Abstract
Mericitabine (RG7128) is a first-in class nucleoside polymerase inhibitor (NPI), which requires intracellular uptake and phosphorylation to two active triphosphates. Mathematical modeling has provided important insights for characterizing HCV-RNA decline and estimating in vivo effectiveness of antiviral agents; however it has not been used to characterize viral kinetics with NPIs. HCV RNA was frequently measured in 32 treatment-experienced patients infected with HCV genotype-1 during and after mericitabine monotherapy for 14 days with 750-mg or 1500-mg administered once (QD) or twice daily (BID). Initial decline of HCV RNA was typically slower than with interferon-alpha or protease inhibitors and 12 patients presented a novel pattern of HCV RNA kinetics characterized by a monophasic viral decline. Viral kinetics could be well fitted by assuming that the effectiveness in blocking viral production gradually increased over time to reach its final value, ε2, consistent with previous accumulation time estimates of intracellular triphosphates. ε2 was high with BID dosing (mean 750-mg and 1500-mg: 98.0% and 99.8%, P=0.018) and significantly higher than in patients treated QD (mean QD vs BID: 90% vs 99%, P<10−7). Virus rebounded rapidly upon drug discontinuation, which was attributed to the elimination of active drug and the subsequent decline of drug effectiveness with mean t1/2=13.9 h in the BID regimens.
Conclusion
The observed slower initial decline likely represents the time needed to accumulate intracellular triphosphates and is consistent with in vitro data. When administered BID, mericitabine reached a high, dose-dependent, final effectiveness in blocking viral production, that rapidly dropped upon treatment cessation. Understanding HCV RNA kinetics with mericitabine could provide valuable insights for combining it with other direct-acting antiviral agents.
Introduction
Chronic hepatitis C virus (HCV) infection has a worldwide prevalence of about 3% (1). Achieving a long-term sustained virologic response (SVR), defined as undetectable HCV RNA in serum 24 weeks after the end of treatment, is the most effective way to either prevent disease progression (2). Treatment outcome with pegylated interferon (peg-IFN) and ribavirin (RBV) administered for 48 weeks, is correlated with HCV genotype and SVR is only achieved in approximately 50% of HCV genotype 1 patients, the most prevalent genotype in western countries (3).
Direct acting antiviral (DAA) agents constitute a new stage in HCV therapy. HCV protease inhibitors improved treatment outcomes when added to peg-IFN/RBV in both treatment-naive and treatment-experienced patients (4–7). However, the benefits of this strategy will remain limited due to safety, tolerability, and convenience limitations associated with peg-IFN. In addition, the development of protease inhibitor resistance, particularly in non-responders to peg-IFN/RBV and patients infected with HCV genotype 1a, will further limit the efficacy of triple combination treatment of a protease inhibitor added to peg-IFN/RBV (8). Thus, there is a continuing need to develop more effective, better tolerated, and more convenient treatment regimens in the form of DAA combinations, as an all-oral interferon sparing treatment, or as quadruple therapy through the addition of two DAA’s to the current peg-IFN/RBV.
Mericitabine (RG7128) is an oral cytidine nucleoside analog prodrug that exhibited strong antiviral effectiveness against the HCV polymerase across all HCV genotypes (9–11), with no evidence of resistance reported in patients treated with mericitabine monotherapy for 14 days (12). Upon entering the hepatocyte, mericitabine is converted to a cytidine monophosphate, which is then further converted to both a cytidine and a uridine triphosphate. Both triphosphate forms are active, with the cytidine form predominating at least early following the initiation of treatment (13).
Viral dynamic modeling has provided valuable insights for quantifying the effects of (peg)-IFN, ribavirin and HCV protease inhibitors and estimating their antiviral effectiveness in vivo (14), but it has not been used to analyze data from nucleoside HCV polymerase inhibitor treatment studies. Here, we analyzed and modeled HCV RNA kinetics from 32 interferon-treatment experienced patients, infected with HCV genotype 1, who were treated for 14 days with 750-mg or 1500-mg doses of mericitabine alone daily (QD) or twice a day (BID). In addition, HCV RNA was frequently measured after the end of the dosing period, which allowed us an opportunity to examine the determinants of the post-treatment viral rebound.
Methods
Patients and treatment
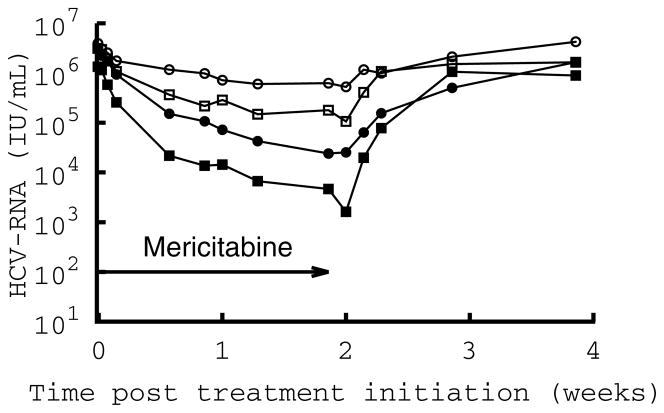
The RG7128 clinical study was a multicenter, observer-blinded, randomized, placebo-controlled study in non-cirrhotic patients chronically infected with HCV genotypes 1 (30-1a; 10-1b) that had previously failed interferon therapy with or without ribavirin. Multiple oral doses of mericitabine were administered for 14 days to 32 HCV-infected patients, split into four cohorts (n=10 patients per cohort with 8 getting drug and 2 taking placebo) with regimens of 750-mg QD, 1500-mg QD, 750-mg BID and 1500-mg BID. Mean changes in HCV RNA per dosing group are displayed in Figure 1.
