Background: RNA interference (RNAi) is a useful tool to know the function of a gene in a cell under any kind of stress.
Results: RNAi-mediated knockdown of genes in dendritic cells identified unreported genes and pathways that regulate its various functions during Mycobacterium tuberculosis infection.
Conclusion: The identified genes could be potential targets in drug and vaccine designing.
Significance: Understanding the role of host factors that regulate priming of immune responses is crucial to study host-pathogen interactions.
Keywords: Bacteria, Host-pathogen Interactions, Immunology, Immunosuppression, Infectious Diseases
Abstract
With rising incidence of acquired drug resistance among life-threatening pathogens, alternative approaches to improve therapy and vaccination have taken center stage. To this end, genome-wide and pathway-specific siRNA libraries are being employed increasingly to identify genes that regulate immune responses against a number of pathogens. In this study using calcium and cysteine protease pathway-specific siRNA libraries, we identified genes that play critical roles in modulating diverse functions of dendritic cells (DCs) during Mycobacterium tuberculosis infection. Knockdown of many of these genes in the two pathways resulted in reduced bacterial burden within DCs. These included genes that regulated activation of transcription factors, ubiquitin-specific peptidases, and genes that are involved in autophagy and neddylation. Knockdown of certain genes increased the expression of IL-12p40 and surface densities of costimulatory molecules in an antigen- and receptor-specific manner. Increased IL-12p40 and costimulatory molecules on DCs also promoted the development of Th1 responses from a Th2 inducing antigen. Furthermore, modulation of autophagy and oxidative burst appeared to be one of the mechanisms by which these genes regulated survival of M. tuberculosis within DCs. Although some genes regulated specific responses, others regulated multiple responses that included IL-12 production, T cell priming, as well as intracellular survival of M. tuberculosis. Further dissection of the mechanisms such as neddylation, by which these genes regulate immune responses, would improve our understanding of host parameters that are modulated during M. tuberculosis infection.
Introduction
Tuberculosis poses an ever-increasing risk during one's lifetime (1–4). The specificity breadth and intensity of immune responses to infection by Mycobacterium tuberculosis is dependent on pathogen derived molecular patterns and host responses (5–7). Elucidation of factors involved in mediating immune responses during different stages of infection, namely latent/asymptomatic, active disease and during chemotherapy remains a prerequisite for the effective control of infection both in terms of vaccine development and drug discovery (8). Multiple sets of interactions between the host and the pathogen at all stages of infection are regulated at various levels culminating in differential phenotypic outcomes. Identification of genes that positively and negatively regulate these interactions would identify factors that shape effective responses (9–11).
Among the antigen presenting cells of the immune system, dendritic cells (DCs)3 are the most potent and act as a bridge between the innate and the acquired arm of the immune system (12). This is attributed largely to their ability to stimulate naïve quiescent T cells, thereby initiating a primary immune response. DC subsets colonize, and are recruited to specific tissues immediately following an antigenic insult, where they initiate divergent immune responses. Depending upon the activation status, DCs initiate either inflammatory or regulatory responses that determine whether a pathogen will be cleared or retained, thus, grossly affecting the survival of the host (13, 14). Although macrophages are the preferred hosts for mycobacteria, it is being increasingly recognized that M. tuberculosis infects DCs as well and DCs are crucial to initiate protective immune responses affecting mycobacterial survival in the host (15, 16). Therefore, regulation of DC function in the context of mycobacterial infection is a key area that needs detailed investigation. Mycobacteria target a number of surface receptors on DCs, e.g. the mannose receptor, CD11b (Mac-1), CD11c and DEC-205, TLR2, TLR4, and TLR9 (8, 17, 18). Some of these receptors are employed by macrophages as well.
Recently, RNAi has emerged as an important genomic tool to carryout large-scale functional studies. The use of siRNA libraries against a specific pathway is a powerful technique to study the effect of that pathway on the function of a set of related genes inside a cell (9–11). Two key pathways that are targeted by M. tuberculosis in DCs (and also macrophages) are the calcium pathway that affects the survival and proinflammatory response generation from DCs (19, 20) and the cysteine protease pathway that largely effect antigen processing and presentation to T cells, thereby modulating priming of T cells early on in the infection process (21). Our own work has also highlighted the role of calcium homeostasis in regulating the survival of M. tuberculosis both in vitro and in vivo (22).
Therefore, in the light of the above, in this study, we elucidated the role of genes of these two pathways in modulating DC function with respect to M. tuberculosis infection by employing pathway-specific siRNA libraries. Our results identify a set of as yet unreported genes that are targeted by M. tuberculosis in modulating the activation and function of DCs with respect to cytokine secretion and proinflammatory T cell responses, anti-defense mechanisms such as autophagy, and reactive oxygen species generation.
EXPERIMENTAL PROCEDURES
Animals
All experiments were conducted following approval from the institutional animal ethics committee. Female BALB/c mice 4–6 weeks of age that were kept in pathogen-free environment were used.
Materials
Fluorescence-tagged antibodies against mouse CD80, CD86, CD54, and CD40 were from BD Biosciences. Recombinant mouse GM-CSF was from R&D Systems (Minneapolis, MN). Antibodies to Beclin-1, ATG5, β-actin, superoxide dismutase 1 (SOD1), siRNAs against mouse genes, and Luminol kits for chemiluminescence detection were purchased from Santa Cruz Biotechnology. Control siRNAs from Santa Cruz Biotechnology (catalog no. sc-37007) was used as a nonspecific control. Pathway-specific siRNA libraries for primary screening were from Dharmacon (Lafayette, CO). siRNAs for the secondary screen were procured from Santa Cruz Biotechnology. ELISA kits were from eBioscience (San Diego, CA). Dichloroflourescin diacetate (DCFH-DA) and FITC-tagged Alexa Fluor 488 were obtained from Molecular Probes (Eugene, OR). Recombinant M. tuberculosis antigens Rv2463 and Rv3416 were expressed and purified as described recently (23). TLR2 ligand Pam3Csk4 was purchased from Invivogen (San Diego, CA). The following reagent was obtained through BEI Resources (NIAID, National Institutes of Health; purified lipoarabinomannan (LAM) from Mycobacterium tuberculosis, strain H37Rv, NR-14848).
Generation of DCs
DCs were differentiated with GM-CSF as described previously (23, 24). Briefly, bone marrow from the tibias and femurs of BALB/c mice were flushed out, and lymphocytes and I-A+ cells were depleted following magnet-assisted cell sorting. Cells were cultured in RPMI 1640 medium containing 10% FCS, 0.05 m 2-mercaptoethanol, 1 mm sodium pyruvate plus 15 ng/ml GM-CSF.
Transfection of DCs with siRNA and Stimulation
For siRNA transfections, 4 × 106/ml bone marrow precursors were transfected with 60 pmol of siRNA for 72 h using the Hiperfect transfection reagent (Qiagen) in OPTIMEM medium (Invitrogen). GM-CSF was added 5 h following transfection, and incubation was continued for 72 h for DC differentiation. Knockdown was verified by RT-PCR, following which, cells were stimulated either with 1 μg/ml Pam3Csk4 or 5 μg/ml mannosylated lipoarabinomannan (manLAM) and/or either with 15 μg/ml Rv2463 or Rv3416 for 24 h. For some experiments, siRNA-transfected DCs were infected with M. tuberculosis H37Rv at 2.5 MOI for indicated times. Cells were processed for monitoring colony-forming units (cfu), reactive oxygen species (ROS) measurement or Western blotting as described below.
Flow Cytometry
Cells were stained for the surface levels of CD80, CD86, CD54, and CD40 using FITC-tagged monoclonal antibodies and analyzed by flow cytometry on FACS Calibur (BD Biosciences) as described previously (23). The data were plotted and analyzed using CellQuest Pro software.
T Cell Enrichment and Processing
Mice were immunized intraperitoneally with chicken egg ovalbumin (50 μg/mouse) for 7 days. Mice were sacrificed, and splenic T cells were enriched by magnet-assisted cell sorting as described previously (25). Briefly, following RBC lysis of spleen homogenates, adherent cells were removed by two rounds of panning over plastic plates. Following this, cells were incubated with anti-CD11c, anti-CD11b, anti-I-Ad, and anti-B220 microbeads to remove contaminating DCs, macrophages, MHCII+ cells, and B lymphocytes, respectively. The purity of the enriched T cells was 98%, as ascertained by surface staining with CD90. The percentage of I-A+ cells was 0.05%. T cells were cocultured with siRNA-transfected and ovalbumin-stimulated DCs for 48 h, and cytokines were measured in culture supernatants.
Measurement of Cytokines
Cytokines in the culture supernatants were measured by employing a sandwich ELISA as described previously (23, 24). The samples were diluted to obtain absorbance in the linear range of the standards.
