Background: The TRAF6-mediated Toll-like receptor (TLR)/IL-1 receptor (IL-1R) pathway is essential for innate immune responses and immune homeostasis.
Results: USP4 deubiquitinates Lys-63-linked polyubiquitination of TRAF6 and thereby prevents the TLR/IL-1R-induced activation of NF-κB and AP-1 transcription factors and subsequent proinflammatory responses.
Conclusion: USP4 plays an essential role in the negative regulation of the TLR/IL-1R signaling-mediated innate immune response.
Significance: USP4 is an attractive new therapeutic target for modulation of innate immune responses.
Keywords: Immunology, Molecular Cell Biology, shRNA, Ubiquitination, Zebrafish, TRAF6, Toll-like Receptor/Interleukin-1 Receptor Signaling, USP4, Innate Immune
Abstract
The Toll-like receptor (TLR)/IL-1 receptor (IL-1R) signaling pathway is essential for innate immune responses and immune homeostasis. Lys-63-polyubiquitinated TRAF6 mediates its downstream signaling activation. In a gain-of-expression screen of 66 different deubiquitinating enzymes, we identified USP4 as a potent negative regulator of TLR/IL-1R signaling and TRAF6-interacting protein. USP4 deubiquitinates TRAF6 and thereby prevents the activation of NF-κB and AP-1 transcription factors and subsequent proinflammatory responses. LPS-treated usp4-depleted zebrafish larvae expressed higher levels of proinflammatory cytokines and were more susceptible to endotoxic challenge. Taken together, our results demonstrate that USP4 plays an essential role in negative regulation of the TLR/IL-1R signaling-mediated innate immune response.
Introduction
The Toll-like receptor (TLR3)/IL-1 receptor (IL-1R) superfamily, which shares a conserved cytoplasmic TLR/IL-1R domain, signals an evolutionarily conserved pathway that is critical for innate immune responses (1–3). After activation by ligand, TLR/IL-1R recruits downstream signaling molecules via the TLR/IL-1R domain and subsequently activates the transcription factors NF-κB and AP-1, which control the expression of key immunoregulatory genes (2). Conserved microbial molecules such as bacterial LPS stimulate innate immune responses by binding to TLRs, particularly on macrophages (1, 4). Upon activation of TLRs, immune cells are triggered to secrete proinflammatory cytokines such as IL-1, IL-6, and TNF, which in turn rapidly amplify inflammatory signaling cascades. Activation of macrophages is a critical initial step in the activation of immune responses. After activation, TLR/IL-1R signaling must be restricted to protect the organism against uncontrolled immune activation and to avoid serious systemic disorders (2, 5–7).
Both the magnitude and duration of TLR/IL-1R-initiated immune responses rely on downstream TRAF6 signaling (8–10). As a mediator for the TLR/IL-1R superfamily, TRAF6 is recruited to the receptor complexes and forms oligomers upon signaling activation. TRAF6 is a RING domain-containing E3 ubiquitin ligase. Oligomerization of TRAF6 activates its ligase activity, leading to Lys-63-linked polyubiquitination of targets, including TRAF6 itself (11). Lys-63 ubiquitin conjugation on TRAF6 recruits TAB2 and activates the TAB2-associated TAK1 kinase (12), which subsequently phosphorylates and activates IκB kinases. IκB kinase then phosphorylates IκBs, leading to degradation of IκB and consequently activation of NF-κB (13–15). In addition, TAK1 can activate the JNK and p38 MAPK family members, which trigger AP-1 activation (16). The fact that TRAF6 knock-out mice have a defect in IL-1β- and LPS-induced activation of NF-κB and JNK suggests an essential role for TRAF6 in immune responses (10).
We assumed that there are multiple negative regulators, especially deubiquitinating enzymes (DUBs), which could be employed by organisms to regulate TRAF6 activation, thereby maintaining immune homeostasis. For this purpose, we screened up to 66 DUBs and identified USP4 (ubiquitin-specific protease 4) as a critical negative regulator of TLR/IL-1R signaling through cleavage of the Lys-63-linked polyubiquitin chain on TRAF6.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
HEK293T, HeLa, and mouse embryonic fibroblasts (MEFs) were cultured in DMEM supplemented with 10% FBS (HyClone) and 100 units/ml penicillin/streptomycin (Invitrogen). Macrophage cell lines RAW264.7 and THP-1 were cultured in RPMI 1640 medium supplemented with 10% FBS. cDNAs encoding FLAG-tagged DUBs were purchased from Addgene. Myc-WT USP4 and Myc-USP4 C311 expression constructs were cloned and verified by DNA sequencing. Expression constructs for FLAG-TRAF6, HA-WT ubiquitin (Ub), HA-K48R Ub, HA-K63R Ub, HA-Lys-48 Ub-only, and HA-Lys-63 Ub-only have been described previously (17). IL-1β, LPS, and MG132 were purchased from Sigma. The antibodies used for immunoprecipitation and immunoblotting were raised against the following proteins: β-actin (A5441, Sigma), c-Myc (a-14, sc-789, Santa Cruz Biotechnology), HA (Y-11, sc-805, Santa Cruz Biotechnology), FLAG (M2, Sigma), USP4 (U0635, Sigma), TRAF6 (H-274, Santa Cruz Biotechnology), Lys-63 linkage-specific Ub (clone HWA4C4, Enzo Life Sciences), phospho-IκBα Ser-32 (2859, Cell Signaling), phospho-IκBα Ser-32/Ser-36 (9246, Cell Signaling), and IκBα (9247, Cell Signaling).
Lentiviral Transduction and Generation of Stable Cell Lines
Lentiviruses were produced by transfecting shRNA-targeting plasmids together with helper plasmids pCMV-VSVG, pMDLg-RRE (group specific antigen/polymerase), and pRSV-REV into HEK293T cells. Cell supernatants were harvested 48 h after transfection and either used to infect cells or stored at −80 °C.
To obtain stable cell lines, cells were infected at low confluence (20%) for 24 h with lentiviral supernatants diluted 1:1 with normal culture medium in the presence of 5 ng/ml Polybrene (Sigma). 48 h after infection, cells were placed under puromycin selection for 1 week and then passaged before use. Puromycin was used at 1 μg/ml to maintain macrophage RAW264.7 and THP-1 cells and at 5 μg/ml to maintain MEF cells.
