Background: A30P α-synuclein is associated with the familial form of Parkinson disease.
Results: The secondary structure and chemical shifts of A30P α-synuclein fibrils are in high agreement with wild-type.
Conclusion: A30P α-synuclein fibrils adopt the same structure as the wild-type.
Significance: This work is an important step toward elucidating the association between the early onset Parkinson disease mutants of α-synuclein fibrils and Parkinson disease pathology.
Keywords: Mutant, Parkinson Disease, Protein Aggregation, Protein Structure, Solid-state NMR
Abstract
α-Synuclein (AS) is associated with both sporadic and familial forms of Parkinson disease (PD). In sporadic disease, wild-type AS fibrillates and accumulates as Lewy bodies within dopaminergic neurons of the substantia nigra. The accumulation of misfolded AS is associated with the death of these neurons, which underlies many of the clinical features of PD. In addition, a rare missense mutation in AS, A30P, is associated with highly penetrant, autosomal dominant PD, although the pathogenic mechanism is unclear. A30P AS fibrillates more slowly than the wild-type (WT) protein in vitro and has been reported to preferentially adopt a soluble, protofibrillar conformation. This has led to speculation that A30P forms aggregates that are distinct in structure compared with wild-type AS. Here, we perform a detailed comparison of the chemical shifts and secondary structures of these fibrillar species, based upon our recent characterization of full-length WT fibrils. We have assigned A30P AS fibril chemical shifts de novo and used them to determine its secondary structure empirically. Our results illustrate that although A30P forms fibrils more slowly than WT in vitro, the chemical shifts and secondary structure of the resultant fibrils are in high agreement, demonstrating a conserved β-sheet core.
Introduction
α-Synuclein (AS)5 is centrally implicated in Parkinson disease (PD) and several other neurodegenerative disorders (1). The unifying feature of the “synucleinopathies” is aggregation and accumulation of AS protein within intracellular inclusions (2). Although these diseases are typically sporadic, several mutations in the SNCA gene encoding AS are associated with familial PD, including single-point mutations A53T (3), A30P (4), and E46K (5), and allele duplication (6) or triplication (7). Although the PD-associated AS mutations are extremely rare, analysis of their pathogenicity could significantly illuminate the mechanisms underlying sporadic disease. The normal function of AS is not precisely known, but it shares conserved structural features with the exchangeable apolipoproteins (8), and several lines of evidence suggest a role for AS in presynaptic vesicle trafficking (9–11). Like the wild-type (WT) protein, all mutant forms of AS are intrinsically unfolded in aqueous solution but adopt an α-helical secondary structure within their N-termini upon binding to phospholipid vesicles or detergent micelles (12, 13). Within A30P, the helical domain is partially disrupted by proline substitution, with a consequent decrease in lipid affinity (12), whereas the A53T and E46K mutants exhibit similar or enhanced lipid binding compared with WT AS (12, 14). Thus, altered lipid affinity is not a unifying phenotype for disease-associated mutants.
One hypothesis to account for the pathogenicity of the PD-related mutations is that they promote pathological AS aggregation. However, A30P AS has been reported to fibrillate more slowly than WT AS in vitro (15) and to populate a soluble, protofibrillar intermediate preferentially, whereas WT readily progresses to mature, insoluble fibrils (16). This has fueled speculation that the mechanism of A30P toxicity may be fundamentally different from that of WT AS. Only recently has tissue from a familial PD patient with the A30P mutation become available for analysis. This individual displayed neuropathology typical of idiopathic PD but with a greater than typical load of insoluble fibrillar aggregates (17). This is a surprising result that strongly implicates fibrillar AS in the pathogenesis of A30P-dependent PD.
A30P and WT AS fibrils have similar morphologies when viewed by low resolution techniques, like electron microscopy (18). Although recent solution NMR studies of quenched hydrogen/deuterium (H/D) exchange have suggested that the A30P mutation does not perturb the location or arrangement of β-strands in WT AS fibrils (19), it is not possible to draw site-specific conclusions regarding structure from H/D exchange experiments alone; for example, some β-sheet regions may be more exposed than others, or protected regions may not exhibit a β-sheet secondary structure. In addition, these indirect measurements rely on low molecular mass samples, which require fibrils to be broken down to smaller units. In contrast, solid-state NMR (SSNMR) is uniquely positioned to obtain atomic resolution structural information of systems (like AS fibrils) that are noncrystalline, insoluble, and of high molecular weight; these results are achieved without altering sample integrity. In this study, we sought to determine whether A30P AS fibrils differ from WT AS fibrils at the atomic level of secondary structure. Our results, based on chemical shift analysis obtained with multidimensional SSNMR experiments, illustrate that A30P and WT AS fibrils are highly similar in secondary structural details.