Figure 1.
Mean decline in HCV RNA per dosing group. Dotted line: 750 mg every day (750 QD); dashed line: 1500 mg every day (1500 QD); dashed-dotted line: 750 mg every 12 h (750 BID); solid line: 1500 mg every 12 h (1500 BID).
Samples for plasma HCV RNA analysis using the Roche Cobas TaqMan (limit of detection <15 IU/mL) were collected at baseline (day 0), 4 hours, 12 hours, and then at day 1, 4, 6, 7, 9, and 13 during treatment and at days 14, 15, 16, 20, and 27 after the end of treatment.
Kinetics of viral decline under treatment using the standard model
The kinetics of viral decline under treatment was modeled using the standard model of HCV kinetics (15), defined by the following set of differential equations:
| (1) |
where T0 represents the density of target cells that can be infected by virus (V), with rate constant β. In the model, infected cells (I) die or lose their infected state with rate constant δ and produce virions at constant rate p per cell. Virions are assumed to be cleared from serum with rate constant c. It is also assumed that the target cell level is constant throughout the study period and remains at its pre-treatment steady state value T0=cδ/pβ (15).
After an initial pharmacologic delay of length t0, therapy was assumed to reduce the rate of viral production per cell from p to p(1−ε), where ε is the drug effectiveness, with ε=1 implying the drug is 100% effective in blocking viral production. Since this model assumes constant effectiveness (CE) under treatment, it has been called the CE model (16).
A model that allows treatment effectiveness to increase with time
We also explored an alternative model where the treatment effectiveness in blocking viral production, ε, can change over time during therapy:
| (2) |
where ε1 and ε2 are the initial and the final values of the treatment effectiveness, respectively, and k defines the rapidity of the change in effectiveness. This function generates an effectiveness that increases with time (assuming ε2>ε1). The use of this function (Eq. 2) combined with the viral dynamics model (Eq. 1) is called the varying effectiveness (VE) model (17). Note that if the initial and the final effectiveness are equal (ε1=ε2) or if the changes in the drug effectiveness are very rapid (k large), the VE model is equivalent to the CE model. In that respect, the CE model is a particular case of the VE model. If k is small, then the rate of viral load decline until ε ≈ ε2 depends on the value of k.
Kinetics of viral increase after the end of treatment
Let tend denote the time when the last dose was taken and t1 the length of the delay until drug effectiveness starts decreasing. For QD and BID regimens tend =13 days and tend =13.5 days, respectively. Assuming that the drug effectiveness is related to the intracellular drug concentration C(t) by an Emax model (18) of the form:
| (3a) |
and assuming that after tend + t1 the concentration of active drug decreases exponentially with rate ke, the drug effectiveness is given by:
| (3b) |
where εmax is the effectiveness at time tend + t1. If the drug is eliminated at the same rate in the circulation and intracellularly, then ke approximates the rate constant of elimination for the parent nucleoside from the systemic circulation. A more general form of the Emax model includes a Hill coefficient n. When a Hill coeffient is included in Eq.(3a), ke in Eq. (3b) is replaced by nke, and the combined parameter nke is then estimated. We find (see Results) that nke is approximately equal to the measured mean drug clearance rate suggesting n=1. Hence we assume n=1 and have omitted the Hill coefficient from further consideration.
Statistical Methods
Model parameters were obtained by a maximum likelihood method using MONOLIX version 3.1 (http://software.monolix.org), a software program based on a stochastic approximation expectation-approximation (SAEM) algorithm (19). After the population parameters and the between-subject variabilities were found the estimated parameters for each individual were deduced using empirical Bayes estimates (20). Thus, all dosing groups were analyzed simultaneously and the parameters have the same distributions, irrespective of the dosing groups. For each parameter, we report the population estimates and standard errors, as well as the first and third quartiles of the individual estimates (when the sample size was large enough). One subject (#92206) did not respond to treatment and therefore was not included in the final analysis.
In order to reduce the number of parameters to be estimated in the VE model, c was fixed to 6 d−1 (15). Moreover, t0 was determined empirically as the last sampling time before the viral load declined by more than 0.2 log10 and did not increase afterward above baseline, or before two consecutive decreasing HCV RNA measurements.
Two covariates were included in the model to study their impact on the viral kinetic parameters. The first covariate was the treatment dosing regimen group. Except for the determination of the final treatment effectiveness (ε2), the QD and BID groups were treated together. Also, we considered a second covariate distinguishing patients having or not a having monotonic viral decline throughout the treatment period. For that purpose we computed for each patient by linear regression the slope, s2, of the HCV RNA measurements between t=4 days and t=13 days, a period typically considered to be part of the second phase of viral decline (Table S1). A t-test was used to assess whether s2 was significantly different than 0. If s2 was not significantly (P>0.1) different than 0, the patient was said to have a flat second phase response. Using this criterion 52% (16/31) of the patients had a flat second phase response, with no difference in distribution among dosing regimen (Table S1).