Western Blotting for Signaling Molecules
At the end of incubation, cells were chilled on ice, washed once with ice-cold PBS, and lysed in buffer containing 10 mm HEPES (pH 7.9), 10 mm KCL, 0.1 mm EDTA, 0.1 m EGTA, 0.5% Nonidet P-40, and 2 μg/ml each of aprotinin, leupeptin, and pepstatin. The suspension was centrifuged at 13,000 rpm for 2 min at 4 °C. The supernatant was designated as the cytoplasmic extract. Twenty micrograms of cytoplasmic extract was resolved on 10% SDS-PAGE and subsequently transferred onto nitrocellulose membrane (Hybond C pure, Amersham Biosciences). The blots were then probed with antibodies to various molecules, followed by HRP-labeled secondary antibodies. Furthermore, a parallel set of samples was run separately on SDS-PAGE and probed for β-actin as loading control. The blots were later developed by chemiluminescence using the luminol reagent.
Measurement of Intracellular Reactive Oxygen Species
Intracellular ROS levels were measured by flow cytometry, as described previously, using the redox-sensitive dye DCFH-DA (23, 26). The nonfluorescent DCFH-DA readily diffuses into the cells where it is hydrolyzed to the polar derivative nonfluorescent dichlorofluorescin, which is oxidized in the presence of H2O2 to the highly fluorescent dichlorofluorescein. Thirty minutes prior to the end of each incubation period, 1 × 106 cells/ml were incubated with 10 μm DCFH-DA in the dark. Cells were thoroughly and quickly washed with pulse spin and immediately acquired for analyses in FACSCalibur (BD Biosciences). The data were plotted and analyzed using CellQuest Pro software.
Confocal Microscopy
2 × 106/ml siRNA-transfected DCs were stimulated with manLAM along with Rv2463 for 4 h. Cells were fixed with 2% paraformaldehyde, permeabilized with 0.1% saponin, and incubated with antibodies against Beclin-1 or ATG5 followed by anti-rabbit FITC-tagged Alexa Fluor 488. Cells were again fixed with 4% paraformaldehyde. Confocal imaging was performed with Nikon A1 laser scan confocal microscope with 60× objective magnification, numerical aperture 1.4, refractive index 1.5, Plan Apo optics equipped with an argon laser, using excitation and emission wavelength of 488 and 525, respectively. Data were analyzed using the NIS Elements AR software. Expression levels were quantified as average of sum intensity of each fluorescent field.
Statistics
A two-tailed Student's t test was carried out to obtain p values. Values of p < 0.05 were considered as significant.
RESULTS
Genes of Calcium Calmodulin and Cysteine Protease Pathways Modulate Survival of M. tuberculosis within DCs
First, we employed pathway-specific libraries to identify genes that regulated the survival of M. tuberculosis inside DCs, in keeping with the now accepted view that in addition to macrophages, M. tuberculosis also targets DCs during primary infection (15). Knockdown of a number of genes in either the calcium calmodulin pathway (supplemental Fig. 1) or cysteine protease pathway (supplemental Fig. 2) significantly modulated the survival of M. tuberculosis in DCs. This included an increase as well a decrease in the bacterial burden within DCs. This indicated that the two pathways contain genes that play positive and negative roles in regulating the survival of mycobacteria within DCs, although knockdown of very few genes in the cysteine protease pathway significantly increased intracellular bacterial loads. To proceed further, we chose to characterize the function of genes whose knockdown inhibited the survival of M. tuberculosis inside DCs because understanding the mechanisms by which these genes inhibited host-mediated responses against M. tuberculosis would identify strategies for either vaccine or drug development. These included calcium/calmodulin-dependent protein kinase II α (Camkiia) that regulates activation of transcription factors NF-κB and NF-AT and the activation of the MAPK pathway (27); proviral integration site 2 (Pim2) reported as an anti-apoptotic protein (28). It also included a number of serine threonine kinases such as SNF-related kinase (Snrk) and testis-specific serine kinase 1 (Stk22a), which regulate chromatin remodeling. These also act as a novel substrate for liver kinase B1 (Lkb1) and are involved in the spermatogenesis and hormone regulation in the testis (29). In addition, knockdown of Prkaa2 (protein kinase AMP-activated, α2), which modulates the activation of K+ channels and Ulk1 (unc51-like kinase 1)-mediated autophagy (30), also decreased bacterial burden. As inhibiting these genes resulted in enhanced killing of M. tuberculosis by DCs, this indicated that M. tuberculosis modulates the activity and/or function of these genes for its survival and regulation of immune responses from DCs. Although the role of calcium in mediating the survival of M. tuberculosis has been well studied, the role of these genes in M. tuberculosis infection has not yet been documented.
Similarly, genes in the cysteine protease pathway, whose knockdown resulted in reduced survival of M. tuberculosis included members of the ubiquitin-specific protease family, e.g. Usp25 (ubiquitin-specific peptidase 25) and Usp9y (ubiquitin-specific peptidase 9Y). Other genes that had a similar effect were Uchl1 (ubiquitin carboxyl-terminal hydrolase L1), which modulates free monomeric ubiquitin levels and has been identified as a novel gene for Parkinson disease (31), Lgmn (legumain), another cysteine endopeptidase that regulates apoptosis (32), and Ctsh (cathepsin H), which plays a crucial role in processing and presentation of antigen peptides to T cells (33).
Interestingly, knockdown of SUMO/Senp8 (sentrin-specific peptidase 8), which plays an important role in neddylation and sumoylation (34) had a significant effect on the survival of M. tuberculosis within DCs. A recent report indicated the role of one such ubiquitin protease during M. tuberculosis infection, wherein some of the PE-polymorphic CG-proteins of M. tuberculosis were found to be resistant to these proteases (35). However, their roles in specifically regulating M. tuberculosis survival have not been reported.
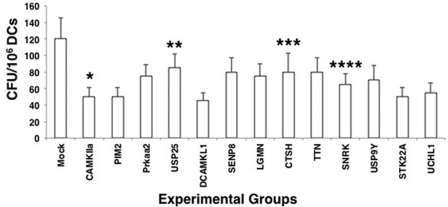
To validate our results, we shortlisted some of the genes from the above two pathways that significantly reduced the survival of M. tuberculosis inside DCs and investigated their ability to modulate M. tuberculosis survival in DCs using siRNAs from a different source. As shown in Fig. 1, knockdown of all of the 13 shortlisted genes reduced the survival of M. tuberculosis by >50%, and therefore, these 13 genes were selected for subsequent experimentation and characterization. Efficiency of knockdown of select genes was ascertained by RT-PCR (supplemental Fig. 3).
FIGURE 1.
Knockdown of select genes of the two pathways in DCs results in decreased survival of M. tuberculosis. DCs were transfected with siRNAs against indicated genes belonging to either the calcium calmodulin pathway or cysteine protease pathway for 72 h, followed by infection with 2.5 MOI M. tuberculosis H37Rv for 48 h. Cell lysates were plated onto 7H11 agar plates, and cfu were monitored after 2–3 weeks. Mock represents knockdown with a control siRNA. Data represent mean ± S.D. of three independent experiments. *, p = 0.012 for Mock versus Camkiia; **, p = 0.017 for MOCK versus Usp25; ***, p = 0.006 for Mock versus Ctsh; and ****, p = 0.016 for Mock versus Snrk. TTN, titin.
DC Genes Differentially Regulate IL-12p40 Production by TLR2 and DC-SIGNR1
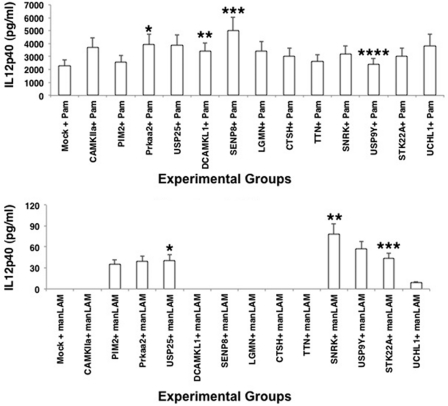
We next carried out a detailed characterization of the functional effects regulated by the validated genes. First, we investigated the ability of these genes to influence the production of IL-12p40 from DCs. We had shown recently that TLR2 induces higher IL-12p40 production in DCs when compared with DC-SIGN (DC-specific ICAM-3 grabbing non-integrin) R1 in the context of M. tuberculosis infection. To this end, we stimulated TLR2 with Pam3Csk4 and DC-SIGNR1 with manLAM as described previously (24). As shown in Fig. 2, knockdown of many genes significantly up-regulated IL-12p40 expression over and above that obtained following TLR2 stimulation. Maximum effects with >2-fold increases were observed upon knockdown of Camkiia, Usp25, Prkaa2, Uchl1, and Senp8. Expectedly, and consistent with our earlier report, DC-SIGNR1 stimulation did not result in any detectable induction of IL-12p40. However, following knockdown of either Pim2 or Prkaa2 or Usp25 or Snrk or Usp9y or Stk22a, a significant induction of IL-12p40, albeit to different levels, was now observed (Fig. 2). These results indicated that the genes that negatively regulated survival of M. tuberculosis also negatively regulated IL-12p40 production, more significantly following stimulation of DC-SIGNR1. Furthermore, some genes such as Usp25 and Snrk mediated IL-12 increase from both receptors, whereas others displayed a receptor specific effect, indicating compartmentalization of functions.
