Abstract
In genetic association studies, due to the varying underlying genetic models, no single statistical test can be the most powerful test under all situations. Current studies show that if the underlying genetic models are known, trend-based tests, which outperform the classical Pearson χ2 test, can be constructed. However, when the underlying genetic models are unknown, the χ2 test is usually more robust than trend-based tests. In this paper, we propose a new association test based on a generalized genetic model, namely the generalized order-restricted relative risks model. Through a Monte Carlo simulation study, we show that the proposed association test is generally more powerful than the χ2 test, and more robust than those trend-based tests. The proposed methodologies are also illustrated by some real SNP datasets.
Key Words: Genetic association, Robust test, Trend test, SNP
1. Introduction
In case-control genome-wide association (GWA) studies, hundreds of thousands of single nucleotide polymorphisms (SNPs) are tested to determine whether they are associated with the common disease of interest. If a SNP is in linkage disequilibrium with a disease locus, it will not be independent of the status of the disease. Although there may exist gene-gene or gene-environment interactions, the first and crucial step in GWA studies is to identify single SNPs that are associated with disease.
For a SNP with two alleles, A and a, which is assumed to be at risk, there are three genotypes: AA, Aa, and aa. Suppose that there are r cases and s controls in the study. In the r cases, there are r0, r1, and r2 affected people with genotypes AA, Aa, and aa, respectively. There are s0, s1, and s2 people with genotypes AA, Aa, and aa, respectively, in the s unaffected controls.
Testing whether there is an association between the genotype and the disease status is equivalent to testing the association in the 2 × 3 contingency table. Pearson's χ2 test with 2 degrees of freedom (df) is one of the most commonly used statistical methods for testing the association in a contingency table. Note that for SNP data, it is reasonable to assume that the relative risk associated with Aa is between the risks associated with AA and aa. However, Pearson's χ2 test does not utilize this feature of the order-restricted risks in the SNP data. On the contrary, the Cochran-Armitage trend test (CATT) was designed to incorporate the trend to increase the detecting power. The CATT with appropriate scores can be more powerful than Pearson's χ2 test if the underlying genetic model is known [1, 2, 3, 4]. In general, the scores used in the CATT for SNP data are (0, x, 1), where x is a number between 0 and 1 and the optimal value of x depends on the true underlying genetic model. For instance, if the genetic models are recessive, additive/multiplicative (log additive), and dominant, the optimal values of x in the CATTs are 0, 0.5 and 1, respectively. The CATTs with optimal scores have been shown to be more powerful than Pearson's χ2 test, provided that the underlying genetic model is known [4].
However, the genetic model is usually unknown in practice and the CATT with a non-optimal score may perform poorly. In other words, the CATT is not as robust as the χ2 test, and it is sensitive to the departure of assumed genetic models. To increase the robustness of CATT, several trend-based methods have been proposed for the situations where the underlying genetic models are unknown [5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17]. For instance, the maxmin efficiency robust test (MERT) by Gastwirth [18, 19], and the maximum of the three optimal CATTs under recessive, additive, and dominant models (MAX3) have been studied [7]. Zheng and Ng [16] also proposed a two-phase procedure (GMS) with the selection of the genetic model based on the data in the first stage and then used the optimal score based on the chosen model for the CATT in the second stage. Although the above CATT-based methods have been shown to be robust compare to the CATT, there are some limitations for these methods when the analytic null distributions are either unavailable or too complicated. Consequently, Monte Carlo or numerical methods are required to compute the p values of these test procedures.
This paper is organized as follows. First, in Section 2.1, we propose a generalized genetic model for the SNP data, namely a generalized order-restricted relative risks (ORRR) model, in which we assume that the two relative risks are monotonically increasing or decreasing. This ORRR model covers a wide range of ideal models. For instance, the recessive, additive, multiplicative and dominant models are special cases of the ORRR model. Then, we propose a statistical test based on the ORRR model in Section 2.2. Moreover, a restricted likelihood ratio test under the ORRR model is also considered in Section 2.2. Since the new test uses the order-restricted property of the relative risks, it is expected to be more powerful than the χ2 test under many situations in general. On the other hand, unlike the CATT, the proposed test does not assume a specific genetic model; it is not sensitive to the misspecification of the underlying genetic models and therefore is more robust than CATT. In Section 3, a Monte Carlo simulation study is used to study the performance of the proposed procedure. We show that our proposed method is more robust than the existing methods and has decent power properties. The proposed methodologies are illustrated using some real SNP data in Section 4. Conclusions are provided in Section 5.
2. Proposed Test Procedure
2.1. A Generalized Genetic Model and Existing Methods
Table 1 gives the data structure of a case-control GWA study. The relative risks of genotypes Aa and aa to AA are defined as:
Table 1.