Lentiviral shRNAs were obtained from Sigma (MISSION® shRNA). Typically, five shRNAs were identified and tested, and the most effective two shRNAs were used for the experiment. We used TRCN0000004040 (shRNA#1) and TRCN0000004041 (shRNA#2) for human USP4 knockdown and TRCN0000030740 (shRNA m#1) and TRCN0000030741 (shRNA m#2) for mouse USP4 knockdown.
Transcription Reporter Assay
HEK293T cells seeded in 24-well plates were transfected by calcium phosphate with the indicated plasmids. 24 h after transfection, cells were untreated or treated with IL-1β (5 ng/ml) or LPS (20 μg/ml) for 8 h and then harvested. Luciferase activities were determined using a PerkinElmer luminometer. The internal transfection control β-gal (30 ng) was used to normalize luciferase activity. Each experiment was performed in triplicate, and the data represent the mean ± S.D. of three independent experiments after normalization to β-gal activity. For all transfections, an empty vector was used as a control to ensure that the same amount of DNA were transfected into cells.
Immunoprecipitation and Immunoblotting
Cells were lysed with 1 ml of lysis buffer (20 mm Tris-HCl (pH 7.4), 2 mm EDTA, 25 mm NaF, and 1% Triton X-100) plus protease inhibitors (Sigma) for 10 min at 4 °C. After centrifugation at 12,000 × g for 15 min, the protein concentration was measured, and equal amounts of lysate were used in immunoprecipitation with different antibodies and protein A-Sepharose (GE Healthcare) for 3 h at 4 °C. Thereafter, the precipitants were washed three times with wash buffer (50 mm Tris-HCl (pH 8.0), 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% SDS), and the immune complexes were eluted with sample buffer containing 1% SDS for 5 min at 95 °C. The immunoprecipitated proteins were then separated by SDS-PAGE. Western blotting was performed with specific antibodies and secondary anti-mouse or anti-rabbit antibodies conjugated to horseradish peroxidase (Amersham Biosciences). Visualization was achieved with chemiluminescence.
In Vivo Ubiquitination Assay
Cells were washed with PBS and lysed in radioimmunoassay buffer (20 mm NaH2PO4, Na2HPO4 (pH 7.4), 150 mm NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, and 1% SDS) supplemented with protease inhibitors and 10 mm N-ethylmaleimide. Lysates were sonicated, boiled at 95 °C for 5 min, diluted with radioimmunoassay buffer containing 0.1% SDS, and centrifuged at 4 °C for 15 min. The supernatant was incubated with a specific antibody and protein A-Sepharose for 3 h at 4 °C. After extensive washing, bound proteins were eluted with 2× SDS sample buffer and separated on a SDS-polyacrylamide gel, followed by immunoblotting.
Real-time Quantitative RT-PCR (qRT-PCR)
Total RNAs were prepared using a NucleoSpin® RNA II kit (BIOKÉ). 1 μg of RNA was reverse-transcribed using a RevertAidTM first-strand cDNA synthesis kit (Fermentas). Real-time PCR was conducted with SYBR Green incorporation (Applied Bioscience) using a StepOne Plus real-time PCR system (Applied Bioscience). All values of the target gene expression levels were normalized to GAPDH. All primers used in qRT-PCR are listed in supplemental Table 1.
Zebrafish Injection
Full-length zebrafish usp4 cDNA was obtained using a pair of primers (zU4full primers listed in supplemental Table 1). mRNA was synthesized using T7 RNA polymerase and the pXt7-usp4 template (18). The morpholino oligonucleotide (MO) sequences (usp4 splicing blocker and mismatch control) are listed in supplemental Table 1. MO efficiency was tested by RT-PCR using exon 4 and 5 primers (zU4 in supplemental Table 1).
Statistical Analysis
Statistical analyses were performed with a two-tailed unpaired t test. p < 0.05 was considered statistically significant.
RESULTS
DUB cDNA Expression Screen Identifies USP4 as Suppressor of IL-1β Signaling and TRAF6-interacting Protein
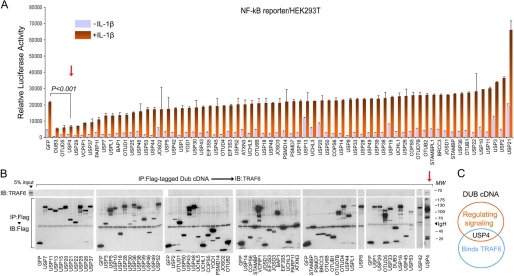
We started this project with a genetic screen to identify DUBs that could mitigate TLR/IL-1R-initiated reporter activity (Fig. 1A). 66 expression plasmids encoding FLAG-tagged human DUBs were individually cotransfected with an NF-κB transcription reporter in HEK293T cells. Cells were treated with a suboptimal dose of IL-1β and then assayed for luciferase activity. From this screen, multiple DUB cDNAs, including usp4, were identified as NF-κB repressors (Fig. 1A). Considering that multiple ubiquitination-related IL-1β-induced signaling steps could be regulated by DUBs, we then focused our attention on identifying specific DUBs for TRAF6. The same collections of DUBs were individually overexpressed in HEK293T cells, and cell lysates were prepared. Blotting the FLAG-DUB immunoprecipitates for endogenous TRAF6 revealed that only ectopic USP4 detectably interacted with endogenous TRAF6 (Fig. 1B). Thus, USP4 was identified both as a suppressor of IL-1β signaling and as a TRAF6-interacting protein.
FIGURE 1.
DUB cDNA screen identifies USP4 as suppressor of IL-1β signaling and TRAF6-interacting protein. A, luciferase activity analysis of the IL-1β (5 ng/ml)-induced NF-κB reporter in HEK293T cells transfected with reporters along with FLAG-tagged DUB cDNA plasmids. FLAG-tagged GFP served as control. The DUBs inhibiting the transcriptional reporter activity by >40% in triplicate (p < 0.05) were considered as candidates for repressors. B, TRAF6 immunoblotting (IB) followed by FLAG immunoprecipitation (IP) of cell lysates from HEK293T cells expressing FLAG-tagged DUB cDNA plasmids. C, diagram of the screening results showed USP4 is a TRAF6-interacting protein that also regulates IL-1β signaling.
