Background: Tablysin-15 from a blood-feeding arthropod binds integrins.
Results: The structure of tablysin-15 reveals a hydrophobic channel in addition to a canonical integrin-binding RGD motif.
Conclusion: Leukotrienes are ligands for the hydrophobic channel.
Significance: Tablysin-15 contains an integrin-binding site and a cavity for scavenging proinflammatory leukotrienes.
Keywords: Calorimetry, Crystal Structure, Integrins, Leukotriene, X-ray Crystallography, CAP Domain, Antigen 5, Disintegrin, Salivary
Abstract
The antihemostatic/antiangiogenic protein tablysin-15 is a member of the CAP (cysteine-rich secretory, antigen 5, and pathogenesis-related 1 protein) superfamily and has been shown to bind the integrins αIIbβ3 and αVβ3 by means of an Arg-Gly-Asp (RGD) tripeptide sequence. Here we describe the x-ray crystal structure of tablysin-15 and show that the RGD motif is located in a novel structural context. The motif itself is contained in a type II β-turn structure that is similar in its conformation to the RGD sequence of the cyclic pentapeptide cilengitide when bound to integrin αVβ3. The CAP domain also contains a hydrophobic channel that appears to bind a fatty acid molecule in the crystal structure after purification from Escherichia coli. After delipidation of the protein, tablysin-15 was found to bind proinflammatory cysteinyl leukotrienes with submicromolar affinities. The structure of the leukotriene E4-tablysin-15 complex shows that the ligand binds with the nonfunctionalized end of the fatty acid chain buried in the hydrophobic pocket, whereas the carboxylate end of the ligand binds forms hydrogen bond/salt bridge interactions with polar side chains at the channel entrance. Therefore, tablysin-15 functions as an inhibitor of integrin function and as an anti-inflammatory scavenger of eicosanoids.
Introduction
Blood-feeding arthropods are responsible for the transmission of a variety of parasitic and viral diseases (1). Transmission is usually tied to the process of feeding where the host skin is pierced by the insect feeding apparatus and a salivary secretion is injected (2). The saliva contains proteins, peptides, and small molecules that inhibit host systems of inflammation and hemostasis (1). In addition to serving as a neutral injection medium for pathogens, the saliva has been found in various cases to either enhance or limit the infection, presumably by modifying the skin environment at the feeding site. Proteins have been identified that antagonize the processes of platelet activation, coagulation, mast cell function, immunity, and vasoconstriction.
Members of the CAP (cysteine-rich secretory, antigen 5 and pathogenesis-related 1 protein) superfamily of proteins, similar to the venom antigen Ves v 5 of the wasp, have been described from saliva of most blood-feeding insects including mosquitoes, sand flies, and triatomine bugs (3–5). The functions of the vast majority of these proteins have not been determined, but recently tablysin-15, a CAP superfamily member from the saliva of the horsefly Tabanus yao, was shown to be a very potent inhibitor of platelet and endothelial cell function (6). The protein binds tightly with integrins αIIbβ3 and αVβ3, thereby inhibiting the adhesion of platelets to fibrinogen and endothelial cells to the matrix protein vitronectin. It can therefore act as an inhibitor of platelet aggregation and angiogenesis, and its efficacy in vivo has been demonstrated in rodent models (6). Tablysin-15 binding with integrins is mediated by an Arg-Gly-Asp (RGD) sequence motif lying near the N terminus of the protein. In snake venom disintegrins, tick salivary proteins, and some endogenous vertebrate proteins, this sequence is known to interact directly with the head structure of the integrin and is the basis for inhibition by these proteins (7, 8). The structures of αVβ3 and αIIbβ3 bound with RGD peptides shows that the motif binds at a cleft between the α and β subunits and makes contact with both (9–12). An inhibitory RGD sequence has not been previously observed in a CAP superfamily member, making tablysin-15 unique as a scaffold for a disintegrin-like motif.
In this study, we describe the three-dimensional structure of tablysin-15 and characterize the structural basis for its inhibitory mechanism. Remarkably, in addition to showing the context of the RGD motif, this structure reveals the presence of an unexpected lipid-binding pocket that accommodates a cysteinyl leukotriene molecule. These eicosanoids are released in the skin by activated mast cells where they cause pain and itching and elicit changes in vascular permeability at the site of an insect bite (13, 14). Salivary proteins having this function have been described from mosquitoes and ticks, but the structural context for the binding site is different (13, 15–17). Ticks bind eicosanoids using proteins from the lipocalin family (17), whereas mosquitoes bind them with odorant-binding protein-like molecules from the family known as D7 (13, 15). The presence of this binding site in tablysin-15 represents the acquisition of a second function by this salivary protein. First, it can act in the skin and vasculature as an inhibitor of platelet aggregation and endothelial cell activation. Second, it can inhibit the proinflammatory effects of cysteinyl leukotrienes that are released from mast cells in the area of the bite.
EXPERIMENTAL PROCEDURES
Materials
l-Arginine, was obtained from Sigma. Leukotrienes C4, D4, and E4 (LTC4, LTD4, and LTE4),3 and arachidonic acid were purchased from Cayman Chemical Company. The selenomethionine media kit (SelenoMet) was obtained from Molecular Dimensions Ltd. Crystallization reagents were obtained from Hampton Research. Native collagen fibrils (type I) from equine tendons used in platelet experiments were purchased from Chrono-Log Corporation. Convulxin was prepared as described previously (18).
Protein Expression, Refolding, and Purification
The cDNA sequence of tablysin-15 was modified by PCR to remove the signal sequence and insert NdeI and XhoI restriction sites to be used for cloning into the vector pET17b. The expression construct was then transformed into the Escherichia coli strain BL21(DE3)pLys-E. Protein was expressed after induction by isopropyl β-d-thiogalactopyranoside at 37 °C and inclusion bodies prepared as described previously (19). The protein was dissolved in 20 mm Tris-HCl, pH 8.0, 6 m guanidinium hydrochloride, 10 mm dithiothreitol and diluted into 8 liter of 20 mm Tris-HCl, pH 7.0, 0.3 m arginine monohydrochloride. After ultrafiltration the protein was purified using size exclusion chromatography on Sephacryl S-100 using 20 mm Tris-HCl, 150 mm NaCl, pH 8.0, followed by cation exchange chromatography on SP-Sepharose. The protein was further purified by additional gel filtration on Superdex 75. For some experiments the protein was also purified by reversed phase HPLC on C4, using a gradient of aqueous acetonitrile in the presence of 0.1% trifluoroacetic acid. The purity and identity of the preparations were assessed by SDS-PAGE.