SNP data in GWA studies
| Genotype: | AA | Aa | aa | Total |
|---|---|---|---|---|
| Case | r0 | r1 | r2 | r |
| Control | s0 | s1 | s2 | s |
| Total | n0 | n1 | n2 | n |
| (1) |
For many genetic models, we can reasonably assume Pr(case|Aa), the disease risk associated with genotype Aa, is between Pr(case|AA) and Pr(case|aa). Specifically, if a is the at-risk allele, the relative risks satisfy: λ1 ≥ 1 and λ2 ≥ λ1 with at least one of the inequalities being strictly greater. If A is the at-risk allele, the relative risks satisfy 1 ≥ λ1 and λ1 ≥ λ2 where at least one of the inequalities is strict. The monotonicity of the relative risks is also known as order-restricted relative risks. Here, a genetic model with order-restricted relative risks is called a generalized ORRR model. We can see that the aforementioned ideal models (assuming the at-risk allele is a), that is, recessive (λ1 = 1, λ2 > λ1), additive (λ1 = (1 + λ2)/2), multiplicative (λ2 1 λ12), and dominant (λ1 = λ2 > 1), are all special cases of the generalized ORRR model.
As mentioned in Section 1, in addition to the χ2 test, existing statistical test procedures, including CATT, MAX3, GMS and MERT, for the null hypothesis that there is no association between disease and the genotype are also considered here. The CATT statistic can be written as [10]:
where (x0, x1, x2,) (0, x, 1).
The statistic for MAX3 is [7]:
The statistic for GMS is [16, 17]:
GMS =
where I is the indicator function, and the Hardy-Weinberg disequilibrium trend test (HWDTT) statistic is given by [20]:
^P = r2/r + r1/(2r))2, ^Q = s2/s-(s2/s + s1(2s))2, and c is a constant and usually chosen as 1.645.
The statistic for MERT is [19]:
2.2. The Proposed Test
The proposed test is designed to detect the alternative hypothesis that the underlying genetic model belongs to the generalized ORRR model. Suppose the allele frequencies for AA, Aa, and aa are p0, p1, p2 for case and q0, q1, and q2 for control, respectively.
Under the null hypothesis that there is no association between disease and the genotype, we have p0 = q0, p1 = q1, and p2 = q2.
Equation 1 can be expressed as
| (2) |
where Pr(AA) = Pr(AA|case)Pr(case) + Pr(AA|control)Pr(control) = kp0 + (1 − k)q0 and k is the disease prevalence. Similarly we have Pr(Aa) = kp1 + (1 − k) q1 and Pr(AA) = kp2 + (1 − k) q2.
Then, equation 1 can be written as
| (3) |
Assuming the at-risk allele is a, the alternative hypothesis we want to test is based on the generalized ORRR genetic model for which the relative risks satisfy λ1 ≥ 1 and λ2 ≥ λ1, with at least one of the inequalities being strict.
From equation 3, we can write λ1 ≥ 1 and λ2 ≥ λ1 as
| (4) |
Let
then detecting the alternative hypothesis is equivalent to detecting that both T1 and T2 are non-negative and at least one of them is strictly greater than 0. Therefore, we propose a statistical test procedure based on T1 and T2.
Given the observed data presented in table 1, the sample estimates of T1 and T2 can be obtained as
| (5) |
respectively. It can be shown that under the null hypothesis of no association (i.e. pi = qi, i = 0, 1, 2), the expected values of and are zeros (i.e. ) and the variance-covariance matrix of and is
where
Since the variance-covariance matrix is unknown, it can be estimated by replacing the pis by their consistent estimators, The estimated variance-covariance matrix can be expressed as
Because is a positive definite square matrix, eigen-decomposition of gives = PDP', where D is a diagonal matrix
d1 and d2 are the eigenvalues of , and the columns of P are the corresponding eigenvectors of which satisfies PP′ = I.
under the null hypothesis, Z1 and Z2 are asymptotically independent standard normally distributed random variables, where
It can be shown that all elements in are non-negative. Therefore, under the alternative hypothesis, we would expect E(Z1) and E(Z2) to be non-negative where at least one of them is strictly positive. Note that if the at-risk allele is A instead of a in table 1 (i.e. column 1 and column 3 are being switched), statistics Z1 and Z2 are still valid with E(Z1) and E(Z2) being non-positive and with at least one of them being strictly negative under the alternative hypothesis.
By taking the order-restricted property of the SNPs data into account, we consider the statistics
| (6) |
where is the cumulative distribution function of the standard normal distribution. The asymptotic distributions of C1 and C2 are as follows.