FIGURE 2.
Knockdown of specific genes results in enhanced IL-12p40 expression from TLR2 and DC-SIGNR1. DCs were transfected with siRNAs against specific genes as in Fig. 1, followed by stimulation with either 1 μg/ml Pam3Csk4 (Pam) or 5 μg/ml manLAM for 24 h. Mock represents knockdown with a control siRNA. IL-12p40 levels in culture supernatants were measured by ELISA. Bars represent mean ± S.D. of three independent experiments. Upper panel, *, p = 0.012 for Mock versus Prkaa2; **, p = 0.006 for Mock versus Dcamkl1; ***, p = 0.013 for Mock versus Senp8; ****, p = 0.03 for Mock versus Usp9y. Lower panel, *, p = 0.016 for Mock versus Usp25; **, p = 0.01 for Mock versus Snrk; ***, p = 0.009 for Mock versus Stk22a.
Specific Genes Modulate M. tuberculosis Antigen-mediated IL-12p40 Production
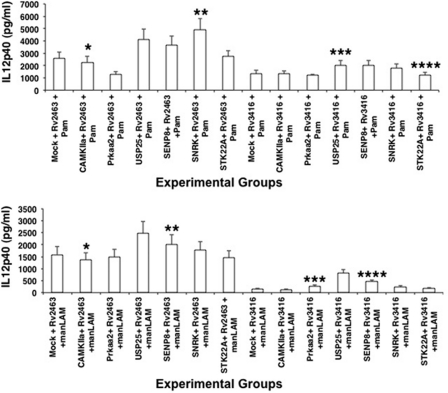
We recently reported the enrichment and functional characterization of M. tuberculosis genes expressed inside macrophages as a function of infection and time. These genes were named as day1 and day5 antigens, based on their expression patterns at 24 and 120 h post-infection, respectively (23). Characterization of these antigens revealed their role in suppression of TLR2 and M. tuberculosis mediated IL-12p40 production, M. tuberculosis mediated expression of surface T cell costimulatory and MHC molecules, surface expression of cytokine receptors, generation of ROS and antigen-specific T cell responses. Furthermore, overexpression of these day1 and day5 antigens also resulted in increased survival of M. tuberculosis inside DCs as well as macrophages. In light of the above results, we therefore investigated the role of the host genes identified in the present study in mediating the effects of the day1 and day5 antigens in some of the above mentioned responses. To this end, we knocked down genes in DCs followed by stimulation with a day1 (Rv2463) or a day5 (Rv3416) antigen along with costimulation of TLR2 and DC-SIGNR1. Levels of IL-12p40 were scored in the supernatants. As shown in Fig. 3, knockdown of genes such as Usp25, Senp8, and Snrk significantly increased the levels of IL-12p40 expression following stimulation of TLR2 and DC-SIGNR1 in the presence of day1 and day5 antigens. In fact, the increase in the levels was more significant with the day1 antigen when compared with day5 antigen, thus indicating a role of these genes in modulating early responses. This also indicated an antigen specific effect of these genes in mediating IL-12p40 production by TLR2. In contrast, no significant up-regulation of IL-12p40 was observed following knockdown of Camkiia, Prkaa2, and Stk22a. These results indicate a differential role for these genes that is both antigen- and receptor-specific. Furthermore, the fact that the increase was observed in the context of day1 antigen stimulation also indicates that these genes play a negative role in mediating early priming of Th1 responses.
FIGURE 3.
Day1 and day5 M. tuberculosis antigens differentially modulate IL-12p40 expression in pathway-specific gene-silenced DCs. Indicated genes were knocked down in DCs followed by stimulation with either 15 μg/ml Rv2463 or Rv3416 for 24 h. Cells were later stimulated with either 1 μg/ml Pam3Csk4 (Pam) or 5 μg/ml manLAM for 24 h. Culture supernatants were screened for the levels of IL-12p40. Mock represents knockdown with a control siRNA. Data represent mean ± S.D. of three independent experiments. Upper panel, *, p = 0.004 for Mock+Rv2463+Pam3Csk4 versus Camkiia+Rv2463+Pam3Csk4; **, p = 0.01 for Mock+Rv2463+Pam3Csk4 versus Snrk+Rv2463+Pam3Csk4; ***, p = 0.017 for Mock+Rv3416+Pam3Csk4 versus Usp25+Rv3416+Pam3Csk4; ****, p = 0.1 for Mock+Rv3416+Pam3Csk4 versus Stk22a+Rv3416+Pam3Csk4. Lower panel, *, p = 0.16 for Mock+Rv2463+manLAM versus Camkiia+Rv2463+manLAM; **, p = 0.009 for Mock+Rv2463+manLAM versus Senp8+Rv2463+manLAM; ***, p = 0.012 for MOCK+Rv3416+manLAM versus Prkaa2+Rv3416+manLAM; ****, p = 0.004 for MOCK+Rv3416+manLAM versus Senp8+Rv3416+manLAM.
Modulation of Costimulatory Molecule Expression by Host Genes
A primary function of a DC is to initiate T cell responses, during which the surface densities of costimulatory molecules play a determinant role. We therefore investigated the ability of three genes (based on their ability to significantly modulate IL-12p40 responses) to modulate the surface densities of key costimulatory molecules on DCs. Our results indicate that these genes differentially influenced the surface densities of key costimulatory molecules. For example, as shown in Table 1, knockdown of Snrk increased the expression of CD80 following stimulation with Rv2463 (day1 antigen) in the context of TLR2 but not DC-SIGNR1. However, no significant changes were observed with Rv3416 (day5 antigen) in the context of either TLR2 or DC-SIGNR1. Similarly, knockdown of Usp25 resulted in increased expression of CD80 and CD54 following stimulation with Rv3416 and DC-SIGNR1. Similar to Usp25, knockdown of Senp8 showed a similar pattern with increased expression of all the costimulatory molecules (CD80, CD86, CD40) with maximum increase in the levels of CD54 following stimulation with Rv3416 and DC-SIGNR1.
TABLE 1.
Knockdown of specific genes modulates surface densities of costimulatory molecules on DCs
The indicated genes were knocked down in DCs with specific siRNAs followed by stimulation with either Pam3Csk4 (Pam) or manLAM along with Rv2463 or Rv3416, as indicated. The surface levels of indicated markers were monitored by flow cytometry. Data are represented as increase (+) or decrease (−) over that observed in control siRNA transfected and similarly stimulated DCs. Number of + or − indicates levels of increase or decrease, respectively. Data are representative of two independent experiments.
| Gene/stimulation | CD80 | CD86 | CD40 | CD54 |
|---|---|---|---|---|
| SNRK | ||||
| Rv2463+Pam | ++ | +/− | +/− | +/− |
| Rv2463+manLAM | +/− | +/− | +/− | +/− |
| Rv3416+Pam | +/− | +/− | +/− | +/− |
| Rv3416+manLAM | + | + | + | +/− |
| USP25 | ||||
| Rv2463+Pam | +/− | +/− | +/− | +/− |
| Rv2463+manLAM | + | +/− | +/− | +/− |
| Rv3416+Pam | − | +/− | +/− | + |
| Rv3416+manLAM | ++ | + | + | +++ |
| SENP8 | ||||
| Rv2463+Pam | + | +/− | +/− | +/− |
| Rv2463+manLAM | +/− | +/− | +/− | +/− |
| Rv3416+Pam | − | +/− | +/− | +/− |
| Rv3416+manLAM | + | + | + | +++ |
Furthermore, the three genes showed an antigen- and receptor-specific modulation of costimulatory molecules. Typically, stimulation of DC-SIGNR1 in the context of Rv3416 a day5 antigen was more effective in increasing the expression of most costimulatory molecules following knockdown of different genes when compared with similar stimulation of TLR2. This indicated that early priming of T cells by the day1 antigen (Rv2463) was regulated by these genes via modulation of IL-12 (as observed in Fig. 3); the late or secondary priming of T cells by the day5 antigen (Rv3416) was regulated at the level of costimulatory molecules by these genes. These results further indicate a role for these genes in influencing immune responses as a function of time and receptor triggering. These observations are consistent with the documentation that during M. tuberculosis infections, TLR2 triggering occurs early on in the infection process and is followed by stimulation of DC-SIGN by soluble manLAM secreted by infected macrophages as the infection proceeds with time (36).