Endogenous USP4 Is Repressor of IL-1β- and LPS-induced NF-κB Signaling
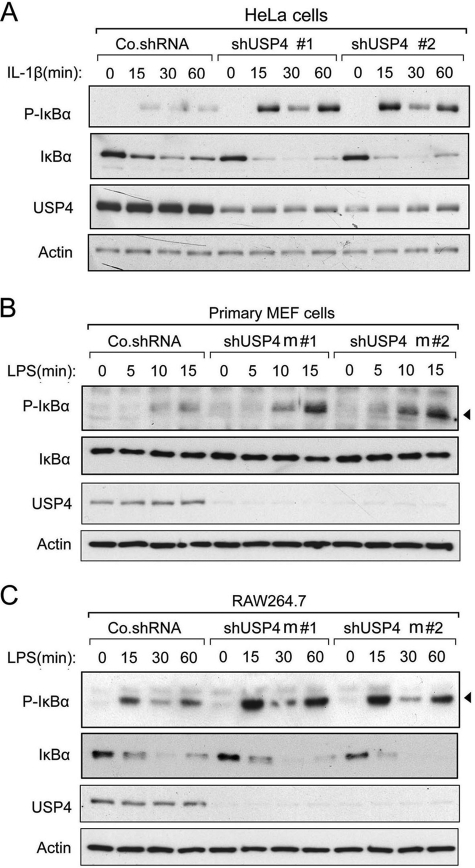
USP4 was originally identified as an ubiquitous nuclear protein with oncogenic potential (19). Later, it was found to efficiently cleave the ubiquitin-proline bond in Ub fusion proteins of different sizes (20). USP4 was reported to cleave both Lys-63- and Lys-48-linked polyubiquitination on targets (21, 22). To address whether USP4 is relevant for endogenous TLR/IL-1R activation, we monitored the IκBα phosphorylation levels induced by IL-1β or LPS. In HeLa cells and primary MEFs, lentivirus-mediated knockdown of USP4 elevated both IL-1β- and LPS-induced phosphorylated IκBα levels (Fig. 2, A and B). Similar observations were detected in macrophages challenged with LPS (Fig. 2C). In line with these results, knockdown of USP also enhanced NF-κB reporter activity in these cells (see Fig. 5, G and I).
FIGURE 2.
USP4 depletion up-regulates IL-1β- and LPS-induced NF-κB signaling. A, immunoblot of control and USP4-depleted (shUSP4m#1 and shUSP4m#2) HeLa cells treated with IL-1β (10 ng/ml) as indicated. Co.shRNA, control non-targeting shRNA. B, immunoblot of control and USP4-depleted MEF cells treated with LPS (1 μg/ml) as indicated. C, immunoblot of control and USP4-depleted RAW264.7 cells treated with LPS (1 μg/ml) as indicated.
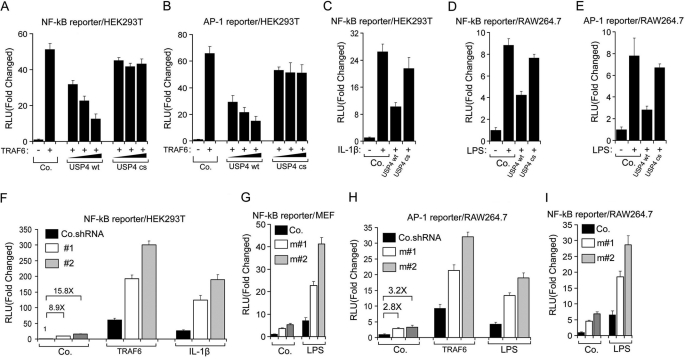
FIGURE 5.
USP4 suppresses TRAF6-mediated TLR/IL-1R signaling. A and B, luciferase activity analysis of NF-κB (A) and AP-1 (B) reporters in HEK293T cells transfected with reporters along with TRAF6, WT USP4, or C311S (cs) mutant USP4 plasmid. Co., control empty vector. RLU, relative luciferase units. C, luciferase activity of NF-κB reporter stimulated with IL-1β (10 ng/ml) in the presence or absence of WT USP4 or C311S USP4 in HEK293T cells. D and E, luciferase activity of the NF-κB (D) and AP-1 (E) reporters stimulated with LPS (1 μg/ml) in the presence or absence of WT USP4 or C311S USP4 in RAW264.7 cells. F, luciferase activity of the NF-κB reporter in HEK293T cells with USP4 depletion (#1 or #2) and stimulated with TRAF6 or IL-β (10 ng/ml). Co.shRNA, control non-targeting shRNA. G, luciferase activity of the NF-κB reporter in MEF cells with USP4 depletion (m#1 or m#2) and stimulated with LPS (1 μg/ml). H, luciferase activity of the AP-1 reporter in RAW264.7 cells with USP4 depletion and stimulated with TRAF6 or LPS (1 μg/ml). I, luciferase activity of the NF-κB reporter in RAW264.7 cells with USP4 depletion and stimulated with LPS (1 μg/ml). For all reporter assays, data represent the mean ± S.D. (n = 3).