Theorem 1
Under the null hypothesis, C1 and C2 are asymptotically χ2 distributed with 4 df.
The proposed test statistic is the maximum of C1 and C2 denoted as
| (7) |
It should be noted that C1 and C2 are not independent. However, the following asymptotic property can lead us to an approximation of the p value associated with statistic W.
Theorem 2
Under the null hypothesis of no association, the survival function of W is asymptotically bounded by
| (8) |
where γ = 1 − χ24(w) and χ24(·) is the cumulative distribution function of the χ2 distribution with 4 df.
Theorem 2 suggests that we can estimate the p value of the test procedure based on W by 2γ, and with small γ, the approximation is very accurate.
Under the ORRR genetic models, some other statistical tests can also be applied. For instance, the restricted likelihood ratio test (RLRT) has been proposed to detect the association for the 2 × k contingency table [21, 22]. For SNP data with k = 3, the RLRT statistic is:
where and , are order-restricted MLEs satisfying
Usually is estimated using the pool adjacent violators algorithm (PAVA) [23], and the above statistic has a weighted χ2 distribution (χ2) under the null hypothesis [24]. Its p value is . For the SNP data, k = 3 and the weights can be estimated using [22] . For association studies, the order (increase or decrease) is usually unknown before we observe the data. According to Barlow et al. [24], we can first compute the p values for both increasing and decreasing alternatives and then compute the overall p value as two times the smaller one.
3. Monte Carlo Simulation Study
A Monte Carlo simulation study is used to study the performance and the power properties of the proposed procedure as well as some existing procedures in the literature. We assume that the rows of case and control in table 1 follow multinomial distributions with probabilities p = (p0, p1, p2) and q = (q0, q1, q2), respectively.
Let
where λ1 and λ2 are the relative risks. Since p0 + p1 + p2 = 1, we have
Therefore, for given qi s, and λ1, λ2, the values of the corresponding pis can be obtained from the above formulas.
For the controls, we first assume Hardy-Weinberg equilibrium (HWE) holds and the minor allele frequencies (MAF) are 0.3 and 0.5. The numbers of cases (r) and controls (s) both equal 2,500 in our simulations. We use different λ1 and λ2 to compare the performance of our proposed method with those of GMS, MERT, MAX3, Pearson's χ2 test, CATT with x = 0.5, and the RLRT. In our simulation study, we use significance level α = 10−5 to reflect the real situation of GWA studies where the total number of SNPs are large and the point-wise significance levels are usually very small. For each setting, we used 1,000,000 realizations to estimate the type I error rates (sizes) and power values of those test procedures. To estimate the p values of MAX3, GMS, and MERT, we used R package ‘Rassoc’ with option ‘asy’ which uses the asymptotic null distribution [17].
Note that under the null hypothesis that λ1 = λ2 = 1, the estimated rejection rates are the estimated type I error rates. These estimated type I error rates of different test procedures are presented in table 2. Figures 1 and 2 plot the estimated rejection rates of different test procedures when HWE holds. We also considered the situations where HWE does not hold for controls. Specifically, we assume the probabilities for genotypes (AA, Aa, aa) are (0.1, 0.3, 0.6) or (0.6, 0.3, 0.1) in controls. Figures 3 and 4 plot the estimated rejection rates for these two settings, respectively.
Table 2.
Estimated type I error rates for the test procedures discussed under different settings
| Setting | W | X2 | MAX3 | GMS | CATT | MERT | RLRT |
|---|---|---|---|---|---|---|---|
| HWE (MAF = 0.3) | 1.2 × 10–5 | 1.3 × 10–5 | 1.4 × 10–5 | 1.4 × 10–5 | 1.3 × 10–5 | 1.5 × 10–5 | 2.3 × 10–5 |
| HWE (MAF = 0.5) | 0.7 × 10–5 | 0.8 × 10–5 | 0.8 × 10–5 | 0.8 × 10–5 | 0.9 × 10–5 | 0.8 × 10–5 | 0.9 × 10–5 |
| q = (0.1, 0.3, 0.6) | 1.2 × 10–5 | 1.1 × 10–5 | 1.0 × 10–5 | 1.0 × 10–5 | 1.1 × 10–5 | 1.4 × 10–5 | 1.6 × 10–5 |
| q = (0.6, 0.3, 0.1) | 0.6 × 10–5 | 1.1 × 10–5 | 0.6 × 10–5 | 0.6 × 10–5 | 1.1 × 10–5 | 0.8 × 10–5 | 0.5 × 10–5 |
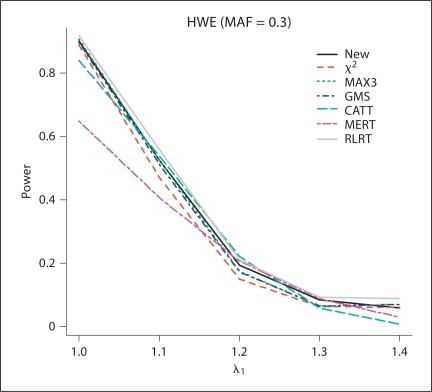
Fig. 1.