A selenomethionine derivative of tablysin was prepared by expression of the plasmid construct described above in BL834(DE3)pLysS grown in SelenoMet media (Molecular Dimensions) as per the manufacturer's instructions. The protein was folded and purified using the methods described above.
Isothermal Titration Calorimetry
Tablysin was prepared for isothermal titration calorimetry experiments by dialysis against 20 mm Tris-HCl 0.15 m NaCl pH 7.4 for 1 h. Binding experiments were performed using a MicroCal VP-ITC instrument. The lipid ligands in ethanol or methyl acetate were dried under a stream of nitrogen in a glass vial; then dissolved in 20 mm Tris-HCl, 0.15 m NaCl, pH 7.4; suspended; vortexed; and sonicated for 8 min. Other ligands were dissolved in 20 mm Tris-HCl, 0.15 m NaCl, pH 7.4. The ligand and protein solutions were degassed prior to each experiment. The assays were performed at 30 °C with successive 10-μl injections. After subtraction of the heats of dilution, the measured heats were converted to enthalpies and analyzed by fitting to a single-site binding model using the Microcal Origin software.
Platelet Aggregation Assays
Platelet-rich plasma was obtained from platelet donors at the Department of Transfusion Medicine, National Institutes of Health. The platelet-rich plasma (100 μl) was diluted with Tyrode's solution containing BSA to a concentration of 200,000–400,000 platelet/μl. Platelet aggregation was measured in a Chrono-Log lumiaggregometer (Chrono-Log Corp.) with stirring at 1,200 rpm at 37 °C. Tablysin was incubated with the platelet preparation for 1 min before adding either collagen at 1.5 μg/ml or convulxin at 75 pm. Platelet aggregation was measured by determining the change in transmittance for 3–5 min.
Smooth Muscle Bioassays
The contraction of the guinea pig ileum in response to LTC4 was measured isotonically with an initial load of 1.0 g of tension. Tissue preparations were bathed in modified Tyrode's solution (with 10 mm HEPES buffer, pH 7.4) during the course of the experiment, and the solutions were oxygenated by bubbling air into the bath. Ileum preparations were preincubated with 5 μm tablysin, and LTC4 was added in 0.1 μm increments until maximum contraction was reached.
Crystallization and Data Collection
Crystals of tablysin-15 and its selenomethionine derivative were produced using the hanging drop vapor diffusion method from 10% PEG 6000, 100 mm citric acid, 13 mm praseodymium (III) acetate hydrate, pH 5.6. The crystals were flash-frozen after a short soak in the crystallization buffer described above, minus the praseodymium (III) acetate hydrate but containing 10% glycerol. To obtain a complex of LTE4 with tablysin-15, a concentrated protein sample was mixed with an equimolar concentration of LTE4. Data were collected at beamline 22-BM at the Southeast Regional Collaborative Access Team, APS, Argonne National Laboratory. Tablysin crystallized in the space group P3121with a single molecule in the asymmetric unit (Table 1). Data sets were collected from the native protein and a selenomethionine derivative, and two sites were located. After building and refinement of the structure, one of these was assigned as a praseodymium ion. Data from the leukotriene E4-tablysin-15 cocrystal was collected using a laboratory source (Rigaku rotating copper anode generator), and reflections were recorded using an R-Axis IV detector.
TABLE 1.
Data collection, phasing and refinement statistics for tablysin-15 and its LTE4 complex
| Crystal | Selenomethionine | Ligand-free | LTE4 |
|---|---|---|---|
| Resolution (Å) | 49.4-1.57 | 32.2-1.65 | 18.2-2.50 |
| Beamline | 22-BM | 22-BM | Lab source |
| Wavelength (Å) | 0.97625 | 0.99999 | 1.54178 |
| Completeness (total/high resolution shell) | 99.7/98.5 | 98.3/99.5 | 99.5/99.8 |
| Average redundancy (total/high resolution shell) | 15.9/12.6 | 7.3/5.2 | 3.7/3.7 |
| Rmerge (total/high resolution shell)a | 6.0/32.7 | 6.2/51.7 | 5.4/14.3 |
| I/σI (total/high resolution shell) | 18.0/7.8 | 11.9/4.4 | 14.5/8.1 |
| Observed reflections | 545,971 | 403,475 | 31,960 |
| Unique reflections | 34,232 | 29,066 | 8,629 |
| Space group | P3121 | P3121 | P3121 |
| Unit cell dimensions (Å) | |||
| a | 69.57 | 69.60 | 69.77 |
| b | 69.57 | 69.60 | 69.77 |
| c | 86.21 | 86.13 | 85.47 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 |
| Phasing statistics | |||
| Number of sitesb | 2 | ||
| Contrast (SHELXE) | 0.60 | ||
| Root mean square deviations | |||
| Bond lengths (Å) | 0.012 | 0.013 | 0.013 |
| Bond angles (°) | 1.04 | 1.02 | 1.25 |
| Ramachandran plot (favored/allowed) | 92.4/100 | 92.3/100 | 89.4/100 |
| Wilson temperature factor | 15.56 | 19.06 | 39.9 |
| Rcryst/Rfreec | 18.5/21.1 | 19.2/22.6 | 20.2/25.3 |
a Rmerge = Σh|Ih − <I>|/Σh<I>, where <I> is the mean intensity of all symmetry-related reflections Ih.
b Anomalous signal from one selenium site and one praseodymium ion site.
c Rcryst = Σ|Fobs − Fcalc|/ΣFobs. Rfree as for Rcryst using a 5% random subset of the data for calculation.
Structure Solution and Refinement
Reflection data were processed using HKL2000 (20) for the selenomethionine and native unliganded structures and Crystal Clear (21) for the LTE4 complex. The structure of tablysin was determined using the single wavelength anomalous dispersion method with data collected at the selenium absorption edge (Table 1). Initial phases were obtained using the HKL2MAP implementation of SHELX C, D, and E, for the location of selenium sites, solvent flattening, and density modification (22). Much of the model was built using Buccaneer (23), and the remainder of the protein backbone structure and the side chains were built manually using the native data set. Cycles of rebuilding and refinement were performed with Coot (24) and REFMAC 5 (25) or Phenix (26) to obtain the final structure of the unliganded protein. Model quality was checked using the MOLPROBITY web server (27). The structural figures presented here were produced with PyMOL (DeLano Scientific). The coordinate and reflection files for the selenomethionine derivative with palmitate modeled in the binding pocket (entry 3U3L), the wild-type protein containing palmitate (entry 3U3N), and the complex with (E,E,Z,Z)-7,9,11,14-eicosatetraenoic acid (entry 3U3U) have been deposited in the RCSB Protein Data Bank.
