Background: ATP-dependent closure of the N-gate is a key step for negative DNA supercoiling by gyrase.
Results: Potassium ions are required for efficient nucleotide-induced N-gate closure and DNA supercoiling but are dispensable for DNA relaxation.
Conclusion: Potassium ions help coordinate the nucleotide cycle to gate movements of gyrase.
Significance: Coordinated conformational changes are crucial for the function of ATP-driven molecular machines in general.
Keywords: ATPases, DNA Gyrase, DNA Topoisomerase, DNA Topology, Fluorescence Resonance Energy Transfer (FRET), GHKL ATPase, Potassium Ions, Negative Supercoiling, Nucleotide-driven Conformational Change
Abstract
DNA gyrase catalyzes ATP-dependent negative supercoiling of DNA by a strand passage mechanism that requires coordinated opening and closing of three protein interfaces, the N-, DNA-, and C-gates. ATP binding to the GyrB subunits of gyrase causes dimerization and N-gate closure. The closure of the N-gate is a key step in the gyrase catalytic cycle, as it captures the DNA segment to be transported and poises gyrase toward strand passage. We show here that K+ ions are required for DNA supercoiling but are dispensable for ATP-independent DNA relaxation. Although DNA binding, distortion, wrapping, and DNA-induced narrowing of the N-gate occur in the absence of K+, nucleotide-induced N-gate closure depends on their presence. Our results provide evidence that K+ ions relay small conformational changes in the nucleotide-binding pocket to the formation of a tight dimer interface at the N-gate by connecting regions from both GyrB monomers and suggest an important role for K+ in synchronization of N-gate closure and DNA-gate opening.
Introduction
Gyrase is a DNA topoisomerase that introduces negative supercoils into plasmid DNA in an ATP-dependent reaction (1) and plays an important role in DNA replication, transcription, and recombination (2). The active unit of gyrase is composed of two GyrA and two GyrB subunits that assemble into an A2B2 heterotetramer. Gyrase forms two cavities, delimited by three protein interfaces termed the N-, DNA-, and C-gates (see Fig. 1A). During ATP-dependent negative supercoiling, these gates open and close in a coordinated manner to allow for strand passage toward negative DNA supercoiling (3–8). The gyrase catalytic cycle begins when a gate DNA (G-segment) is bound at the DNA-gate and distorted (7). Interaction of DNA flanking the G-segment with the C-terminal domains (CTDs)2 causes the CTDs to move upward and sideways (9), and complete wrapping of DNA leads to narrowing of the N-gate formed by the GyrB subunits (8). The N-gate acts as an ATP-dependent clamp (10): ATP binding to the ATPase domains of the GyrB subunits induces GyrB dimerization and N-gate closure (8, 11–13), leading to the trapping of the transport DNA (T-segment). N-gate closure and DNA-gate opening appear to be coupled, with possible contributions from the T-segment (10). After passage of the T-segment through the gap in the G-segment, the G-segment is religated, the T-segment leaves the enzyme through the C-gate formed by GyrA (14, 15), and the N-gate reopens (8, 12). Our recent results provide evidence for a bidirectional communication between the N- and DNA-gates of gyrase (7, 8) that coordinates their movements in the supercoiling cycle.
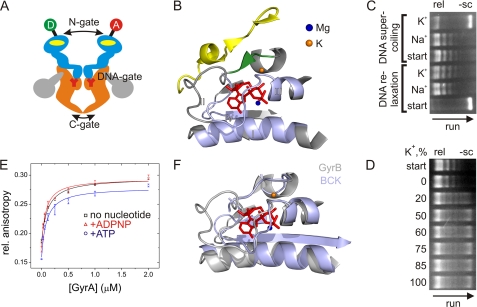
FIGURE 1.