The estimated power values of the different test procedures when HWE (MAF = 0.3) holds for controls with λ1 = 1.0, 1.1, 1.2, 1.3, 1.4, and λ2 = 1.4.
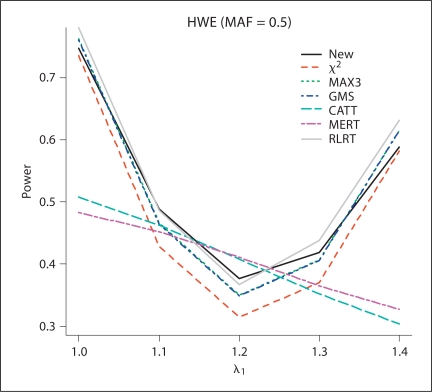
Fig. 2.
The estimated power values of the different test procedures when HWE (MAF = 0.5) holds for controls with λ1 = 1.0, 1.1, 1.2, 1.3, 1.4 and λ2 = 1.4.
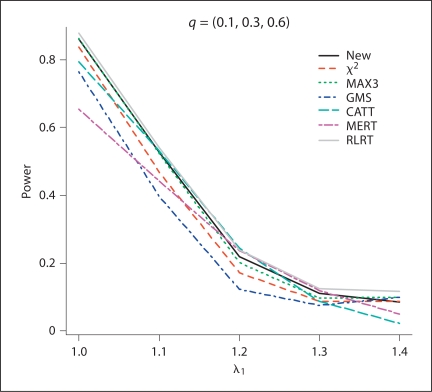
Fig. 3.
The estimated power values of the different test procedures when probabilities for genotypes (AA, Aa, aa) are (0.1, 0.3, 0.6) in controls with λ1 = 1.0, 1.1, 1.2, 1.3, 1.4, and λ2 = 1.4.
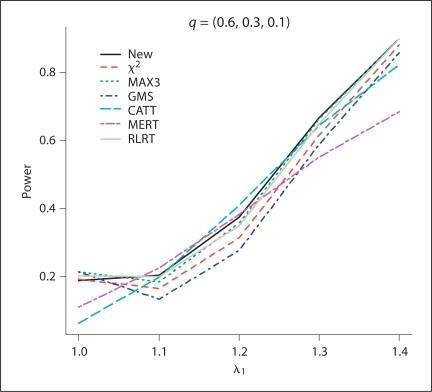
Fig. 4.
The estimated power values of the different test procedures when probabilities for genotypes (AA, Aa, aa) are (0.6, 0.3, 0.1) in controls with λ1 = 1.0, 1.1, 1.2, 1.3, 1.4, and λ2 = 1.4.
From the simulation results, for all of the methods except for RLRT, the methods control type I error rates quite well. In general, RLRT and our proposed method have similar powers. However, our simulation study shows that sometimes RLRT had inflated type I error rates. For example, when HWE holds and MAF = 0.3, the estimated size using RLRT was 2.3 × 10−5, which is statistically significantly different than the nominal level 1 × 10−5 at significance level 0.05. The situation can be even worse when MAF is smaller. If we assume HWE holds and MAF = 0.2 and 0.1, the estimated sizes from RLRT were 2.5 × 10−5, and 2.8 × 10−5, respectively. Also we can see that MAX3 and GMS have similar performances, while CATT with score x = 0.5 and MERT have power values close to each other.
For additive models (λ1 = 1.2, λ2 = 1.4), MERT and CATT are usually more powerful than other methods, as expected. However, when the true genetic model is dominant (λ1 = λ2 = 1.4) or recessive (λ1 = 1, λ2 = 1.4), MERT and CATT perform much worse than other methods. This indicates that MERT and CATT are sensitive to the underlying genetic models and therefore they are not robust. In contrast, MAX3 and GMS perform much better than MERT and CATT for dominant and recessive models; while they both have low power values for additive models. Under models other than recessive, additive, and dominant (i.e. λ1 = 1.1, λ2 = 1.4 and λ1 = 1.3, λ2 = 1.4), the proposed test and RLRT have among the three largest power values. Furthermore, if our proposed method is not the most powerful test for a given situation, it always has power value close to the largest one (usually the second or the third largest in power values). This indicates that our proposed method is robust in the sense that it has comparable power under different situations considered in the simulation study. This robustness property is one of the merits of the proposed method because the underlying genetic models are seldom known in practice.
