Background: Tandem L27 domains are important for multidomain proteins to assemble into supramolecular complexes for cell polarity regulation.
Results: Tandem L27 domain-mediated tripartite Patj/Pals1/Mals2 and DLG1/CASK/Mals2 complexes form in a mutually independent assembly mode.
Conclusion: The mutually independent assembly mode may be a novel mechanism for tandem L27 domain-mediated, tripartite complex formation.
Significance: These findings reveal the distinct mechanism of tandem L27 domain-mediated assembly of obligate supramolecular complexes.
Keywords: Analytical Ultracentrifugation, Cell Polarity, Crystal Structure, Protein Assembly, Scaffold Proteins, Heterotrimer, Tandem L27 Domains, Tripartite Complex
Abstract
The assembly of supramolecular complexes in multidomain scaffold proteins is crucial for the control of cell polarity. The scaffold protein of protein associated with Lin-7 1 (Pals1) forms a complex with two other scaffold proteins, Pals-associated tight junction protein (Patj) and mammalian homolog-2 of Lin-7 (Mals2), through its tandem Lin-2 and Lin-7 (L27) domains to regulate apical-basal polarity. Here, we report the crystal structure of a 4-L27 domain-containing heterotrimer derived from the tripartite complex Patj/Pals1/Mals2. The heterotrimer consists of two cognate pairs of heterodimeric L27 domains with similar conformations. Structural analysis and biochemical data further show that the dimers assemble mutually independently. Additionally, such mutually independent assembly of the two heterodimers can be observed in another tripartite complex, Disks large homolog 1 (DLG1)/calcium-calmodulin-dependent serine protein kinase (CASK)/Mals2. Our results reveal a novel mechanism for tandem L27 domain-mediated, supramolecular complex assembly with a mutually independent mode.
Introduction
Establishment of cell polarity is indispensable for most eukaryotic cell functions. The asymmetric distribution of a set of evolutionarily conserved cell polarity proteins or lipids is required for establishing and maintaining cell polarization (1, 2). Many of these conserved proteins are multidomain scaffold proteins, and they often interact with each other to assemble supramolecular protein complexes, thereby mediating the fundamental process for cell polarity (3, 4). The L27 domain, initially identified in the Caenorhabditis elegans Lin-2 and Lin-7 proteins (5), exists in many scaffold proteins and is involved in assembling essential supramolecular protein complexes that play critical roles in cell polarity (6–11).
Pals1, also known as membrane-associated palmitoylated protein 5 (MPP5),3 belongs to the membrane-associated guanylate kinase (MAGUK) family and contains two tandem L27 domains called L27N and L27C, a PDZ domain, an SH3 domain, and a guanylate kinase (GuK) domain (see Fig. 1A). Patj contains one N-terminal L27 domain that mediates binding to the L27N domain in Pals1 followed by 10 PDZ domains (see Fig. 1A) (10). Mammalian homolog of Lin-7 (Mals) has three genes (Mals1, Mals2, and Mals3) in mammals, and each encodes a small protein containing an L27 domain, which binds the L27C domain of Pals1, and a PDZ domain (see Fig. 1A) (6, 12, 13). Loss of Mals destabilizes Pals1, leading to tight junction defects (14). When the Mals genes were genetically disrupted in a mouse model, Patj and Pals1 were lost from the apical surface, suggesting that L27 domain-mediated, tripartite Patj/Pals1/Mals complex assembly is essential for apical-basal polarity (15, 16).
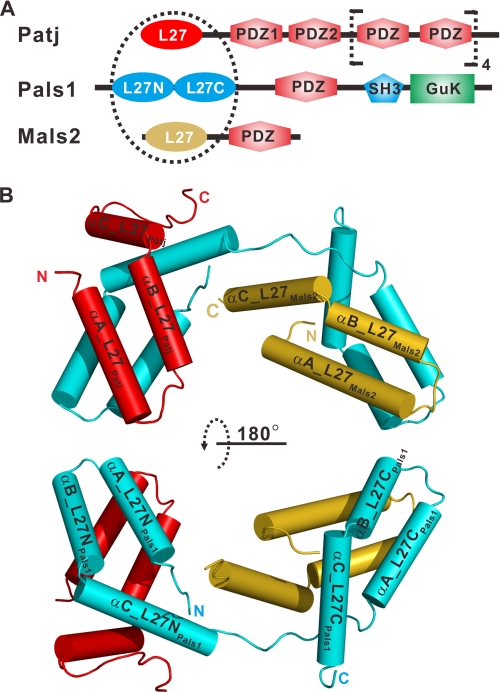
FIGURE 1.
Structure of L27 heterotrimer. A, schematic of the domain organization of rat Patj, human Pals1, and mouse Mals2. The dashed circle indicates the tripartite complex consisting of four L27 domains. The L27 domains of Patj (L27Patj), Pals1 ((L27N-L27C)Pals1), and Mals2 (L27Mals2) are shown in red, cyan, and gold, respectively. GuK, guanylate kinase. B, graphic representation of the overall structure of the heterotrimer colored as in A. Two α-helices formed by artificial linkers are not shown in the figure for clarity.
The structural basis of cognate L27 domain heterodimer assembly has been studied extensively by NMR and x-ray crystallography and has previously led to the proposal of a unified model of symmetric L27 homotetramers (dimer of heterodimers) (17–20). Recently, the structure of a tandem L27 domain-mediated tripartite L27DLG1/(L27N-L27C)MPP7/L27Mals3 complex showed the asymmetric, cooperative assembly of a heterotrimer consisting of two cognate pairs of heterodimeric L27 domains (21).