RESULTS
Overall Structure of Tablysin-15
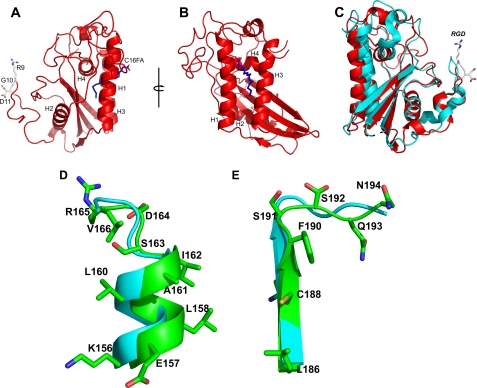
Database searches with the tablysin-15 sequence revealed similarity to the CAP superfamily, which contains the cysteine-rich secretory proteins, the wasp venom antigen 5 (Ves v 5), and pathogenesis-related 1 proteins (3, 28). Consistent with this, tablysin-15 has an overall structure that is also similar to Ves v 5 and other members of the CAP superfamily (3, 29–31) (Fig. 1). The structure is made up of a four-stranded antiparallel β-sheet sandwiched between two helical regions (Fig. 1, A and B). On one side of the sheet lies a cluster of three α-helices with the two most exterior helices (H1 and H3) being parallel to one another and perpendicular to the third (H4). A fourth short helical segment is also present on this side of the β-sheet. On the opposite side of the β-sheet, a single α-helix (H2) is present (Fig. 1). The structure is stabilized by five disulfide bonds.
FIGURE 1.
The structure of tablysin-15. A, ribbon diagram showing the CAP domain structure of tablysin-15 with α-helices H1–H4 labeled. The RGD motif is shown in stick representation with carbon colored white, nitrogen colored blue, and oxygen colored red. Palmitic acid in the hydrophobic pocket of the molecule is also shown in stick representation with carbon colored blue and oxygen colored red. B, rotation of the molecule in A ∼90 degrees around the axis shown. C, superposition of Ves v 5 (cyan) and tablysin-15 (red). The RGD motif of tablysin-15 is shown as in A. D and E, superposition of the CAP1 and CAP2 motif region of Ves v 5 (cyan) with tablysin-15 (green). Side chains are only shown for the tablysin-15 structure. D shows the CAP1 motif region, and E shows the CAP2 motif region.
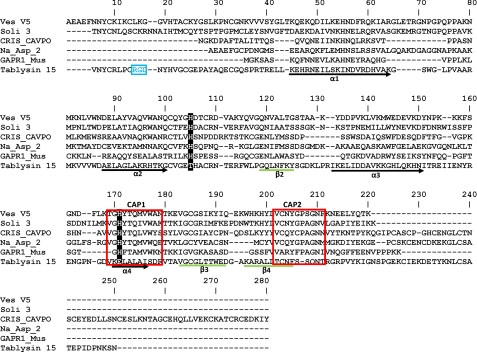
Tablysin-15 is unusual in its lack of strong conservation in two sequence motifs characteristic of the CAP superfamily as described in the PROSITE database (28). The CAP1 motif has the sequence [GDER][HR][FYWH][TVS][QA][LIVM][LIVMA]WXX[STN], and CAP2 has the sequence [LIVMFYH][LIVMFY]XC[NQRHS]YX[PARH]X[GL]N[LIVMFYWDN] (Figs. 1, D and E, and 2). An alignment of tablysin-15 with other CAP domain sequences showed that only Ile-160 at position 7 of the CAP1 motif is conserved (Fig. 2). The CAP2 motif is completely conserved through Asn-187 at position 5 but is less highly conserved in the final seven residues. Despite this divergence in sequence, the structures of the protein backbones in the CAP1 motif in helix H4 and the CAP2 motif in β-sheet 4 and the C-terminal coiled region are quite similar in tablysin-15 and Ves v 5 (Fig. 1, D and E). Two histidine residues, including one in CAP1, have been shown to coordinate a bound zinc ion in the snake venom CAP domain protein natrin and human glioma pathogenesis-related protein 1 (32, 33). These residues are widely conserved in CAP domain proteins, including Ves v 5, but are not present in tablysin-15, indicating a loss of this metal-binding site (Fig. 2).
FIGURE 2.
Amino acid sequence alignment of tablysin-15 with taxonomically diverse CAP domain proteins. α-Helical segments are indicated as black arrows, and β-strands are indicated by green lines. The RGD motif is colored in blue. The PROSITE motifs CAP1 and CAP2 are bordered in red. The histidine positions forming the conserved metal-binding site in CAP domain proteins are highlighted in black. Ves V5, Ves v 5, Vespula vulgaris (Insecta); Soli 3, Sol i 3, Solenopsis invicta (Insecta); Na_Asp_2, Na-ASP-2, Necator americanus (Chromadorea); CRIS_CAPVO, CRISP-2, Cavia porcellus (Mammalia); GAPR1_Mus, Golgi-associated plant pathogenesis-related protein 1, Mus musculus (Mammalia).
The C-terminal segment of tablysin-15 is longer than that of Ves v 5 and is similar in conformation to the hinge region between the CAP domain and the CRISP domain of the calcium channel blocker triflin (3, 31). CRISP domain proteins normally contain two disulfide bonds in the hinge region (28), and one of these disulfides (linking Cys-209 and Cys-220) is conserved in tablysin-15. The second disulfide in the hinge region of triflin is not conserved, nor is the first disulfide bond stabilizing the ion channel regulator region of the CRISP domain. In tablysin-15, the C terminus is made up of a coil region that runs along the surface of the CAP domain rather than folding into a separate domain structure.