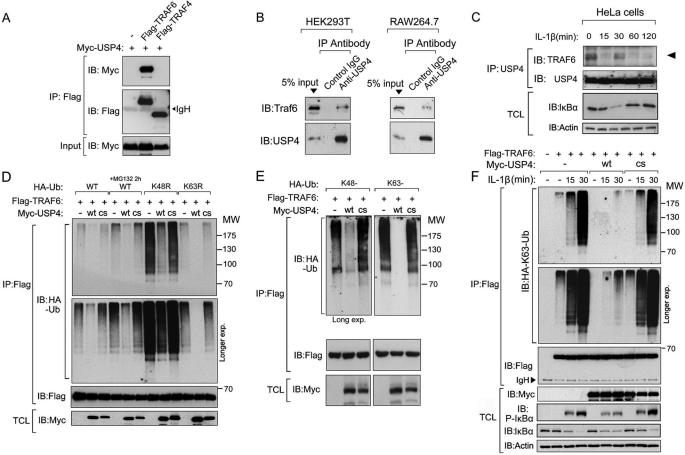
Endogenous USP4 Interacts with TRAF6 and Deubiquitinates TRAF6
We next confirmed that USP4 and TRAF6 physically interact with each other. Co-immunoprecipitation and Western blotting of transfected 293T cell lysates showed that FLAG-tagged TRAF6, but not TRAF4, interacted with Myc-tagged USP4 (Fig. 3A). Domain mapping in TRAF6 and USP4 showed that the USP domain in USP4 mediated its binding to TRAF6. Within TRAF6, the TRAF domain, but not the Ring finger or zinc finger domain, could bind to USP4 (supplemental Fig. 1). Moreover, in co-immunoprecipitation assays with lysates of HEK293T, RAW264.7, and other non-transfected cells, endogenous USP4 was detected in complex with endogenous TRAF6 (Fig. 3B and data not shown). Importantly, the binding of USP4 to TRAF6 was found to be regulated by ligand stimulation, as it was reduced after 15 min, increased after 30 min, and again reduced after 60 min, thereby showing the opposite pattern as that of phosphorylation of IκBα (Fig. 3C and compare with Fig. 2A; also see “Discussion”). We next asked if USP4 serves as a DUB for TRAF6. To address this, we first analyzed the ubiquitylation pattern of TRAF6. HEK293T cells were transfected with expression plasmids encoding HA-tagged wild-type Ub or mutants (K48R and K63R), FLAG-tagged TRAF6 proteins were then affinity-purified, and their ubiquitylation pattern were visualized by immunoblotting against HA-ubiquitin. Polyubiquitylation appeared as a major modification of TRAF6, and it could be stabilized by MG132 treatment (Fig. 3D). Both the K48R and K63R Ub mutants were found to be conjugated to TRAF6, indicating that multiple types of polyubiquitination occur on TRAF6 (Fig. 3D). Ectopic expression of wild-type USP4, but not the catalytically inactive USP4 mutant (C311S) (23), inhibited TRAF6 polyubiquitylation (Fig. 3D). Crucially, when we used Ub mutants that form only Lys-48 or Lys-63 polyubiquitin chains, USP4 inhibited the conjugation of both chains on TRAF6 in a deubiquitinating activity-dependent manner (Fig. 3E). As an essential step for TLR/IL-1R signaling transduction, the Lys-63-linked polyubiquitin chain on TRAF6 mediated downstream cascade activation (14). As monitored by ubiquitination assay, IL-1β ligand-induced HA-Lys-63 Ub conjugation on FLAG-TRAF6 was inhibited by wild-type USP4, but not by the inactive mutant (C311S) (Fig. 3F).
FIGURE 3.
USP4 associates with TRAF6 and deubiquitinates Lys-63-linked polyubiquitination of TRAF6. A, immunoprecipitation (IP) and immunoblot (IB) analysis of cell lysates from HEK293T cells expressing FLAG-TRAF6 and Myc-USP4. FLAG-TRAF4, which does not interact with Myc-USP4, was employed as a negative control. B, endogenous association between USP4 and TRAF6 as measured by immunoblot detection of TRAF6 upon USP4 immunoprecipitation in both HEK293T and RAW264.7 cells. C, endogenous interaction of TRAF6 with USP4 after IL-1β (10 ng/ml) stimulation in HeLa cells. TCL, total cell lysate. D and E, immunoprecipitation and immunoblot analysis of cell lysates from HEK293T cells expressing WT or mutant HA-Ub and FLAG-TRAF6 in the presence or absence of wild-type (wt) or mutant C311S (cs) Myc-USP4. F, immunoblot of HA-Lys-63-linked polyubiquitination of TRAF6 in HeLa cells expressing HA-Lys-63-linked Ub along with wild-type or C311S mutant Myc-USP4 after IL-1β (10 ng/ml) stimulation for indicated times.
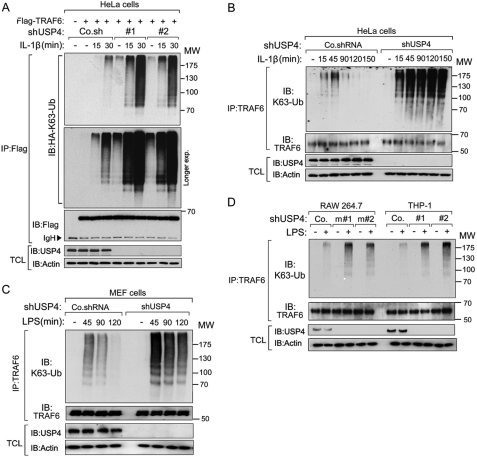
USP4 Depletion Enhances Lys-63-linked Polyubiquitination of TRAF6
Importantly, loss of endogenous USP4 enhanced basal as well as IL-1β-induced Lys-63-linked polyubiquitylation on FLAG-TRAF6 (Fig. 4A), indicating that endogenous USP4 is a potent regulator of TRAF6 activity. When we examined in a time course the IL-1β-induced endogenous TRAF6 Lys-63 polyubiquitin chain, we observed an enhancement of both magnitude and duration in USP4-depleted cells compared with control cells (Fig. 4B). In line with our previous findings for IL-1β stimulation, LPS-induced TRAF6 Lys-63-linked polyubiquitination was also elevated in USP4-depleted MEFs, macrophages (RAW264.7), and human monocytic cells (THP-1) (Fig. 4, C and D).
FIGURE 4.