EXPERIMENTAL PROCEDURES
Protein Sample Preparation
Natural abundance and uniformly 13C,15N-labeled WT and A30P full-length, monomer samples were expressed and purified as described previously (20). Briefly, recombinant protein was expressed in Escherichia coli BL21(DE3), while grown in minimal medium supplemented with 13C,15N BioExpress (Cambridge Isotopes). Purification was performed by thermal lysis, hydrophobic interaction, and size exclusion chromatography resulting in high yield (>40 mg/liter). The sample purity was confirmed by gel electrophoresis and mass spectrometry. To verify that the mutation was present, the plasmid was sequenced and 1H-15N heteronuclear single-quantum correlation (HSQC) spectra of the purified monomer samples were acquired (see supplemental Fig. S1).
Thioflavin T Fluorescence
Solutions of monomeric WT and A30P AS (1 mm, 10 mm phosphate buffer, 2.7 mm KCl, 137 mm NaCl, pH 7.4) were filtered, and fibril formation was measured by monitoring Thioflavin T (15 μm, Sigma-Aldrich) fluorescence using established protocols (21). Control wells were prepared to account for light scatter and possible quenching. 96-well plates were incubated at 37 °C and agitated for 16 min prior to each reading with 4 min of no agitation. Seven replicates were performed for both A30P and WT AS.
Electron Microscopy
WT and A30P AS fibril samples were treated with Karnovsky's fixative and negatively stained with 2% ammonium molybdate (w/v). Samples were applied on Formvar carbon-coated grids (300 mesh) and were viewed with a Hitachi H600 transmission electron microscope, operating at 75 kV.
Solution NMR Spectroscopy
Solution NMR spectroscopy is discussed in the supplemental text.
Solid-state NMR Spectroscopy
An initial solution of monomeric, natural abundance A30P AS (1 mm protein, 50 mm phosphate buffer, pH 7.5, 0.02% azide, and 0.1 mm EDTA) was filtered with a 0.22-μm syringe filter. This solution was incubated with shaking (200 rpm) at 37 °C for 3 weeks to produce mature fibrils. These fibrils were then used to seed future uniformly 13C,15N-labeled A30P AS fibrils. After the allotted time, fibril solutions were washed, dried, packed into 3.2-mm standard or thin wall rotors (Varian, Fort Collins, CO), and rehydrated with 36% (m/v) water according to previously described protocols (22).
A 14.1-Tesla (600 MHz, 1H frequency) Varian Infinity Plus spectrometer equipped with a 3.2-mm T3 Varian BalunTM 1H-13C-15N MAS probe, in triple resonance mode, was utilized to perform all SSNMR experiments. Experiments employed tangent ramped cross-polarization (23) and SPINAL-64 (24, 25) 1H decoupling with a field strength of ∼75 kHz during evolution and acquisition periods. For three-dimensional 15N-13C-13C and 13C-15N-13C correlation experiments, band-selective SPECIFIC cross-polarization (26) was utilized for heteronuclear polarization transfer between 15N and 13C and DARR (27) mixing for 13C homonuclear polarization transfer. All experiments were acquired under 13.3-kHz MAS and at a cooling gas temperature of 10 °C with 90 standard cubic feet per hour flow, resulting in an actual sample temperature of 18 ± 5 °C. The adamantane downfield peak was assumed to resonate at 40.48 ppm as an external chemical shift reference (28). The two- and three-dimensional spectra used for chemical shift assignments are listed in supplemental Table S1.
Data were processed with back linear prediction applied to the direct dimension. Zero filling, Lorentzian-to-Gaussian apodization and/or cosine bells were applied for each dimension before Fourier transformation using NMRPipe (29). Peak picking, assignments, and peak heights were obtained with SPARKY software (30) using the approximation of Gaussian line shapes for peak integration.
RESULTS AND DISCUSSION
A30P AS Fibrillates More Slowly than WT
WT and A30P AS solutions were prepared, as described previously (20), to monitor fibril formation with Thioflavin T fluorescence. An increase in lag time indicates a decrease in the fibrillation rate, as shown for A30P compared with WT (Fig. 1a). Our data show WT AS to propagate fibrils after 24 h and A30P after 72 h. These results are in agreement with those of Li et al. (31) and Meuvis et al. (32). Although A30P and WT fibrillate at different rates, a comparison of electron micrographs of mature fibrils, formed after 3 weeks of incubation, exhibit no distinguishable changes in the fibril morphology upon mutation (Fig. 1b).
FIGURE 1.