More details on the fitting method and statistical analysis of the model are given in the supplementary materials.
Results
Analysis of the changes in viral load using the standard model of viral dynamics
We first fitted the data using the standard (CE) model of viral dynamics (Eq. 1;3a–b) (Table 1). The mean value estimated for c was surprisingly small (0.98 d−1) with no significant differences between the dosing groups (QD vs BID, P=0.84). Interestingly, even the maximal value for c estimated in our sample (3.42 d−1) was lower than what has been typically found with IFN-based therapy (6 d−1) (15, 21). How to understand this result? A closer analysis of the early viral kinetics induced by mericitabine reveals that the initial rate of viral decline (in the first days after treatment initiation) is much slower than what was previously seen with IFN (21). Although the CE model attributes this slow decline to a low rate of viral clearance, this interpretation is dubious as the rate of viral clearance is a physiological quantity and, consequently, c should not depend on the antiviral strategy.
Table 1.
Viral kinetic parameter estimates obtained using the CE model.
| c [d−1] | ε QD 750mg | ε QD 1500mg | ε BID 750mg | ε BID 1500mg | δ [d−1] | t1[d] | ke [d−1] QD | ke [d−1] BID | AIC | |
|---|---|---|---|---|---|---|---|---|---|---|
| Population parameters | 0.98 | 0.80 | 0.90 | 0.98 | 0.997 | 0.019 | 0.16 | 0.67 | 1.49 | 295.9 |
| SE1 | 0.14 | 0.095 | 0.044 | 0.012 | 0.0015 | 0.0066 | 0.079 | 0.012 | 0.028 |
Then, what other factor may explain the slow initial rate of viral decline? A mathematical analysis of the CE model reveals that the initial viral decline should be approximately linear with slope cε. Thus, assuming c is as high as what was found during IFN-based therapy, a modest initial viral decline can be explained by an initially modest treatment effectiveness, consistent with the conversion and accumulation of intracellular nucleoside triphosphates that occurs with nucleoside analogues (13). Therefore, we studied the possibility of a gradual increase of mericitabine antiviral effectiveness over time by fitting the viral load data using the VE model (Eq. 2). A comparison of the model fit to each individual patient’s viral load data is given in the Supplemental Material (Fig. S1). Interestingly, the VE model had a lower Akaike information criterion value (22) than the CE model, and thus provided better fits to the data, even after correcting for the additional numbers of parameter involved in this model. Since the VE model gave a better fit than the CE model, we only discuss in the following the results provided by the VE model.
In the VE model, the initial antiviral effectiveness of mericitabine increased upon the initiation of dosing with characteristic rate k, so that it reached half of the final drug effectiveness, ε2, in time ln2/k. Mericitabine’s final drug effectiveness, ε2 was high with BID dosing (mean 750-mg and 1500-mg: 98% and 99.8%, P=0.018) and significantly higher than in patients treated QD (mean QD vs BID: 90% vs 99%, P<10−7).
Possibly due to small sample sizes, no difference in the initial antiviral effectiveness, ε1, was found between the dosing groups (QD vs BID, P=0.40) or with the pattern of viral decline. Consistent with the intracellular pharmacokinetics, the estimated value of ε1 was low in the vast majority of patients (mean 0.38), and probably reflects the minimal antiviral effectiveness needed to generate a discernable viral decline. The differences in HCV RNA decline between the four dosing regimens could be only explained by differences in the final drug effectiveness in blocking viral production, ε2, and in the rate at which the effectiveness was increasing over time, k (Table 2). Since the two QD groups did not show a significantly different effectiveness (mean ε2=0.86 with 750-mg QD vs mean ε2=0.94 with 1500-mg QD, P=0.20), they were treated together as a single group (mean±standard error (SE) ε2=0.90±0.050). The antiviral effectiveness was significantly higher in the two BID groups compared to the QD groups (mean±SE ε2 for 750-mg BID was 0.98±0.0091, P=0.0056; while for 1500-mg group it was 0.998±0.0012, P<10−7). The rapidity of the change in treatment effectiveness, measured by the parameter k, was significantly higher in flat vs non-flat second phase responders (P<10−9). Also, this parameter was associated with the treatment regimen (QD vs BID, P=0.017), which indicates that, for a similar final effectiveness, patients given the BID regimens built up effectiveness faster than patients in the QD regimens. Patients receiving a BID regimen reached 90% and 99% of the estimate final effectiveness with a mean time of 2.9 and 6.5 days, respectively (Table S1 and Fig. 2).
Table 2.