Position and Structure of RGD Motif
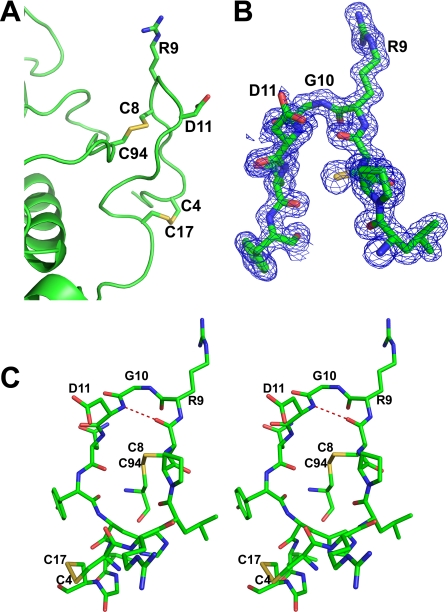
Inhibition of integrins αIIbβ3 and αVβ3 by tablysin-15 is dependent on the presence of an RGD sequence motif located near the N terminus of the protein (Figs. 1A, 2, and 3). The RGD motif lies in an N-terminal extension from the CAP domain and forms a tongue-like structure that allows it to insert into the cleft in the integrin head group between the α and β subunits in a manner similar to that seen in the structure of cilengitide a cyclic RGD pentapeptide bound with integrin αVβ3 (Figs. 1A and 3) (12). The sequence occurs at the apex of a type II β-turn (34) that contains a hydrogen bond between the carbonyl oxygen of Cys-8 and the amide group of Asp-11 (Fig. 3). The structure of the loop is further stabilized by two disulfide bonds. Cys-8 immediately precedes Arg-9, the first residue of the RGD motif, and is disulfide linked to Cys-94 (Fig. 3A). The second disulfide bond links Cys-4 and Cys-17. Both of these disulfides are conserved in Ves v 5 (Fig. 2), and the backbone conformation of the loop is also quite similar in the two proteins (Fig. 1). The extended N terminus of the CAP domain that contains the RGD motif is not present in related proteins from nematodes and mammals (28). Tight turns stabilized by disulfide bonds are also found in the small disintegrin proteins from snake venoms such as trimestatin and flavoridin (7).
FIGURE 3.
Structure of the loop of tablysin-15 containing the RGD motif. A, ribbon diagram of tablysin-15 highlighting the RGD sequence and the two disulfide bonds stabilizing the RGD loop. Carbon is shown in green, nitrogen is in blue, oxygen is in red, and sulfur is in yellow. B, electron density (blue, contoured at 1 σ) covering the RGD motif. The structure is colored as in A. C, stereo view of the RGD-containing loop showing hydrogen binding of the β-turn. Hydrogen bonds are shown as dashed red lines, and atoms are colored as described for A.
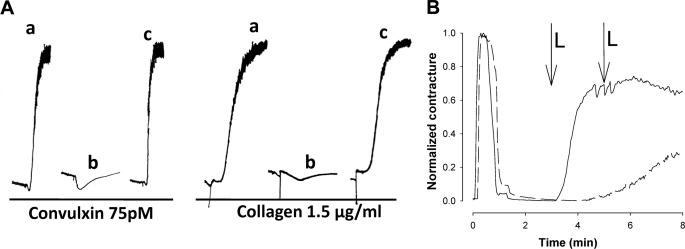
To verify the RGD motif as the inhibitory feature of tablysin-15, we mutated the motif to Ala-Gly-Ala. The mutant protein exhibited a loss of anti-platelet aggregation activity after stimulation by collagen or the GPVI agonist convulxin, indicating that binding of αIIbβ3 is responsible for all of the observed activity of tablysin-15 (Fig. 4A).
FIGURE 4.
Physiological activities of tablysin-15. A, the RGD motif and inhibition of collagen and convulxin-induced platelet aggregation. Aggregometer traces are measured from stirred platelet suspensions treated with convulxin and collagen at the concentrations shown. a, agonist in the absence of tablysin-15; b, agonist in the presence of 85 nm tablysin-15; c, agonist in the presence of 100 nm of the RGD9–11:AGA site-directed mutant tablysin-15. B, inhibition of LTC4-induced contraction of a preparation of guinea pig ileum by tablysin-15. Solid line, no tablysin-15 added. Dashed line, tablysin-15 added to 5 μm. In both experiments, histamine was first added at time 0 to induce a contraction. The preparation was then washed with fresh Tyrode's solution resulting in relaxation. At the 3- and 5-min time points (marked L), LTC4 was added to 100 and 200 nm concentrations, respectively. The first addition of LTC4 caused maximal contraction in the absence of tablysin-15. A second addition of LTC4 resulted in a slow contraction in the presence of LTC4.
Tablysin-15 Contains Fatty Acid-binding Pocket
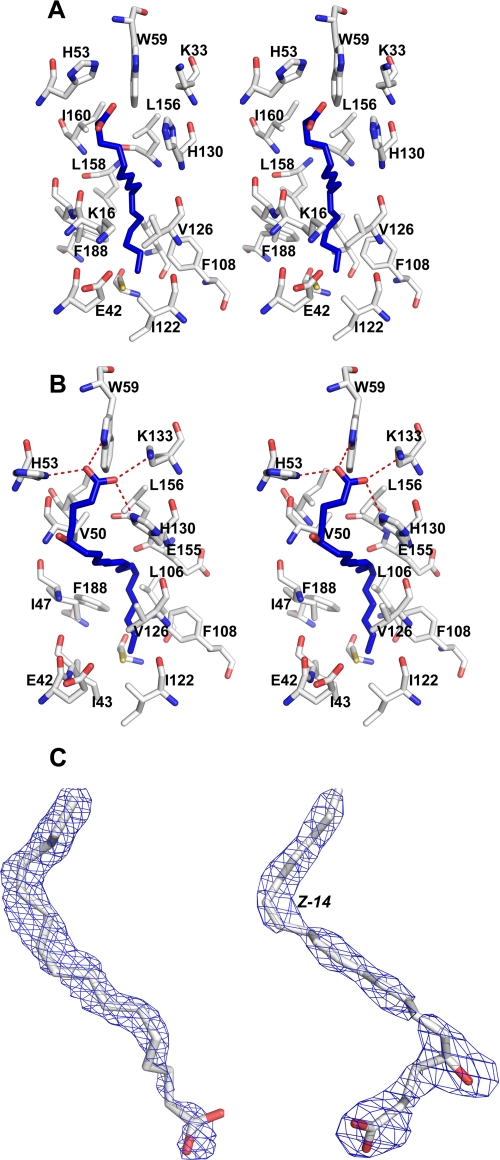
Surprisingly, in addition to the RGD sequence, the tablysin-15 structure was found to contain a hydrophobic pocket or channel located between helices 1, 3, and 4 that also includes one face of the β-sheet (Figs. 1 and 5). The pocket is solvent-accessible with an entrance flanked by the side chains of His-53, His-130, and Trp-59 and is similar to the eicosanoid-binding pocket found in the salivary D7 proteins of mosquitoes (13, 15). The interior of the channel is lined with primarily hydrophobic residues including Val-50, Val-126, Val-54, Leu-156, Ile-47, Ile-43, Ile-122, Leu-106, Leu-184, and Phe-108 (Fig. 5).