DNA gyrase and influence of K+ and Na+ ions on topoisomerase activities and DNA binding. A, schematic representation of gyrase and the N-, DNA-, and C-gates. The active enzyme consists of two GyrA (orange and gray) and two GyrB (light blue) subunits. The yellow ovals indicate ATP-binding sites in GyrB, and Y represents the active site tyrosine in GyrA. The GyrA CTDs are depicted in gray. The circles indicate the attachment sites for donor (D) and acceptor (A) dyes at the N-gate. B, ATP-binding site in GyrB (Protein Data Bank code 1EI1, GyrB dimer in complex with ADPNP). The conserved GHKL motifs (I–IV) are depicted in light blue. The Mg2+ ion coordinating the phosphates is shown as a blue sphere, and the K+ ion is depicted in orange in the location where it is found in the protein kinase BCK (Protein Data Bank code 1GJV; see F). The K+ ion connects the α-helix contacting the phosphates of the bound nucleotide with the short β-strand (green) that forms a β-sheet with an α-strand from the second GyrB monomer (yellow). C, DNA relaxation (rel) and negative supercoiling (−sc) of DNA by gyrase in the presence of Na+ or K+. The arrow indicates the direction of the gel run. DNA relaxation (no ATP; start indicates negatively supercoiled DNA used as a substrate) is independent of K+, but negative DNA supercoiling (with ATP; start indicates relaxed DNA used as a substrate) requires K+ ions. D, dependence of negative DNA supercoiling on the K+ concentration. All reactions were performed at a constant ionic strength (100 mm, NaCl/KCl mixture) with 20 nm GyrA, 80 nm GyrB, 20 nm pUC18, and 1.5 mm ATP, varying the KCl concentration. The arrow indicates the direction of the gel run. The relaxed plasmid used as a substrate is shown in the start row. Sodium ions are not inhibitory for the supercoiling reaction. E, anisotropy titrations of an Alexa Fluor 488- and Alexa Fluor 546-labeled 60-bp DNA (7) in buffer containing Na+ with GyrA (8 μm GyrB added to form active gyrase). Kd values for the gyrase-DNA complex were 99 ± 12 nm (no nucleotide), 73 ± 16 nm (ATP), and 59 ± 11 nm (ADPNP). Error bars denote S.D. from three independent experiments. F, superposition of GyrB (gray; Protein Data Bank code 1EI1, GyrB dimer in complex with ADPNP) and BCK (light blue; Protein Data Bank code 1GJV), demonstrating the similar structures of the nucleotide-binding sites.
ATP binding and hydrolysis are essential for DNA supercoiling. The ATPase site is located in the N-terminal domain of GyrB (11, 12). GyrB belongs to the GHKL (GyrB-Hsp90-histidine/serine protein kinases-MutL) phosphotransferase superfamily (reviewed in Ref. 16). Members of this family share a catalytic site for ATP hydrolysis formed by four conserved sequence motifs. The ATP-binding pocket formed by motifs I, II, and IV is covered by a flexible lid formed by motif III (see Fig. 1B) (16). ATP hydrolysis requires the presence of an Mg2+ ion in the ATP-binding pocket, and a K+ ion is required for ATP binding and hydrolysis in the rat branched-chain α-keto acid dehydrogenase kinase (BCK) (see Fig. 1F) (17). K+ increases the thermodynamic stability of GyrB by stabilizing the ATPase domain (18). It was shown previously that Micrococcus luteus gyrase requires K+ ions for DNA supercoiling (19). We have noted that K+ ions are also required for DNA supercoiling by Bacillus subtilis gyrase (7). In this work, we investigated the role of monovalent ions in the gyrase supercoiling cycle. We show that K+ ions are required for ATP-dependent negative supercoiling. DNA binding, DNA distortion, and DNA-induced N-gate narrowing do not require the presence of K+. ATP binding and hydrolysis are also only mildly affected in the absence of K+. In contrast, nucleotide-dependent closure of the N-gate is hampered in the absence of K+, pointing to a role of potassium ions in stabilizing the dimer interface in the closed N-gate. DNA relaxation by gyrase does not depend on the presence of K+ ions, rationalizing the previous observation that the N-gate is not required for this reaction (20).
EXPERIMENTAL PROCEDURES
Site-directed Mutagenesis, Protein Production, and Purification
To investigate the role of the K+ ions in conformational changes of the gyrase N-gate, we used a GyrBA fusion protein in which the coding region of GyrB (amino acids 1–638) was fused to the coding region of GyrA (amino acids 1–821) with the short linker coding for the peptide GAP (8). Mutants GyrBA_E17C and GyrBA_S7C were generated, purified, and labeled with Alexa Fluor 488 (donor) and Alexa Fluor 546 (acceptor) as described previously (8). Donor/acceptor-labeled proteins showed topoisomerase activity within 2-fold of wild-type gyrase (8).
DNA Substrates and Fluorescence Anisotropy Titrations
As a model for a gate DNA, a 60-bp DNA substrate with a contained preferred cleavage site for B. subtilis gyrase in the center (21) was prepared from the complementary strands (Purimex) as described (7). The Kd value of complexes between GyrB2GyrA2 and the 60-bp DNA was determined in fluorescence anisotropy titrations with 10 nm Alexa Fluor 488/Alexa Fluor 546-labeled 60-bp DNA in 50 mm Tris-HCl (pH 7.5), 100 mm KCl or NaCl, and 10 mm MgCl2 at 37 °C using the fluorescence of Alexa Fluor 546 as a probe, and data were analyzed using the solution of the quadratic equation (Equation 1) as described previously (7, 8),
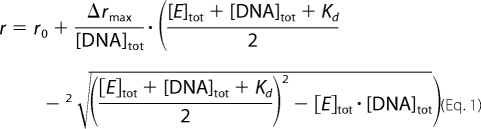 |
where r0 is the anisotropy of free DNA, Δrmax is the amplitude, [E]tot is the total enzyme concentration, and [DNA]tot is the total DNA concentration. The GyrB concentration was 8 μm. ATP (Pharma-Waldhof GmbH) and ADPNP (Jena Bioscience) were added at concentrations of 2 mm.