USP4 depletion increases ligand-induced Lys-63-linked polyubiquitination of TRAF6. A, immunoblot (IB) of Lys-63-linked polyubiquitination of TRAF6 in HeLa cells expressing HA-Lys-63-linked Ub along with USP4 depletion (shUSP4#1 or shUSP4#2) after IL-1β (10 ng/ml) stimulation for the indicated times. Co.sh, control non-targeting shRNA. IP, immunoprecipitation; TCL, total cell lysate. B, immunoblot of endogenous Lys-63-linked polyubiquitination of TRAF6 in control or USP4-depleted HeLa cells upon IL-1β (10 ng/ml) stimulation for the indicated times. Co.shRNA, control non-targeting shRNA. C, immunoblot of endogenous Lys-63-linked polyubiquitination of TRAF6 in control and USP4-depleted MEF cells upon LPS (1 μg/ml) stimulation for the indicated times. D, immunoblot of endogenous Lys-63-linked polyubiquitination of TRAF6 in control and USP4-depleted RAW264.7 or THP-1 cells (shUSP4m#1 or shUSP4m#2 for RAW264.7 and shUSP4#1 or shUSP4#2 for THP-1) after LPS (1 μg/ml) stimulation for 2 h.
USP4 Suppresses TRAF6-mediated TLR/IL-1R Signaling
Lys-63 autoubiquitination of TRAF6 promotes activation of its downstream transcription factors AP-1 and NF-κB, which in turn induce proinflammatory cytokine gene expression. We assessed the effects of wild-type and mutant USP4 on AP-1 and NF-κB activation using transcriptional reporter assays and real-time qPCR analysis for target gene expression. Expression of TRAF6 in HEK293T cells substantially increased AP-1 and NF-κB reporter activity, whereas expression of USP4 inhibited this increase in a dose- and deubiquitinating enzyme activity-dependent manner (Fig. 5, A and B). IL-1β stimulation increased NF-κB reporter activity in 293T cells, whereas ectopic expression of wild-type USP4, but not the corresponding C311S mutant, inhibited this activation (Fig. 5C). Similarly, the activation of AP-1 and NF-κB in macrophages by LPS challenge was inhibited by wild-type USP4, but not by the C311S mutant (Fig. 5, D and E). In line with this, shRNA-mediated depletion of endogenous USP4 augmented TRAF6-induced AP-1 and NF-κB activation and IL-1β-stimulated NF-κB activation (Fig. 5, F and H). USP4 depletion also elevated LPS-induced AP-1 and NF-κB activation (Fig. 5, G–I).
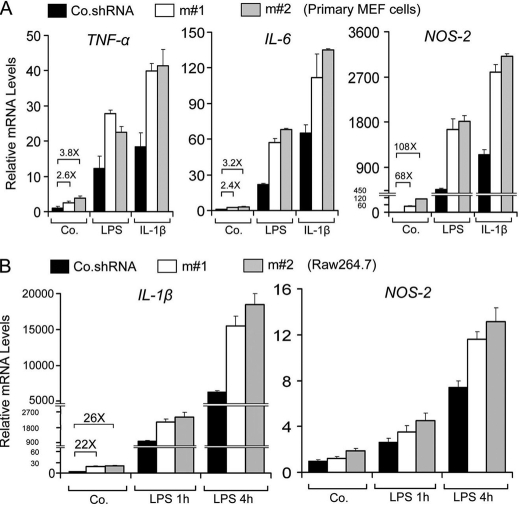
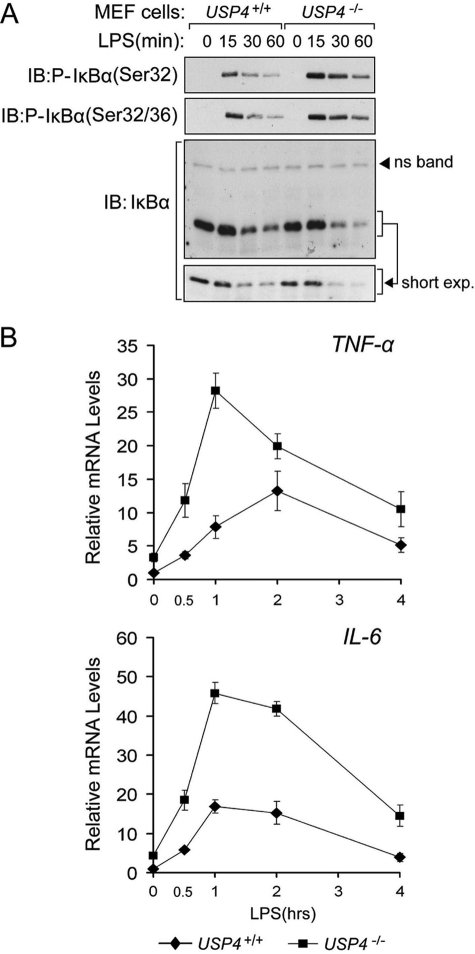
We then measured the effect of USP4 on the expression of NF-κB- and AP-1-dependent target genes (24). Lentivirus shRNA-mediated USP4 knockdown enhanced LPS-and IL-1β-induced TNF, IL-6, and NOS-2 (Fig. 6A and data not shown). Moreover, USP4-depleted macrophages showed enhanced LPS-induced levels of IL-1β and NOS-2 (Fig. 6B and data not shown). Furthermore, compared with wild-type MEFs, USP4−/− MEFs expressed higher levels of phosphorylated IκBα and TNF and IL-6 mRNAs in response to LPS (Fig. 7).
FIGURE 6.
USP4 depletion up-regulates TLR/IL-1R downstream genes. A, qRT-PCR analysis of target genes of TLR/IL-1R signaling in primary MEF cells treated with or without LPS (1 μg/ml) or IL-1β (10 ng/ml) for 4 h. B, qRT-PCR analysis of target genes of IL-1R/TLR signaling in RAW264.7 cells treated with LPS (100 ng/ml) for the indicated times. Data represent the mean ± S.D. of triplicates and are representative of at least two independent experiments (A and B). Co.shRNA, control non-targeting shRNA; m#1 and m#2, shRNAs for mouse USP4; Co., control empty vector.
FIGURE 7.
Elevated TLR/IL-1R signaling responses in USP4 knock-out MEF cells. A, immunoblot (IB) of control and USP4 knock-out MEF cells treated with LPS (1 μg/ml) as indicated. ns, nonspecific. B, qRT-PCR analysis of cytokine gene induction of LPS (1 μg/ml) in control and USP4 knock-out MEF cells.