In vitro fibrillation conditions provide microscopically well ordered A30P AS fibrils that form slower than WT. a, average fibril formation assay of (red triangles) and A30P AS fibrils (blue circles) monitored by Thioflavin T fluorescence. Error bars were determined from seven replicates for each. b, comparison of the electron micrographs of WT (left) and A30P AS (right) fibrils. 13C′ (c), 13CA (d), and 13CB (e) chemical shift plots between two individual batches of A30P AS fibrils. f, 13C-13C two-dimensional with 50-ms DARR mixing of A30P AS fibrils. Overlaid expansions of WT (red) on to A30P (blue) AS fibrils for the Thr/Ser (CB-C′) (g) and Thr/Ser (CB-CA) (h) regions.
A30P AS Fibril Morphology Is Highly Reproducible
SSNMR has proven to be a useful structural biology technique in exploring the structure and dynamics of amyloid fibrils, like AS fibrils (33–35). Certain amino acids (i.e. Gly, Ala, Thr, Ser, Ile, and Pro) are distinctively identifiable by their unique chemical shift patterns and are highly sensitive to secondary structure (36–40). Therefore, to evaluate slight variations between one fibril batch to the next, 13C-13C two-dimensional spectra with 50-ms DARR (27) mixing were acquired of three different batches of A30P AS fibrils, prepared as described previously for uniformly 13C,15N-labeled WT AS fibrils (20, 22). The linear regression analysis of two individual batches showed R2 values of 0.996, 0.999, and 0.999 for 13C′, 13CA and 13CB, respectively (Fig. 1, c–e). Independent batches also exhibit average chemical shift variations of less than 0.2 ppm. Our A30P fibril samples are microscopically well ordered, evidenced by the narrow heterogeneous line widths, averaging 0.2 ppm (Fig. 1f). Many of the spectral fingerprints of A30P AS fibrils are identical to those of WT; for example, the highly resolved Thr and Ser regions, shown in Fig. 1, g and h.
A30P AS Fibril Chemical Shift Assignments
SSNMR heteronuclear (15N-13C) and homonuclear (13C-13C) two-dimensional experiments with longer DARR mixing times were acquired on uniformly 13C,15N-labeled A30P AS fibrils to detect intra- and interresidue correlations and confirm pairwise assignments. Some signal patterns could be assigned immediately, but the majority of correlation patterns were degenerate due to the presence of sequential, imperfect repeats (KTKEGV) in the AS sequence. Accordingly, three-dimensional experiments (supplemental Table S1) were acquired to obtain unambiguous, site-specific chemical shift assignments.
15N-13C-13C three-dimensional experiments detect a common nitrogen frequency to associate two neighboring residues (the i and i−1 residues). Additionally, 13C-15N-13C three-dimensional experiments can be used to gain an added common frequency such as 13CA or 13C′ in the second dimension. For example, a CAN(CO)CX starts with 13C polarization on 13CA nuclei and is transferred to 15N nuclei; this is followed by a polarization transfer from 15N to 13C′ nuclei (where no chemical shift evolution takes place) using specific cross-polarization (26). Once these cross-polarization transfers have occurred, the resulting polarization on the 13C′ nucleus is transferred through space to the side chain 13C nuclei using the DARR mixing scheme. Using these three multidimensional experiments, we conducted chemical shift assignments of sequential residues using well established techniques for making de novo site-specific chemical shift assignments (22, 43–49). Fig. 2 illustrates how the backbone walk method is used to assign seven consecutive residues in A30P AS fibrils. Sequential backbone assignments were achieved for the stretches of Val40-Val49, Gly51-Val55, and Glu57-Asp98 (supplemental Figs. S2 and S3). A total of 63 unique de novo resonance assignments were possible for A30P AS fibrils (supplemental Table S2) without relying upon the WT AS chemical shift lists.
FIGURE 2.
Multidimensional spectra with high sensitivity and resolution allowed for the chemical shift assignments of A30P AS fibrils. Left, backbone walk schematic. Right, illustration of backbone connectivity through the NCACX (red), NCOCX (blue), and CAN(CO)CX (purple) spectra of residues Val71-Asn65 for A30P AS fibrils. All spectra were acquired with 50-ms DARR mixing and processed with 0.5 ppm of line broadening in each dimension.
The resolution and sensitivity of A30P AS fibril spectra were in some instances better than those observed with WT fibrils; for example, a number of signals (corresponding to residues Glu57, Glu61, and Asp98) were detected in the loop regions that were not evident in comparable spectra of WT AS fibrils. In previous studies of the WT AS fibrils, residues Tyr39 to Lys43, Lys58 to Lys60, Gln62, and Lys97 (highlighted in supplemental Table S2) could only be assigned using data acquired with 13C-sparsely labeled samples (22), which provide higher resolution data compared with uniformly 13C-labeled samples (50). For A30P, these residues were detected and assigned using uniformly 13C-labeled samples, which we attribute to technical improvements in the data collection and potential differences in the dynamics of the loop regions. Some of these signals show slight chemical shift perturbations relative to WT (see below), consistent with small conformational differences and/or chemical exchange effects.