Figures 1–4 clearly show that the proposed methods have highest or close to highest powers for all the situations considered in the simulations. It should be noticed that for figures 1 and 2, when the estimated power values for MAX3 and GMS are very close to each other, the differences between the two lines for these two methods are not appreciable. From our simulation study, we observe that the power values of some methods not only depend on the genotypic frequencies, but also the genetic model. For example, in figure 2, except for CATT and MERT, the power values of all other methods decrease when λ1 increases for λ1 < 1.2 (λ1 = 1.2 is the additive model) and the power values increase when λ1 increases for λ1 > 1.2.
4. Numerical Illustrations
In this section, we apply our proposed method with others to some real SNPs reported from four GWA studies with 100,000–500,000 SNPs for age-related macular degeneration (AMD) [25], two cancer studies [26, 27], and a hypertension study [28]. The datasets are summarized in table 3, which were taken from [17].
Table 3.
Datasets for numerical illustrations (adapted from [17])
| GWA study SNP ID | Case | Control | ||||
|---|---|---|---|---|---|---|
| AA | Aa | aa | AA | Aa | aa | |
| AMD | ||||||
| rs380390 | 50 | 35 | 11 | 6 | 25 | 19 |
| rs1329428 | 2 | 24 | 68 | 5 | 29 | 14 |
| Prostate cancer | ||||||
| rs1447295 | 25 | 283 | 864 | 10 | 218 | 929 |
| rs6983267 | 223 | 598 | 351 | 301 | 579 | 277 |
| rs7837688 | 27 | 283 | 861 | 11 | 206 | 939 |
| Breast cancer | ||||||
| rs10510126 | 10 | 180 | 955 | 14 | 272 | 854 |
| rs12505080 | 50 | 477 | 608 | 99 | 408 | 628 |
| rs17157903 | 18 | 316 | 777 | 26 | 220 | 862 |
| rs1219648 | 250 | 543 | 352 | 170 | 538 | 433 |
| rs7696175 | 187 | 605 | 353 | 249 | 496 | 396 |
| rs2420946 | 242 | 546 | 357 | 165 | 537 | 440 |
| Hypertension | ||||||
| rs2820037 | 40 | 587 | 1,325 | 72 | 684 | 2,180 |
| rs6997709 | 118 | 716 | 1,116 | 237 | 1,201 | 1,500 |
| rs7961152 | 416 | 963 | 570 | 492 | 1,448 | 992 |
| rs11110912 | 67 | 647 | 1,237 | 83 | 804 | 2,049 |
| rs1937506 | 113 | 742 | 1,097 | 244 | 1,205 | 1,484 |
| rs2398162 | 111 | 624 | 1,205 | 194 | 1,121 | 1,608 |
Table 4 reports the p values for each SNP from the different methods. It can be observed that when the genetic model is between recessive and dominant (Z1 > 0 and Z2 > Z1, or Z1 < Z2 and Z2 < 0), the proposed method has similar p values with those from CATT, which is usually more powerful than other methods. However, when the genetic model is close to recessive or dominant, CATT performs poorly, but GMS, MAX3 and our proposed method have similar p values and better than CATT. This indicates that GMS, MAX3 and our proposed method perform similar under this situation. For SNPs having large absolute values but with different signs of Z1 and Z2, their genetic models do not belong to the generalized genetic model. Under those situations, Pearson's χ2 test has the smallest p value as expected, since it is more robust than any other method, while CATT and MERT have large p values. The p values of the proposed method are similar to those from GMS and MAX3. Note that the p values from RLRT are usually similar to or smaller than those from our proposed test. However, we found that ten out of the seventeen estimated MAF values from controls are less than 0.3; these small p values may be due to the liberal nature of the RLRT for highly unbalanced data as mentioned before.
Table 4.