In this study, we solved the crystal structure of the L27 domain heterotrimer from the tripartite complex Patj/Pals1/Mals2. The structure of the L27 domain complex, together with data derived from various biochemical studies, establishes a novel, symmetric, and mutually independent assembly mode of heterotrimer formation mediated by tandem L27 domains in the specific tripartite Patj/Pals1/Mals2 complex. Additionally, we showed that another specific tripartite DLG1/CASK/Mals2 complex is also formed via tandem L27 domain-mediated, symmetric, and mutually independent assembly.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
To construct a single-chain fusion protein of the L27Patj/(L27N-L27C)Pals1/L27Mals2 complex (supplemental Fig. S1A and molecule 1 in supplemental Fig. S1C), DNA fragments corresponding to the L27 domain of rat Patj (residues 1–68), the tandem L27 domains of human Pals1 (residues 119–232), and the L27 domain of mouse Mals2 (residues 3–66) were amplified by PCR and linked with two rhinovirus 3C protease-cleavable segments (Leu-Glu-Val-Leu-Phe-Gln-Gly-Pro), and a triglycine cassette (GGG) was inserted before the second 3C protease-cleavable segment. The single open reading frame was cloned into the pET32a vector (Novagen, San Diego, CA), in which the S-tag and the thrombin recognition site were replaced with a sequence encoding a tobacco etch virus (TEV) protease-cleavable segment (Glu-Asn-Leu-Tyr-Phe-Gln-Ser).
BL21(DE3) CodonPlus Escherichia coli cells harboring the expression plasmid were grown in LB medium at 37 °C until the A600 reached 0.6 and then induced with 0.3 mm isopropyl-β-d-thiogalactoside at 16 °C for ∼16–18 h. After centrifugation at 5000 rpm for 15 min, E. coli cells were resuspended in T50N500I5 buffer (50 mm Tris-HCl, pH 7.9, 500 mm NaCl, and 5 mm imidazole) supplemented with 1 mm phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, and 1 μg/ml antipain. The cells were then lysed by sonication. After the lysates had been centrifuged at 18,000 rpm for 30 min, the supernatant was loaded onto a nickel-nitrilotriacetic acid-agarose column (Qiagen, Valencia, CA) that was equilibrated with T50N500I5 buffer. The nickel-nitrilotriacetic acid column was washed with 3 column volumes of T50N500I5 buffer. The His6-tagged protein was eluted with T50N500I5 buffer containing 500 mm imidazole. The eluted proteins were digested with TEV protease overnight to remove the N-terminal His6 tag and then loaded onto a HiLoad 26/60 Superdex 200 size-exclusion column (GE Healthcare) and eluted with T50N50E1D1 buffer (50 mm Tris-HCl, pH 8.0, 50 mm NaCl, 1 mm EDTA, and 1 mm DTT) at a flow rate of 2.5 ml/min. Each fraction of the column eluate was 5 ml. The single-chain fusion protein peak of the L27Patj/(L27N-L27C)Pals1/L27Mals2 complex was identified by SDS-PAGE, and the corresponding fractions were pooled and concentrated to 90 mg/ml for crystallization trials. To obtain the three separate chains of the L27Patj/(L27N-L27C)Pals1/L27Mals2 complex (supplemental Fig. S2A), 3C protease was added to the single-chain fusion protein to cleave the covalent linker. A Se-Met-substituted version of the L27Patj/(L27N-L27C)Pals1/L27Mals2 single-chain fusion protein was produced following the same protocol that was used for the wild-type protein, with the exception that methionine auxotroph E. coli B834 (DE3) cells and LeMaster medium were used to express the recombinant protein.
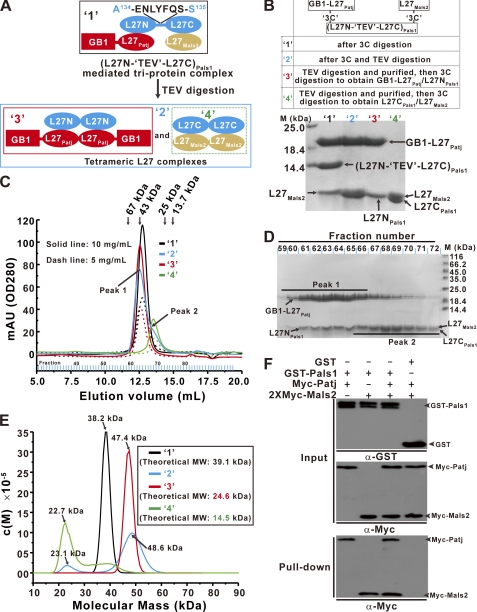
For the GB1-L27Patj/(L27N-L27C)Pals1/27Mals2 single-chain fusion protein (molecule 2 in supplemental Fig. S1C), a B1 domain of streptococcal protein G (GB1) tag was fused to the N terminus of molecule 1, and a TEV protease-cleavable segment (Glu-Asn-Leu-Tyr-Phe-Gln-Ser) was inserted between amino acids Ala134 and Ser135 in the linker region between the tandem L27 domains of (L27N-L27C)Pals1. These tandem L27 domains were designated (L27N-TEV-L27C)Pals1. Protein expression and purification were performed using methods similar to those used for the single-chain fusion protein of the L27Patj/(L27N-L27C)Pals1/L27Mals2 complex mentioned above. To prepare protein sample for biochemical analysis, the GB1-L27Patj/(L27N-27C)Pals1/L27Mals2 single-chain fusion protein was digested with 3C protease to obtain three separate chains for the GB1-L27Patj/(L27N-TEV-L27C)Pals1/L27Mals2 complex (complex 1 in Fig. 4, A and B). To obtain the complexes of GB1-L27Patj/L27NPals1 (complex 3 in Fig. 4, A and B) and L27CPals1/L27Mals2 (complex 4 in Fig. 4, A and B), the single-chain fusion protein was first digested with TEV protease and then loaded onto a Mono Q column (GE Healthcare) to individually obtain each covalently linked GB1-L27Patj-L27NPals1 and L27CPals1-L27Mals2 complex. The GB1-L27Patj-L27NPals1 and the L27CPals1-L27Mals2 complexes were identified by SDS-PAGE according to their different molecular masses. Each complex was pooled and then digested with 3C protease to cleave the covalent linker. It was noted that L27NPals1, L27CPals1, and L27Mals2 migrated as band on 15% Tricine-SDS-PAGE gels because of their similar molecular masses (7633.7, 7178.1, and 7452.5 Da, respectively) (see Fig. 4, B and D).