FIGURE 5.
Structure of the hydrophobic binding pocket of tablysin-15. A, stereo view of the protein pocket containing a molecule of palmitic acid. Protein carbon atoms are shown in white, oxygen atoms are red, and nitrogen atoms are blue. The ligand carbon atoms are shown in blue, and oxygen atoms are in red. B, stereo view of the protein pocket containing a molecule of (E,E,Z,Z)-7,9,11,14-eicosatetraenoic acid. Coloring is the same as in A. Hydrogen bonds are shown as dashed red lines. C, electron density (2Fo − Fc) covering the ligands in the structure with no added ligand (left) and after cocrystallization with (E,E,Z,Z)-7,9,11,14-eicosatetraenoic acid (right). Carbon atoms are colored white, and oxygen atoms are red. Electron density is colored blue. For orientation, the cis-14 unsaturation is labeled in (E,E,Z,Z)-7,9,11,14-eicosatetraenoic acid.
Electron density not accounted for by the protein was observed in the pocket in both the native and selenomethionine structures. The unexplained electron density enters the pocket between the side chains of His-53, His-130, and Trp-59 and makes a 90° bend near the interior end of the channel (Fig. 5C). The tubular shape of the density is reminiscent of that seen in crystal structures of eicosanoid complexes of the mosquito D7 proteins (15) and can probably be attributed to a fatty acid molecule derived from the E. coli expression host that is bound during the refolding process. When palmitic acid, a 16-carbon saturated fatty acid, was modeled into the binding site, continuous electron density was observed in the weighted 2Fo − Fc map of the refined structure contoured at 0.8 σ (Fig. 5, A and C). In this model, the ω-end of the hydrocarbon chain is inserted into the hydrophobic pocket (Fig. 5A), and the carboxyl group lies near the entrance.
Tablysin-15 Binds Cysteinyl Leukotrienes
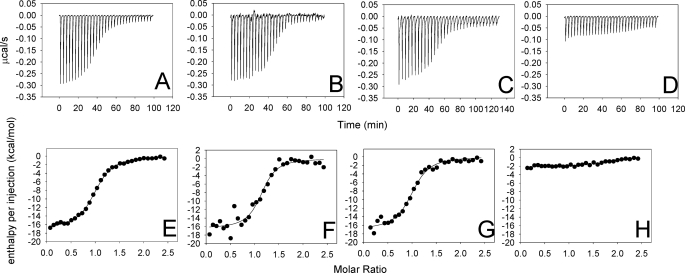
The presence of a possible fatty acid bound in the hydrophobic pocket suggested to us that tablysin-15 may function as a scavenger of bioactive lipids including leukotrienes or thromboxanes. When a panel of eicosanoid compounds was screened using isothermal titration calorimetry, only the cysteinyl leukotrienes LTC4, LTD4, and LTE4 elicited significant heats of binding (Fig. 6). The reaction stoichiometries estimated by fitting the data to a single-site binding model indicated that one molecule of ligand was binding per molecule of protein (Table 2). The observed binding affinities were small relative to the leukotriene-binding D7 proteins of mosquito saliva (13, 15), suggesting that the unknown ligand observed in the crystal structures may compete with the leukotriene for its binding site, thereby lowering the apparent affinity for the added ligand. To remove the bound lipid, we further purified the protein using reversed phase liquid chromatography with a gradient of aqueous acetonitrile as the eluent. This delipidated protein bound cysteinyl leukotrienes with at least 4-fold higher affinity and an increase in the magnitude of the observed binding enthalpy of ∼2-fold (Table 2 and Fig. 6 and supplemental Fig. S1). Conversely, the 20-carbon fatty acid arachidonic acid did not bind significantly with tablysin-15 (Fig. 6 and Table 2).
FIGURE 6.
Binding of eicosanoid compounds with tablysin-15 as measured by isothermal titration calorimetry. A–D, heats measured on injection of eicosanoids. A, LTC4; B, LTD4; C, LTE4; D, arachidonic acid. E–H, enthalpies per injection calculated from A–D. E, LTC4; F, LTD4; G, LTE4; H, arachidonic acid. Solid circles represent the measured enthalpies, whereas the solid line represents the best fit to a single-site binding model. Thermodynamic parameters derived from this curve fitting are given in Table 2. All of the experiments were performed with a ligand (syringe) concentration of 50 μm and a protein (cell) concentration of 5 μm, with the exception of LTE4, which had a ligand concentration of 40 μm and a protein concentration of 4 μm.
TABLE 2.
Thermodynamic parameters for eicosanoid binding with tablysin-15
| Ligand | ΔH | Kd | 1N | TΔS |
|---|---|---|---|---|
| kcal/mol | nm | mol of ligand bound/mol of protein | kcal/mol | |
| LTC4 | −16.9 ± 0.1 | 178.6 ± 10.5 | 0.99 | −7.6 |
| LTD4 | −16.4 ± 0.6 | 105.4 ± 35.6 | 1.13 | −6.8 |
| LTE4 | −16.9 ± 0.4 | 133.3 ± 23.2 | 1.00 | −7.4 |
| Arachidonic acid | −2.1 ± 0.1 | >10,000 |
LTC4, LTD4, and LTE4 differ from each other in the structure of the peptide substituent at C-6. LTC4 is the product secreted by mast cells and contains a glutathione moiety conjugated with the eicosanoid fatty acid chain through a thioether linkage. LTC4 is metabolized extracellularly to LTD4 and LTE4 by successive cleavages of the peptide substituent to produce cysteinylglycine (LTD4) and cysteine (LTE4) moieties (35). All three products are active in stimulating inflammatory responses in the skin (14). Like the mosquito D7 proteins, tablysin-15 bound the three cysteinyl leukotrienes with similar affinity, despite the fact that they differ substantially in the structure of the peptide (13, 15) (Fig. 6 and Table 2). This suggests that only the fatty acid portion of the molecule makes important binding contacts with the protein and that the protein would be effective at sequestering all three compounds at the feeding site.
The ability of tablysin-15 (reversed phase HPLC purified) to block LTC4-induced smooth muscle contraction was tested using a preparation of guinea pig ileum (Fig. 4B). The addition of 5 μm tablysin-15 abrogated contraction of the ileum induced by 100 nm LTC4, indicating that the protein effectively sequestered the ligand and prevented the activation of CysLT receptors (Fig. 4B). The further addition of LTC4 to a concentration of 200 nm produced a detectable but reduced level of contraction.