DNA Supercoiling
Negatively supercoiled pUC18 was purified from transformed Escherichia coli XL1-Blue (Promega midiprep system). Relaxed plasmid was prepared from negatively supercoiled pUC18 using gyrase in the absence of ATP. If not indicated otherwise, DNA relaxation and supercoiling activities were assayed as described (8) with 20 nm negatively supercoiled or relaxed pUC18, 200 nm GyrA, and 800 nm GyrB in 50 mm Tris-HCl (pH 7.5), 100 mm KCl or NaCl, and 10 mm MgCl2. For supercoiling reactions, buffers were supplemented with 1.5 mm ATP. Reactions were stopped after 5 min by adding 0.5% SDS and 25 mm EDTA. Products were analyzed on a 1% agarose gel.
Steady-state ATPase Activity
ATPase activity was measured in a coupled enzymatic assay as described (22) with 200 nm (50 nm in the presence of pUC18) GyrBA in 50 mm Tris-HCl (pH 7.5), 100 mm KCl or NaCl (or mixtures), 10 mm MgCl2, and 4 mm ATP. If not indicated otherwise, the pUC18 concentration was 100 nm. Experimental results were analyzed according to Michaelis-Menten kinetics using Equation 2,
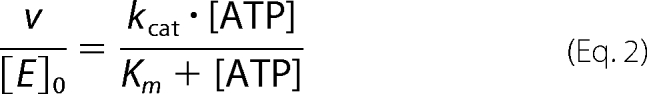 |
where [ATP] is the ATP concentration, Km is the Michaelis constant, [E]0 is the total GyrBA concentration, kcat is the turnover number, and v is the rate of product formation.
Analytical Size Exclusion Chromatography
The oligomeric state of GyrB was analyzed on a SuperdexTM 200 10/300 GL size exclusion column (GE Healthcare) equilibrated in 50 mm Tris-HCl (pH 7.5), 200 mm KCl or NaCl, 10 mm MgCl2, and 2 mm β-mercaptoethanol at room temperature as described (22). Samples of 50 μm GyrB in buffer containing K+ or Na+ were preincubated in the absence or presence of 5 mm ADPNP at 37 °C for 15 min before each run.
Single-molecule FRET (smFRET) Experiments
smFRET experiments with 50 pm donor/acceptor-labeled GyrBA or DNA (concentration of donor fluorophore) (7) were performed at 37 °C in 50 mm Tris-HCl (pH 7.5), 100 mm KCl or NaCl, and 10 mm MgCl2 using a home-built confocal microscope as described (7, 8, 23). Buffers were treated with active charcoal to reduce background fluorescence. Measured fluorescence intensities from bursts of >100 photons were corrected for background, for cross-talk from donor fluorescence into the acceptor channel and vice versa, for different detection efficiencies and quantum yields of donor and acceptor, and for direct excitation of the donor as described (23) using previously reported correction parameters for GyrBA_E17C and GyrBA_S7C (8) and the 60-bp DNA (7). We have previously validated our correction procedure and have shown that it yields corrected FRET efficiencies that reflect correct intermolecular distances (see Ref. 24 for a detailed description).
RESULTS
ATP-dependent DNA Supercoiling by Gyrase Requires Potassium Ions
We have noted previously that DNA supercoiling by B. subtilis gyrase is sensitive to the presence of K+ ions (7–9) and therefore supplemented all buffers with 100 mm KCl. To investigate the effect of K+ ions systematically, we first monitored the relaxation and supercoiling activities of gyrase in the presence of K+ and Na+ (Fig. 1C). DNA relaxation occurred in both the presence of K+ and Na+. In contrast, ATP-dependent DNA supercoiling strictly required K+ and did not occur in the presence of Na+. When we performed the DNA supercoiling reaction at constant ionic strength (100 mm cations, mixture of NaCl and KCl) but varied the fraction of K+ ions from 0 to 100%, supercoiled DNA appeared at concentrations of 20 mm KCl and above (Fig. 1D), indicating that Na+ is not inhibitory for DNA supercoiling by gyrase.