FIGURE 4.
Tandem L27 domains are important for L27Patj/(L27N-L27C)Pals1/L27Mals2 heterotrimer assembly. A, schematic representation of the GB1-L27Patj/(L27N-TEV-L27C)Pals1/L27Mals2 heterotrimeric complex and the two tetrameric L27 domain complexes formed after digestion of the GB1-L27Patj/(L27N-TEV-L27C)Pals1/L27Mals2 complex with TEV protease. Extension of the linker between the tandem L27 domains with a TEV protease cleavage segment (ENLYFQS) was designated (L27N-TEV-L27C)Pals1. B, four complexes are represented by 1–4 and were prepared according to the summary in the top panel. Bottom panel, 15% Tricine-SDS-PAGE showing each purified complex. C, size-exclusion chromatography of each indicated protein was carried out on a Superose 12 column at two concentrations. mAU, milliabsorbance units. D, after treatment with TEV protease, the digestion mixture formed by the GB1-L27Patj/(L27N-TEV-L27C)Pals1/L27Mals2 complex was fractionated and analyzed by 15% Tricine-SDS-PAGE. E, analytical ultracentrifugation SV experiments measuring the molecular mass of each protein. The inset shows the theoretical molecular weight (MW) of each protein. F, GST pulldown experiments using the Patj/Pals1/Mals2 ternary complex and either Patj/Pals1 or Pals1/Mals2 binary complex formation. Input represents 1% of the cell lysis used in the assay.
For the L27DLG1/(L27N-3C-L27C)CASK/L27Mals2 single-chain fusion protein (molecule 3 in supplemental Fig. S1C), the DNA sequences encoding the L27 domain of rat DLG1 (residues 1–65), tandem L27 domains of mouse CASK (residues 329–460), and L27 domain of mouse Mals2 (residues 2–78) were amplified by PCR. The three fragments were fused into a single open reading frame using two human thrombin-cleavable segments (Leu-Val-Pro-Arg-Gly-Ser). Furthermore, a 3C protease-cleavable segment was inserted between the amino acids Ile412 and Arg413 in the linker region between the tandem L27 domains of (L27N-L27C)CASK. These tandem L27 domains are designated (L27N-3C-L27C)CASK. The single-chain DNA sequence was cloned into an in-house-modified version of the pET32a vector in which the S-tag was removed. The L27DLG1/(L27N-3C-L27C)CASK/L27Mals2 fusion protein was produced by following a protocol similar to that used for the single-chain fusion protein of the L27Patj/(L27N-L27C)Pals1/L27Mals2 complex. Finally, the single-chain fusion protein was digested with thrombin to form the three separate chains of the L27DLG1/(L27N-3C-L27C)CASK/L27Mals2 complex.
Analytical Ultracentrifugation
Sedimentation velocity (SV) and sedimentation equilibrium (SE) experiments were performed in a Beckman Coulter XL-I analytical ultracentrifuge (Beckman Coulter) using double-sector or six-channel centerpieces and sapphirine windows. Before the experiments, the proteins were transferred to buffer containing 50 mm PBS, pH 7.4, 100 mm NaCl, and 1 mm EDTA by Superose 12 10/300 GL column (GE Healthcare). Proteins at absorbances of 0.6 and 1.2 at 280 nm were loaded into double-sector cells for SV experiments, which were conducted at 42,000 rpm and 10 °C and with absorbance detected at 280 nm. For SE experiments, data were collected at 4,000, 22,000, and 27,000 rpm and 4 °C by interference detection using six-channel cells. The concentrations of the proteins were ∼27 and 40 μm. The buffer composition (density and viscosity) and protein partial specific volume (V-bar) were obtained sing the SEDNTERP program (available through the Boston Biomedical Research Institute). The SV and SE data were analyzed using the SEDFIT and SEDPHAT programs (22, 23), respectively.
Crystallization and Data Collection
The wild-type L27Patj/(L27N-L27C)Pals1/L27Mals2 protein complex in the single-chain fusion form (90 mg/ml in T50N50E1D1 buffer) was crystallized using sitting drop vapor diffusion equilibrated with a reservoir solution of 2.8 m sodium acetate trihydrate, pH 7.0. Crystals were grown over 1 month at 20 °C and directly flash-frozen in liquid nitrogen. The Se-Met-substituted crystals were produced in the same manner as the wild-type crystals. Diffraction data sets were collected using beamline BL17U1 at the Shanghai Synchrotron Radiation Facility (SSRF) and processed using the HKL2000 software (24). Both wild-type and Se-Met-substituted crystals belonged to the space group P6122. The wild-type crystals diffracted to 2.05 Å, with unit cell dimensions of a = 145.2 Å, b = 145.2 Å, and c = 202.5Å. Single anomalous data were collected for Se-Met-substituted crystals at the selenium peak wavelength. Se-Met-substituted crystals diffracted to 2.7 Å, with unit cell dimensions of a = 145.1 Å, b = 145.1 Å, and c = 202.4 Å.