A30P α-Synuclein Forms Fibrils That Are Structurally Similar to Wild Type
We applied linear regression analysis to the complete set of WT (22) and A30P AS fibril chemical shifts (Fig. 3). The chemical shifts of 13CA and 13CB, which report primarily upon secondary structure, showed a high agreement (R2 values of 0.999 and 0.998, respectively) and confirm nearly identical secondary structures between A30P and WT AS fibrils. The 15N and 13C' chemical shifts exhibit R2 values of 0.998 and 0.991, respectively, which are consistent with modest perturbations in hydrogen bonding and electrostatics upon mutation (51, 52).
FIGURE 3.
Comparison of the chemical shift assignments of A30P and WT AS fibrils (22) demonstrates that the fibril is mostly unchanged upon A30P mutation. 15N (a), 13CA (b), 13C′ (c) and 13CB (d) chemical shift plots of WT versus A30P AS fibrils are shown. Residues that differ by more than 0.5 ppm are labeled (open circles).
As demonstrated previously (53), the dipolar-based CANCO experiment produces a correlation for the most rigid residues in a given sample. Amyloid fibrils contain a rigid core, where dipolar-based experiments transfer polarization with high efficiency, and mobile regions, where polarization transfer is inefficient in dipolar-based experiments (34, 54). Thus, the signal intensities report qualitatively on rigidity. Sixty-one correlations were identified in the CANCO spectrum of A30P AS fibrils, of a possible maximum of 139 backbone pair correlations from 140 residues. Of the correlations detected in the CANCO spectra, 91% were unambiguously assigned, as described in Fig. 2. Fig. 4c demonstrates the trend in signal intensity by residue number for A30P AS fibrils. The region with the greatest intensity is found for residues 68–94, which includes the most hydrophobic stretch, 71–82 (55). Signals from mobile regions, such as the termini, are not observed in these spectra, due to the low efficiency of dipolar-mediated polarization transfer for mobile residues.
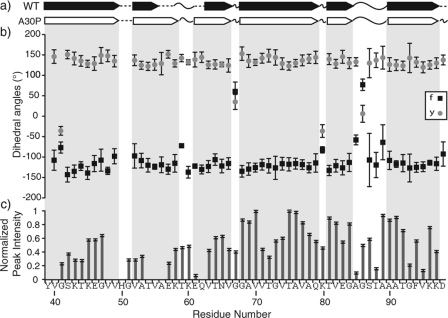
FIGURE 4.
Comparison of the secondary structures between WT (22) and A30P AS fibrils demonstrates that the fibril core is mostly unchanged upon A30P mutation. a, representation of the secondary structure of WT and A30P AS fibrils based on TALOS+ analysis (β-strands, arrows; turn or loop, curved lines; not predicted, dashed line). TALOS+ predicted backbone dihedral angles φ (black squares) and ψ (gray circles), with error bars based on the 10 best TALOS+ data base matches (b) and the normalized peak heights from CANCO as a function of residue number for A30P (c).
SSNMR detects differences in chemical environments with high sensitivity (37–40, 56). This allows for the identification of amino acids and prediction of backbone dihedral angles (φ and ψ) and secondary structure with the TALOS+ program, illustrated in Fig. 4, a and b (38, 57). Our results pertaining to A30P AS fibrils indicate a highly similar β-sheet secondary structure relative to the WT (22) (Fig. 4a). This is consistent with the localization of A30 outside the stable β-sheet core for the WT fibril structure, thus exerting no major effect on the β-sheet secondary structure of the core upon mutation (22).
The improvement in data quality of A30P AS fibrils allowed for the detection of additional residues and extended empirical determination of secondary structure compared with the WT in the Val55-Val63 region. In addition, when the chemical shifts of this region were compared between A30P and WT AS fibrils, perturbations greater than 0.5 ppm for Val55, Lys58, Lys60, and Val63 were observed; this variation significantly exceeds the batch-to-batch variations in individual sample preparations (∼0.2 ppm). These localized chemical shift perturbations support the idea that this region somehow interacts with residue 30 in the AS fibril structure. We envision three possible scenarios. (i) The region Val55-Val63 is proximate to A30 in the folded WT structure; these interactions are modified as a direct result of the A30P mutation. (ii) The region Val55-Val63 of one molecule is proximate to A30 of a neighboring molecule, due to the quaternary arrangement of the fibril; such interactions would also change as a direct result of the A30P mutation. (iii) The region Val55-Val63 interacts with the N-terminal domain of the WT fibrils, and the mutation A30P disrupts the organization of this N-terminal domain; thus the perturbations in the region Val55-Val63 would be an indirect consequence of the mutation. These three scenarios are not mutually exclusive.