Knockdown of Genes Induce Th1 Responses by Th2-promoting Antigen
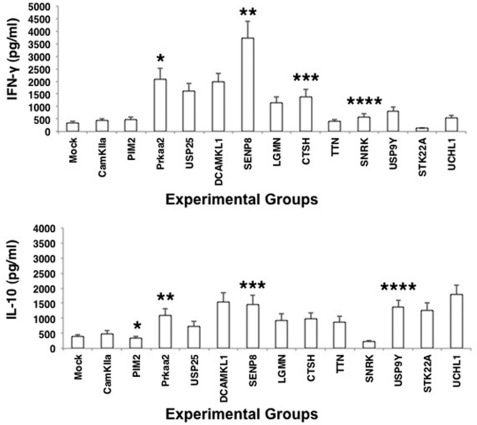
Keeping the above results in mind, we next investigated the quality of T cell responses from ovalbumin-stimulated DCs following knockdown of the genes. Ovalbumin is an antigen that is known to induce Th2 responses and is a typical antigen widely used in mouse models of asthma (37). Because a potent Th1 response is a prerequisite and a marker for protective responses during M. tuberculosis infections, we investigated whether knockdown of the genes would skew the ovalbumin-induced Th2 response to a Th1 response. To this end, genes were knocked down in DCs followed by stimulation with ovalbumin for 24 h. The DCs were then co-cultured with T cells enriched from ovalbumin-primed mice. The levels of IFN-γ and IL-10 were scored in the supernatants 48 h later. As shown in Fig. 4, ovalbumin induced a typical Th2 response with higher levels of IL-10 when compared with IFN-γ. However, knockdown of Prkaa2, Senp8, Dcamkl1, and Usp25 (and to some extent knockdown of Snrk and Ctsh) significantly skewed the Th2 response to a Th1 response by increasing the ratio of IFN-γ over IL-10. However, knockdown of Camkiia, Pim2, Ttn, and Uchl1 had no significant effect, whereas knockdown of Stk22a had a marginal decrease in IFN-γ levels, indicating that not all genes have the ability to influence all responses. Nevertheless, these results were consistent with the role of these genes in influencing IL-12p40 levels from TLR2 and DC-SIGNR1. The results further indicated that these genes played a negative role in the priming of Th1 responses from infected or antigen-stimulated DCs during M. tuberculosis infections and inhibiting these genes could indeed potentiate Th1 responses.
FIGURE 4.
Knockdown of Prkaa2, Usp25, and Senp8 induces Th1 responses from ovalbumin-stimulated DCs. Indicated genes were knockdown in DCs followed by stimulation with 15 μg/ml ovalbumin for 24 h followed by co-culture for 48 h with T cells enriched from ovalbumin-primed mice. Culture supernatants were screened for the levels of IFN-γ and IL-10. Mock represents knockdown with a control siRNA. Data represent the mean ± S.D. of three independent experiments. Upper panel, *, p = 0.013 for Mock versus Prkaa2; **, p = 0.01 for Mock versus Senp8; ***, p = 0.013 for Mock versus Ctsh; ****, p = 0.014 for Mock versus Snrk. Lower panel, *, p = 0.012 for Mock versus Pim2; **, p = 0.02 for Mock versus Prkaa2; ***, p = 0.014 for Mock versus Senp8; ****, p = 0.008 for Mock versus Usp9y.
Genes of Calcium and Cysteine Protease Pathways Increase Autophagy in DCs
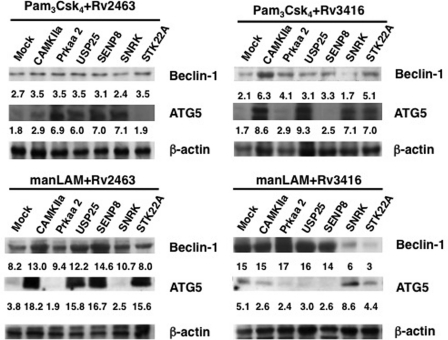
Of late, autophagy has emerged as a key defense mechanism employed by infected cells to clear intracellular pathogens (38, 39). A recent report elucidated the role of autophagy during M. tuberculosis infection, wherein promoting autophagy by rapamycin increased the vaccine potential of Bacillus Calmette-Guerin (40). In addition, as mentioned above, Prkaa2 is reported to modulate autophagic responses (30). Therefore, to investigate whether knockdown of the genes would modulate autophagic responses in DCs to regulate M. tuberculosis survival, we monitored the expression levels of some autophagic markers. Following silencing with specific genes, we stimulated either TLR2 or DC-SIGNR1 along with either the day1 or day5 antigen and scored for the expression levels of ATG5 and Beclin-1. As shown in Fig. 5, knockdown of Camkiia, Prkaa2, Usp25, Senp8, or Stk22a followed by stimulation with TLR2 and day1 antigen, resulted in increased expression of Beclin-1, whereas the expression of ATG5 was increased following knockdown of Prkaa2, Usp25, Senp8, and Snrk, and marginally by Camkiia but not by Stk22a. Interestingly, knockdown of the above genes followed by stimulation of TLR2 along with the day5 antigen showed a different pattern. Although knockdown of only Camkiia or Stk22a significantly increased the expression of both Beclin-1 and ATG5, knockdown of the other genes had specific effects on either one of the autophagic markers. For example, although knockdown of Prkaa2 increased the expression of only Beclin-1 with no effect on the expression of ATG5, a reverse pattern for observed in the case of knockdown of either Usp25 and Snrk, wherein no significant changes were observed with respect to Beclin-1 levels, but a significant increase in the levels of ATG5 were observed. Similarly, knockdown of all genes (except Stk22a) increased the expression levels of Beclin-1 following by stimulation with DC-SIGNR1 along with the day1 antigen. On the other hand, knockdown of only four genes, namely Camkiia, Usp25, Senp8, and Stk22a, significantly increased the expression of ATG5.
FIGURE 5.
Knockdown of genes induces autophagy from TLR2 and DC-SIGNR1. DCs were transfected with siRNAs against indicated genes and later stimulated with either 15 μg/ml Rv2463 or Rv3416 for 24 h, followed by stimulation with either 1 μg/ml Pam3Csk4 (Pam) or 5 μg/ml manLAM for 24 h. Cytoplasmic extracts were Western blotted for Beclin-1 and ATG5 expression. Mock represents knockdown with a control siRNA. Numbers below the Beclin-1 or ATG5 blots indicate relative intensities of the bands. Data from one of three experiments are shown.
In contrast, stimulation of DC-SIGNR1 and the day5 antigen either had no effect on Beclin-1 expression following knockdown of most genes, whereas its levels were reduced below basal levels following knockdown of Snrk and Stk22a. Interestingly, the knockdown of only Snrk increased ATG5 levels, with no effects upon knockdown of the other genes. These results reiterate the receptor- and antigen-specific effects in the regulation of autophagy by these genes. For example, a reverse effect of Snrk was observed in the case of stimulation of DC-SIGNR1 with the day5 antigen, wherein a polar effect was evident with respect to regulation of the expression of two genes in the autophagic pathway. Similar opposite effects were seen in the case of Stk22a with respect to ATG5 expression in the context of TLR2 stimulation along with the day1 and the day5 antigen. Taken together, the results in Fig. 5, display an interesting pattern, wherein specific genes in the calcium and cysteine protease pathway regulate the expression levels of different genes in the autophagic pathway, depending on the type of receptor that is stimulated by diverse antigens. Overall, the knockdown of these genes had a positive effect on autophagy, albeit at different levels, indicating a net negative role during mounting of autophagic responses during M. tuberculosis infection. This could reflect the redundancy that is observed for most processes in biological systems following a stress response in the form of pathogenic insult or abiotic pressures.
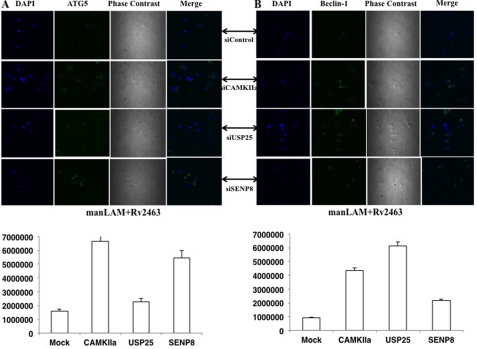
We also monitored the expression of Beclin-1 and ATG5 in select groups by confocal microscopy. To this end, we selected Camkiia, Usp25, and Senp8 because knockdown of these genes significantly increased the expression of both Beclin-1 and ATG5 following stimulation of DC-SIGNR1 with the day1 antigen. As shown in Fig. 6, and consistent with the results in Fig. 5, knockdown of either Camkiia or Senp8 or Usp25 maximally increased the expression of Beclin-1 upon costimulation of day1 antigen (Rv2463) with DC-SIGNR1.
FIGURE 6.