Structure Determination and Refinement
The HKL2MAP program (25) was used to search eight selenium sites in one asymmetric unit cell. The initial phases were then calculated by PHENIX software (26). Model building and refinement were performed using COOT (27) and PHENIX (28). After the initial main-chain model was built, the wild-type data were applied to carry out iterative refinement to assign all side chains. The final structure had an Rcryst value of 18.0% and an Rfree value of 22.1%. The Ramachandran plot generated by the program PROCHECK (29) shows that 95.0% residues are in their most favored regions, 4.6% residues are in additional allowed regions, 0.4% residues are in generously allowed regions, and no residue is in disallowed regions. Detailed data collection and refinement statistics are summarized in supplemental Table S1. All figures were made with the PyMOL program (30).
Glutathione S-Transferase(GST) Pulldown Assays
GST-L27Patj, GST-Mals2, and GST-Pals1 fusion proteins were expressed in E. coli BL21 (DE3) CodonPlus cells and purified using a glutathione-Sepharose 4B column (GE Healthcare) and a Superdex 200 size-exclusion column. Transfected HEK293T cells were lysed using 500 μl of ice-cold cell lysis buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 10% glycerol, 1% Nonidet P-40, 1 mm phenylmethylsulfonyl fluoride, 1 μg/ml leupeptin, and 1 μg/ml antipain) and cleared by centrifugation at 13,000 rpm for 20 min at 4 °C. Soluble fractions were incubated with GST fusion proteins at 4 °C for 2 h. Glutathione-Sepharose 4B beads (GE Healthcare) were then added for further incubation at 4 °C for 2 h. The beads were washed with cell lysis buffer and boiled in SDS sample buffer. The prepared samples were separated by SDS-PAGE and analyzed using Western blots.
Co-immunoprecipitation
HEK293T cells were plated at ∼6.5 × 106 cells in 10-cm dishes. The following day, 70–80% confluent cells were transfected with the indicated amounts and various combinations of plasmids. At 24 h after transfection, HEK293T cells were lysed and cleared by centrifugation at 13,000 rpm for 20 min at 4 °C. The supernatants were then incubated with anti-GFP antibody (1 μg) at 4 °C for 2 h. The immune complexes were immobilized on protein A/G agarose beads (Pierce) for an additional 2 h. The resin was washed three times with cell lysis buffer and eluted with SDS sample buffer. Samples were then subjected to Western blot analysis.
RESULTS
Assembly of L27 Domain Heterotrimer
To identify a suitable L27 domain complex for crystallization, several constructs from the same or different species were designed and tested for protein expression and purification. After extensive crystal screening, high-quality crystals were obtained only with the construct containing the L27 domain of rat Patj (L27Patj), the tandem L27 domains of human Pals1 ((L27N-L27C)Pals1), and the L27 domain of mouse Mals2 (L27Mals2). L27Patj, (L27N-L27C)Pals1, and L27Mals2 were fused into a single polypeptide by two 3C protease-cleavable segments. An additional GGG cassette was inserted before the second 3C protease cleavage segment (supplemental Fig. S1, A and C). This technique, which has been used extensively in the determination of L27 domain structures by our laboratory and others, does not alter the global structure of the complexes (17, 19–21). The purified L27Patj/(L27N-L27C)Pals1/L27Mals2 complex, both as a single-chain fusion and as three separate chains, eluted as a single peak from an analytical gel filtration column with a molecular mass corresponding to that of the heterotrimer (supplemental Fig. S2, A–C). Analytical ultracentrifugation confirmed that the L27Patj/(L27N-L27C)Pals1/L27Mals2 complex assembled into a heterotrimer with a molecular mass of ∼30.4 and ∼32.3 kDa for complexes with and without covalent linkers, respectively (supplemental Fig. S2, D–G). We conclude that the L27Patj/(L27N-L27C)Pals1/L27Mals2 complex forms a heterotrimer containing four L27 domains.
Structure of L27 Domain Heterotrimer
The structure of the L27 domain heterotrimer was determined using single-wavelength anomalous dispersion. The Fourier map calculated from the initial single-wavelength anomalous dispersion phases showed that the four molecules of the L27Patj/(L27N-L27C)Pals1/L27Mals2 complex were present in one asymmetric unit (supplemental Fig. S3). The four molecules were similar in structure, with a root mean square deviation of less than 0.61 Å for the 245 Cα atoms. Therefore, we discuss only molecule A (colored red in supplemental Fig. S3) in the following section.
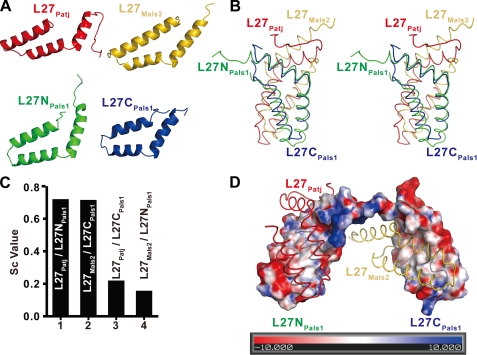
The overall structure of the L27Patj/(L27N-L27C)Pals1/L27Mals2 complex consists of two cognate pairs of L27 domain heterodimers (L27Patj/L27NPals1 and L27CPals1/L27Mals2) connected by the hinge region between L27NPals1 and L27CPals1 (Fig. 1B and supplemental Fig. S1B). Each L27 domain had three α-helices, although they share only 5–14% amino acid sequence identity (Figs. 1B and 2A and supplemental Fig. S4). The superimposition of the heterodimeric L27Patj/L27NPals1 onto L27CPals1/L27Mals2 showed that these two heterodimers have similar conformations (Fig. 2B). The buried surface area was calculated using the AREAIMOL program (31) and was 2512.4 Å2 for the L27Patj/L27NPals1 heterodimer and 2471.6 Å2 for the L27CPals1/L27Mals2 heterodimer, indicating that these two dimers pack tightly. Furthermore, the calculation of the surface complementary value (32) showed that the L27NPals1 and L27CPals1 domains are complementary to the L27Patj and L27Mals2 domains, respectively, but cannot be exchanged (Fig. 2C), indicating that each cognate pair of L27 heterodimers (L27Patj/L27NPals1 and L27CPals1/L27Mals2) is an integral structural unit for each L27 heterodimer assembly.