ATP Hydrolysis by Gyrase Is Similar in Presence of K+ and Na+ Ions
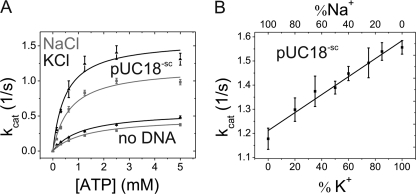
To pinpoint the effect of K+ on DNA supercoiling, we next performed steady-state ATPase activity experiments in the presence of K+ and Na+ (Fig. 2A). The intrinsic ATPase activity was slightly higher in the presence of K+ (kcat = 0.59 ± 0.03 s−1) compared with kcat = 0.46 ± 0.02 s−1 in the presence of Na+. In the presence of Na+, the turnover number increased by 2.6-fold when plasmid DNA was added to kcat = 1.19 ± 0.07 s−1. In the presence of K+, DNA-stimulated ATP hydrolysis occurred with kcat = 1.58 ± 0.13 s−1, corresponding to a 2.7-fold stimulation. Overall, DNA-stimulated ATP hydrolysis was thus slightly faster in the presence of K+, but the effects were small, and the DNA stimulation of ATP hydrolysis was similar for both ions. Km values of gyrase for ATP were also independent of the ion present in both the absence (1.3 ± 0.2 mm with Na+ and 1.3 ± 0.2 mm with K+) and presence (0.66 ± 0.13 mm with Na+ and 0.5 ± 0.2 mm with K+) of DNA. Although there was a trend toward lower Km values in the presence of K+, the differences were again small, and the effects of K+ on ATP binding appeared to be negligible.
FIGURE 2.
Effect of K+ and Na+ ions on steady-state ATPase activity of gyrase. A, dependence of the steady-state ATPase rate on the ATP concentration. The Km values in the presence of Na+ (gray) or K+ (black) are virtually identical without DNA (1.3 ± 0.2 mm with Na+ and 1.3 ± 0.2 mm with K+) and in the presence of supercoiled plasmid DNA (0.66 ± 0.13 mm with Na+ and 0.5 ± 0.2 mm with K+). There was no significant stimulation of the ATPase activity by potassium ions without DNA (kcat = 0.46 ± 0.02 s−1 for Na+ and 0.59 ± 0.03 s−1 for K+) or in the presence of 100 nm pUC18 (kcat = 1.19 ± 0.07 s−1 for Na+ and 1.58 ± 0.13 s−1 for K+). Error bars denote S.D. from three independent experiments. B, steady-state ATPase activity as a function of K+/Na+ buffer composition in the presence of 80 nm pUC18 plasmid DNA. The low ATPase activity at 100 mm NaCl increased linearly with increasing K+ content. Error bars denote S.D. from three independent experiments.
To further elucidate the role of K+ ions in the ATPase activity, we performed ATPase assays at constant ionic strength (100 mm cations) but varied the fraction of K+ ions from 0 to 100% (Fig. 2B). The kcat values show a linear increase with increasing K+/Na+ ratio. At higher K+ concentrations, the maximum effect on DNA supercoiling was achieved, indicating that saturation of gyrase with K+ occurs within this concentration range. Thus, the linear dependence indicates that binding of one K+ ion (to one of the two GyrB subunits) is sufficient for DNA supercoiling.
DNA Binding and Distortion Are Independent of K+
To quantify the effect of K+ on DNA binding to gyrase, we determined the Kd values of gyrase-DNA complexes in fluorescence anisotropy titrations of a 60 bp-DNA (7, 8). This DNA serves as a model gate DNA for DNA gyrase (7) and elicits a conformational change of the CTDs similar to plasmid DNA (9), justifying its use as a model to investigate interactions of gyrase with DNA at the beginning of the supercoiling cycle. The Kd values of the gyrase-DNA complex in the presence of Na+ ions were 99 ± 12 nm (no nucleotide), 73 ± 16 nm (in the presence of 2 mm ATP), and 59 ± 11 nm (in the presence of 2 mm non-hydrolysable ATP analog ADPNP). The Kd values were in the same low nanomolar range as determined previously in the presence of K+ ions (7), demonstrating that DNA binding to gyrase is independent of K+ ions.
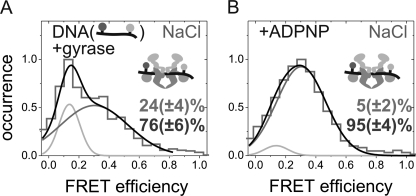
Using smFRET experiments and a donor/acceptor-labeled 60-bp DNA substrate, we previously reported that DNA bound at the DNA-gate occurs in two conformations when K+ ions are present (7). A high FRET state corresponds to DNA slightly distorted from B-form geometry, and a low FRET state corresponds to DNA severely distorted from B-form geometry (7). The severe distortion of the DNA bound at the DNA-gate requires the presence of the CTDs of gyrase (8). Here, we tested if the DNA distortion is possible in the presence of Na+ ions in smFRET experiments with the donor/acceptor-labeled 60-bp DNA (Fig. 3A). The FRET histograms of DNA bound to gyrase show two populations, one with a low FRET efficiency of ∼0.14 and one with a higher FRET efficiency of ∼0.34. The histograms can be described by two Gaussian distributions, yielding a population of 24 ± 4% for the severely distorted DNA (low FRET). This value is similar to the fraction of severely distorted DNA in the presence of K+ (7), confirming further that the interaction of gyrase with DNA does not require K+ ions. Thus, the DNA-gate appears to be assembled and fully functional in the absence of K+ ions.