Knockdown of Camkiia or Usp25 or Senp8 increases antigen- and receptor-mediated Beclin-1 and ATG5 expression. A, DCs were transfected with siRNAs against the indicated genes, followed by costimulation with 5 μg/ml manLAM and 15 μg/ml Rv2463 for 4 h. Cells were incubated with antibody to Beclin-1 (A) and followed by staining with anti-rabbit FITC-tagged Alexa Fluor 488 (B). Images were captured under a confocal microscope. Bars below each panel represent the expression levels quantified as average of sum intensity of each fluorescent point by using NIS Elements AR software. siControl represents knockdown with a control siRNA. Data from one of three experiments are shown.
Key Genes in Two Pathways Modulate Oxidative Burst in DCs
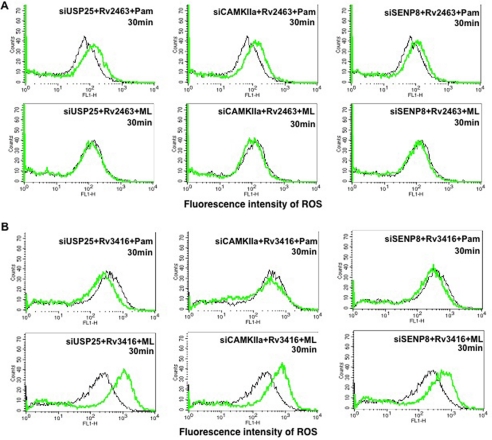
We had earlier reported the role of ROS in mediating the survival of mycobacteria inside DCs (26). Furthermore, we had established a positive correlation between intracellular calcium influx and ROS generation in DCs. Increased intracellular calcium and ROS levels reduced the survival of mycobacteria in DCs (26). Therefore, we monitored ROS levels following knockdown of a few genes followed by specific antigen and receptor stimulation. As shown in Fig. 7, an interesting pattern was observed with respect to modulation of ROS by antigens and receptors. In the case of day1 antigen (Rv2463), knockdown of the three genes increased ROS levels by TLR2, whereas a reverse effect was observed upon stimulation of DC-SIGNR1 (Fig. 7A). In contrast, in the case of the day5 antigen (Rv3416), knockdown of the three genes increased ROS levels by DC-SIGNR1, whereas a reverse effect was observed upon stimulation of TLR2 (Fig. 7B). These results indicate that with the early antigen (Rv2463), these genes played a negative role in ROS induction from TLR2, whereas a positive role was observed following DC-SIGNR1 stimulation. Conversely, with the late antigen (Rv3416), these genes played a positive role in ROS induction from TLR2 and a negative role upon DC-SIGNR1 stimulation. A similar pattern was obtained following knockdown of Snrk and Stk22a and to an extent with Prkaa2 (supplemental Fig. 4). These results also point to a differential activation status of these genes during a time-dependent antigenic stimulation of different receptors. Furthermore, these results add support to our earlier report wherein we proposed a complementary role for these antigens as a function of infection and time that constantly work toward keeping immune responses suppressed (23). These results indicated that modulating the oxidative burst could be one of the mechanisms by which these genes regulate survival of M. tuberculosis in DCs.
FIGURE 7.
Day1 and day5 antigens differentially regulate ROS generation in gene-silenced DCs upon stimulation of TLR2 or DC-SIGNR1. DCs were transfected with siRNAs against indicated genes, followed by stimulation with either Rv2463 (A) or Rv3416 (B) along with either 1 μg/ml Pam3Csk4 (Pam) or 5 μg/ml manLAM (ML) for the indicated times. 30 min prior to the incubation period, cells were loaded with 10 μm DCFH-DA. At the end of the incubation period, cells were quickly and thoroughly washed with culture medium and immediately analyzed for ROS levels by flow cytometry. Mock represents transfection with control siRNAs. Black lines depict ROS levels in DCs transfected with control siRNAs. Green lines depict DCs transfected with gene-specific siRNAs. Data from one of three experiments are shown.
Key Genes Regulate Autophagy in M. tuberculosis-infected DCs
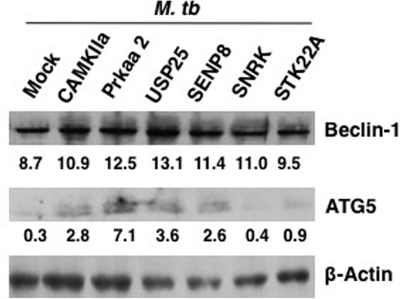
We extended the above observations with infection of DCs with live virulent M. tuberculosis. Specific genes were knocked down with specific siRNAs followed by infection with M. tuberculosis H37Rv for 24 h. First, we monitored the effect of knockdown of five genes in modulating autophagic responses in M. tuberculosis-infected DCs. As shown in Fig. 8, knockdown of Camkiia, Prkaa2, Usp25, Senp8, and Snrk increased the levels of Beclin-1, whereas knockdown of Stk22a had a marginal effect. On the other hand, knockdown of Camkiia, Prkaa2, Usp25, and Senp8 (to an extent Stk22a) significantly increased ATG5 expression in M. tuberculosis infected DCs, whereas knockdown of SNRK had no effect. The fact that the increase in the levels of Beclin-1 was not as dramatic as observed following stimulation with day1 or day5 antigens, and TLR2 or DC-SIGNR1 could be attributed to the fact that during M. tuberculosis infection multiple receptors would be stimulated with multiple surface antigens, in contrast to stimulation of specific receptors (TLR2 or DC-SIGNR1) with specific antigens (Rv2463 or Rv3416). The cross-talk between signals emanating from multiple receptors would indeed have bearings on the levels of gene expression of the autophagic pathway. Nevertheless, the fact that there was an increase in the levels of both Beclin-1 and ATG5 by one gene or the other indicates that these genes played a net negative role in regulating autophagy during M. tuberculosis infection.
FIGURE 8.
Knockdown of select genes induces autophagy in M. tuberculosis-infected DCs. DCs were transfected with siRNAs against indicated genes, followed by infection with 2.5 MOI M. tuberculosis (M. tb) H37Rv for 24 h. Cytoplasmic extracts were Western blotted for Beclin-1 and ATG5 expression. MOCK represents knockdown with a control siRNA. Numbers below the Beclin-1 or ATG5 blots indicate relative intensities of the bands. Data from one of two experiments are shown.
Key Genes Regulate ROS Generation and SOD1 Levels in M. tuberculosis-infected DCs
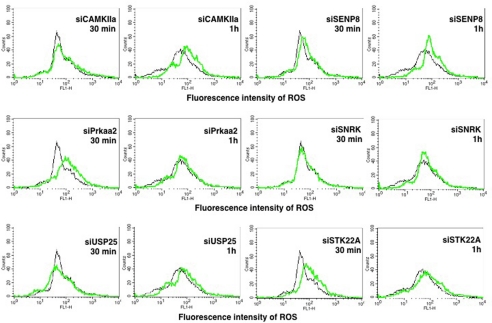
We next monitored the levels of ROS in M. tuberculosis DCs following knockdown of the above genes. As shown in Fig. 9, individual knockdown of all of the genes increased ROS levels in infected cells albeit with different kinetics and to different extents. Although the data in Fig. 7 indicated that an increase in ROS levels upon stimulation of either TLR2 or DC-SIGNR1 with a day1 or the day5 antigen was observed within 30 min, the kinetics of increase in ROS levels were different with respect to M. tuberculosis infection. Although knockdown of Senp8 or Camkiia or Usp25 increased ROS levels 1-h post infection, knockdown of Prkaa2 or Stk22a increased ROS levels within 30 min. On the other hand, knockdown of Snrk had no significant effect. These results once again reiterate that these genes played a negative role in regulating oxidative burst during M. tuberculosis infection, and the delayed kinetics observed in some genes could be attributed to stimulation of multiple receptors as against individual receptors as argued above for autophagy.
FIGURE 9.
Knockdown of select gens enhances ROS generation in M. tuberculosis-infected DCs. DCs were transfected with siRNAs against indicated genes and later infected with 2.5 MOI M. tuberculosis H37Rv for indicated times. 30 min prior to the incubation period, cells were loaded with 10 μm DCFH-DA. At the end of the incubation period, cells were quickly and thoroughly washed with culture medium and fixed with 4% paraformaldehyde for 2 h. Cells were washed once again with culture medium and immediately analyzed for ROS levels by flow cytometry. Mock represents transfection with control siRNAs. Black lines depict ROS levels in DCs transfected with control siRNAs. Green lines depict DCs transfected with gene-specific siRNAs. Data from one of two experiments are shown.