FIGURE 2.
Binding specificity in L27 domain heterotrimer. A, graphic representation of individual L27 domains. L27Patj, L27NPals1, L27CPals1, and L27Mals2 are shown in red, green, blue, and gold, respectively. B, stereo view of the superimposition of the two heterodimers. C, bar graph of surface complementary (Sc) values of heterodimeric, L27 domain complexes. D, electrostatic surface representation of (L27N-L27C)Pals1. Positive, negative, and neutral charges are shown in red, blue, and white, respectively. The graphic representation of L27Patj and L27Mals2 is shown in red and gold, respectively.
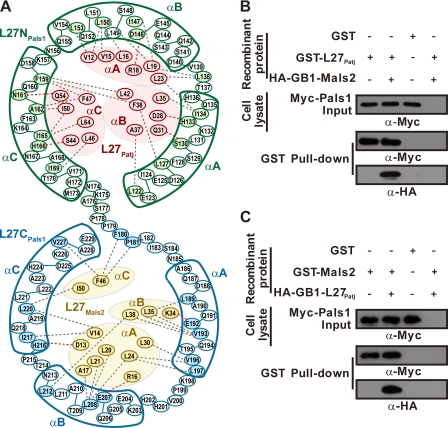
Although there are similarities in conformation and buried surface area, when assembled in this heterotrimer complex, L27Patj/L27NPals1 and L27CPals1/L27Mals2 do not interact with each other (Fig. 3A), indicating that each L27 heterodimer is formed mutually independently. This result contrasts sharply with previous findings that two L27 heterodimers are asymmetric and cooperative in the L27DLG1/(L27N-L27C)MPP7/L27Mals3 heterotrimeric complex (21). To verify our findings, we performed a GST pulldown assay. Our results showed that L27Patj bound Pals1 with similar affinities in the absence or presence of Mals2 (Fig. 3B) and vice versa (Fig. 3C).
FIGURE 3.
Dissection of L27 heterotrimer assembly. A, schematic illustration of detailed, noncovalent interactions within the interface between L27Patj and L27NPals1 of one heterodimer and L27CPals1 and L27Mals2 of the other heterodimer. Residues from L27Patj, L27NPals1, L27CPals1, and L27Mals2 are shown in red, green, blue, and gold ellipses, respectively. Residues involved in the interactions are indicated in the corresponding color. Hydrophobic interactions and hydrogen bonds between residues are designated by gray and red dashed lines, respectively. B and C, GST pulldown experiments with L27Patj/Pals1 (B) or Mals2/Pals1 binary complexes (C) and L27Patj/Pals1/Mals2 ternary complex formation (B and C). Precipitated proteins were visualized by first blotting with anti-Myc antibody and then stripping the membrane and immunoblotting with anti-HA antibodies. Input represents 1% of the cell lysis used in the assay.
Surface electrostatic analysis suggested that the assembly of the L27Patj/L27NPals1 and L27CPals1/L27Mals2 heterodimers is mediated primarily by extensive hydrophobic interactions inside the core of the complex (Fig. 2D). In addition to hydrophobic interactions, several hydrogen bonds between charged residues are involved in the assembly of the two L27 heterodimers. Detailed interactions in each L27 heterodimer (L27Patj/L27NPals1 and L27CPals1/L27Mals2) of the L27Patj/(L27N-L27C)Pals1/L27Mals2 complex are shown in Fig. 3A. The residues participating in the interactions are evolutionarily conserved according to structure-based sequence alignment (supplemental Fig. S5).
L27 domains of the same homologous protein from different species are usually conserved. The amino acid sequence identity of L27Patj among mouse, rat, and human is very high (supplemental Fig. S5A). Within the structural region of L27Patj (residues Gln11–Ser56), only four residues are divergent over 46 amino acids, and these four residues are not involved in binding to L27NPals1. Within the structural region of the tandem L27 domains of Pals1 (residues Leu122–Glu229) (supplemental Fig. S5B), there are nine different residues over 108 amino acids across mouse, rat, and human, and these residues are also not involved in interactions between each cognate pair of L27 domains except for one residue (Val169 versus Ile169, which are similar hydrophobic residues). The amino acid sequences of L27Mals2 from mouse, rat, and human are completely conserved (supplemental Fig. S5C). Taken together, we conclude that the 4-L27 domain-containing heterotrimer from the same species should have a very similar structure to that determined in this study with rat Patj, human Pals1, and mouse Mals2. Furthermore, several structures of cognate pair L27 domains from mixed species have been determined (18–20).
Specificity of Heterotrimer Assembly
To test the role of the tandem L27 domains in heterotrimeric L27Patj/(L27N-L27C)Pals1/L27Mals2 complex formation, a TEV protease cleavage segment (ENLYFQS) was inserted between amino acids Ala134 and Ser135 in the linker region between the L27 domains of (L27N-L27C)Pals1 (referred to as (L27N-TEV-L27C)Pals1 below) (Fig. 4A and supplemental Fig. S1C). This linker region mutant (L27N-TEV-L27C)Pals1 can also form a heterotrimer with GB1-tagged L27Patj (GB1-L27Patj) and L27Mals2, as indicated by the elution of the purified protein complex as a single peak from size-exclusion chromatography with a molecular mass corresponding to a heterotrimer (Fig. 4C, black line). Analytical ultracentrifugation confirmed that the GB1-L27Patj/(L27N-TEV-L27C)Pals1/L27Mals2 complex assembled into a heterotrimer with a molecular mass of ∼38.2 kDa (Fig. 4E, black line). Interestingly, when the GB1-L27Patj/(L27N-TEV-L27C)Pals1/L27Mals2 complex was digested with TEV protease, the resulting protein mixture eluted as two peaks from size-exclusion chromatography (Fig. 4C, cyan line). More detailed biochemical analysis revealed that each peak represented a tetrameric L27 complex containing the cognate pairs GB1-L27Patj/L27NPals1 and L27CPals1/L27Mals2 (Fig. 4, B–E). Taken together, these data reveal that the tandem L27 domains are essential for L27Patj/(L27N-L27C)Pals1/L27Mals2 ternary complex assembly. Furthermore, a full-length, tripartite Patj/Pals1/Mals2 complex could be assembled by tandem L27 domains in a GST pulldown assay in vitro and a co-immunoprecipitation assay in vivo (Fig. 4F and supplemental Fig. S6).