Viral kinetic parameter estimates obtained using the VE model.
| ε1 | k [d−1] QD flat | k [d−1] QD non-flat | k [d−1] BID flat | k [d−1] BID non-flat | ε2 QD 750mg | ε2 QD 1500mg | ε2 BID 750mg | ε2 BID 1500mg | δ [d−1] | t1[d] | ke [d−1] QD | ke [d−1] BID 750mg | ke [d−1] BID 1500mg | AIC | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Population parameters | 0.38 | 1.06 | 0.23 | 2.03 | 0.43 | 0.86 | 0.94 | 0.98 | 0.998 | 0.023 | 0.37 | 0.55 | 0.77 | 1.57 | 241.9 |
| SE | 0.11 | 0.34 | 0.086 | 0.42 | 0.092 | 0.089 | 0.040 | 0.0081 | 0.0015 | 0.0056 | 0.08 | 0.10 | 0.19 | 0.54 | |
| Q1-Q32 | 0.30–0.61 | 0.02–0.04 | 0.3–0.5 | 0.4–0.6 |
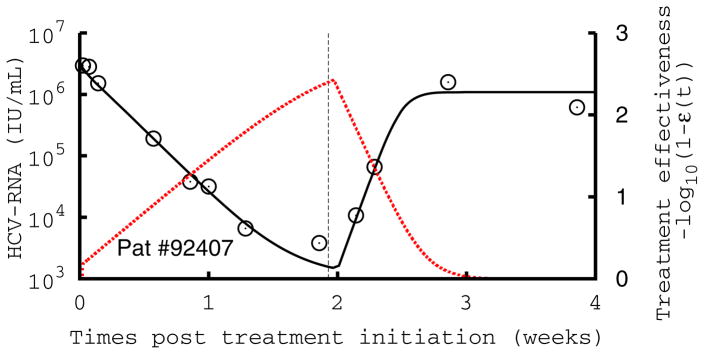
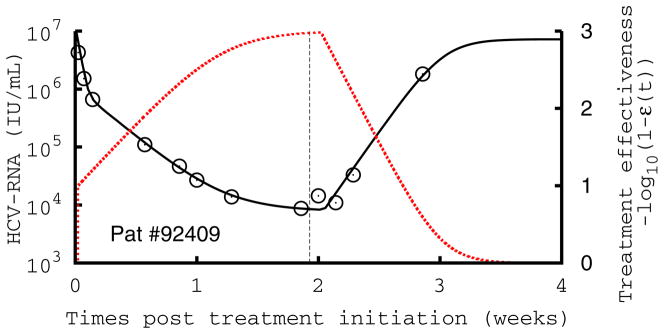
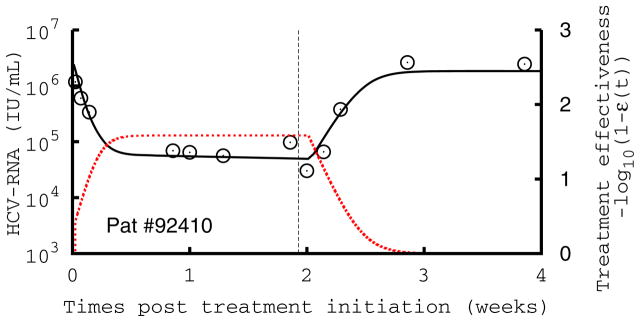
Figure 2.
Comparison of measured HCV RNA (circle) and best-fit prediction by the viral kinetic model during the 14-days of treatment with mericitabine monotherapy followed by 14 days of follow-up in three patients treated with 1500 mg BID. The number in each figure indicates the subject number, and the dotted red line the treatment effectiveness. The vertical line indicates the time of treatment cessation. These patients are displayed because they exhibit a monophasic decline (#92407), a biphasic type of decline with a significant increase in antiviral effectiveness in the first week of treatment (#92409), and a flat second phase of viral decline (#92410), where the increase in antiviral effectiveness is rapid, but flattens out and remains stable during the second phase of antiviral decline. Parameters used are given in Table S1.
In patients that needed additional time to reach high levels of antiviral effectiveness, there was a slower initial rate of viral decline, with 12 patients exhibiting a single phase of monotonic decline, rather than the characteristic two or three phases of viral decline usually observed with IFN or protease inhibitor therapy (14) (Table S1). Figure 2 displays the three patterns of viral kinetics observed in this study: a monophasic decline (Fig. 2A), or a biphasic decline, characterized by a rapid first phase lasting for 1–4 days followed by a slower second phase, with a slope that was either significantly greater than zero (Fig. 2B) or flat (Fig. 2C). The mean value estimated for δ was 0.023 d−1 with no differences across dosing groups (P=0.30)or patterns of decline (P=0.20).
After the dosing period, the viral load returned to its pretreatment value rapidly. We estimated a mean delay, t1, of 0.37 days before treatment effectiveness began declining. In our model, the loss in drug effectiveness was assumed to be exponential with an estimated mean rate, ke, of 0.55 d−1 and 1.20 d−1 in the QD and BID dosing groups, respectively (P=0.0005), with the rate being significantly larger in the 1500 mg BID group than in the 750 mg BID group (ke=1.57 d−1 vs 0.77 d−1, P=0.015).