Furthermore, our SSNMR results demonstrate that the N-terminus of A30P is more dynamic than the fibril core, based upon the absence of N-terminal signals in the dipolar spectra. Perhaps significantly, Goedert and co-workers report that the truncation of the N-terminal domain of AS causes an increase in the fibrillation lag time of WT AS (42). This demonstrates that the intact N-terminal domain plays a role in promoting fibrillation and suggests a mechanism whereby disruption of the N-terminal domain (via the A30P mutation) might inhibit fibrillation kinetics without altering the fibril structure. These observations are consistent with the hypothesis that the A30P mutation affects the susceptibility to form fibrils in vitro, hence the propensity to form more oligomeric species than mature fibrils (31).
Although the propensity to fibrillize more slowly in vitro is well documented for A30P, a recent report of neuropathology in a familial PD patient with the A30P mutation demonstrated a greater than typical load of insoluble fibrillar aggregates as compared with sporadic PD patients (17). This seeming paradox remains to be explained, although it is plausible that relative fibrillation kinetics might differ significantly in a cellular environment. The A30P mutation decreases lipid affinity and disrupts the structure of the lipid-bound protein relative to WT (12), which could alter the trafficking, localization, and turnover of A30P or its interactions with chaperone molecules. Elucidation of the cellular factors that modulate AS fibrillation are a subject of ongoing research.
CONCLUSIONS
We have performed a site-specific comparison between the secondary structures of full-length WT and A30P AS fibrils. Chemical shift assignments and the empirically determined secondary structure demonstrate that A30P adopts the wild-type fibril structure, despite the fact that A30P forms fibrils more slowly. Our results show that the A30P mutation does not substantially perturb the resulting AS fibril secondary structure at an atomic level.
Supplementary Material
Acknowledgments
We thank Kem Winter and Shin Lee for assisting with sample preparation and Andrew J. Nieuwkoop for technical assistance with NMR data collection.
This work was supported, in whole or in part, by National Institutes of Health Grant R01-GM073770.

This article contains supplemental Figs. S1–S3, Tables S1 and S2, text, and additional references.
The chemical shift assignments for A30P AS fibrils have been deposited in the Biological Magnetic Resonance Data Bank (BMRB) under accession number 17214.
- AS
- α-synuclein
- PD
- Parkinson disease
- SSNMR
- solid-state NMR
- DARR
- dipolar assisted rotational resonance
- HSQC
- heteronuclear single-quantum correlation.
REFERENCES
- 1. Goedert M. (2001) α-Synuclein and neurodegenerative diseases. Nat. Rev. Neurosci. 2, 492–501 [DOI] [PubMed] [Google Scholar]
- 2. Galvin J. E., Lee V. M., Trojanowski J. Q. (2001) Synucleinopathies: clinical and pathological implications. Arch. Neurol. 58, 186–190 [DOI] [PubMed] [Google Scholar]
- 3. Polymeropoulos M. H., Lavedan C., Leroy E., Ide S. E., Dehejia A., Dutra A., Pike B., Root H., Rubenstein J., Boyer R., Stenroos E. S., Chandrasekharappa S., Athanassiadou A., Papapetropoulos T., Johnson W. G., Lazzarini A. M., Duvoisin R. C., Di Iorio G., Golbe L. I., Nussbaum R. L. (1997) Mutation in the α-synuclein gene identified in families with Parkinson's disease. Science 276, 2045–2047 [DOI] [PubMed] [Google Scholar]
- 4. Krüger R., Kuhn W., Müller T., Woitalla D., Graeber M., Kösel S., Przuntek H., Epplen J. T., Schöls L., Riess O. (1998) Ala30Pro mutation in the gene encoding α-synuclein in Parkinson's disease. Nat. Genet. 18, 106–108 [DOI] [PubMed] [Google Scholar]
- 5. Zarranz J. J., Alegre J., Gómez-Esteban J. C., Lezcano E., Ros R., Ampuero I., Vidal L., Hoenicka J., Rodriguez O., Atarés B., Llorens V., Gomez Tortosa E., del Ser T., Muñoz D. G., de Yebenes J. G. (2004) The new mutation, E46K, of α-synuclein causes Parkinson and Lewy body dementia. Ann. Neurol. 55, 164–173 [DOI] [PubMed] [Google Scholar]