Structure of Tablysin-15-LTE4 Complex
After delipidation by reversed phase HPLC, tablysin-15 was crystallized in the presence of equimolar LTE4. The data were collected to a resolution of 2.5 Å, and the position of the bound ligand was examined. Although LTE4 was bound at less than full occupancy, electron density surrounding the ligand was visible and differed from that in the structure without added ligands and allowed us to model the eicosanoid chain ((E,E,Z,Z)-7,9,11,14-eicosatetraenoic acid) in the ligand-binding pocket (Fig. 5). The conformation of the ligand is similar to that of the bound fatty acid seen in the native structure, with the ω-end of the two molecules occupying similar positions. The major difference lies in the appearance of a bend in the fatty acid chain occurring at carbons 5 and 6 that allows the binding site to accommodate the additional two carbon atoms of LTE4 (Fig. 5). A break in the electron density around C7 is visible, but the carboxyl end of the chain can be seen to extend between the side chains of His-53, Trp-59, His-130, and Lys-133 where it can form hydrogen bonding/salt bridge interactions with all of these, although the imidazole group of His-53 shows disorder. These interactions induce a shift of ∼1.5 Å in the position of the loop containing Trp-59, putting it in position to form a hydrogen bond with the carboxyl group. The hydroxyl group of LTE4 does not interact directly with the protein, and the cysteinyl moiety is apparently disordered.
DISCUSSION
Tablysin-15 has been previously shown to facilitate blood feeding by inhibiting the aggregation of platelets through binding with integrin αIIbβ3 (6). It has also proven to be a potent antagonist of integrin αVβ3, inhibiting microvascular endothelial cell adhesion at low nanomolar levels (6). The structure presented here shows the location and conformation of an inhibitory RGD motif in the context of a novel structural scaffold, the CAP domain. Additionally we found the protein to contain a novel eicosanoid-binding site located in the center of the CAP domain that appears to act in scavenging cysteinyl leukotrienes at the site of blood feeding. A second function can now be assigned to the protein: that of an anti-inflammatory agent acting to ameliorate the effects of mast cell-derived cysteinyl leukotrienes produced in response to the presence of salivary protein antigens in the skin.
The RGD motif has long been known as a ligand for integrin receptors and is present in snake venom disintegrins, as well as endogenous vertebrate proteins including fibronectin, vitronectin, fibrinogen, von Willebrand factor, and activated protein C where it serves in cellular adhesion processes (7). The details of the interaction of RGD peptides with integrins are shown in the published crystal structures of αVβ3 in complex with the cyclic pentapeptide cilengitide and αIIbβ3 in complex with various ligands (10, 12). Cilengitide binds αVβ3 in a crevice made up of the β-propeller domain of the αV chain and the βA domain from the β3 chain. The arginine side chain forms salt bridge interactions with two aspartate residues in the β-propeller domain, whereas the aspartate side chain of the ligand forms a larger network of salt bridge/hydrogen bonding interactions with residues of the β3 chain including a Mg2+ ion at the MIDAS site in the βA domain (12). In αIIbβ3, interactions with the RGD motif are similar, but the distance between elements of the binding site is larger because of a deeper, broader crevice in the αIIb chain relative to αV (10). This is reflected in the structures of αIIbβ3-selective ligands that tend to have larger distances between the cationic (Arg) and anionic (Asp) contact points. The αIIbβ3 binding sequence in the γ subunit of fibrinogen contains a KQAGD sequence in which the lysine residue performs the function of arginine in RGD, whereas the aspartate plays the same role. This motif is too large to be accommodated at the αVβ3 binding site (9, 36, 37).
In tablysin-15 the loop containing the RGD motif is stabilized by two disulfide bonds and exhibits a conformation similar to comparable loops of structurally characterized venom disintegrins such as trimestatin and flavoridin (38, 39). The type II β-turn is well ordered, producing a compact loop that gives the RGD motif dimensions very similar to those of cilengitide. This may explain the apparently higher affinity of tablysin-15 for αVβ3 than for αIIbβ3 as estimated from cellular adhesion assays (6). It is interesting to note that superposition of the RGD tripeptide sequences in flavoridin and tablysin-15 results in very little spatial overlap in the remainder of the protein. This suggests that integrin-ligand interactions outside of the RGD motif must be quite different in the two types of disintegrin molecules.
Inhibition of the proinflammatory and prohemostatic effects of eicosanoids and biogenic amines is an important function of the salivary secretions of blood feeders. Histamine and serotonin released by mast cells leads to swelling, itching, and pain responses that can interfere with feeding and prevent the uptake of a complete blood meal (13, 15). Mast cells also secrete cysteinyl leukotrienes that elicit wheal and flare responses in the skin (14). Thromboxanes, catecholamines, and serotonin released from platelets promote platelet aggregation and cause vasoconstrictive responses. Removal of these mast cell and platelet products from the bite site is a common inhibitory mechanism to facilitate feeding, and the saliva of most groups of blood feeders contain proteins that bind some or all of these compound types. As a group, salivary proteins that act by scavenging effector molecules rather than blocking their cellular targets have been given the name kratagonists (5). Until now, salivary proteins performing these functions have been restricted to three families, the lipocalins, the D7s and the “yellow proteins” (19, 40–42). With tablysin-15, a fourth family, the CAP domain proteins, has been added to the list of known kratagonists. Blood feeding has evolved separately in the major taxa of blood feeders (5), and it is not surprising that through a process of convergent evolution, different protein scaffolds have been adapted to perform the same function.
The leukotriene-binding pocket of tablysin-15 lying between helices H1, H3, and H4 is formed with little displacement of its helical elements when compared with Ves v 5 (3). Rather, differences in side chain structure along the interface between helices H1 and H3 produce the pocket. In other CAP domain structures, including Ves v 5 and the fire ant venom antigen Sol i 3, the space between these α-helices is filled, and no channel leading to the protein surface exists (Fig. 7) (3, 30). The structure of the binding pocket and ligand binding mode are similar to those reported for mosquito D7 proteins where the nonfunctionalized end of the fatty acid chain inserts deep into the hydrophobic pocket, whereas the carboxylate end of the chain lies near the entrance to the channel where it forms hydrogen bonding or salt bridge interactions with polar residues at the protein surface (13, 15). Like the D7s, the peptide substituent of the leukotriene is not well ordered, suggesting that it does not form extensive polar contacts with the protein molecule (13, 15). This is reflected in the similar binding affinities for LTC4, LTD4, and LTE4.
FIGURE 7.
