Background: Nod factor perception (NFP) protein is a plant, lysin motif receptor-like kinase.
Results: Disulfide bridges that connect the three extracellular lysin motifs and the intracellular dead-kinase domain are essential for NFP function.
Conclusion: Post-translational modifications are required for NFP folding, trafficking, and functioning.
Significance: Structural information will help to determine NFP biochemical function.
Keywords: Disulfide, Glycosylation, Plant, Post-translational Modification, Receptor Structure-Function, N-Glycans, Nodulation, Rhizobium, Symbiosis
Abstract
The lysin motif receptor-like kinase, NFP (Nod factor perception), is a key protein in the legume Medicago truncatula for the perception of lipochitooligosaccharidic Nod factors, which are secreted bacterial signals essential for establishing the nitrogen-fixing legume-rhizobia symbiosis. Predicted structural and genetic analyses strongly suggest that NFP is at least part of a Nod factor receptor, but few data are available about this protein. Characterization of a variant encoded by the mutant allele nfp-2 revealed the sensitivity of this protein to the endoplasmic reticulum quality control mechanisms, affecting its trafficking to the plasma membrane. Further analysis revealed that the extensive N-glycosylation of the protein is not essential for biological activity. In the NFP extracellular region, two CXC motifs and two other Cys residues were found to be involved in disulfide bridges, and these are necessary for correct folding and localization of the protein. Analysis of the intracellular region revealed its importance for biological activity but suggests that it does not rely on kinase activity. This work shows that NFP trafficking to the plasma membrane is highly sensitive to regulation in the endoplasmic reticulum and has identified structural features of the protein, particularly disulfide bridges involving CXC motifs in the extracellular region that are required for its biological function.
Introduction
The establishment of the nitrogen-fixing symbiosis between legume plants and rhizobia bacteria requires a molecular dialog between the host and the microsymbiont. Secretion of rhizobial lipochitooligosaccharidic signals, called Nod factors (NFs),2 and their perception by the host plant are necessary for activating the plant root nodule organogenesis and infection programs, and these factors are also involved in the partner specificity between the legume species and the bacterial strains (1, 2). Successful interaction leads to development of infected root nodules (nodulation) in which the bacteria fix dinitrogen.
Genetic analysis has identified genes involved in NF perception and signal transduction. NFP of Medicago truncatula (3) and its orthologues NFR5 of Lotus japonicus and SYM10 of pea (4, 5) are required for all NF responses and for nodulation and infection. Another gene, LYK3, is also required for nodulation and infection in M. truncatula, whereas in L. japonicus, a similar gene, NFR1, is required, like NFR5, for all NF responses (5, 6).
NFP, LYK3, and their orthologues are receptor-like kinases (RLKs), belonging to the lysin motif (LysM-RLK) class. LysM-RLKs occur in all plants, and phylogenetic analysis has defined two subfamilies, which have been called the LYR and LYK families (3). In M. truncatula, there are at least 17 LysM-RLKs, and NFP and LYK3 are members of the LYR and LYK subfamilies, respectively. Like all RLKs (7), the LysM-RLKs are type I membrane proteins with a predicted signal peptide and an extracellular region, followed by a single transmembrane-spanning helix and an intracellular region (IR) exhibiting homology to a protein kinase. The lack of certain highly conserved kinase features and the lack of in vitro kinase activity of the IR of NFP (3) and NFR5 (8) suggest that these proteins and indeed many LYR proteins may be part of the large number of plant RLKs with “dead kinases” (9). In contrast, LYK3 and NFR1 have active kinases, which are necessary for their symbiotic roles (3, 8, 10).
All of the LysM-RLKs are predicted to encode proteins with three LysM domains in their extracellular regions, which are separated by characteristic CXC motifs (where X represents any amino acid) in the interdomain spacer regions. A similar structure also occurs in related LysM receptor-like proteins (LYM proteins) that lack an IR (11). LysM domains are protein motifs of about 40 amino acids (AA), which were first described in bacterial autolysins but indeed are found in many eukarya and bacteria proteins, often in association with other domains (12). Only in plants are they associated with kinase domains (12). LysM domains are implicated in the binding of GlcNAc-containing molecules. However, although genetic analysis implicates the symbiotic LysM-RLKs of legumes in binding of lipochitooligosaccharidic NFs (1, 13, 14), this has not yet been demonstrated biochemically.
We previously showed that NFP is highly N-glycosylated (15), and of the two mutant alleles of NFP that have been identified (3), nfp-1 appears to be a null allele, whereas nfp-2 bears a mutation (S67F) located in a putative N-glycosylation site in the first LysM domain. In addition, the presence of the highly conserved pairs of CXC motifs between the LysM domains in LysM-RLKs suggests that they may play a structural role in the protein, perhaps through the formation of disulfide (S-S) bridges.