- 6. Chartier-Harlin M. C., Kachergus J., Roumier C., Mouroux V., Douay X., Lincoln S., Levecque C., Larvor L., Andrieux J., Hulihan M., Waucquier N., Defebvre L., Amouyel P., Farrer M., Destée A. (2004) α-Synuclein locus duplication as a cause of familial Parkinson's disease. Lancet 364, 1167–1169 [DOI] [PubMed] [Google Scholar]
- 7. Singleton A. B., Farrer M., Johnson J., Singleton A., Hague S., Kachergus J., Hulihan M., Peuralinna T., Dutra A., Nussbaum R., Lincoln S., Crawley A., Hanson M., Maraganore D., Adler C., Cookson M. R., Muenter M., Baptista M., Miller D., Blancato J., Hardy J., Gwinn-Hardy K. (2003) α-Synuclein locus triplication causes Parkinson's disease. Science 302, 841–841 [DOI] [PubMed] [Google Scholar]
- 8. George J. M., Jin H., Woods W. S., Clayton D. F. (1995) Characterization of a novel protein regulated during the critical period for song learning in the zebra finch. Neuron 15, 361–372 [DOI] [PubMed] [Google Scholar]
- 9. Darios F., Ruipérez V., López I., Villanueva J., Gutierrez L. M., Davletov B. (2010) α-Synuclein sequesters arachidonic acid to modulate SNARE-mediated exocytosis. EMBO Rep. 11, 528–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nemani V. M., Lu W., Berge V., Nakamura K., Onoa B., Lee M. K., Chaudhry F. A., Nicoll R. A., Edwards R. H. (2010) Increased expression of α-synuclein reduces neurotransmitter release by inhibiting synaptic vesicle reclustering after endocytosis. Neuron 65, 66–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Scott D. A., Tabarean I., Tang Y., Cartier A., Masliah E., Roy S. (2010) A pathologic cascade leading to synaptic dysfunction in α-synuclein-induced neurodegeneration. J. Neurosci. 30, 8083–8095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Perrin R. J., Woods W. S., Clayton D. F., George J. M. (2000) Interaction of human α-synuclein and Parkinson's disease variants with phospholipids: structural analysis using site-directed mutagenesis. J. Biol. Chem. 275, 34393–34398 [DOI] [PubMed] [Google Scholar]
- 13. Fredenburg R. A., Rospigliosi C., Meray R. K., Kessler J. C., Lashuel H. A., Eliezer D., Lansbury P. T. (2007) The impact of the E46K mutation on the properties of α-synuclein in its monomeric and oligomeric states. Biochemistry 46, 7107–7118 [DOI] [PubMed] [Google Scholar]
- 14. Choi W., Zibaee S., Jakes R., Serpell L. C., Davletov B., Crowther R. A., Goedert M. (2004) Mutation E46K increases phospholipid binding and assembly into filaments of human α-synuclein. FEBS Lett. 576, 363–368 [DOI] [PubMed] [Google Scholar]
- 15. Li J., Uversky V. N., Fink A. L. (2001) Effect of familial Parkinson's disease point mutations A30P and A53T on the structural properties, aggregation, and fibrillation of human α-synuclein. Biochemistry 40, 11604–11613 [DOI] [PubMed] [Google Scholar]
- 16. Conway K. A., Lee S. J., Rochet J. C., Ding T. T., Williamson R. E., Lansbury P. T. (2000) Acceleration of oligomerization, not fibrillization, is a shared property of both α-synuclein mutations linked to early onset Parkinson's disease: implications for pathogenesis and therapy. Proc. Natl. Acad. Sci. U.S.A. 97, 571–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Seidel K., Schöls L., Nuber S., Petrasch-Parwez E., Gierga K., Wszolek Z., Dickson D., Gai W. P., Bornemann A., Riess O., Rami A., Den Dunnen W. F., Deller T., Rüb U., Krüger R. (2010) First appraisal of brain pathology owing to A30P mutant α-synuclein. Ann Neurol 67, 684–689 [DOI] [PubMed] [Google Scholar]
- 18. van Raaij M. E., Segers-Nolten I. M., Subramaniam V. (2006) Quantitative morphological analysis reveals ultrastructural diversity of amyloid fibrils from α-synuclein mutants. Biophys. J. 91, L96–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cho M. K., Kim H. Y., Fernandez C. O., Becker S., Zweckstetter M. (2011) Conserved core of amyloid fibrils of wild-type and A30P mutant α-synuclein. Protein Sci. 20, 387–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kloepper K. D., Woods W. S., Winter K. A., George J. M., Rienstra C. M. (2006) Preparation of α-synuclein fibrils for solid-state NMR: expression, purification, and incubation of wild-type and mutant forms. Protein Expression Purification 48, 112–117 [DOI] [PubMed] [Google Scholar]