p values for real SNP data from the various methods and observed statistics Z1 and Z2 from the proposed method
| SNP | X2 | MAX3 | GMS | CATT | MERT | RLRT | Z1 | Z2 | W |
|---|---|---|---|---|---|---|---|---|---|
| rs380390 | 1.8 × 10–6 | 1.0 × 10–6 | 2.0 × 10–6 | 3.1 × 10–7 | 3.9 × 10–7 | 3.1 × 10–7 | –4.04 | –3.11 | 9.0 × 10–7 |
| rs1329428 | 3.6 × 10–6 | 1.0 × 10–6 | 1.0 × 10–6 | 8.7 × 10–7 | 8.5 × 10–6 | 9.2 × 10–7 | 1.92 | 4.60 | 2.0 ×10–6 |
| rs1447295 | 1.9 × 10–4 | 8.3 × 10–5 | 8.1 × 10–5 | 4.5 × 10–5 | 6.0 × 10–5 | 4.7 × 10–5 | –2.46 | –3.32 | 8.4 × 10–5 |
| rs6983267 | 3.5 × 10–5 | 3.0 × 10–5 | 2.9 × 10–5 | 7.9 × 10–6 | 7.4 × 10–6 | 1.5 × 10–5 | 3.67 | 2.66 | 1.5 × 10–5 |
| rs7837688 | 1.6 × 10–5 | 5.0 × 10–6 | 6.0 × 10–6 | 2.7 × 10–6 | 8.3 × 10–6 | 3.5 × 10–6 | −2.51 | −3.98 | 6.9 × 10–6 |
| rs10510126 | 3.7 ×10–6 | 1.0 × 10–6 | 3.0 × 10–6 | 1.4 × 10–6 | 1.7 × 10–4 | 8.2 × 10–7 | 0.78 | 4.94 | 2.9 × 10–6 |
| rs12505080 | 1.8 10–5 | 7.4 × 10–5 | 8.3 × 10–5 | 0.32 | 0.039 | 5.3 × 10–5 | 4.27 | –1.90 | 2.4 × 10–4 |
| rs17157903 | 9.9 × 10–6 | 5.3 × 10–5 | 4.8 × 10–5 | 6.3 × 10–4 | 0.058 | 3.8 × 10–5 | 1.31 | –4.62 | 5.0 × 10–5 |
| rs1219648 | 7.5 × 10–6 | 4.0 × 10–6 | 7.0 × 10–6 | 1.8 × 10–6 | 1.4 × 10–6 | 2.9 × 10–6 | –3.92 | –2.87 | 3.2 × 10–6 |
| rs7696175 | 1.6 × 10–5 | 2.1 × 10–3 | 1.9 × 10–3 | 0.59 | 0.40 | 1.8 × 10–3 | 3.77 | –2.80 | 1.7 × 10–3 |
| rs2420946 | 8.8 × 10–6 | 7.0 × 10–6 | 4.0 × 10–6 | 1.9 × 10–6 | 1.5 × 10–6 | 3.5 × 10–6 | –3.82 | –2.95 | 3.7 × 10–6 |
| rs2820037 | 7.7 × 10–7 | 1.0 × 10–6 | 2.0 × 10–6 | 5.8 × 10–5 | 0.013 | 2.3 × 10–6 | 1.03 | –5.21 | 2.9 × 10–6 |
| rs6997709 | 4.4 × 10–5 | 2.8 × 10–5 | 1.4 × 10–5 | 7.9 × 10–6 | 1.9 × 10–5 | 1.4 × 10–5 | 2.43 | 3.77 | 1.9 × 10–5 |
| rs7961152 | 3.0 × 10–5 | 1.4 ×10–5 | 1.6 ×10–5 | 7.4 ×10–6 | 6.0 ×10–6 | 1.4 ×10–5 | –3.68 | –2.70 | 1.3 ×10–5 |
| rs11110912 | 1.9 × 10–5 | 5.0 ×10–6 | 1.9 ×10–5 | 9.2 ×10–6 | 2.2 ×10–4 | 5.8 ×10–6 | –1.08 | –4.53 | 1.3 ×10–5 |
| rs1937506 | 4.5 ×10–5 | 3.4 ×10–5 | 2.6 ×10–5 | 9.2 ×10–6 | 8.5 ×10–6 | 1.3 ×10–5 | 3.13 | 3.19 | 1.9 ×10–5 |
| rs2398162 | 5.7 ×10–6 | 1.0 ×10–6 | 2.0 ×10–6 | 7.9 ×10–6 | 1.2 ×10–4 | 1.7 ×10–6 | 1.02 | 4.81 | 4.0 ×10–6 |
In these illustrations, we can also see that the two observed statistics Z1 and Z2 can be used to determinate the genetic model and the at-risk allele. For example, for SNP rs380390 in table 4, Z1 = −4.04, Z2 = −3.11, since both Z1 and Z2 are negative, the at-risk allele is A instead of a; the genetic model should be neither recessive nor dominant, but between the two since the absolute values of Z1 and Z2 are much larger than 1. From table 4, we also see that some SNPs have Z1 and Z2 with different signs. Three out of six SNPs from the breast cancer data fall into this category. This situation deserves special attention. It is possible that the underlying genetic models are over- or under-dominant. But it is also possible that this happened merely due to chance or something else, such as population substructure, which needs further investigation. The breast cancer SNP data in table 3 were taken from the Nurses’ Health Study (NHS) and three additional studies have been conducted by the authors [26]. The p values for SNP rs17157903 from the other three studies are 0.72, 0.49 and 0.92, respectively, using the χ2 test. Therefore the association between the SNP rs17157903 and breast cancer needs to be validated by future studies.