USP4 Functions as in Vivo Negative Regulator of TLR/IL-1R Signaling
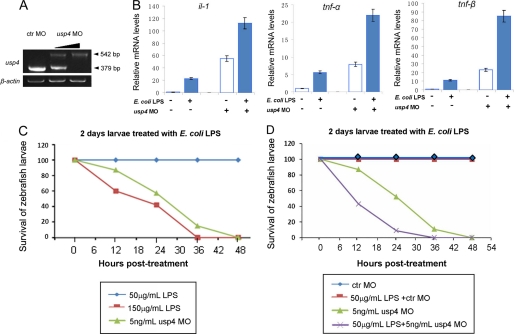
To further verify the in vivo involvement of USP4 in TLR signaling, we compared LPS-driven cytokine expression in control and usp4-depleted zebrafish larvae. For this purpose, we used a splicing blocker against zebrafish usp4 that keeps its target mRNA in the premature form, resulting in a 542-bp PCR product from cDNA of usp4 morphants, in contrast to the 379-bp fragment from cDNA of control MO-injected embryos (Fig. 8A). Analysis of mRNA isolated from zebrafish challenged with LPS showed that usp4-depleted zebrafish produced more TNF-α, TNF-β, and IL-1β mRNAs than control zebrafish (Fig. 8B). Because endotoxin shock as induced by Escherichia coli LPS is mediated in part by TNF, we analyzed the survival of control and morphant zebrafish upon culturing in LPS-containing medium. Because early treatment can cause nonspecific embryonic death, we chose 2-day larvae (48 h post-fertilization) as the starting point and first defined the lethal and sublethal dose in our system (25). 150 μg/ml LPS-containing medium kept normal larvae partly alive in the first 18 h of treatment, but almost all of them had died after 36 h (Fig. 8C). In contrast, all normal larvae survived in 50 μg/ml LPS medium until 7 days post-fertilization. We therefore used 150 μg/ml LPS as the lethal concentration and 50 μg/ml LPS as the sublethal concentration. Next, we analyzed the effect of usp4 depletion in combination with a sublethal dose of LPS. As shown in Fig. 8D, control MO-injected zebrafish larvae were not affected by this dose at all, but only a partial amount of usp4-depleted zebrafish survived 10 h after treatment. The fact that the usp4-depleted zebrafish were more susceptible to LPS challenge indicates that usp4 functions as an in vivo negative regulator of TLR/IL-1R signaling.
FIGURE 8.
USP4 depletion promotes LPS-induced death of zebrafish larvae. A, RT-PCR of the efficiency of usp4 splicing blocker MO. The lower dose of injection was 2 ng/ml, and the higher dose was 5 ng/ml. B, qRT-PCR analysis of target genes of IL-1/TNF signaling in zebrafish larvae treated with different combinations of usp4 MO and LPS. C and D, different survival ratio of usp4 morphants, LPS-treated larvae, and their combination. All data were calculated from three independent experiments.
DISCUSSION
As an important part of the host defense system, TLR/IL-1R signaling needs to be tightly controlled to sustain immune homeostasis and to avoid detrimental responses (26). Uncontrolled activation of the TLR/IL-1R system leads to unrestrained innate immune responses and results in serious systemic disorders such as septic shock and rheumatoid arthritis. To avoid inappropriate overactivation, multiple negative regulators are engaged to serve this purpose. To further elucidate the immune signaling network and to identify novel regulators of the TLR/IL-1R system, in this study, we focused on DUBs. As a member of the USP family of DUBs, USP4 was identified as a suppressor of IL-1β-initiated signaling and as a TRAF6-binding partner. In multiple cell types, endogenous USP4 was shown to associate with TRAF6 and to inhibit IL-1β- and LPS-induced IκBα phosphorylation. Thus, USP4 was expected to function through interacting and targeting TRAF6 deubiquitination. In vivo ubiquitination assays confirmed this hypothesis; USP4 inhibited both basal and ligand-induced Lys-63-linked polyubiquitination of TRAF6 in both macrophages and other cells. Subsequently, we demonstrated that USP4 plays a critical role in inhibiting NF-κB- and AP-1-dependent transcription and proinflammatory cytokine expression. USP15, which is highly similar to USP4 (27), did not bind to TRAF6 or regulate TRAF6 signaling (Fig. 1). Consistent with this result, few changes were observed in polyubiquitinated TRAF6 when USP15 was overexpressed (data not shown). Notably, an enhanced inflammatory response and a reduced threshold to lethal endotoxin challenge were obtained in zebrafish larvae lacking endogenous usp4. Thus, by specifically cleaving the Lys-63-linked polyubiquitin chain of TRAF6, USP4 controls TLR/IL-1R signaling, thereby contributing to the maintenance of immune homeostasis both in vitro and in vivo.
In the past decade, multiple regulators have been reported to modulate TLR/IL-1R signaling (14, 28), including DUBs such as CYLD and A20, which could target multiple TNF receptor family members to block NF-κB activation (29–35). Moreover, a recent study demonstrated that USP4 can deubiquitinate TAK1, a downstream target of TRAF6 (36). While our manuscript was in preparation, another study was published that confirmed the role of USP4 in TRAF6 regulation in vitro (37). This suggests that USP4 can regulate NF-κB activation on at least two distinct points, providing a “double check” for TLR/IL-1R-induced NF-κB activation to avoid excessive damage to the host. Our data do not exclude the possibility that there are other targets of USP4 in the TLR/IL-1R signaling pathway.
Another aspect that deserves discussion is the role of USP4 in the reversal of TRAF6 Lys-48-linked polyubiquitylation. At the biochemical level, TRAF6 ubiquitylation has so far been investigated almost exclusively in the context of Lys-63-linked polyubiquitylation and signal activation, yet it is unclear how this can be reconciled with the apparent protein stability-related Lys-48 ubiquitin chain conjugation on TRAF6. USP4 can also operate on the pool of Lys-48-linked polyubiquitylated TRAF6, which could be relevant to regulation of TRAF6 stability under certain experimental conditions. However, endogenous TRAF6 expression remains largely unchanged upon USP4 ectopic expression or depletion. Therefore, whether USP4 affects TRAF6 protein stability under certain biological condition requires further investigation.