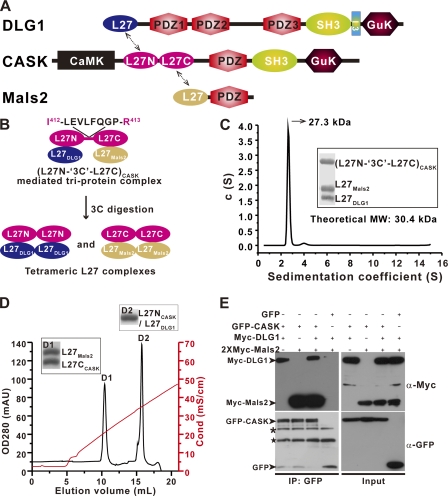
In mammals, several other scaffold proteins contain two tandem L27 domains (supplemental Fig. S7). To further investigate tandem L27 domain-mediated, supramolecular complex assembly, we examined the tandem L27 domains of CASK in more detailed studies. CASK, which, like Pals1, contains two tandem L27 domains interacts with DLG1 through the cognate pair L27DLG1/L27NCASK and with Mals2 through the cognate pair L27CCASK/L27Mals2 to form a tripartite DLG1/CASK/Mals2 cell polarity complex (Fig. 5A) (7, 15, 17, 19, 33). To investigate the role of CASK tandem L27 domains in the assembly of the DLG1/CASK/Mals2 ternary complex, we used strategies similar to those applied to Patj/Pals1/Mals2. A 3C protease cleavage segment (LEVLFQGP) was inserted between amino acids Ile412 and Arg413 in the linker region between the L27 domains of (L27N-L27C)CASK (referred to as (L27N-3C-L27C)CASK below) (Fig. 5B and supplemental Fig. S1C). The purified three separated chains of the L27DLG1/(L27N-3C-L27C)CASK/L27Mals2 complex assembled into a heterotrimer with a molecular mass of ∼27.3 kDa as determined by analytical ultracentrifugation (Fig. 5C). After 3C protease digestion, the resulting L27DLG1/(L27N-3C-L27C)CASK/L27Mals2 complex protein mixture was loaded onto an ion-exchange column, and we obtained two tetrameric L27 domain complexes for each cognate pair of L27DLG1/L27NCASK and L27CCASK/L27Mals2 (Fig. 5, B and D) (17, 19). Taken together, these data reveal that the tandem L27 domains of CASK are essential for L27DLG1/(L27N-L27C)CASK/L27Mals2 ternary complex assembly. Additionally, a full-length tripartite DLG1/CASK/Mals2 complex could be assembled by tandem L27 domains in a co-immunoprecipitation assay in vivo (Fig. 5E).
FIGURE 5.
L27DLG1/(L27N-L27C)CASK/L27Mals2 heterotrimer formation requires the tandem L27 domains of CASK. A, schematic domain organization of rat DLG1, mouse CASK, and mouse Mals2. Four L27 domains (dashed arrows) mediate tripartite complex formation. The L27 domains from DLG1 (L27DLG1), the tandem L27 domains of CASK ((L27N-L27C)CASK), and the L27 domain of Mals2 (L27Mals2) are shown in blue, magenta, and gold, respectively. B, schematic representation of the L27DLG1/(L27N-3C-L27C)CASK/L27Mals2 heterotrimeric complex and the two tetrameric L27 domain complexes formed by 3C protease digestion of the L27DLG1/(L27N-3C-L27C)CASK/L27Mals2 complex. Extension of the linker between the tandem L27 domains with a 3C protease cleavage segment (LEVLFQGP) is designated as (L27N-3C-L27C)CASK. C, c(S) distribution from an SV experiment of the L27DLG1/(L27N-3C-L27C)CASK/L27Mals2 complex. The inset shows the purified complex used in the SV experiment. MW, molecular weight. D, Mono Q chromatogram profile of the L27DLG1/(L27N-3C-L27C)CASK/L27Mals2 complex after 3C protease digestion. L27DLG1/L27NCASK and L27CCASK/L27Mals2 complexes eluted in the D1 and D2 peaks, respectively. mAU, milliabsorbance units; OD, optical density; Cond (mS/cm), Conductance (milliSiemens per centimeters). E, co-immunoprecipitation (IP) experiments with DLG1/CASK/Mals2 ternary complex and DLG1/CASK or CASK/Mals2 binary complex formation. Input represents 1% of the cell lysis used in the assay. The black asterisk represents nonspecific protein bands, and the black star represents the GFP antibody heavy chain.
DISCUSSION
A bioinformatic survey in mammals reveals that L27 domains from various scaffold proteins are categorized into two subfamilies: type A and type B (supplemental Fig. S7) (17). Not only are all L27 monomers unable to specifically self-associate, they cannot interact with distinct monomers from within the same type and can form heterodimers only with monomers from a different type (7, 17, 33). Previous studies (17, 19) have shown that the symmetric L27 homotetramers (dimer of heterodimers) is a unified assembly mode for cognate pairs of type A/type B L27 domain complexes.