5. Conclusions
In GWA studies, since the underlying genetic models are usually unknown, choosing a powerful statistic test is desirable. There is no single test performing uniformly better than the other competitors, and most of the existing methods may suffer from serious power lost under some models. Through Monte Carlo simulations and the study of real SNP data, we have seen that our proposed method is more robust and powerful than existing methods in many situations.
The robustness of the proposed test is expected since, unlike the CATT test, it does not require a complete genetic model specification except that we assume the model belongs to a generalized ORRR model. Therefore it is not very sensitive to model mis-specification. Meanwhile, the proposed test correctly incorporates the property of the monotonicity of the relative risks for SNP data in GWA studies which results in power gains. Moreover, based on the simulation results, we observed that when the genetic models are not one of the perfect models (i.e. recessive, additive, and dominant models), the proposed test usually has the highest or second highest power. In real world applications, the perfect models may be rare if not impossible; hence the proposed method is certainly preferable. Finally, through simulation (data not shown) and real data (see table 4), when the genetic model is not ORRR, e.g. over- or under-dominant, the proposed method has reasonable power. Beside the robustness of the proposed method, another advantage of the proposed method is that the p value can be easily approximated with very high accuracy. Although sometimes RLRT can also be applied to generalized ORRR models, it should be used with caution as it inflates type I error rates when the data are highly unbalanced (e.g. HWE with small MAF).
Since some of SNPs are highly correlated due to linkage disequilibrium, the p values obtained from individual SNPs are also correlated. Traditional multiple tests correction methods, such as the Bonferroni procedure, are not appropriate. One may choose instead to use the recently proposed method which is based on the concept of effective number [29].
Appendix
Proof of Theorem 1
For large sample sizes, which are usually available for GWA studies, we can assume Z1 and Z2 are independently and identically distributed as standard normal, so that Φ(Z1) and Φ(Z2) are independently and identically distributed uniformly between 0 and 1. Therefore, according to Fisher [30], C1 is χ2 distributed with df = 4. Similarly, C2 is χ2 distributed with df = 4.
Proof of Theorem 2
For a large sample size, from Theorem 1, C1 and C2 are both χ2 distributed with df = 4. Using the concept of associated random variables by Esary et al. [31] and Theorem 2 by Owen [32], we have
Acknowledgements
The first author would like to thank for the support from the NIH grant (UL1 RR024148) awarded to the University of Texas Health Science Center at Houston. The authors would also like to thank Professor Wayne Woodward for his suggestions which resulted in a much improved version of the manuscript.
References
- 1.Armitage P. Tests for linear trends in proportions and frequencies. Biometrics. 1955;11:375–386. [Google Scholar]
- 2.Balding DJ. A tutorial on statistical methods for population association studies. Nat Rev Genet. 2006;7:781–791. doi: 10.1038/nrg1916. [DOI] [PubMed] [Google Scholar]
- 3.Cochran W. Some methods for strengthening the common chi-square tests. Biometrics. 1954;10:417–451. [Google Scholar]
- 4.Zheng G, Freidlin B, Gastwirth JL. Comparison of robust tests for genetic association using case-control studies. In: Rojo J, editor. Optimality: The Second Erich L. Lehmann Symposium, May 19–22, 2004, Rice University. IMS Lecture Notes – Monograph Series. vol 49. Beachwood: Institute of Mathematical Statistics; 2006. pp. 253–265. [Google Scholar]
- 5.Chen Z, Zheng G. Exact robust tests for detecting candidate-gene association in case-control trio design. J Data Sci. 2005;3:19–33. [Google Scholar]
- 6.Freidlin B, Podgor MJ, Gastwirth JL. Efficiency robust tests for survival or ordered categorical data. Biometrics. 1999;55:883–886. doi: 10.1111/j.0006-341x.1999.00883.x. [DOI] [PubMed] [Google Scholar]
- 7.Freidlin B, Zheng G, Li Z, Gastwirth JL. Trend tests for case-control studies of genetic markers: power, sample size and robustness. Hum Hered. 2002;53:146–152. doi: 10.1159/000064976. [DOI] [PubMed] [Google Scholar]