- 21. Conway K. A., Harper J. D., Lansbury P. T., Jr. (2000) Fibrils formed in vitro from α-synuclein and two mutant forms linked to Parkinson's disease are typical amyloid. Biochemistry 39, 2552–2563 [DOI] [PubMed] [Google Scholar]
- 22. Comellas G., Lemkau L. R., Nieuwkoop A. J., Kloepper K. D., Ladror D. T., Ebisu R., Woods W. S., Lipton A. S., George J. M., Rienstra C. M. (2011) Structured regions of α-synuclein fibrils include the early onset Parkinson's disease mutation sites. J. Mol. Biol. 411, 881–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hediger S., Meier B. H., Kurur N. D., Bodenhausen G., Ernst R. R. (1994) NMR cross-polarization by adiabatic passage through the Hartmann-Hahn condition (APHH). Chem. Phys. Lett. 223, 283–288 [Google Scholar]
- 24. Fung B. M., Khitrin A. K., Ermolaev K. (2000) An improved broadband decoupling sequence for liquid crystals and solids. J. Magn. Reson. 142, 97–101 [DOI] [PubMed] [Google Scholar]
- 25. Comellas G., Lopez J. J., Nieuwkoop A. J., Lemkau L. R., Rienstra C. M. (2011) Straightforward, effective calibration of SPINAL-64 decoupling results in the enhancement of sensitivity and resolution of biomolecular solid-state NMR. J. Magn. Reson. 209, 131–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Baldus M., Petkova A. T., Herzfeld J., Griffin R. G. (1998) Cross polarization in the tilted frame: assignment and spectral simplification in heteronuclear spin systems. Mol. Phys. 95, 1197–1207 [Google Scholar]
- 27. Takegoshi K., Nakamura S., Terao T. (2001) 13C-1H dipolar-assisted rotational resonance in magic-angle spinning NMR. Chem. Phys. Lett. 344, 631–637 [Google Scholar]
- 28. Morcombe C. R., Zilm K. W. (2003) Chemical shift referencing in MAS solid state NMR. J. Magn. Reson. 162, 479–486 [DOI] [PubMed] [Google Scholar]
- 29. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 30. Goddard T. D., Kneller D. G. (2006) SPARKY 3.106, University of California, San Francisco [Google Scholar]
- 31. Li J., Uversky V. N., Fink A. L. (2002) Conformational behavior of human α-synuclein is modulated by familial Parkinson's disease point mutations A30P and A53T. Neurotoxicology 23, 553–567 [DOI] [PubMed] [Google Scholar]
- 32. Meuvis J., Gerard M., Desender L., Baekelandt V., Engelborghs Y. (2010) The conformation and the aggregation kinetics of α-synuclein depend on the proline residues in its C-terminal region. Biochemistry 49, 9345–9352 [DOI] [PubMed] [Google Scholar]
- 33. Jaroniec C. P., MacPhee C. E., Astrof N. S., Dobson C. M., Griffin R. G. (2002) Molecular conformation of a peptide fragment of transthyretin in an amyloid fibril. Proc. Natl. Acad. Sci. U.S.A. 99, 16748–16753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Siemer A. B., Arnold A. A., Ritter C., Westfeld T., Ernst M., Riek R., Meier B. H. (2006) Observation of highly flexible residues in amyloid fibrils of the HET-s prion. J. Am. Chem. Soc. 128, 13224–13228 [DOI] [PubMed] [Google Scholar]
- 35. Wasmer C., Lange A., Van Melckebeke H., Siemer A. B., Riek R., Meier B. H. (2008) Amyloid fibrils of the HET-s(218–289) prion form a β-solenoid with a triangular hydrophobic core. Science 319, 1523–1526 [DOI] [PubMed] [Google Scholar]
- 36. Wishart D. S., Watson M. S., Boyko R. F., Sykes B. D. (1997) Automated 1H and 13C chemical shift prediction using the BioMagResBank. J. Biomol. NMR 10, 329–336 [DOI] [PubMed] [Google Scholar]
- 37. Wishart D. S., Sykes B. D. (1994) Chemical shifts as a tool for structure determination. Methods Enzymol. 239, 363–392 [DOI] [PubMed] [Google Scholar]
- 38. Cornilescu G., Delaglio F., Bax A. (1999) Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13, 289–302 [DOI] [PubMed] [Google Scholar]
- 39. Wishart D. S., Sykes B. D. (1994) The 13C chemical-shift index: a simple method for the identification of protein secondary structure using 13C chemical-shift data. J. Biomol. NMR 4, 171–180 [DOI] [PubMed] [Google Scholar]
- 40. Oldfield E. (2002) Chemical shifts in amino acids, peptides, and proteins: from quantum chemistry to drug design. Annu. Rev. Phys. Chem. 53, 349–378 [DOI] [PubMed] [Google Scholar]
- 41. Deleted in proof.