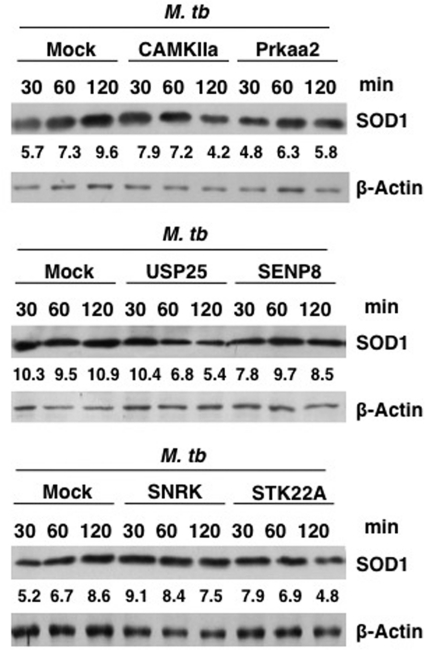
Because generation of ROS is often associated with modulations in the levels of the ROS quencher SOD1, we monitored SOD1 levels in M. tuberculosis-infected DCs following knockdown of the genes. As shown in Fig. 10, infection with M. tuberculosis increased SOD1 levels with time, indicating down-modulation of oxidative burst. However, knockdown of most genes resulted in a significant decrease in SOD1 levels. With respect to the MOCK control, knockdown of Camkiia or Usp25 resulted in a decrease of SOD1 levels by 56 and 50%, respectively. Likewise, knockdown of Prkaa2 and Stk22a brought down SOD1 levels by 40 and 44%, respectively. On the other hand, knockdown of SENP8 and SNRK resulted in a modest decrease of SOD1 expression levels by 22 and 13%, respectively, with respect to the MOCK control. These results indicate that although all the genes analyzed in effect decreased SOD1 levels, they varied in their kinetics in down-modulating SOD1 levels. For example, although knockdown of genes such as Camkiia, Usp25, and Stk22a down-modulated SOD1 levels at later times post-infection (2 h), knockdown of genes such as Prkaa2 and Senp8 had an early effect with respect to down-regulation of SOD1 as their effects were evident within 30 min post-infection. Knockdown of Snrk although showed a modest decrease at 2 h, but the fact that a downward trend was observed indicates a late regulation of SOD1 post-infection by this gene. These results are not only in agreement with the data in Fig. 9, wherein knockdown of most genes enhanced ROS levels, but also reiterate the complex network of regulation that is associated with the generation of ROS, which is interlinked with SOD1 expression by genes in the calcium and cysteine protease pathways. Overall, the results obtained with live M. tuberculosis infections confirm the negative role of the identified genes in regulating immune responses during tuberculosis infection.
FIGURE 10.
Knockdown of genes decreases SOD1 expression from M. tuberculosis-infected DCs. DCs were transfected with siRNAs against indicated genes, followed by infection with 2.5 MOI M. tuberculosis (M. tb) H37Rv for the indicated times. Cytoplasmic extracts were Western blotted for SOD1 expression. Mock represents knockdown with a control siRNA. Numbers below the SOD1 blots indicate relative intensities of the bands. Data from one of two experiments are shown.
DISCUSSION
With the emergence of drug resistance in pathogenic strains, including M. tuberculosis, and the increasing risk and failures associated with multi-drug-resistant and extensively drug-resistant TB, together with the protracted development of new and effective drugs against these strains, research priorities have begun to focus on the abilities of the host machinery to combat long lasting and resurgent infections. To this end, several attempts have been made to understand the role of host genes in mediating protection as also the regulation of immune responses against these pathogens (9–11). The survival of M. tuberculosis in the host cell is facilitated through a dynamic interplay of cellular and intracellular immune responses. These include down-regulation and inhibition of antigen presentation, cellular activation pathways, and the activation of bacterial responses (24, 41). Furthermore, several reports point toward a role of M. tuberculosis antigens in mediating suppressive responses on host cells. For example, the 19-kDa lipoprotein inhibits IFN-γ-mediated responses in macrophages via TLR2 (42). Over the years, we have also highlighted the role of M. tuberculosis antigens, specifically CFP-10, in modulating DC functions (22–24, 43, 44).
Several studies have demonstrated the utility of RNAi-based screens, which use siRNA libraries to identify genes that play crucial roles that shape functional and phenotypic outcomes (45) and to identify potential drug targets (46). For example, siRNA libraries against the apoptotic pathway have been used to identify several genes regulating cancer (47). Furthermore, siRNA libraries have been used in systematic and cost-effective genome-wide loss-of-function screens with the aim of assessing the role of specific genes in neoplastic phenotypes and the rapid identification of novel drug targets (46). The apoptotic pathway library has been used to identify genes active during apoptosis, proliferation, and cell cycle (48). MAPK pathway-specific siRNA libraries have been used to study cell cycle regulation (49). Using similar approaches, the role of phosphatases that regulate NF-κB activation has been elucidated (50). Recently, genes regulating the level of ploidy in HeLa cells were profiled using genome-scale RNAi profiling and >2000 genes were identified to regulate ploidy in cancerous cells (51). A recent report has also identified genes in human macrophages that regulate the survival of M. tuberculosis wherein inhibiting xenophagic pathways and the formation of autophagosomes as an increase in the levels of LC3 II upon the silencing of some target genes was observed (52).
In light of the above reports, we investigated the roles of genes that mediate immune responses to M. tuberculosis via DCs. To that end, we restricted our studies to investigating the roles of genes involved in specific pathways, namely the calcium calmodulin pathway and cysteine protease pathway. These pathways were chosen based on reports, including ours, on the role of calcium in modulating immune responses to M. tuberculosis from DCs and macrophages (22) and the important role of antigen presentation in stimulating T cell responses (12–14). In addition, calcium plays a determinant role in the generation of proinflammatory responses (19, 20) and also regulates the survival of M. tuberculosis in macrophages and DCs (22).
Using siRNA libraries to these pathways, we initially investigated the role of these genes in mediating survival of M. tuberculosis inside DCs. This was carried out to shortlist crucial genes that could be pursued for further characterization. A set of 13 genes was identified whose knockdown resulted in a significant reduction in cfus. Interestingly, many of the genes have not been reported to regulate immune responses to M. tuberculosis, although these genes have been reported to influence various immune related responses. For example, TLR stimulation of DCs result in the activation of Camkiia and it is also involved in the regulation and distribution of MHCII levels in DCs (53).
PIM-2, an anti-apoptotic protein (28, 54), is expressed at high levels in acute promyelocytic leukemia patients (55). Usp25, a ubiquitin-specific protease enzyme, is involved in deubiquitination process that has an essential role in regulating various cellular pathways, cell cycle regulation, transcription etc. Dcamkl1 (doublecortin and CaM kinase-like-1) is expressed in post-mitotic neurons (56) and is known as a putative stem cell marker. Senp8 is a NEDD8-specific protease that has an important role in deneddylation of cullin, a substrate for neddylation (34). In addition to playing a role during apoptosis, Lgmn, an asparaginyl endopeptidase, is overexpressed in many human solid tumors, breast, colon, and prostate cancer and sparsely expressed in normal tissues (32). Snrk is reported as a novel substrate for liver kinase B1 (29). It also controls angioblast populations in the lateral plate mesoderm in zebrafish (57). Usp9y is one of the major Y-linked spermatogenesis genes, which acts as a fine tuner to improve efficiency of human spermatogenesis (58).
As reported by others, several responses observed with live infections can be reproduced by stimulating immune cells such as DCs or macrophages with M. tuberculosis antigens (22, 42–44). To this end, we also recently reported the role of antigens expressed inside of M. tuberculosis-infected macrophages as a function of infection and time in modulating M. tuberculosis-mediated responses to DCs and macrophages (23).
Therefore, in the next set of experiments, we investigated the ability of these genes to modulate M. tuberculosis antigen-specific responses from DCs. To that end, again based on our earlier report (24), we focused on the roles of two receptors, TLR2 and DC-SIGNR1, which induce contrasting responses with respect to M. tuberculosis (59). Taken together, the results indicate that knockdown of many of these genes resulted in enhanced proinflammatory cytokine responses, (as observed by increased levels of IL-12p40) from both TLR2 and to some extent DC-SIGNR1. This was further corroborated with increases in the surface densities of costimulatory molecules, as well as induction of Th1 responses from an antigen known to favor Th2 responses. These results emphasize an as yet unknown function of these genes in regulating immune responses to M. tuberculosis.
Following this, we next investigated the mechanism involved in regulation of M. tuberculosis survival inside DCs as mediated by the identified genes. To that end, our results indicated that they regulate autophagy and the generation of oxidative bursts. The results indicate that knockdown of most of the genes are able to increase the expression of Beclin-1 and ATG5, autophagic markers upon TLR2 and DC-SIGNR1 stimulation with day1 and day5 M. tuberculosis antigens. Likewise, the generation levels of ROS also showed an enhanced effect following knockdown of genes upon stimulation of TLR2 and DC-SIGNR1 with day1 and day5 M. tuberculosis antigens. Interestingly, ROS was regulated by these genes in a manner that was reflective of time-based responses. This indicated differential activation of these genes at different times post-infection when antigens secreted from infected macrophages would counter DCs that are recruited to the sites of infection and modulate their function as proposed in our earlier report (23).