USP4 has the capability to cleave free Lys-63 polyubiquitin chains (38). The free polyubiquitin chains synthesized by TRAF6 are direct activators for TAK1 (39). Thus, another target(s) of USP4 might be the free polyubiquitin chains that are synthesized by TRAF6.
The fact that USP4-TRAF6 association is regulated by ligand stimulation emphasizes the physiological relevance of this interaction. USP4-TRAF6 association was clearly detected in the absence of ligand, but upon stimulation, USP4 transiently and partially dissociated from TRAF6. We noticed that at the 15-min time point, when USP4-TRAF6 complex levels are low, the IL-1β-initiated phospho-IκBα levels reached a peak (shown in Fig. 2A). The continuous USP4-TRAF6 association in the absence of ligand indicates that USP4 is required to keep signaling in the suppressed state. This is consistent with observations that USP4 depletion alone already up-regulated to certain level the downstream signaling (Figs. 5 and 6). The ligand-regulated dynamic USP4-TRAF6 interaction indicates the existence of a self-regulatory machinery. Further studies are required to elucidate how the cell manages to achieve such tight regulation.
Combined, the in vitro and in vivo evidence elucidates critical functions of USP4 in the termination of TRAF6-mediated, TLR/IL-1R-induced NF-κB and AP-1 activation, indicating that USP4 restricts the amplifying cascade of innate immune responses at both the initial and subsequent stages. Moreover, as an in vivo regulator of TLR/IL-1R signaling, USP4 is an attractive new therapeutic target for the modulation of innate immune responses.
Supplementary Material
Acknowledgments
We are grateful to our colleagues for helpful discussions and technical assistance. We thank Drs. Xiongbin Lu and Maréne Landström for reagents.
This work was supported by Netherlands Organization for Scientific Research Grant VICI 016.066.606, the Centre for Biomedical Genetics (to P. t. D.), and National Nature Science Foundation of China Grants 30900748 and 31171388 (to H. H.).

This article contains supplemental Fig. 1 and Table 1.
- TLR
- Toll-like receptor
- IL-1R
- IL-1 receptor
- DUB
- deubiquitinating enzyme
- MEF
- mouse embryonic fibroblast
- Ub
- ubiquitin
- qRT-PCR
- quantitative RT-PCR
- MO
- morpholino oligonucleotide.
REFERENCES
- 1. Takeda K., Kaisho T., Akira S. (2003) Toll-like receptors. Annu. Rev. Immunol. 21, 335–376 [DOI] [PubMed] [Google Scholar]
- 2. Akira S., Takeda K. (2004) Toll-like receptor signaling. Nat. Rev. Immunol. 4, 499–511 [DOI] [PubMed] [Google Scholar]
- 3. Dunne A., O'Neill L. A. (2003) The interleukin-1 receptor/Toll-like receptor superfamily: signal transduction during inflammation and host defense. Sci. STKE 2003, re3. [DOI] [PubMed] [Google Scholar]
- 4. Janeway C. A., Jr., Medzhitov R. (2002) Innate immune recognition. Annu. Rev. Immunol. 20, 197–216 [DOI] [PubMed] [Google Scholar]
- 5. Akira S., Takeda K., Kaisho T. (2001) Toll-like receptors: critical proteins linking innate and acquired immunity. Nat. Immunol. 2, 675–680 [DOI] [PubMed] [Google Scholar]
- 6. Barton G. M., Medzhitov R. (2003) Toll-like receptor signaling pathways. Science 300, 1524–1525 [DOI] [PubMed] [Google Scholar]
- 7. Akira S. (2003) Toll-like receptor signaling. J. Biol. Chem. 278, 38105–38108 [DOI] [PubMed] [Google Scholar]
- 8. Bradley J. R., Pober J. S. (2001) Tumor necrosis factor receptor-associated factors (TRAFs). Oncogene 20, 6482–6491 [DOI] [PubMed] [Google Scholar]
- 9. Chung J. Y., Park Y. C., Ye H., Wu H. (2002) All TRAFs are not created equal: common and distinct molecular mechanisms of TRAF-mediated signal transduction. J. Cell Sci. 115, 679–688 [DOI] [PubMed] [Google Scholar]
- 10. Lomaga M. A., Yeh W. C., Sarosi I., Duncan G. S., Furlonger C., Ho A., Morony S., Capparelli C., Van G., Kaufman S., van der Heiden A., Itie A., Wakeham A., Khoo W., Sasaki T., Cao Z., Penninger J. M., Paige C. J., Lacey D. L., Dunstan C. R., Boyle W. J., Goeddel D. V., Mak T. W. (1999) TRAF6 deficiency results in osteopetrosis and defective interleukin-1, CD40, and LPS signaling. Genes Dev. 13, 1015–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang C., Deng L., Hong M., Akkaraju G. R., Inoue J., Chen Z. J. (2001) TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature 412, 346–351 [DOI] [PubMed] [Google Scholar]
- 12. Arbibe L., Mira J. P., Teusch N., Kline L., Guha M., Mackman N., Godowski P. J., Ulevitch R. J., Knaus U. G. (2000) Toll-like receptor 2-mediated NF-κB activation requires a Rac1-dependent pathway. Nat. Immunol. 1, 533–540 [DOI] [PubMed] [Google Scholar]
- 13. Scherer D. C., Brockman J. A., Chen Z., Maniatis T., Ballard D. W. (1995) Signal-induced degradation of IκBα requires site-specific ubiquitination. Proc. Natl. Acad. Sci. U.S.A. 92, 11259–11263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Deng L., Wang C., Spencer E., Yang L., Braun A., You J., Slaughter C., Pickart C., Chen Z. J. (2000) Activation of the IκB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103, 351–361 [DOI] [PubMed] [Google Scholar]
- 15. Chen Z., Hagler J., Palombella V. J., Melandri F., Scherer D., Ballard D., Maniatis T. (1995) Signal-induced site-specific phosphorylation targets IκBα to the ubiquitin-proteasome pathway. Genes Dev. 9, 1586–1597 [DOI] [PubMed] [Google Scholar]