FIGURE 3.
Conformations of G-segment DNA bound to DNA-gate of gyrase in presence of Na+. A, smFRET histogram for a phosphorothiolate-modified 60-bp DNA carrying donor and acceptor fluorophores flanking the cleavage site (7) bound to gyrase. The high (76%) and low (24%) FRET species correspond to a slightly and severely distorted conformation of bound DNA described previously (7). The distribution of these two conformers is similar to that observed previously in the presence of K+ (7). B, smFRET histograms after ADPNP addition to the sample in A. The fraction of the low FRET species was reduced (to 5%), following behavior observed previously in the presence of K+ (7).
K+ Is Dispensable for DNA-induced Narrowing of N-gate but Is Required for Stable Nucleotide-induced N-gate Closure
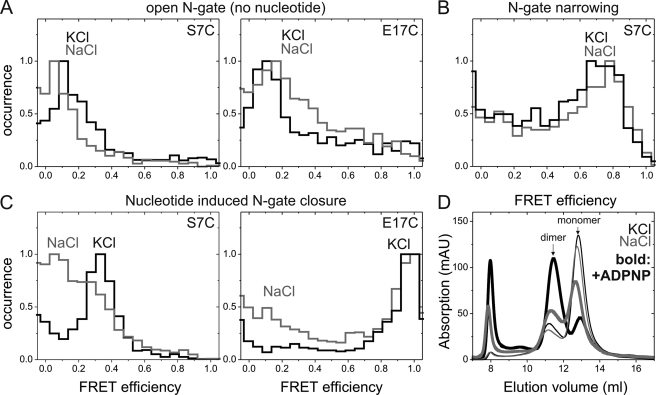
We have previously shown that binding of DNA at the DNA-gate and complete wrapping around the CTDs trigger a narrowing of the N-gate (8). To avoid dissociation of GyrB subunits and formation of incomplete complexes containing only one GyrB subunit under single-molecule conditions with picomolar concentrations of the labeled species, we used a GyrBA fusion protein to investigate the effect of K+ on N-gate conformation. As reported previously (8), GyrBA labeled at position 7 (GyrBA_S7C) or 17 (GyrBA_E17C) flanking the N-gate shows FRET histograms with a unimodal distribution of FRET efficiencies around EFRET = 0.1 in the presence of K+. In the presence of Na+, similar histograms were obtained (Fig. 4A), demonstrating that the N-gate is in a similar open conformation independent of the identity of the ion present.
FIGURE 4.
Effect of K+ on N-gate conformation. A, smFRET histograms of N-gate labeled GyrBA_S7C (left) and GyrBA_E17C (right) in KCl (black) or NaCl (gray) buffer. FRET efficiencies were similar and correspond to the open conformation of the N-gate for both of the buffers. B, smFRET histogram of GyrBA_S7C in the presence of pUC18 and K+ (black) or Na+ (light gray). The increase in FRET efficiencies corresponds to DNA-induced N-gate narrowing and was virtually identical in the presence of K+ and Na+. C, smFRET histograms of GyrBA_S7C (left) and GyrBA_E17C (right) after addition of ADPNP. The FRET efficiency increased due to N-gate closure in the presence of K+, but less efficient closure of the N-gate was observed in the presence of Na+, although the relative populations of gyrase with open and closed N-gates were different for the two mutants. Thus, K+ ions are not required for DNA-induced N-gate narrowing but are required for stable nucleotide-induced N-gate closure. D, oligomeric states of GyrB in the absence (thin lines) or presence (thick lines) of ADPNP. Without nucleotide, the elution profiles are independent of the ion present (K+, black lines; Na+ ions, gray lines), and the predominant peak eluting at 13 ml corresponds to monomeric GyrB. Quantitative analysis of the elution profiles with Gaussian distributions yielded 28% dimer (Na+) and 25% dimer (K+). In the presence of ADPNP, the second peak corresponding to the GyrB dimer was slightly more populated in the presence of Na+ (42% dimer) and became dominant in the presence of K+ (69% dimer). These results confirm that K+ ions are required for stable nucleotide-induced GyrB dimerization and efficient N-gate closure.