N-Glycosylation and S-S bridges are post-translational modifications that occur in many eukaryotic proteins exposed to the extracellular medium (secreted and surface membrane-anchored or transmembrane proteins, such as NFP). S-S bridges also occur in prokaryotes but to a lesser extent than in eukaryotes. N-Glycosylation and S-S bridges have been reported to play many roles, including facilitating the folding, trafficking and function of the protein as well as protecting it from an extracellular medium, which, in plants, is acidic and rich in proteases (16). These post-translational modifications have recently been shown to be important for trafficking to the plasma membrane (PM) and/or the functioning of several plant RLKs of the leucine-rich repeat (LRR-RLKs) class (17). Both modifications occur in the endoplasmic reticulum (ER). S-S bridges are formed between two Cys residues, due to the favorable redox condition and with the help of protein disulfide isomerases. N-Glycosylation, initially with mannose-rich N-glycans, occurs on Asn residues at specific sites, which consist of the consensus sequence Asn-X-Ser/Thr, where X is any AA except Pro and is mediated by the oligosaccharyltransferase complex (16). The N-glycans then undergo cycles of trimming and elongation, which lead to dissociation/association with the ER lumenal lectin chaperones calnexin/calreticulin (18). N-Glycans are further modified in the Golgi apparatus to give complex N-glycans, which in plants are often fucosylated on the proximal GlcNAc residue (19). In addition to calnexin/calreticulin, BIP and its partners are other ER non-lectin lumenal chaperones that bind hydrophobic exposed domains of unfolded proteins. All together, the lumenal and the cytosolic chaperones retain the nascent proteins in the ER until they are correctly folded. Long retention of unfolded proteins increases the probability for the protein to be targeted for degradation by the ER-associated degradation pathway or eventually by the vacuole (20). This represents the ER quality control (ER-QC).
The aim of this work was to identify the structural features of NFP required for its activity. To this end, we performed a large scale structure function analysis, using a combination of genetic complementation and biochemical approaches. In particular, we examined the role of post-translational modifications in the extracellular region and conserved residues in the intracellular region. The work presented here contributes to our understanding of the structure and regulation of plant LysM-RLKs in signal transduction and to the functioning of NFP in nodulation.
EXPERIMENTAL PROCEDURES
Cloning and Plant Transformation
A cloning vector was built containing an expression cassette consisting of a HindIII site, the cauliflower mosaic virus 35S promoter (Pro35S) or the NFP promoter, −1137 to −1 bp before ATG (ProNFP), a BglII site, the coding region of NFP fused in frame to a protein tag, an EcoRI site, the Nos terminator, and a SmaI site. The protein tags used were the yellow fluorescent protein, sYFP2 (21), monomeric red fluorescent protein (RFP) (22), and 3×FLAG (Sigma). A construction of NFP deleted from its intracellular region and fused to monomeric RFP (NFPΔIR-RFP) contained amino acids 1–283 of NFP. Point mutations were introduced using the QuikChange mutagenesis kit (Stratagene) using the NFP-RFP cloning vector for the mutations in the NFP extracellular region. The NFP-3×FLAG cloning vector was used for mutations in the NFP IR. The expression cassettes were then transferred to the binary plasmid pBin+ using HindIII and SmaI. Plasmids containing PMA4-GFP (23), HDEL-GFP (24), or HVR-ROP-mTurquoise (25, 26) are as described.
The resulting plasmids were introduced into Agrobacterium rhizogenes (ARqua1) or into Agrobacterium tumefaciens (LBA4404) by electroporation. M. truncatula kanamycin-resistant roots were produced on nfp-1 plantlets (3), essentially as described (27). After 4 weeks on agar plates supplemented with 20 μg/ml kanamycin, the composite plants were transferred to growth pouches. One week later, the root systems were inoculated with Sinorhizobium meliloti (strain 2011). Nodules were counted at 10 and 14 days postinoculation (dpi), and the root systems were then harvested and stored at −80 °C for future immunoblotting analysis. Nicotiana tabacum BY2 cells were transformed by co-culturing for 3 days at 25 °C in dark without shaking, several dilutions of A. tumefaciens suspensions at 1 A600 (10–200 μl) with 4 ml of 4-day-old BY2 cells (weekly subcultured). Bacteria and plant cells were previously washed and resuspended in BY2 cell culture medium (4.4 g/liter Murashige and Skoog salts, 30 g/liter sucrose, 0.2 g/liter KH2PO4, 5 mg/liter thiamine, 0.2 mg/liter 2,4-dichlorophenoxyacetic acid, pH 5.4) supplemented with 100 nm acetosyringone. Cells were then washed three times and plated and grown at 25 °C in the dark on solid BY2 cell culture medium (0.8% agar) supplemented with 400 mg/liter carbenicillin and 100 mg/liter kanamycin until a calli appeared and then cultured in BY2 cell culture medium with only kanamycin. Protoplasts were transformed as described (28).