Collectively, our results indicate the identification of genes that play a significant role in modulating immune responses to M. tuberculosis from DCs. Genes such as Usp25, Snrk, and Senp8 displayed broad negative regulation with respect to proinflammatory responses from DCs as well as regulation of M. tuberculosis survival. Furthermore, many of the genes identified in this report acted in a specific manner that are indicative of fine tuning of immune responses during infection and the ability of M. tuberculosis to modulate the activation status of these genes for immune evasion. Interestingly, Senp8 is involved in neddylation of various intracellular molecules (60). The role of neddylation during M. tuberculosis infection has not been reported. Some of the above results were extended to experimentation with live M. tuberculosis infections and similar results were obtained. Most genes down-regulated autophagy and ROS during live infections thereby adding support to the above observations. This indicated that these genes played a negative role during specific antigenic stimulation as well as whole bacterial infection. Therefore, delineating the mechanisms employed by these genes in regulating some of these responses could increase our understanding on the roles played by these genes in the pathogenesis of M. tuberculosis.
Supplementary Material
This work was supported by grants from the Department of Biotechnology, Ministry of Science and Technology, Government of India (to K. N.).

This article contains supplemental Figs. 1–4.
- DC
- dendritic cell
- DCFH-DA
- dichloroflourescin diacetate
- DC-SIGNR1
- DC-SIGN-related molecule 1
- TLR
- Toll-like receptor
- manLAM
- mannosylated LipoArabinoMannan
- ROS
- reactive oxygen species
- MOI
- multiplicity of infection
- SNRK
- SNF-related kinase
- USP25
- ubiquitin-specific peptidase 25
- SOD1
- superoxide dismutase 1.
REFERENCES
- 1. World Health Organization (2009) World Health Organization Fact Sheet, Geneva, Switzerland [Google Scholar]
- 2. Young D. (2009) Animal models of tuberculosis. Eur. J. Immunol. 39, 2011–2014 [DOI] [PubMed] [Google Scholar]
- 3. Ottenhoff T. H. (2009) Overcoming the global crisis: “Yes, we can,” but also for TB … ? Eur. J. Immunol. 39, 2014–2020 [DOI] [PubMed] [Google Scholar]
- 4. Fine P. E. (1995) Variation in protection by BCG: Implications of and for heterologous immunity. Lancet 346, 1339–1345 [DOI] [PubMed] [Google Scholar]
- 5. Hess J., Schaible U., Raupach B., Kaufmann S. H. (2000) Exploiting the immune system: Toward new vaccines against intracellular bacteria. Adv. Immunol. 75, 1–88 [DOI] [PubMed] [Google Scholar]
- 6. Manabe Y. C., Bishai W. R. (2000) Latent Mycobacterium tuberculosis persistence, patience, and winning by waiting. Nat. Med. 6, 1327–1329 [DOI] [PubMed] [Google Scholar]
- 7. Foote S. (1999) Mediating immunity to mycobacteria. Nat. Genet. 21, 345–346 [DOI] [PubMed] [Google Scholar]
- 8. Flynn J. L., Chan J. (2001) Immunology of tuberculosis. Annu. Rev. Immunol. 19, 93–129 [DOI] [PubMed] [Google Scholar]
- 9. Ganesan A. K., Ho H., Bodemann B., Petersen S., Aruri J., Koshy S., Richardson Z., Le L. Q., Krasieva T., Roth M. G., Farmer P., White M. A. (2008) Genome-wide siRNA-based functional genomics of pigmentation identifies novel genes and pathways that impact melanogenesis in human cells. PLoS Genet. 4, e1000298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Luo J., Emanuele M. J., Li D., Creighton C. J., Schlabach M. R., Westbrook T. F., Wong K. K., Elledge S. J. (2009) A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell 137, 835–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cullen L. M., Arndt G. M. (2005) Genome-wide screening for gene function using RNAi in mammalian cells. Immunol. Cell Biol. 83, 217–223 [DOI] [PubMed] [Google Scholar]
- 12. Steinman R. M. (1999) Dendritic cells in Fundamental Immunology (Paul William E., ed) pp. E47–72, Lippincott-Raven Publishers [Google Scholar]
- 13. Banchereau J., Steinman R. M. (1998) Dendritic cells and the control of immunity. Nature 392, 245–252 [DOI] [PubMed] [Google Scholar]
- 14. Reis e Sousa C. (2001) Dendritic cells as sensors of infection. Immunity 14, 495–498 [DOI] [PubMed] [Google Scholar]
- 15. Tian T., Woodworth J., Sköld M., Behar S. M. (2005) In vivo depletion of CD11c+ cells delays the CD4+ T cell response to Mycobacterium tuberculosis and exacerbates the outcome of infection. J. Immunol. 175, 3268–3272 [DOI] [PubMed] [Google Scholar]
- 16. Pecora N. D., Fulton S. A., Reba S. M., Drage M. G., Simmons D. P., Urankar-Nagy N. J., Boom W. H., Harding C. V. (2009) Mycobacterium bovis BCG decreases MHC-II expression in vivo on murine lung macrophages and dendritic cells during aerosol infection. Cell. Immunol. 254, 94–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hemmi H., Takeuchi O., Kawai T., Kaisho T., Sato S., Sanjo H., Matsumoto M., Hoshino K., Wagner H., Takeda K., Akira S. (2000) A Toll-like receptor recognizes bacterial DNA. Nature 408, 740–745 [DOI] [PubMed] [Google Scholar]
- 18. Bodnar K. A., Serbina N. V., Flynn J. L. (2001) Fate of Mycobacterium tuberculosis within murine dendritic cells. Infect. Immun. 69, 800–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Noble A., Truman J. P., Vyas B., Vukmanovic-Stejic M., Hirst W. J., Kemeny D. M. (2000) The balance of protein kinase C and calcium signaling directs T cell subset development. J. Immunol. 164, 1807–1813 [DOI] [PubMed] [Google Scholar]
- 20. Malik Z. A., Thompson C. R., Hashimi S., Porter B., Iyer S. S., Kusner D. J. (2003) Cutting edge: Mycobacterium tuberculosis blocks Ca2+ signaling and phagosome maturation in human macrophages via specific inhibition of sphingosine kinase. J. Immunol. 170, 2811–2815 [DOI] [PubMed] [Google Scholar]
- 21. Hsing L. C., Rudensky A. Y. (2005) The lysosomal cysteine proteases in MHC class II antigen presentation. Immunol. Rev. 207, 229–241 [DOI] [PubMed] [Google Scholar]
- 22. Gupta S., Salam N., Srivastava V., Singla R., Behera D., Khayyam K. U., Korde R., Malhotra P., Saxena R., Natarajan K. (2009) Voltage-gated calcium channels negatively regulate protective immunity to Mycobacterium tuberculosis. PLoS One 4, e5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gupta D., Sharma S., Singhal J., Satsangi A. T., Antony C., Natarajan K. (2010) Suppression of TLR2-induced IL-12, reactive oxygen species, and inducible nitric oxide synthase expression by Mycobacterium tuberculosis antigens expressed inside macrophages during the course of infection. J. Immunol. 184, 5444–5455 [DOI] [PubMed] [Google Scholar]
- 24. Srivastava V., Manchanda M., Gupta S., Singla R., Behera D., Das G., Natarajan K. (2009) Toll-like receptor 2 and DC-SIGNR1 differentially regulate suppressors of cytokine signaling 1 in dendritic cells during Mycobacterium tuberculosis infection. J. Biol. Chem. 284, 25532–25541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Srivastava V., Vashishta M., Gupta S., Singla R., Singla N., Behera D., Natarajan K. (2011) Suppressors of cytokine signaling inhibit effector T cell responses during Mycobacterium tuberculosis infection. Immunol. Cell Biol. 89, 786–791 [DOI] [PubMed] [Google Scholar]
- 26. Sinha A., Singh A., Satchidanandam V., Natarajan K. (2006) Impaired generation of reactive oxygen species during differentiation of dendritic cells (DCs) by Mycobacterium tuberculosis secretory antigen (MTSA) and subsequent activation of MTSA-DCs by mycobacteria results in increased intracellular survival. J. Immunol. 177, 468–478 [DOI] [PubMed] [Google Scholar]
- 27. Nguyen A., Chen P., Cai H. (2004) Role of CaMKII in hydrogen peroxide activation of ERK1/2, p38 MAPK, HSP27, and actin reorganization in endothelial cells. FEBS Lett. 572, 307–313 [DOI] [PubMed] [Google Scholar]
- 28. Yan B., Zemskova M., Holder S., Chin V., Kraft A., Koskinen P. J., Lilly M. (2003) The PIM-2 kinase phosphorylates BAD on serine 112 and reverses BAD-induced cell death. J. Biol. Chem. 278, 45358–45367 [DOI] [PubMed] [Google Scholar]