Donor/acceptor-labeled GyrBA_S7C showed the largest difference in FRET efficiencies between open and narrowed N-gates (8) and is thus suitable to distinguish these conformations. When we monitored the conformation of the N-gate upon DNA binding in the presence of Na+ instead of K+, the FRET efficiency increased from 0.1 to 0.7 (Fig. 4B), similar to the value observed previously for the gyrase-DNA complex in the presence of KCl. Thus, DNA-induced N-gate narrowing occurs independently of K+ ions. In addition, these results further support that the interaction of gyrase with DNA at the DNA-gate is quantitatively and qualitatively similar in the presence and absence of K+ ions.
In the presence of ADPNP, the N-gate of gyrase closes (8) due to dimerization of the GyrB subunits (11, 22). When K+ ions are present, complete closure of the N-gate is observed as an increase in the mean FRET efficiency to 0.34 for GyrBA_S7C and to 0.97 for GyrBA_E17C (Fig. 4C) (8). These FRET efficiencies are in agreement with the distances expected from the structure of dimeric GyrB (11). When we performed the same experiment in the presence of Na+, a significant fraction of the molecules maintained a low FRET efficiency characteristic of the open N-gate after ADPNP addition, although the fraction of gyrase-ADPNP complexes with an open N-gate was different for the two mutants (Fig. 4C).
To independently confirm that nucleotide-induced N-gate closure requires K+ ions, we performed size exclusion chromatography experiments in the presence of K+ or Na+ ions (Fig. 4D) (22). GyrB eluted mainly as a monomer in the presence of Na+ (22) or K+ (Fig. 4D), with an additional small peak that corresponded to GyrB dimers. Addition of ADPNP to GyrB in the presence of K+ ions (Fig. 4D) led to the efficient formation of GyrB dimers, whereas little dimer formation was observed in the presence of Na+ (Fig. 4D) (22). These results confirm that K+ ions are required for efficient closure of the N-gate in response to nucleotide binding and suggest a stabilization of the N-gate interface by K+ ions.
The smFRET experiments indicate that intramolecular signaling from the DNA-gate to the N-gate (linked by DNA binding and distortion to N-gate narrowing) (Fig. 4B) is functional without K+ present. To finally test for communication from the N-gate to the DNA bound at the DNA-gate, we monitored the effect of nucleotide binding on gate DNA conformation by adding ADPNP to the gyrase-DNA complex containing donor/acceptor-labeled DNA (Fig. 3B). Addition of ADPNP reduced the fraction of severely distorted DNA in both the presence (7) and absence (Fig. 3B) of K+, indicating that K+ is dispensable for the communication from the N-gate back to the DNA-gate, even though N-gate closure cannot occur efficiently.
DISCUSSION
In this work, we have shown that potassium ions are required for the supercoiling activity of the B. subtilis gyrase. Although DNA binding, DNA distortion, and DNA-induced N-gate closure are not sensitive to the type of monovalent ions present, efficient nucleotide-induced N-gate closure depends on the presence of potassium ions. K+ has been demonstrated to increase the thermodynamic stability of the GyrB ATPase domain (18), and a potassium ion is present in the catalytic site of other members of the GHKL family (25). Although we cannot exclude that GyrB contains additional binding sites for monovalent ions, it seems likely that the K+ ion present in the ATP-binding site is responsible for the observed effect of K+ on N-gate closure. GyrB dimerization is a prerequisite for ATP hydrolysis to occur (12), although we have previously shown that the rate of ATP hydrolysis already increases with the fraction of gyrase with a narrowed N-gate (8). Gyrase contains two ATP-binding sites, but the ATP hydrolysis rate shows a linear dependence on the concentration of K+, suggesting that the presence of one K+ ion is sufficient for nucleotide-induced GyrB dimerization and N-gate closure, in line with the observation that ATP bound to one of the subunits is sufficient to trigger GyrB dimerization and strand passage (13, 26). (If two K+ ions were required, a dependence on the square of the K+ concentration would be expected.) The effects of K+ ions on the ATPase activity are small, suggesting that the bound potassium may contribute to a downstream effect required for strand passage. DNA binding at the DNA-gate leads to N-gate narrowing in the absence of K+, indicating that intramolecular communication from the DNA-gate to the N-gate is a K+-independent process. Similarly, the previously observed effect of ADPNP binding on the conformation of DNA bound at the DNA-gate occurs in the absence of K+. These findings suggest that the change in DNA conformation upon ADPNP binding is a step separate from and prior to N-gate closure and demonstrate that at least partial communication from the N-gate to the DNA-gate is possible without K+. We have previously provided evidence that N-gate closure poises gyrase for DNA-gate opening and strand passage toward negative supercoiling (8), in line with the so-called double-lock rule put forward by Roca (27, 28). The results shown here illustrate that K+ ions play an important role in coordinating these movements. K+ ions are dispensable for DNA relaxation, corroborating the notion that DNA relaxation may be catalyzed by a distinct mechanism.