Under physiological conditions, the L27 domain often forms a tandem L27 domain-mediated heterotrimer to achieve its biological functions as a cell polarity regulator (10, 15, 34). A previous study of a 4-L27 domain-containing heterotrimer from a tripartite DLG1/MPP7/Mals3 complex showed that the heterodimer of L27CMPP7/L27Mals3 has multiple contacts with L27DLG1 of the L27DLG1/L27NMPP7 heterodimer (21), suggesting that the assembly of the L27CMPP7/L27Mals3 heterodimer promotes the recruitment of L27DLG1 and further facilitates the L27DLG1/L27NMPP7 heterodimer formation. In support of this idea, a prior study showed that the association of DLG1 with MPP7 required the prior formation of a complex between MPP7 and Mals3 (34). Thus, these studies revealed that the two cognate pairs of L27 heterodimers assemble into heterotrimers in a tandem L27 domain-mediated cooperative mode. It was reported that MPP3(DLG3) and MPP2(DLG2), two Mals-binding proteins, bind DLG1/synapse-associated protein 97 (SAP97) and require both the L27N and the L27C domains of MPP3 and MPP2, respectively (35), indicating that these heterotrimers may also form through the tandem L27 domain-mediated cooperative assembly mode.
In sharp contrast to this observation, the structure reported here illustrates that within the L27Patj/(L27N-L27C)Pals1/L27Mals2 heterotrimeric complex, there is no interaction between the cognate pairs of L27 heterodimers (L27Patj/L27NPals1 and L27CPals1/L27Mals2), indicating that each prototypical L27 heterodimer is formed independently (Figs. 1 and 3). A similar situation seems to exist in the tandem L27 domain-mediated heterotrimeric assembly of L27DLG1/(L27N-L27C)CASK/L27Mals2 complex (Fig. 5). Furthermore, our biochemical results indicate that the tandem L27 domains of Pals1 and CASK are indispensable for the tripartite assembly of the Patj/Pals1/Mals2 and DLG1/CASK/Mals2 complexes, respectively (Figs. 4 and 5). Together, our results suggest a novel mechanism for tandem L27 domain-mediated supramolecular complex formation by a mutually independent mode in tripartite Patj/Pals1/Mals2 and DLG1/CASK/Mals2 complexes.
The L27N domain-binding partners of MPP4 and Pals2 (MPP6) have not yet been identified, and the mechanism of L27 domain-mediated tripartite complex assembly mechanism must be uncovered to fully elucidate their biological functions. Nevertheless, although the assembly mode of heterotrimeric structure is divergent, the 4-L27 domain-containing heterotrimeric structure may represent a general assembly mode for tandem L27 domain-mediated obligate tripartite complexes. This distinct mechanism of tandem L27 domain-mediated assembly of heterotrimeric structures may, in part, reveal the correct assembly mode of L27 domain scaffold proteins during the organization of specific supramolecular protein complexes. This hypothesis remains to be tested by further functional studies in vivo. Such tandem L27 domain-mediated multimeric scaffolds provide various nucleation platforms for the organization of suprasignaling complexes that have been implicated in a wide range of elementary cellular processes, including asymmetric cell division, cell polarity control, and the regulation of cytoskeletal dynamics and signal transduction.
Supplementary Material
Acknowledgments
We are grateful to the staff at the beamline BL17U1 of the Shanghai Synchrotron Radiation Facility (SSRF).
This work was funded by the 973 Program (Grant 2009CB825504), the Natural Science Foundation of China (NSFC) (Grants 31140029, 31100527, and 31170684), and the Fundamental Research Funds for the Central Universities (Grants 65011621 and 65020241).

This article contains supplemental Table S1 and Figs. S1–S7.
The atomic coordinates and structure factors (code 3UIT) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- MPP5
- membrane-associated palmitoylated protein 5
- CASK
- calmodulin-dependent serine protein kinase
- TEV
- tobacco etch virus
- SV
- sedimentation velocity
- SE
- sedimentation equilibrium
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1. Macara I. G. (2004) Par proteins: partners in polarization. Curr. Biol. 14, R160–R162 [PubMed] [Google Scholar]
- 2. Mellman I., Nelson W. J. (2008) Coordinated protein sorting, targeting, and distribution in polarized cells. Nat. Rev. Mol. Cell Biol. 9, 833–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pawson T., Nash P. (2003) Assembly of cell regulatory systems through protein interaction domains. Science 300, 445–452 [DOI] [PubMed] [Google Scholar]
- 4. Funke L., Dakoji S., Bredt D. S. (2005) Membrane-associated guanylate kinases regulate adhesion and plasticity at cell junctions. Annu. Rev. Biochem. 74, 219–245 [DOI] [PubMed] [Google Scholar]
- 5. Doerks T., Bork P., Kamberov E., Makarova O., Muecke S., Margolis B. (2000) L27, a novel heterodimerization domain in receptor targeting proteins Lin-2 and Lin-7. Trends Biochem. Sci. 25, 317–318 [DOI] [PubMed] [Google Scholar]
- 6. Butz S., Okamoto M., Südhof T. C. (1998) A tripartite protein complex with the potential to couple synaptic vesicle exocytosis to cell adhesion in brain. Cell 94, 773–782 [DOI] [PubMed] [Google Scholar]
- 7. Lee S., Fan S., Makarova O., Straight S., Margolis B. (2002) A novel and conserved protein-protein interaction domain of mammalian Lin-2/CASK binds and recruits SAP97 to the lateral surface of epithelia. Mol. Cell Biol. 22, 1778–1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bachmann A., Schneider M., Theilenberg E., Grawe F., Knust E. (2001) Drosophila Stardust is a partner of Crumbs in the control of epithelial cell polarity. Nature 414, 638–643 [DOI] [PubMed] [Google Scholar]