- 8.Gonzalez JR, Carrasco JL, Dudbridge F, Armengol L, Estivill X, Moreno V. Maximizing association statistics over genetic models. Genet Epidemiol. 2008;32:246–254. doi: 10.1002/gepi.20299. [DOI] [PubMed] [Google Scholar]
- 9.Kwak M, Joo J, Zheng G. A robust test for two-stage design in genome-wide association studies. Biometrics. 2009;65:1288–1295. doi: 10.1111/j.1541-0420.2008.01187.x. [DOI] [PubMed] [Google Scholar]
- 10.Sasieni PD. From genotypes to genes: doubling the sample size. Biometrics. 1997;53:1253–1261. [PubMed] [Google Scholar]
- 11.Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S, Balkau B, Heude B, Charpentier G, Hudson TJ, Montpetit A, Pshezhetsky AV, Prentki M, Posner BI, Balding DJ, Meyre D, Polychronakos C, Froguel P. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 12.Slager SL, Schaid DJ. Case-control studies of genetic markers: power and sample size approximations for Armitage's test for trend. Hum Hered. 2001;52:149–153. doi: 10.1159/000053370. [DOI] [PubMed] [Google Scholar]
- 13.Song K, Elston RC. A powerful method of combining measures of association and Hardy-Weinberg disequilibrium for fine-mapping in case-control studies. Stat Med. 2006;25:105–126. doi: 10.1002/sim.2350. [DOI] [PubMed] [Google Scholar]
- 14.Wang K, Sheffield VC. A constrained-likelihood approach to marker-trait association studies. Am J Hum Genet. 2005;77:768–780. doi: 10.1086/497434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng G, Freidlin B, Li Z, JL G. Choice of scores in trend tests for case-control studies of candidate gene associations. Biometrical J. 2003;45:335–348. [Google Scholar]
- 16.Zheng G, Ng HKT. Genetic model selection in two-phase analysis for case-control association studies. Biostatistics. 2008;9:391–399. doi: 10.1093/biostatistics/kxm039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zang Y, Fung WK, Zheng G. Simple algorithms to calculate the asymptotic null distributions of robust tests in case-control genetic association studies in R. J Stat Software. 2010;33:1–24. [Google Scholar]
- 18.Gastwirth JL. On robust procedures. J Am Stat Assoc. 1966;61:929–948. [Google Scholar]
- 19.Gastwirth JL. The use of maximin efficiency robust tests in combining contingency tables and survival analysis. J Am Stat Assoc. 1985;80:380–384. [Google Scholar]
- 20.Song K, Elston RC. A powerful method of combining measures of association and Hardy-Weinberg disequilibrium for fine mapping in case-control studies. Stat Med. 2006;25:105–126. doi: 10.1002/sim.2350. [DOI] [PubMed] [Google Scholar]
- 21.Agresti A, Coull BA. The analysis of contingency tables under inequality constraints. J Stat Pplan Inference. 2002;107:45–73. [Google Scholar]
- 22.Silvapulle MJ, Sen PK. Constrained Statistical Inference: Inequality, Order, and Shape Restrictions. Hoboken, NJ: John Wiley & Sons; 2005. [Google Scholar]
- 23.Ayer M, Brunk H, Ewing GM, Reid W, Silverman E. An empirical distribution function for sampling with incomplete information. Ann Math Stat. 1955:641–647. [Google Scholar]
- 24.Barlow R, Bartholomew D, Bremner J, Brunk H. Statistical Inference under Order Restrictions. New York: Wiley; 1972. [Google Scholar]
- 25.Klein RJ, Zeiss C, Chew EY, Tsai JY, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, Bracken MB, Ferris FL, Ott J, Barnstable C, Hoh J. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hunter DL, Kraft P, Jacobs KB, Cox DG, Yeager N, Hankinson SE, Wacholder S, Wang Z, Welch R, et al. A genome-wide association study identifies alleles in FGFR2 associated with risk of sporadic postmenpausal breast cancer. Nat Genet. 2007;39:870–874. doi: 10.1038/ng2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yeager M, Orr N, Hayes R, Jacobs K, Kraft P, Wacholder S, Minichiello M, Fearnhead P, Yu K, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 28.The Wellcome Trust Case Control Consortium: Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Z, Liu Q. A new approach to account for the correlations among single nucleotide polymorphisms in genome-wide association studies. Hum Hered. 2011;72:1–9. doi: 10.1159/000330135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fisher RA, editor. Statistical Methods for Research Workers. Edinburgh: Oliver and Boyd; 1932. [Google Scholar]
- 31.Esary JD, Proschan F, Walkup DW. Association of random variables, with applications. Ann Math Stat. 1967;38:1466–1474. [Google Scholar]
- 32.Owen AB. Karl Pearson's meta-analysis revisited. Ann Stat. 2009;37:3867–3892. [Google Scholar]