- 16. Häcker H., Tseng P. H., Karin M. (2011) Expanding TRAF function: TRAF3 as a tri-faced immune regulator. Nat. Rev. Immunol. 11, 457–468 [DOI] [PubMed] [Google Scholar]
- 17. Sorrentino A., Thakur N., Grimsby S., Marcusson A., von Bulow V., Schuster N., Zhang S., Heldin C. H., Landström M. (2008) The type I TGF-β receptor engages TRAF6 to activate TAK1 in a receptor kinase-independent manner. Nat. Cell Biol. 10, 1199–1207 [DOI] [PubMed] [Google Scholar]
- 18. Hyatt T. M., Ekker S. C. (1998) Vectors and techniques for ectopic gene expression in zebrafish. Methods Cell Biol. 59, 117–126 [DOI] [PubMed] [Google Scholar]
- 19. Gupta K., Chevrette M., Gray D. A. (1994) The Unp proto-oncogene encodes a nuclear protein. Oncogene 9, 1729–1731 [PubMed] [Google Scholar]
- 20. Gilchrist C. A., Gray D. A., Baker R. T. (1997) A ubiquitin-specific protease that efficiently cleaves the ubiquitin-proline bond. Journal of Biological Chemistry 272, 32280–32285 [DOI] [PubMed] [Google Scholar]
- 21. Song E. J., Werner S. L., Neubauer J., Stegmeier F., Aspden J., Rio D., Harper J. W., Elledge S. J., Kirschner M. W., Rape M. (2010) The Prp19 complex and the Usp4Sart3 deubiquitinating enzyme control reversible ubiquitination at the spliceosome. Genes Dev. 24, 1434–1447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang X., Berger F. G., Yang J., Lu X. (2011) USP4 inhibits p53 through deubiquitinating and stabilizing ARF-BP1. EMBO J. 30, 2177–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wada K., Tanji K., Kamitani T. (2006) Oncogenic protein UnpEL/Usp4 deubiquitinates Ro52 by its isopeptidase activity. Biochem. Biophys. Res. Commun. 339, 731–736 [DOI] [PubMed] [Google Scholar]
- 24. Pahl H. L. (1999) Activators and target genes of Rel/NF-κB transcription factors. Oncogene 18, 6853–6866 [DOI] [PubMed] [Google Scholar]
- 25. Novoa B., Bowman T. V., Zon L., Figueras A. (2009) LPS response and tolerance in the zebrafish (Danio rerio). Fish Shellfish Immunol. 26, 326–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Liew F. Y., Xu D., Brint E. K., O'Neill L. A. (2005) Negative regulation of Toll-like receptor-mediated immune responses. Nat. Rev. Immunol. 5, 446–458 [DOI] [PubMed] [Google Scholar]
- 27. Komander D., Clague M. J., Urbé S. (2009) Breaking the chains: structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol. 10, 550–563 [DOI] [PubMed] [Google Scholar]
- 28. Xia X., Cui J., Wang H. Y., Zhu L., Matsueda S., Wang Q., Yang X., Hong J., Songyang Z., Chen Z. J., Wang R. F. (2011) NLRX1 negatively regulates TLR-induced NF-κB signaling by targeting TRAF6 and IKK. Immunity 34, 843–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kovalenko A., Chable-Bessia C., Cantarella G., Israël A., Wallach D., Courtois G. (2003) The tumor suppressor CYLD negatively regulates NF-κB signaling by deubiquitination. Nature 424, 801–805 [DOI] [PubMed] [Google Scholar]
- 30. Trompouki E., Hatzivassiliou E., Tsichritzis T., Farmer H., Ashworth A., Mosialos G. (2003) CYLD is a deubiquitinating enzyme that negatively regulates NF-κB activation by TNFR family members. Nature 424, 793–796 [DOI] [PubMed] [Google Scholar]
- 31. Brummelkamp T. R., Nijman S. M., Dirac A. M., Bernards R. (2003) Loss of the cylindromatosis tumor suppressor inhibits apoptosis by activating NF-κB. Nature 424, 797–801 [DOI] [PubMed] [Google Scholar]
- 32. Boone D. L., Turer E. E., Lee E. G., Ahmad R. C., Wheeler M. T., Tsui C., Hurley P., Chien M., Chai S., Hitotsumatsu O., McNally E., Pickart C., Ma A. (2004) The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat. Immunol. 5, 1052–1060 [DOI] [PubMed] [Google Scholar]
- 33. Tavares R. M., Turer E. E., Liu C. L., Advincula R., Scapini P., Rhee L., Barrera J., Lowell C. A., Utz P. J., Malynn B. A., Ma A. (2010) The ubiquitin-modifying enzyme A20 restricts B cell survival and prevents autoimmunity. Immunity 33, 181–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee E. G., Boone D. L., Chai S., Libby S. L., Chien M., Lodolce J. P., Ma A. (2000) Failure to regulate TNF-induced NF-κB and cell death responses in A20-deficient mice. Science 289, 2350–2354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shembade N., Ma A., Harhaj E. W. (2010) Inhibition of NF-κB signaling by A20 through disruption of ubiquitin enzyme complexes. Science 327, 1135–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fan Y. H., Yu Y., Mao R. F., Tan X. J., Xu G. F., Zhang H., Lu X. B., Fu S. B., Yang J. (2011) USP4 targets TAK1 to down-regulate TNFα-induced NF-κB activation. Cell Death Differ. 18, 1547–1560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Xiao N., Li H., Luo J., Wang R., Chen H., Chen J., Wang P. (2012) Ubiquitin-specific protease 4 (USP4) targets TRAF2 and TRAF6 for deubiquitination and inhibits TNFα-induced cancer cell migration. Biochem. J. 441, 978–986 [DOI] [PubMed] [Google Scholar]
- 38. Zhao B., Schlesiger C., Masucci M. G., Lindsten K. (2009) The ubiquitin-specific protease 4 (USP4) is a new player in the Wnt signaling pathway. J. Cell. Mol. Med. 13, 1886–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Xia Z. P., Sun L., Chen X., Pineda G., Jiang X., Adhikari A., Zeng W., Chen Z. J. (2009) Direct activation of protein kinases by unanchored polyubiquitin chains. Nature 461, 114–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.