What is the possible role of K+ ions in DNA supercoiling? Although K+ is not present in the GyrB crystal structures, comparison with the GHKL kinase BCK (Fig. 1F) (17) suggests that K+ connects the α-helix that is part of motif III with a short adjacent β-strand (Fig. 1B). This β-strand in turn forms a two-stranded β-sheet with two residues (Lys-11 and Val-12 in E. coli) from the second GyrB monomer (11, 29). These residues are part of the 14-amino acid GyrB N-terminal arms that are intertwined in the GyrB dimer in the presence of ADPNP (29). The arrangement thus suggests a stabilizing role of K+ in the closed N-gate by aligning a secondary structure element connecting regions from both monomers. It has recently been suggested that ATP binding is required by type II topoisomerases to prevent double-strand breaks due to dissociation of gyrase during the catalytic cycle (30). From the putative interactions outlined above and the requirement of K+ ions for secure nucleotide-induced closure of the N-gate, it is conceivable that K+ ions contribute to the stability of the closed N-gate and thereby allow for disruption of the interactions stabilizing the DNA-gate and for DNA-gate opening and strand passage. Notably, the α-helix containing motif III is close to the phosphates of the bound nucleotide. A central role of K+ in relaying small conformational changes in the nucleotide-binding pocket and in synchronizing the nucleotide cycle with DNA-gate movement could be envisaged.
Acknowledgments
We thank Diana Blank, Ines Hertel, Jessica Guddorf, and Andreas Schmidt for excellent technical assistance.
This work was supported by the Swiss National Science Foundation (National Center of Competence in Research Nanoscale Sciences).
- CTD
- C-terminal domain
- BCK
- rat branched-chain α-keto acid dehydrogenase kinase
- ADPNP
- 5′-adenylyl β,γ-imidodiphosphate
- smFRET
- single-molecule FRET.
REFERENCES
- 1. Gellert M., Mizuuchi K., O'Dea M. H., Nash H. A. (1976) DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc. Natl. Acad. Sci. U.S.A. 73, 3872–3876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Corbett K. D., Schoeffler A. J., Thomsen N. D., Berger J. M. (2005) The structural basis for substrate specificity in DNA topoisomerase IV. J. Mol. Biol. 351, 545–561 [DOI] [PubMed] [Google Scholar]
- 3. Williams N. L., Maxwell A. (1999) Probing the two-gate mechanism of DNA gyrase using cysteine cross-linking. Biochemistry 38, 13502–13511 [DOI] [PubMed] [Google Scholar]
- 4. Williams N. L., Maxwell A. (1999) Locking the DNA-gate of DNA gyrase: investigating the effects on DNA cleavage and ATP hydrolysis. Biochemistry 38, 14157–14164 [DOI] [PubMed] [Google Scholar]
- 5. Kampranis S. C., Bates A. D., Maxwell A. (1999) A model for the mechanism of strand passage by DNA gyrase. Proc. Natl. Acad. Sci. U.S.A. 96, 8414–8419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Williams N. L., Howells A. J., Maxwell A. (2001) Locking the ATP-operated clamp of DNA gyrase: probing the mechanism of strand passage. J. Mol. Biol. 306, 969–984 [DOI] [PubMed] [Google Scholar]
- 7. Gubaev A., Hilbert M., Klostermeier D. (2009) The DNA-gate of Bacillus subtilis gyrase is predominantly in the closed conformation during the DNA supercoiling reaction. Proc. Natl. Acad. Sci. U.S.A. 106, 13278–13283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gubaev A., Klostermeier D. (2011) DNA-induced narrowing of the gyrase N-gate coordinates T-segment capture and strand passage. Proc. Natl. Acad. Sci. U.S.A. 108, 14085–14090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lanz M. A., Klostermeier D. (2011) Guiding strand passage: DNA-induced movement of the gyrase C-terminal domains defines an early step in the supercoiling cycle. Nucleic Acids Res. 39, 9681–9694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tingey A. P., Maxwell A. (1996) Probing the role of the ATP-operated clamp in the strand passage reaction of DNA gyrase. Nucleic Acids Res. 24, 4868–4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wigley D. B., Davies G. J., Dodson E. J., Maxwell A., Dodson G. (1991) Crystal structure of an N-terminal fragment of the DNA gyrase B protein. Nature 351, 624–629 [DOI] [PubMed] [Google Scholar]