- 9. Hong Y., Stronach B., Perrimon N., Jan L. Y., Jan Y. N. (2001) Drosophila Stardust interacts with Crumbs to control polarity of epithelia but not neuroblasts. Nature 414, 634–638 [DOI] [PubMed] [Google Scholar]
- 10. Roh M. H., Makarova O., Liu C. J., Shin K., Lee S., Laurinec S., Goyal M., Wiggins R., Margolis B. (2002) The Maguk protein, Pals1, functions as an adapter, linking mammalian homologs of Crumbs and Discs Lost. J. Cell Biol. 157, 161–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lemmers C., Médina E., Delgrossi M. H., Michel D., Arsanto J. P., Le Bivic A. (2002) hINADl/PATJ, a homolog of Discs Lost, interacts with Crumbs and localizes to tight junctions in human epithelial cells. J. Biol. Chem. 277, 25408–25415 [DOI] [PubMed] [Google Scholar]
- 12. Kamberov E., Makarova O., Roh M., Liu A., Karnak D., Straight S., Margolis B. (2000) Molecular cloning and characterization of Pals, proteins associated with mLin-7. J. Biol. Chem. 275, 11425–11431 [DOI] [PubMed] [Google Scholar]
- 13. Jo K., Derin R., Li M., Bredt D. S. (1999) Characterization of MALS/Velis-1, -2, and -3: a family of mammalian LIN-7 homologs enriched at brain synapses in association with the postsynaptic density-95/NMDA receptor postsynaptic complex. J. Neurosci. 19, 4189–4199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Straight S. W., Pieczynski J. N., Whiteman E. L., Liu C. J., Margolis B. (2006) Mammalian lin-7 stabilizes polarity protein complexes. J. Biol. Chem. 281, 37738–37747 [DOI] [PubMed] [Google Scholar]
- 15. Olsen O., Funke L., Long J. F., Fukata M., Kazuta T., Trinidad J. C., Moore K. A., Misawa H., Welling P. A., Burlingame A. L., Zhang M., Bredt D. S. (2007) Renal defects associated with improper polarization of the CRB and DLG polarity complexes in MALS-3 knockout mice. J. Cell Biol. 179, 151–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Srinivasan K., Roosa J., Olsen O., Lee S. H., Bredt D. S., McConnell S. K. (2008) MALS-3 regulates polarity and early neurogenesis in the developing cerebral cortex. Development 135, 1781–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Feng W., Long J. F., Fan J. S., Suetake T., Zhang M. (2004) The tetrameric L27 domain complex as an organization platform for supramolecular assemblies. Nat. Struct. Mol. Biol. 11, 475–480 [DOI] [PubMed] [Google Scholar]
- 18. Li Y., Karnak D., Demeler B., Margolis B., Lavie A. (2004) Structural basis for L27 domain-mediated assembly of signaling and cell polarity complexes. EMBO J. 23, 2723–2733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Feng W., Long J. F., Zhang M. (2005) A unified assembly mode revealed by the structures of tetrameric L27 domain complexes formed by mLin-2/mLin-7 and Patj/Pals1 scaffold proteins. Proc. Natl. Acad. Sci. U.S.A. 102, 6861–6866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Petrosky K. Y., Ou H. D., Löhr F., Dötsch V., Lim W. A. (2005) A general model for preferential hetero-oligomerization of LIN-2/7 domains: mechanism underlying directed assembly of supramolecular signaling complexes. J. Biol. Chem. 280, 38528–38536 [DOI] [PubMed] [Google Scholar]
- 21. Yang X., Xie X., Chen L., Zhou H., Wang Z., Zhao W., Tian R., Zhang R., Tian C., Long J., Shen Y. (2010) Structural basis for tandem L27 domain-mediated polymerization. FASEB J. 24, 4806–4815 [DOI] [PubMed] [Google Scholar]
- 22. Schuck P. (2000) Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and Lamm equation modeling. Biophys. J. 78, 1606–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schuck P. (2003) On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation. Anal. Biochem. 320, 104–124 [DOI] [PubMed] [Google Scholar]
- 24. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 25. Pape T., Schneider T. R. (2004) HKL2MAP: a graphical user interface for macromolecular phasing with SHELX programs. J. Appl. Crystallogr. 37, 843–844 [Google Scholar]
- 26. Zwart P. H., Afonine P. V., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., McKee E., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Storoni L. C., Terwilliger T. C., Adams P. D. (2008) Automated structure solution with the PHENIX suite. Methods Mol. Biol. 426, 419–435 [DOI] [PubMed] [Google Scholar]
- 27. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 30. DeLano W. L. (2010) The PyMOL Molecular Graphics System, version 1.3r1, Schrödinger, LLC, New York [Google Scholar]
- 31. Lee B., Richards F. M. (1971) The interpretation of protein structures: estimation of static accessibility. J. Mol. Biol. 55, 379–400 [DOI] [PubMed] [Google Scholar]
- 32. Lawrence M. C., Colman P. M. (1993) Shape complementarity at protein/protein interfaces. J. Mol. Biol. 234, 946–950 [DOI] [PubMed] [Google Scholar]
- 33. Harris B. Z., Venkatasubrahmanyam S., Lim W. A. (2002) Coordinated folding and association of the LIN-2, -7 (L27) domain: An obligate heterodimerization involved in assembly of signaling and cell polarity complexes. J. Biol. Chem. 277, 34902–34908 [DOI] [PubMed] [Google Scholar]
- 34. Bohl J., Brimer N., Lyons C., Vande Pol S. B. (2007) The stardust family protein MPP7 forms a tripartite complex with LIN7 and DLG1 that regulates the stability and localization of DLG1 to cell junctions. J. Biol. Chem. 282, 9392–9400 [DOI] [PubMed] [Google Scholar]
- 35. Karnak D., Lee S., Margolis B. (2002) Identification of multiple binding partners for the amino-terminal domain of synapse-associated protein 97. J. Biol. Chem. 277, 46730–46735 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.