Discussion
All patients treated with mericitabine were characterized by a relatively slow viral decline in the first 4 days of treatment compared to rates previously observed in treatment naïve patients during daily IFN-based therapy (15) and during therapy with NS3 inhibitors (17), non-nucleoside NS5B inhibitors (23) or NS5A inhibitors (24). However, monotherapy with these agents are limited due to the rapid emergence of viral resistance, which was not observed following 14 days of mericitabine. Whereas patients treated with these other classes of agents typically exhibit a biphasic viral decline (14), about 40% of the patients in our study presented a novel pattern of viral decline, that we identified as “monophasic”, i.e., characterized by a slow first phase of viral decline that extended throughout the 14 days of treatment. We showed that a model assuming mericitabine’s main mode of action was to reduce the rate of virus production, with an effectiveness that increases over time, could describe the data well. The observation that the pyrimidine nucleotide PSI-7977 induces a more rapid first phase of viral decline (25) than mericitabine, even though the active species of PSI-7977, a uridine triphosphate, is the same uridine triphosphate produced by mericitabine (26) suggests that the first phosphorylation may be the step limiting the rapid build up of mericitabine’s antiviral effectiveness (13).
The estimated final treatment effectiveness was strongly associated with the drug regimen, and the BID regimens had a final effectiveness in blocking viral production (mean 750-mg and 1500-mg: 98% and 99.8%, respectively, P=0.018), significantly higher than the QD regimens (mean 99% and 90%, P<10−7). How fast the antiviral effectiveness built up was also drug regimen dependent, and we predicted that 12/16 patients in the BID regimens reached 90% of their final antiviral effectiveness by day 4 (Table S1).
In all patients, the second phase slope of viral decline was modest. This was attributed, in our model, to a low intrinsic rate of loss of infected cells, δ, which might be causally related to the fact that these patients had previously failed IFN-based therapy. However, other interpretations are possible. In models that allows target cell levels to vary, there exists a certain patient-specific antiviral effectiveness level that needs to be exceeded or the virus will not be eliminated (27). In this case, the second phase slope of viral decline will be minimal and will not reflect the loss rate of infected cells (27). From that perspective, the fact that our study population consisted of patients that had previously failed peg-IFN/RBV suggests they may have had a high critical effectiveness, due for instance to more advanced disease with a higher baseline proportion of infected cells (28).
By fitting HCV RNA kinetics after treatment cessation, we could estimate parameters related to the drug pharmacodynamics. We estimated that after drug withdrawal the drug effectiveness, after a delay of 0.37 days, declined with a mean half-life in the QD and BID regimens of 30.2 h and 13.9 h, respectively. Since RG7128 requires intracellular uptake and phosphorylation to two active species, a cytidine triphosphate and a uridine triphosphate, with intracellular half-lives of ~5 h and 38 h, respectively (13), our estimate tends to support that the uridine triphosphate form contributes to maintaining some antiviral effectiveness for a day or two after treatment cessation.
Several assumptions and limitations of our model need to be addressed. The assumption of a gradual increase in antiviral effectiveness that explains the initially slow decrease in viral load still needs to be validated, even though it is supported by the observation that the active forms of mericitabine in vitro take ~48 hours to accumulate to steady-state triphosphate levels (13). It is noteworthy that ribavirin, which needs to be phosphorylated to its mono-, di- and tri-phosphate analogs, when given as monotherapy also induces a monophasic viral decline consistent with the variable effectivenss assumption (29). Second our model does not distinguish between the cytidine and the uridine triphosphates, which could have slightly different potencies and are expected to accumulate at different rates. Third, it is hard to precisely estimate ε1, ε2 and δ as they have overlapping effects on the viral load decline. At least one additional sampling measurement between days 1 and 4 would be necessary to estimate more precisely the initial antiviral effectiveness, ε1. However, the fact that the CE and the VE models provided very similar estimates of ε and δ (see Tables 1 & 2) is an indication that these parameters were precisely estimated, and consequently that infected cell loss/death may be playing a minor role in the overall viral load decline. Lastly, for the sake of parameter identifiability, the target cell level was assumed constant throughout the study period. The kinetics of HCV RNA rebound after the end of treatment may be affected by the increased availability of target cells (30) and hence our estimates of the rate at which antiviral effectiveness decays after the end of treatment may not be as reliable as we would wish.
Recent developments in viral dynamic modeling have emphasized the interplay between the kinetics of intracellular viral RNA (vRNA) and the extracellular viral kinetics measured by serum levels of HCV RNA (31). Within the context of such models it has been shown that the initial rate of decline of serum HCV RNA is proportional to the ability of drug to block the late stages of virion production (i.e., assembly/secretion) (32). If a drug does not block virion assembly/secretion, there may be release of preformed virions during the first phase of viral decline that masks the intrinsic plasma HCV clearance rate (32). Thus the slow initial viral decline observed with mericitabine may reflect that blocking NS5B has only a minimal effect on blocking virus assembly/secretion into the circulation. However, even if a minimal effect in blocking virus assembly/secretion is taken into account using a model that incorporates intracellular events (32), a gradual decrease in the virus production rate, as in the VE model, is still required to fit the data (not shown). On the other hand the action of the RNA dependent RNA polymerase is needed for the production of negative-strand RNA intermediates and thus mericitabine may block the formation of replication complexes (RC). Guedj and Neumann (31) proposed a model that takes into account the kinetics of RC and showed that blocking RC formation leads to the progressive depletion of RC with a rapidity that depends on the level of blockage and the intrinsic turnover rate of RC. However, if only extracellular levels of virus are measured, the contribution of this mechanism to the progressive reduction in virus production can not be distinguished from an increase in antiviral effectiveness.