- 12. Ali J. A., Jackson A. P., Howells A. J., Maxwell A. (1993) The 43-kilodalton N-terminal fragment of the DNA gyrase B protein hydrolyzes ATP and binds coumarin drugs. Biochemistry 32, 2717–2724 [DOI] [PubMed] [Google Scholar]
- 13. Ali J. A., Orphanides G., Maxwell A. (1995) Nucleotide binding to the 43-kilodalton N-terminal fragment of the DNA gyrase B protein. Biochemistry 34, 9801–9808 [DOI] [PubMed] [Google Scholar]
- 14. Roca J., Wang J. C. (1994) DNA transport by a type II DNA topoisomerase: evidence in favor of a two-gate mechanism. Cell 77, 609–616 [DOI] [PubMed] [Google Scholar]
- 15. Roca J., Berger J. M., Harrison S. C., Wang J. C. (1996) DNA transport by a type II topoisomerase: direct evidence for a two-gate mechanism. Proc. Natl. Acad. Sci. U.S.A. 93, 4057–4062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Dutta R., Inouye M. (2000) GHKL, an emergent ATPase/kinase superfamily. Trends Biochem. Sci. 25, 24–28 [DOI] [PubMed] [Google Scholar]
- 17. Machius M., Chuang J. L., Wynn R. M., Tomchick D. R., Chuang D. T. (2001) Structure of rat BCKD kinase: nucleotide-induced domain communication in a mitochondrial protein kinase. Proc. Natl. Acad. Sci. U.S.A. 98, 11218–11223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sissi C., Marangon E., Chemello A., Noble C. G., Maxwell A., Palumbo M. (2005) The effects of metal ions on the structure and stability of the DNA gyrase B protein. J. Mol. Biol. 353, 1152–1160 [DOI] [PubMed] [Google Scholar]
- 19. Liu L. F., Wang J. C. (1978) Micrococcus luteus DNA gyrase: active components and a model for its supercoiling of DNA. Proc. Natl. Acad. Sci. U.S.A. 75, 2098–2102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fu G., Wu J., Liu W., Zhu D., Hu Y., Deng J., Zhang X. E., Bi L., Wang D. C. (2009) Crystal structure of DNA gyrase B′ domain sheds lights on the mechanism for T-segment navigation. Nucleic Acids Res. 37, 5908–5916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bashkirov V. I., Zvingila D. J. (1991) Sequence specificity of Bacillus subtilis DNA gyrase in vivo. Genetica 85, 3–12 [DOI] [PubMed] [Google Scholar]
- 22. Göttler T., Klostermeier D. (2007) Dissection of the nucleotide cycle of B. subtilis DNA gyrase and its modulation by DNA. J. Mol. Biol. 367, 1392–1404 [DOI] [PubMed] [Google Scholar]
- 23. Theissen B., Karow A. R., Köhler J., Gubaev A., Klostermeier D. (2008) Cooperative binding of ATP and RNA induces a closed conformation in a DEAD box RNA helicase. Proc. Natl. Acad. Sci. U.S.A. 105, 548–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hilbert M., Kebbel F., Gubaev A., Klostermeier D. (2011) eIF4G stimulates the activity of the DEAD box protein eIF4A by a conformational guidance mechanism. Nucleic Acids Res. 39, 2260–2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hu X., Machius M., Yang W. (2003) Monovalent cation dependence and preference of GHKL ATPases and kinases. FEBS Lett. 544, 268–273 [DOI] [PubMed] [Google Scholar]
- 26. Kampranis S. C., Maxwell A. (1998) Hydrolysis of ATP at only one GyrB subunit is sufficient to promote supercoiling by DNA gyrase. J. Biol. Chem. 273, 26305–26309 [DOI] [PubMed] [Google Scholar]
- 27. Roca J. (2009) Topoisomerase II: a fitted mechanism for the chromatin landscape. Nucleic Acids Res. 37, 721–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Roca J. (2004) The path of the DNA along the dimer interface of topoisomerase II. J. Biol. Chem. 279, 25783–25788 [DOI] [PubMed] [Google Scholar]
- 29. Brino L., Urzhumtsev A., Mousli M., Bronner C., Mitschler A., Oudet P., Moras D. (2000) Dimerization of Escherichia coli DNA gyrase B provides a structural mechanism for activating the ATPase catalytic center. J. Biol. Chem. 275, 9468–9475 [DOI] [PubMed] [Google Scholar]
- 30. Bates A. D., Berger J. M., Maxwell A. (2011) The ancestral role of ATP hydrolysis in type II topoisomerases: prevention of DNA double-strand breaks. Nucleic Acids Res. 39, 6327–6339 [DOI] [PMC free article] [PubMed] [Google Scholar]