Tunicamycin Treatment
Leaf discs of A. tumefaciens-infiltrated Nicotiana benthamiana leaves were incubated by flotation for 20 h on 10 μm tunicamycin (stock 5 mm tunicamycin (Sigma) in DMSO, diluted in water) prior to microscopy analysis. Leaf discs were then stored at −80 °C before immunoblotting analysis.
PNGase F Treatment
PNGase F treatment was performed on microsomal fractions or on denatured total extracts from transgenic roots. For fractionation, frozen roots were ground for 30 s with a 4-mm metal bead in 2-ml tubes. The powder was diluted in 500 μl of 250 mm sorbitol, 50 mm Tris-HCl, pH 8.0, 2 mm EDTA, 0.6% polyvinylpolypyrrolidone, 5 mm DTT, protease inhibitors (1 mm phenylmethylsulfonyl fluoride and 1 mg/ml each of leupeptin, aprotinin, antipain, chymostatin, and pepstatin; Sigma). 350 μl of 0.8-mm glass beads were added, and samples were reground for 90 s. Samples were centrifuged for 5 min at 10,000 × g, and then the supernatant was recentrifuged for 30 min at 100,000 × g. For denaturation of total extracts, transgenic roots were solubilized in gel loading buffer (see below) and diluted in water. Proteins were then precipitated by 10% trichloroacetic acid (TCA). The pellets were washed twice with 90% acetone. Microsomal fractions and TCA pellets were resuspended by sonication in 45 μl of 25 mm Tris-HCl, pH 7.0. Either 5 μl of PNGase F (Roche Applied Science) or 5 μl of 50% glycerol were added. The samples were incubated for 30 min at 37 °C prior to immunoblotting.
Cysteine Labeling
Microsomal fractions were prepared from 10 g of transgenic N. benthamiana leaves as described (29) and resuspended in 2 ml of denaturing buffer (6 m guanidinium HCl, 0.1 m Tris-HCl, pH 8.0). All steps were performed under nitrogen. The samples were divided in two; 100 μl of freshly prepared 1 m DTT or water were added before a 1-h incubation at room temperature. Microsomal fractions were then diluted, pelleted, washed, and finally resuspended (450 μl) in denaturing buffer. 50 μl of maleimide-PEG2-biotin (Pierce; 10 mm in DMSO) was added before a 1-h incubation at room temperature. Microsomal fractions were then diluted, pelleted, and resuspended (200 μl) in pull-down buffer (150 mm NaCl, 25 mm Tris-Cl, pH 7.5, 10% glycerol). 200 μl of pull-down buffer with 0.5% dodecyl maltoside (Alexis Biochemicals) were added. Samples were incubated for 10 min at 4 °C, diluted 2.5-fold with pull-down buffer, and centrifuged for 15 min at 165,000 × g. 15 μl of magnetic RFP trap (Chromotek) were added to the supernatant before a 1-h incubation at 4 °C. Beads were washed twice in pull-down buffer before analysis of the proteins by immunoblotting.
Immunopurification
200 mg of N. benthamiana transgenic leaves frozen and crushed were ground for 30 s with two 3-mm metal beads in 2-ml tubes. The powder was diluted in 400 μl of pull-down buffer with 0.5% dodecyl maltoside and shaken for 5 min at 4 °C. Samples were centrifuged for 5 min at 5,000 × g. The supernatant was centrifuged for 30 min at 100,000 × g. The supernatant was diluted 5-fold with pull-down buffer, and 10 μl of magnetic RFP trap (Chromotek) were added before a 30-min incubation at 4 °C. Beads were washed once in pull-down buffer with 0.1% dodecyl maltoside before analysis of the proteins by immunoblotting.
Immunoblotting
Protein expression in transgenic roots was followed by analysis of root total extracts (20 mg of roots for 30 μl of gel loading buffer). Other samples were prepared as described above. All samples were solubilized in gel loading buffer (2% SDS, 80 mm Tris-Cl, pH 6.8, 15% glycerol), heated for 5 min at 95 °C, and centrifuged for 5 min at 16,000 × g. After separation by SDS-PAGE on 8% (w/v) acrylamide gels, proteins were transferred onto nitrocellulose membranes. RFP fusions were detected using the serum from a rabbit immunized with an Escherichia coli expressed His6-tagged mCherry protein, at a dilution of 1:5,000, YFP fusions were detected using rabbit anti-GFP (Clontech) at 1:5,000, and BIP was detected using mouse anti-Hsc70 (1D9; Stressgen) at 1:1,000, followed by peroxidase-linked anti-rabbit antibodies (Millipore) or peroxidase-linked anti-mouse antibodies (Pierce). Peroxidase-linked goat anti-biotin antibodies (Sigma) were used at 1:1,000. Bovine serum albumen was used for membrane saturation and incubation of anti-biotin antibodies. Peroxidase activity was revealed by chemiluminescence using the Immobilon kit (Millipore). Molecular weights were estimated by comparison with the PageRuler prestained protein ladder (Fermentas).