How the viral kinetics results in the present hard-to treat patients would translate to treatment naïve patients is unknown, as are the effects to be seen when mericitabine is given in combination with peg-IFN/RBV or other direct-acting antivirals. However, interim results from a large cohort of treatment naïve patients receiving mericitabine and peg-IFN/RBV (PROPEL study) showed that >80% of patients had undetectable HCV RNA in all cohorts receiving the 12 week triple regimen (33). Further, 91% of genotype 1 and 4 patients receiving 1000 mg mericitabine BID and Peg/RBV for 24 weeks had undetectable HCV RNA in the JUMP-C trial (34).
Because nucleoside analogues appear to have a high barrier to resistance, they are very attractive as part of IFN-free combination therapy. Indeed, when mericitabine was combined with the HCV protease inhibitor danoprevir for 13 days, a 5-log decrease in HCV RNA was achieved, in the highest dosed treatment arms (35). During monotherapy with NS3/4 protease inhibitors early treatment-resistance viral strains rapidly emerge that lead to viral rebound (8). Nonetheless, when danoprevir was used in combination with mericitabine or Peg/RBV the viral kinetics were similar (35) with no evidence of treatment-emergent resistance. This may indicate that the viral kinetics of protease-inhibitor based combination regimens may be primarily driven by the more potent protease inhibitor, with mericitabine or Peg/RBV acting to prevent the emergence of protease resistance.
To summarize, our viral dynamic analysis predicts that mericitabine administered BID achieves a high (final) antiviral effectiveness of 0.98 or greater. Our prediction of a high antiviral effectiveness together with the lack of clinical resistance to mericitabine (12, 34, 35) support the idea that mericitabine administed BID offers a valuable candidate for IFN-sparing DAA combination regimens.
Supplementary Material
Figure S1. Comparison of measured HCV RNA (circle) and best-fit prediction by the viral kinetic model during the 14-days of treatment with mericitabine followed by 14 days of follow-up. Patients were dosed at (a) 750 mg every day (750QD), (b) 1500 mg every day (1500 QD), (c) 750 mg every 12 h (750 BID), (d) 1500 mg every 12 h (1500 BID). The number above each figure indicates subject number. The vertical line indicates the time of treatment cessation.
a) 750 QD
b) 1500 QD
c) 750 BID
d) 1500 BID
Table S1 Individual parameters
Acknowledgments
This work was performed under the auspices of the U.S. Department of Energy under contract DE-AC52-06NA25396, and supported by NIH grants RR006555, P20-RR018754, AI065256 and AI028433, by the National Science Foundation under Grant No. NSF PHY05-51164 and by the University of Illinois Walter Payton Liver Center GUILD.
Footnotes
SE: standard error
Q1-Q3: first and third quartiles of the distribution of the individual predicted parameters
Author Contributions: JG, HD, ASP made the analysis, ES and PS provided the data, all authors wrote the manuscript
References
- 1.World Health Organization. Hepatitis C Fact sheet No 164. Revised October 2000. http://www.who.int/mediacentre/factsheets/fs164/en/index.html.
- 2.Cardoso AC, Moucari R, Figueiredo-Mendes C, Ripault MP, Giuily N, Castelnau C, Boyer N, et al. Impact of peginterferon and ribavirin therapy on hepatocellular carcinoma: incidence and survival in hepatitis C patients with advanced fibrosis. J Hepatol. 2010;52:652–657. doi: 10.1016/j.jhep.2009.12.028. [DOI] [PubMed] [Google Scholar]
- 3.Awad T, Thorlund K, Hauser G, Mabrouk M, Gluud C. Peginterferon alpha-2a is associated with higher sustained virological response than peginterferon alfa-2b in chronic hepatitis C: a systematic review of randomized trials. Hepatology. 2010;51:1176–1184. doi: 10.1002/hep.23504. [DOI] [PubMed] [Google Scholar]
- 4.McHutchison JG, Everson GT, Gordon SC, Jacobson IM, Sulkowski M, Kauffman R, McNair L, et al. Telaprevir with peginterferon and ribavirin for chronic HCV genotype 1 infection. N Engl J Med. 2009;360:1827–1838. doi: 10.1056/NEJMoa0806104. [DOI] [PubMed] [Google Scholar]
- 5.McHutchison J, Manns M, Muir A, Terrault N, Jacobson I, Afdhal N, Heathcote E, et al. Telaprevir for previously treated chronic HCV infection. N Engl J Med. 2010;362:1292. doi: 10.1056/NEJMoa0908014. [DOI] [PubMed] [Google Scholar]