Space-filling model of the LTE4-tablysin-15 complex. A, (E,E,Z,Z)-7,9,11,14-eicosatetraenoic acid with carbon atoms shown as white and oxygen as red inserted into the hydrophobic channel of tablysin-15 with carbon atoms shown as cyan, oxygen as red, and nitrogen as blue. B, for comparison, the region of Ves v 5 corresponding to the ligand-binding pocket of tablysin-15 showing a lack of any similar channel.
We have established here that tablysin-15 has evolved two distinct functions that act to inhibit hemostasis and inflammation during feeding. The protein binds β3 integrins by means of its RGD motif and also binds proinflammatory and vasoactive cysteinyl leukotrienes in a novel hydrophobic binding channel. In addition to the αIIbβ3 binding motif in its γ subunit, fibrinogen possesses an RGD motif that interacts with αVβ3. It is thought that the latter interaction functions in stabilization of the thrombus by favoring attachment to the endothelium (9, 36). The binding of both αVβ3 and αIIbβ3 by tablysin-15 may be adaptive in that these interactions would potentially block binding of fibrinogen to both the platelet and the vessel wall, thereby disrupting thrombosis in two ways. Binding with αVβ3 may also increase the effectiveness of the leukotriene binding of tablysin-15 by favoring retention of the protein at the feeding site via tethering by αVβ3, where it can more efficiently scavenge cysteinyl leukotrienes.
Supplementary Material
Acknowledgments
We thank D. Garboczi and A. Gittis for discussions and R. Hearn for technical assistance. We also thank the staff of the Southeast Regional Collaborative Access Team, Advanced Photon Source, Argonne National Laboratory for assistance with x-ray data collection.
This work was supported, in whole or in part, by the National Institutes of Health intramural research program of NIAID. This work was also supported in part by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract W-31-109-Eng-38.

This article contains supplemental Fig. S1.
The atomic coordinates and structure factors (codes 3U3N and 3U3U) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- LTC4
- leukotriene C4
- LTD4
- leukotriene D4
- LTE4
- leukotriene E4.
REFERENCES
- 1. Ribeiro J. M., Francischetti I. M. (2003) Role of arthropod saliva in blood feeding: Sialome and post-sialome perspectives. Annu. Rev. Entomol. 48, 73–88 [DOI] [PubMed] [Google Scholar]
- 2. Ribeiro J. M. (1995) Blood-feeding arthropods. Live syringes or invertebrate pharmacologists? Infect. Agents Dis. 4, 143–152 [PubMed] [Google Scholar]
- 3. Henriksen A., King T. P., Mirza O., Monsalve R. I., Meno K., Ipsen H., Larsen J. N., Gajhede M., Spangfort M. D. (2001) Major venom allergen of yellow jackets, Ves v 5. Structural characterization of a pathogenesis-related protein superfamily. Proteins 45, 438–448 [DOI] [PubMed] [Google Scholar]
- 4. Ribeiro J. M., Andersen J., Silva-Neto M. A., Pham V. M., Garfield M. K., Valenzuela J. G. (2004) Exploring the sialome of the blood-sucking bug Rhodnius prolixus. Insect Biochem. Mol. Biol. 34, 61–79 [DOI] [PubMed] [Google Scholar]
- 5. Ribeiro J. M., Mans B. J., Arcà B. (2010) An insight into the sialome of blood-feeding Nematocera. Insect Biochem. Mol. Biol. 40, 767–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ma D., Xu X., An S., Liu H., Yang X., Andersen J. F., Wang Y., Tokumasu F., Ribeiro J. M., Francischetti I. M., Lai R. (2011) A novel family of RGD-containing disintegrins (Tablysin-15) from the salivary gland of the horsefly Tabanus yao targets αIIbβ3 or αVβ3 and inhibits platelet aggregation and angiogenesis. Thromb. Haemost. 105, 1032–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Calvete J. J., Marcinkiewicz C., Monleón D., Esteve V., Celda B., Juárez P., Sanz L. (2005) Snake venom disintegrins. Evolution of structure and function. Toxicon 45, 1063–1074 [DOI] [PubMed] [Google Scholar]
- 8. Wermelinger L. S., Geraldo R. B., Frattani F. S., Rodrigues C. R., Juliano M. A., Castro H. C., Zingali R. B. (2009) Integrin inhibitors from snake venom. Exploring the relationship between the structure and activity of RGD-peptides. Arch. Biochem. Biophys. 482, 25–32 [DOI] [PubMed] [Google Scholar]
- 9. Springer T. A., Zhu J., Xiao T. (2008) Structural basis for distinctive recognition of fibrinogen γC peptide by the platelet integrin alphaIIbbeta3. J. Cell Biol. 182, 791–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xiao T., Takagi J., Coller B. S., Wang J. H., Springer T. A. (2004) Structural basis for allostery in integrins and binding to fibrinogen-mimetic therapeutics. Nature 432, 59–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xiong J. P., Stehle T., Diefenbach B., Zhang R., Dunker R., Scott D. L., Joachimiak A., Goodman S. L., Arnaout M. A. (2001) Crystal structure of the extracellular segment of integrin αVβ3. Science 294, 339–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xiong J. P., Stehle T., Zhang R., Joachimiak A., Frech M., Goodman S. L., Arnaout M. A. (2002) Crystal structure of the extracellular segment of integrin αVβ3 in complex with an Arg-Gly-Asp ligand. Science 296, 151–155 [DOI] [PubMed] [Google Scholar]
- 13. Alvarenga P. H., Francischetti I. M., Calvo E., Sá-Nunes A., Ribeiro J. M., Andersen J. F. (2010) The function and three-dimensional structure of a thromboxane A2/cysteinyl leukotriene-binding protein from the saliva of a mosquito vector of the malaria parasite. PLoS Biol. 8, e1000547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Soter N. A., Lewis R. A., Corey E. J., Austen K. F. (1983) Local effects of synthetic leukotrienes (LTC4, LTD4, LTE4, and LTB4) in human skin. J. Invest. Dermatol. 80, 115–119 [DOI] [PubMed] [Google Scholar]