- 29. Jaleel M., McBride A., Lizcano J. M., Deak M., Toth R., Morrice N. A., Alessi D. R. (2005) Identification of the sucrose non-fermenting related kinase SNRK, as a novel LKB1 substrate. FEBS Lett. 579, 1417–1423 [DOI] [PubMed] [Google Scholar]
- 30. Lee J. W., Park S., Takahashi Y., Wang H. G. (2010) The association of AMPK with ULK1 regulates autophagy. PLoS One 5, e15394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Snapinn K. W., Larson E. B., Kawakami H., Ujike H., Borenstein A. R., Izumi Y., Kaji R., Maruyama H., Mata I. F., Morino H., Oda M., Tsuang D. W., Yearout D., Edwards K. L., Zabetian C. P. (2011) The UCHL1 S18Y polymorphism and Parkinson disease in a Japanese population. Parkinsonism Relat. Disord. 17, 473–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gawenda J., Traub F., Lück H. J., Kreipe H., von Wasielewski R. (2007) Legumain expression as a prognostic factor in breast cancer patients. Breast Cancer Res. Treat. 102, 1–6 [DOI] [PubMed] [Google Scholar]
- 33. Barrett A. J., Kirschke H. (1981) Cathepsin B, cathepsin H, and cathepsin L. Methods Enzymol. 80, 535–561 [DOI] [PubMed] [Google Scholar]
- 34. Gan-Erdene T., Nagamalleswari K., Yin L., Wu K., Pan Z. Q., Wilkinson K. D. (2003) Identification and characterization of DEN1, a deneddylase of the ULP family. J. Biol. Chem. 278, 28892–28900 [DOI] [PubMed] [Google Scholar]
- 35. Koh K. W., Lehming N., Seah G. T. (2009) Degradation-resistant protein domains limit host cell processing and immune detection of mycobacteria. Mol. Immunol. 46, 1312–1318 [DOI] [PubMed] [Google Scholar]
- 36. van Kooyk Y., Geijtenbeek T. B. (2003) DC-SIGN: Escape mechanism for pathogens. Nat. Rev. Immunol. 3, 697–709 [DOI] [PubMed] [Google Scholar]
- 37. Yadav U. C., Naura A. S., Aguilera-Aguirre L., Ramana K. V., Boldogh I., Sur S., Boulares H. A., Srivastava S. K. (2009) Aldose reductase inhibition suppresses the expression of Th2 cytokines and airway inflammation in ovalbumin-induced asthma in mice. J. Immunol. 183, 4723–4732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Virgin H. W., Levine B. (2009) Autophagy genes in immunity. Nat. Immunol. 10, 461–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Deretic V., Levine B. (2009) Autophagy, immunity, and microbial adaptations. Cell Host Microbe 5, 527–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jagannath C., Lindsey D. R., Dhandayuthapani S., Xu Y., Hunter R. L., Jr., Eissa N. T. (2009) Autophagy enhances the efficacy of BCG vaccine by increasing peptide presentation in mouse dendritic cells. Nat. Med. 15, 267–276 [DOI] [PubMed] [Google Scholar]
- 41. Koul A., Herget T., Klebl B., Ullrich A. (2004) Interplay between mycobacteria and host signaling pathways. Nat. Rev. Microbiol. 2, 189–202 [DOI] [PubMed] [Google Scholar]
- 42. Noss E. H., Pai R. K., Sellati T. J., Radolf J. D., Belisle J., Golenbock D. T., Boom W. H., Harding C. V. (2001) Toll-like receptor 2-dependent inhibition of macrophage class II MHC expression and antigen processing by 19-kDa lipoprotein of Mycobacterium tuberculosis. J. Immunol. 167, 910–918 [DOI] [PubMed] [Google Scholar]
- 43. Latchumanan V. K., Singh B., Sharma P., Natarajan K. (2002) Mycobacterium tuberculosis antigens induce the differentiation of dendritic cells from bone marrow. J. Immunol. 169, 6856–6864 [DOI] [PubMed] [Google Scholar]
- 44. Salam N., Gupta S., Sharma S., Pahujani S., Sinha A., Saxena R. K., Natarajan K. (2008) Protective immunity to Mycobacterium tuberculosis infection by chemokine and cytokine conditioned CFP-10 differentiated dendritic cells. PLoS One. 3, e2869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Moffat J., Sabatini D. M. (2006) Building mammalian signaling pathways with RNAi screens. Nat. Rev. Mol. Cell Biol. 7, 177–187 [DOI] [PubMed] [Google Scholar]
- 46. Ito M., Kawano K., Miyagishi M., Taira K. (2005) Genome-wide application of RNAi to the discovery of potential drug targets. FEBS Lett. 579, 5988–5995 [DOI] [PubMed] [Google Scholar]
- 47. Ovcharenko D., Kelnar K., Johnson C., Leng N., Brown D. (2007) Genome-scale microRNA and small interfering RNA screens identify small RNA modulators of TRAIL-induced apoptosis pathway. Cancer Res. 67, 10782–10788 [DOI] [PubMed] [Google Scholar]
- 48. Alenzi F. Q. (2004) Links between apoptosis, proliferation and the cell cycle. Br. J. Biomed. Sci. 61, 99–102 [DOI] [PubMed] [Google Scholar]
- 49. Su B., Karin M. (1996) Mitogen-activated protein kinase cascades and regulation of gene expression. Curr. Opin. Immunol. 8, 402–411 [DOI] [PubMed] [Google Scholar]
- 50. Li S., Wang L., Berman M. A., Zhang Y., Dorf M. E. (2006) RNAi screen in mouse astrocytes identifies phosphatases that regulate NF-κB signaling. Mol. Cell 24, 497–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kittler R., Pelletier L., Heninger A. K., Slabicki M., Theis M., Miroslaw L., Poser I., Lawo S., Grabner H., Kozak K., Wagner J., Surendranath V., Richter C., Bowen W., Jackson A. L., Habermann B., Hyman A. A., Buchholz F. (2007) Genome-scale RNAi profiling of cell division in human tissue culture cells. Nat. Cell Biol. 9, 1401–1412 [DOI] [PubMed] [Google Scholar]
- 52. Kumar D., Nath L., Kamal M. A., Varshney A., Jain A., Singh S., Rao K. V. (2010) Genome-wide analysis of the host intracellular network that regulates survival of Mycobacterium tuberculosis. Cell 140, 731–743 [DOI] [PubMed] [Google Scholar]
- 53. Connolly S. F., Kusner D. J. (2007) The regulation of dendritic cell function by calcium signaling and its inhibition by microbial pathogens. Immunol. Res. 39, 115–127 [DOI] [PubMed] [Google Scholar]
- 54. Fox C. J., Hammerman P. S., Cinalli R. M., Master S. R., Chodosh L. A., Thompson C. B. (2003) The serine/threonine kinase Pim-2 is a transcriptionally regulated apoptotic inhibitor. Genes Dev. 17, 1841–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Agrawal-Singh S., Koschmieder S., Gelsing S., Stocking C., Stehling M., Thiede C., Thoennissen N. H., Köhler G., Valk P. J., Delwel R., Mills K., Bäumer N., Tickenbrock L., Hansen K., Berdel W. E., Müller-Tidow C., Serve H. (2010) Pim2 cooperates with PML-RARα to induce acute myeloid leukemia in a bone marrow transplantation model. Blood 115, 4507–4516 [DOI] [PubMed] [Google Scholar]
- 56. Lin P. T., Gleeson J. G., Corbo J. C., Flanagan L., Walsh C. A. (2000) DCAMKL1 encodes a protein kinase with homology to doublecortin that regulates microtubule polymerization. J. Neurosci. 20, 9152–9161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pramanik K., Chun C. Z., Garnaas M. K., Samant G. V., Li K., Horswill M. A., North P. E., Ramchandran R. (2009) Dusp-5 and Snrk-1 coordinately function during vascular development and disease. Blood 113, 1184–1191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Krausz C., Degl'Innocenti S., Nuti F., Morelli A., Felici F., Sansone M., Varriale G., Forti G. (2006) Natural transmission of USP9Y gene mutations: A new perspective on the role of AZFa genes in male fertility. Hum. Mol. Genet. 15, 2673–2681 [DOI] [PubMed] [Google Scholar]
- 59. Kaufmann S. H., Schaible U. E. (2003) A dangerous liaison between two major killers: Mycobacterium tuberculosis and HIV target dendritic cells through DC-SIGN. J. Exp. Med. 197, 1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Soucy T. A., Smith P. G., Milhollen M. A., Berger A. J., Gavin J. M., Adhikari S., Brownell J. E., Burke K. E., Cardin D. P., Critchley S., Cullis C. A., Doucette A., Garnsey J. J., Gaulin J. L., Gershman R. E., Lublinsky A. R., McDonald A., Mizutani H., Narayanan U., Olhava E. J., Peluso S., Rezaei M., Sintchak M. D., Talreja T., Thomas M. P., Traore T., Vyskocil S., Weatherhead G. S., Yu J., Zhang J., Dick L. R., Claiborne C. F., Rolfe M., Bolen J. B., Langston S. P. (2009) An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458, 732–736 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.