- 42. Zibaee S., Jakes R., Fraser G., Serpell L. C., Crowther R. A., Goedert M. (2007) Sequence determinants for amyloid fibrillogenesis of human α-synuclein. J. Mol. Biol. 374, 454–464 [DOI] [PubMed] [Google Scholar]
- 43. Huang L., McDermott A. E. (2008) Partial site-specific assignment of a uniformly 13C,15N-enriched membrane protein, light-harvesting complex 1 (LH1), by solid state NMR. BBA-Bioenergetics 1777, 1098–1108 [DOI] [PubMed] [Google Scholar]
- 44. Pauli J., Baldus M., van Rossum B., de Groot H., Oschkinat H. (2001) Backbone and side chain 13C and 15N signal assignments of the α-spectrin SH3 domain by magic angle spinning solid-state NMR at 17.6 teslas. ChemBioChem 2, 272–281 [DOI] [PubMed] [Google Scholar]
- 45. Böckmann A., Lange A., Galinier A., Luca S., Giraud N., Juy M., Heise H., Montserret R., Penin F., Baldus M. (2003) Solid state NMR sequential resonance assignments and conformational analysis of the 2×10.4 kDa dimeric form of the Bacillus subtilis protein Crh. J. Biomol. NMR 27, 323–339 [DOI] [PubMed] [Google Scholar]
- 46. Igumenova T. I., Wand A. J., McDermott A. E. (2004) Assignment of the backbone resonances for microcrystalline ubiquitin. J. Am. Chem. Soc. 126, 5323–5331 [DOI] [PubMed] [Google Scholar]
- 47. Marulanda D., Tasayco M. L., Cataldi M., Arriaran V., Polenova T. (2005) Resonance assignments and secondary structure analysis of E. coli thioredoxin by magic angle spinning solid-state NMR spectroscopy. J. Phys. Chem. B 109, 18135–18145 [DOI] [PubMed] [Google Scholar]
- 48. Pintacuda G., Giraud N., Pierattelli R., Böckmann A., Bertini I., Emsley L. (2007) Solid-state NMR spectroscopy of a paramagnetic protein: assignment and study of human dimeric oxidized Cu(II)-Zn(II) superoxide dismutase (SOD). Angew. Chem. Int. Ed. Engl. 46, 1079–1082 [DOI] [PubMed] [Google Scholar]
- 49. Li Y., Berthold D. A., Gennis R. B., Rienstra C. M. (2008) Chemical shift assignment of the transmembrane helices of DsbB, a 20-kDa integral membrane enzyme, by 3D magic-angle spinning NMR spectroscopy. Protein Sci. 17, 199–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Castellani F., van Rossum B., Diehl A., Schubert M., Rehbein K., Oschkinat H. (2002) Structure of a protein determined by solid-state magic-angle-spinning NMR spectroscopy. Nature 420, 98–102 [DOI] [PubMed] [Google Scholar]
- 51. Schmidt H. L., Sperling L. J., Gao Y. G., Wylie B. J., Boettcher J. M., Wilson S. R., Rienstra C. M. (2007) Crystal polymorphism of protein GB1 examined by solid-state NMR spectroscopy and x-ray diffraction. J. Phys. Chem. B 111, 14362–14369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wylie B. J., Schwieters C. D., Oldfield E., Rienstra C. M. (2009) Protein structure refinement using 13C α-chemical shift tensors. J. Am. Chem. Soc. 131, 985–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Helmus J. J., Surewicz K., Surewicz W. K., Jaroniec C. P. (2010) Conformational flexibility of Y145Stop human prion protein amyloid fibrils probed by solid-state nuclear magnetic resonance spectroscopy. J. Am. Chem. Soc. 132, 2393–2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Helmus J. J., Surewicz K., Nadaud P. S., Surewicz W. K., Jaroniec C. P. (2008) Molecular conformation and dynamics of the Y145Stop variant of human prion protein in amyloid fibrils. Proc. Natl. Acad. Sci. U.S.A. 105, 6284–6289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Giasson B. I., Murray I. V., Trojanowski J. Q., Lee V. M. (2001) A hydrophobic stretch of 12 amino acid residues in the middle of α-synuclein is essential for filament assembly. J. Biol. Chem. 276, 2380–2386 [DOI] [PubMed] [Google Scholar]
- 56. Wishart D. S., Sykes B. D., Richards F. M. (1991) Relationship between nuclear magnetic resonance chemical shift and protein secondary structure. J. Mol. Biol. 222, 311–333 [DOI] [PubMed] [Google Scholar]
- 57. Shen Y., Delaglio F., Cornilescu G., Bax A. (2009) TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 44, 213–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.