- 15. Calvo E., Mans B. J., Ribeiro J. M., Andersen J. F. (2009) Multifunctionality and mechanism of ligand binding in a mosquito antiinflammatory protein. Proc. Natl. Acad. Sci. U.S.A. 106, 3728–3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mans B. J., Ribeiro J. M. (2008) Function, mechanism and evolution of the moubatin-clade of soft tick lipocalins. Insect Biochem. Mol. Biol. 38, 841–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mans B. J., Ribeiro J. M. (2008) A novel clade of cysteinyl leukotriene scavengers in soft ticks. Insect Biochem. Mol. Biol. 38, 862–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Francischetti I. M., Saliou B., Leduc M., Carlini C. R., Hatmi M., Randon J., Faili A., Bon C. (1997) Convulxin, a potent platelet-aggregating protein from Crotalus durissus terrificus venom, specifically binds to platelets. Toxicon 35, 1217–1228 [DOI] [PubMed] [Google Scholar]
- 19. Andersen J. F., Francischetti I. M., Valenzuela J. G., Schuck P., Ribeiro J. M. (2003) Inhibition of hemostasis by a high affinity biogenic amine-binding protein from the saliva of a blood-feeding insect. J. Biol. Chem. 278, 4611–4617 [DOI] [PubMed] [Google Scholar]
- 20. Minor W., Cymborowski M., Otwinowski Z., Chruszcz M. (2006) HKL-3000. The integration of data reduction and structure solution–From diffraction images to an initial model in minutes. Acta Crystallogr. D. Biol. Crystallogr. 62, 859–866 [DOI] [PubMed] [Google Scholar]
- 21. Pflugrath J. W. (1999) The finer things in X-ray diffraction data collection. Acta Crystallogr. D. Biol. Crystallogr. 55, 1718–1725 [DOI] [PubMed] [Google Scholar]
- 22. Schneider T. R., Sheldrick G. M. (2002) Substructure solution with SHELXD. Acta Crystallogr. D. Biol. Crystallogr. 58, 1772–1779 [DOI] [PubMed] [Google Scholar]
- 23. Cowtan K. (2006) The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D. Biol. Crystallogr. 62, 1002–1011 [DOI] [PubMed] [Google Scholar]
- 24. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D. Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D. Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 26. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lovell S. C., Davis I. W., Arendall W. B., 3rd, de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., Richardson D. C. (2003) Structure validation by Cα geometry. φ, ψ, and Cβ deviation. Proteins 50, 437–450 [DOI] [PubMed] [Google Scholar]
- 28. Gibbs G. M., Roelants K., O'Bryan M. K. (2008) The CAP superfamily. Cysteine-rich secretory proteins, antigen 5, and pathogenesis-related 1 proteins–Roles in reproduction, cancer, and immune defense. Endocr. Rev. 29, 865–897 [DOI] [PubMed] [Google Scholar]
- 29. Asojo O. A., Goud G., Dhar K., Loukas A., Zhan B., Deumic V., Liu S., Borgstahl G. E., Hotez P. J. (2005) X-ray structure of Na-ASP-2, a pathogenesis-related-1 protein from the nematode parasite, Necator americanus, and a vaccine antigen for human hookworm infection. J. Mol. Biol. 346, 801–814 [DOI] [PubMed] [Google Scholar]
- 30. Padavattan S., Schmidt M., Hoffman D. R., Markovi-Housley Z. (2008) Crystal structure of the major allergen from fire ant venom, Sol i 3. J. Mol. Biol. 383, 178–185 [DOI] [PubMed] [Google Scholar]
- 31. Shikamoto Y., Suto K., Yamazaki Y., Morita T., Mizuno H. (2005) Crystal structure of a CRISP family Ca2+-channel blocker derived from snake venom. J. Mol. Biol. 350, 735–743 [DOI] [PubMed] [Google Scholar]
- 32. Asojo O. A., Koski R. A., Bonafé N. (2011) Structural studies of human glioma pathogenesis-related protein 1. Acta Crystallogr D Biol Crystallogr 67, 847–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wang Y. L., Kuo J. H., Lee S. C., Liu J. S., Hsieh Y. C., Shih Y. T., Chen C. J., Chiu J. J., Wu W. G. (2010) Cobra CRISP functions as an inflammatory modulator via a novel Zn2+- and heparan sulfate-dependent transcriptional regulation of endothelial cell adhesion molecules. J. Biol. Chem. 285, 37872–37883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chou K. C. (2000) Prediction of tight turns and their types in proteins. Anal. Biochem. 286, 1–16 [DOI] [PubMed] [Google Scholar]
- 35. Boyce J. A. (2005) Eicosanoid mediators of mast cells. Receptors, regulation of synthesis, and pathobiologic implications. Chem. Immunol. Allergy 87, 59–79 [DOI] [PubMed] [Google Scholar]
- 36. Cheresh D. A., Berliner S. A., Vicente V., Ruggeri Z. M. (1989) Recognition of distinct adhesive sites on fibrinogen by related integrins on platelets and endothelial cells. Cell 58, 945–953 [DOI] [PubMed] [Google Scholar]
- 37. Smith J. W., Ruggeri Z. M., Kunicki T. J., Cheresh D. A. (1990) Interaction of integrins αvβ3 and glycoprotein IIb-IIIa with fibrinogen. Differential peptide recognition accounts for distinct binding sites. J. Biol. Chem. 265, 12267–12271 [PubMed] [Google Scholar]
- 38. Fujii Y., Okuda D., Fujimoto Z., Horii K., Morita T., Mizuno H. (2003) Crystal structure of trimestatin, a disintegrin containing a cell adhesion recognition motif RGD. J. Mol. Biol. 332, 1115–1122 [DOI] [PubMed] [Google Scholar]
- 39. Senn H., Klaus W. (1993) The nuclear magnetic resonance solution structure of flavoridin, an antagonist of the platelet GP IIb-IIIa receptor. J. Mol. Biol. 232, 907–925 [DOI] [PubMed] [Google Scholar]
- 40. Mans B. J., Calvo E., Ribeiro J. M., Andersen J. F. (2007) The crystal structure of D7r4, a salivary biogenic amine-binding protein from the malaria mosquito Anopheles gambiae. J. Biol. Chem. 282, 36626–36633 [DOI] [PubMed] [Google Scholar]
- 41. Ribeiro J. M., Walker F. A. (1994) High affinity histamine-binding and antihistaminic activity of the salivary nitric oxide-carrying heme protein (nitrophorin) of Rhodnius prolixus. J. Exp. Med. 180, 2251–2257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xu X., Oliveira F., Chang B. W., Collin N., Gomes R., Teixeira C., Reynoso D., My Pham V., Elnaiem D. E., Kamhawi S., Ribeiro J. M., Valenzuela J. G., Andersen J. F. (2011) Structure and function of a “yellow” protein from saliva of the sand fly Lutzomyia longipalpis that confers protective immunity against Leishmania major infection. J. Biol. Chem. 286, 32383–32393 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.