- 6.Bacon BR, Gordon SC, Lawitz E, Marcellin P, Vierling JM, Zeuzem S, Poordad F, et al. Boceprevir for previously treated chronic HCV genotype 1 infection. N Engl J Med. 2011;364:1207–1217. doi: 10.1056/NEJMoa1009482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Poordad F, McCone J, Jr, Bacon BR, Bruno S, Manns MP, Sulkowski MS, Jacobson IM, et al. Boceprevir for untreated chronic HCV genotype 1 infection. N Engl J Med. 2011;364:1195–1206. doi: 10.1056/NEJMoa1010494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pawlotsky JM. Treatment failure and resistance with direct acting antiviral drugs against hepatitis C virus. Hepatology. 2011;53:1742–1751. doi: 10.1002/hep.24262. [DOI] [PubMed] [Google Scholar]
- 9.Reddy R, Rodriguez-Torres M, Gane E, Robson R, Lalezari J, Everson GT, DeJesus E, et al. Antiviral activity, pharmacokinetics, safety, and tolerability of R7128, a novel nucleoside HCV RNA polymerase inhibitor, following multiple, ascending, oral doses in patients with HCV genotype 1 infection who have failed prior interferon therapy. Hepatology. 2007;46 (Suppl 1):862A. [Google Scholar]
- 10.Gane EJ, Rodriguez-Torres M, Nelson DR, Jacobson IM, McHutchison JG, Jeffers L, Beard A, et al. Antiviral activity of the HCV nucleoside polymerase inhibitor R7128 in HCV genotype 2 and 3 prior non-responders: interim results of R7128 1500mg BID with PEG-IFN and ribavirin for 28 days. Hepatology. 2008;48:1024A. [Google Scholar]
- 11.Rodriguez-Torres M, Lalezari J, Gane EJ, De Jesus E, Nelson DR, Everson GT, Jacobsen I, et al. Potent antiviral response to the HCV nucleoside polymerase inhibitor R7128 for 28 days with peg-IFN and ribavirin: subanalysis by race/ethnicity, weight, and HCV genotype. Hepatology. 2008;48:1160A. [Google Scholar]
- 12.Le Pogam S, Seshaadri A, Ewing A, Kang H, Kosaka A, Yan J, Berrey M, et al. RG7128 alone or in combination with pegylated interferon- 2a and ribavirin prevents hepatitis C virus (HCV) replication and selection of resistant variants in HCV-infected patients. J Infect Dis. 2010;202:1510–1519. doi: 10.1086/656774. [DOI] [PubMed] [Google Scholar]
- 13.Ma H, Jiang WR, Robledo N, Leveque V, Ali S, Lara-Jaime T, Masjedizadeh M, et al. Characterization of the metabolic activation of hepatitis C virus nucleoside inhibitor -d-2 -Deoxy-2 -fluoro-2 -C-methylcytidine (PSI-6130) and identification of a novel active 5 -triphosphate species. J Biol Chem. 2007;282:29812. doi: 10.1074/jbc.M705274200. [DOI] [PubMed] [Google Scholar]
- 14.Guedj J, Rong L, Dahari H, Perelson A. A perspective on modelling hepatitis C virus infection. J Viral Hepat. 2010;17:825–833. doi: 10.1111/j.1365-2893.2010.01348.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Neumann AU, Lam NP, Dahari H, Gretch DR, Wiley TE, Layden TJ, Perelson AS. Hepatitis C viral dynamics in vivo and the antiviral efficacy of interferon-alpha therapy. Science. 1998;282:103–107. doi: 10.1126/science.282.5386.103. [DOI] [PubMed] [Google Scholar]
- 16.Shudo E, Ribeiro RM, Talal AH, Perelson AS. A hepatitis C viral kinetic model that allows for time-varying drug effectiveness. Antivir Ther. 2008;13:919–926. [PubMed] [Google Scholar]
- 17.Guedj J, Perelson AS. HCV RNA declines during antiviral therapy with a second phase rate that increases with drug effectiveness: implications for treatment duration. Hepatology. 2011;53:1801–1808. doi: 10.1002/hep.24272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holford N, Sheiner LB. Kinetics of pharmacologic response. Pharmacol Ther. 1982;16:143–166. doi: 10.1016/0163-7258(82)90051-1. [DOI] [PubMed] [Google Scholar]
- 19.Kuhn E, Lavielle M. Maximum likelihood estimation in nonlinear mixed effects models. Comput Stat Data Anal. 2005;49:1020–1038. [Google Scholar]
- 20.Pinheiro J, Bates D. Mixed-Effects Models in S and S-PLUS. Springer Verlag; 2000. [Google Scholar]
- 21.Adiwijaya B, Hare B, Caron P, Randle J, Neumann A, Reesink H, Zeuzem S, et al. Rapid decrease of wild-type hepatitis C virus on telaprevir treatment. Antivir Ther. 2009;14:591–595. [PubMed] [Google Scholar]
- 22.Akaike H. A new look at the statistical model identification. IEEE Trans Automatic Control. 1974;19:716–723. [Google Scholar]
- 23.Wagner F, Thompson R, Kantaridis C, Simpson P, Troke PJF, Jagannatha S, Neelakantan S, et al. Antiviral activity of the hepatitis C virus polymerase inhibitor filibuvir in genotype 1–infected patients. Hepatology. 2011;54:50–59. doi: 10.1002/hep.24342. [DOI] [PubMed] [Google Scholar]
- 24.Dahari H, Guedj J, Rong L, Nettles RE, Perelson AS. Higher hepatitis C virus (HCV) clearance rates during treatment with direct acting agents compared to interferon-alpha. Hepatology. 2010;52:121A. [Google Scholar]
- 25.Lawitz E, Rodriguez-Torres M, Denning J, Cornpropst M, Clemons D, Mcnair L, Berrey M, et al. Once daily dual-nucleotide combination of PSI-938 and PSI-977 provides 94% HCV RNA<lod at day 14: first purine/pyrimidine clinical combination data (the NUCLEAR study) J Hepatol. 2011;54:S543. [Google Scholar]
- 26.Furman PA, Wang P, Niu C, Bao D, Symonds W, Nagarathnam D, Steuer HM, et al. PSI-7851: a novel liver-targeting nucleotide prodrug for the treatment of hepatitis C. Hepatology. 2009;48:1161A. [Google Scholar]
- 27.Dahari H, Shudo E, Cotler SJ, Layden TJ, Perelson AS. Modelling hepatitis C virus kinetics: the relationship between the infected cell loss rate and the final slope of viral decay. Antivir Ther. 2009;14:459–464. [PMC free article] [PubMed] [Google Scholar]
- 28.Dahari D, Layden-Almer JE, Kallwitz E, Ribeiro RM, Cotler SJ, Layden TJ, Perelson AS. A mathematical model of hepatitis C virus dynamics in patients with high baseline viral loads or advanced liver disease. Gastroenterology. 2009;136:1402–1409. doi: 10.1053/j.gastro.2008.12.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mihm U, Welker M, Teuber G, Wedemeyer H, Berg T, Breske A, Zeuzem S, et al. HCV viral kinetics during ribavirin monotherapy: results of a randomized partially double bling placebo controlled clinical trial. J Hepatol. 2011;54:S185–S185. [Google Scholar]
- 30.Dahari H, Lo A, Ribeiro RM, Perelson AS. Modeling hepatitis C virus dynamics: Liver regeneration and critical drug efficacy. J Theor Biol. 2007;247:371–381. doi: 10.1016/j.jtbi.2007.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guedj J, Neumann AU. Understanding hepatitis C viral dynamics with direct-acting antiviral agents due to the interplay between intracellular replication and cellular infection dynamics. J Theor Biol. 2010;267:330–340. doi: 10.1016/j.jtbi.2010.08.036. [DOI] [PubMed] [Google Scholar]
- 32.Dahari D, Guedj J, Rong L, Nettles RE, Cotler SJ, Layden JE, Perelson AS. New insights into the mechanisms of action of interferon-alpha and BMS-790052: a multi-scale mathematical modeling approach J. Hepatol. 2011;54:S474. [Google Scholar]
- 33.Jensen D, Wedemeyer H, Herring R, Ferenci P, Ma M, Zeuzem S. High rates of early viral response, promising safety profile and lack of resistance-related breakthrough in HCV GT 1/4 patients treated with RG7128 plus PegIFN alfa-2a (40KD)/RBV: planned week 12 interim analysis from the PROPEL study. Hepatology. 2010;52:A81. [Google Scholar]
- 34.Pockros PJ, Jensen D, Tsai N, Taylor RM, Ramji A, Cooper C, Dickson R, et al. First SVR data with the nucleoside analogue polymerase inhibitor mericitabine (RG7128) combined with peginterferon/ribavirin in treatment-naive HCV G1/4 patients: interim analysis from the JUMP-C trial. J Hepatol. 2011;54:S538. [Google Scholar]
- 35.Gane E, Roberts S, Stedman C, Angus P, Ritchie B, Elston R, Ipe D, et al. Oral combination therapy with a nucleoside polymerase inhibitor (RG7128) and danoprevir for chronic hepatitis C genotype 1 infection (INFORM-1): a randomised, double-blind, placebo-controlled, dose-escalation trial. The Lancet. 2010;376:1467–1475. doi: 10.1016/S0140-6736(10)61384-0. [DOI] [PubMed] [Google Scholar]
- 36.Guedj J, Thiébaut R, Commenges D. Maximum likelihood estimation in dynamical models of HIV. Biometrics. 2007;63:1198–1206. doi: 10.1111/j.1541-0420.2007.00812.x. [DOI] [PubMed] [Google Scholar]
- 37.Wu H, Ding AA, DeGruttola V. Estimation of HIV dynamic parameters. Statistics in medicine. 1998;17:2463–2485. doi: 10.1002/(sici)1097-0258(19981115)17:21<2463::aid-sim939>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 38.Samson A, Lavielle M, Mentre F. Extension of the SAEM algorithm to leftcensored data in nonlinear mixed-effects model: Application to HIV dynamics model. Comput Statist Data Anal. 2006;51:1562–1574. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Comparison of measured HCV RNA (circle) and best-fit prediction by the viral kinetic model during the 14-days of treatment with mericitabine followed by 14 days of follow-up. Patients were dosed at (a) 750 mg every day (750QD), (b) 1500 mg every day (1500 QD), (c) 750 mg every 12 h (750 BID), (d) 1500 mg every 12 h (1500 BID). The number above each figure indicates subject number. The vertical line indicates the time of treatment cessation.
a) 750 QD
b) 1500 QD
c) 750 BID
d) 1500 BID
Table S1 Individual parameters