Immunocytolabeling
Root segments transformed with the Pro35S:NFP-RFP construct (or the vector construct) were fixed in 4% formaldehyde, 0.1 m phosphate, 0.1% Triton X-100 buffer, pH 7.4, for 30 min at room temperature followed by subsequent fixation for 60 min in fresh fixing medium without Triton X-100. After rinsing, the specimens were embedded in low melting point wax (30) and cut into 10-μm-thick sections, which were deposited on stick-on-coated slides and dewaxed prior to the immunolocalization procedure. RFP immunolocalization was performed on sections by treating them with anti-RFP antibody diluted 1:20,000 as the primary antibody and goat anti-rabbit HRP (Santa Cruz Biotechnology, Inc.) diluted 1:2,500 as a secondary antibody according to the tyramide signal amplification plus fluorescein system (PerkinElmer Life Sciences). After staining, sections were treated according to Ref. 31.
Microscopy
Fluorescence of all cells was imaged using a Leica SP2 confocal microscope except for that of the protoplasts, imaged using a Zeiss CLSM510 microscope.
qRT-PCR
Biological samples analyzed and qRT-PCR analyses were performed as described (32).
In Silico Analysis
Alignments were made by ClustalW and corrected manually. Transmembrane domains were predicted by TMHMM, and signal peptide was predicted by signalP.
Molecular Modeling
Coordinates of the three LysM domains, previously built from the NMR structure of a LysM domain from E. coli through homology modeling techniques (15), were herein used to build the whole NFP extracellular region (192 AA, from Ile-32 to Pro-223). Loops between domains were generated by using the Biopolymer module of Sybyl X (Tripos Inc.). The same module was used for the generation of the mutated NFP S67F model. Hydrogen atoms were added, and partial atomic charges were derived from the AMBER force field (33). Titratable groups were considered in their standard protonation state at neutral pH. Minimization cycles were then carried out by first optimizing the orientation of hydrogen atoms and side chains. The whole structure was then fully optimized. Energy minimization cycles were performed within the AMBER force field (33). The permittivity was set as a distance-dependent function, and a Powell-type minimizer was used (34). Connolly surfaces were then computed by employing the MOLCAD program, mapping the lipophilic potential properties (35). Images were created within the SybylX graphical environment (Tripos Inc.).
RESULTS
NFP Localizes at Plasma Membrane
Putative NF receptors are expected to localize to the PM, where they may encounter NFs being produced by extracellular rhizobia. To study NFP localization and trafficking, we made YFP, RFP, and 3×FLAG fusions to the C terminus of NFP and transformed wild type (WT) and the nfp-1 (a predicted null allele) mutant of M. truncatula using the A. rhizogenes transformation system. Using the native promoter (ProNFP), we have not been able to detect specific fluorescence (above the autofluorescence level) from the YFP- or RFP-tagged NFP proteins in roots and nodules, although endogenous NFP is expressed in both tissues as shown using a ProNFP:GUS fusion (3) and by NFP mRNA quantification by qRT-PCR (supplemental Fig. S1). We thus tested whether the strong and constitutive promoter (Pro35S) could be used and found that all three tagged NFP fusions expressed from this promoter led to complementation of nfp-1 for nodulation, suggesting that the use of Pro35S and the presence of a C-terminal tag do not compromise the biological function of NFP; indeed, comparison of the 3×FLAG constructs showed that the Pro35S allows a stronger nodulation of M. truncatula than the ProNFP (supplemental Fig. S2). However, still no specific fluorescence (above the autofluorescence level) could be observed from the Pro35S:NFP-YFP or -RFP fusions in M. truncatula roots.
In contrast, the fluorescent NFP fusions were easily detectable when expressed in heterologous systems: cowpea leaf protoplasts, N. tabacum BY2 cells (supplemental Fig. S3), and N. benthamiana leaves (Fig. 1A). In all three systems, localization at the cell periphery was observed. In transformed cowpea protoplasts, the fluorescent fusion of NFP showed co-localization with the PM marker (supplemental Fig. S3A). In N. benthamiana leaf epidermal cells, localization of the protein evolved with time following infiltration with A. tumefaciens containing the construction. Early observations, 36 hpi, revealed NFP fluorescence mainly around the nucleus and in a cortical network resembling the ER (supplemental Fig. S3B). At 72 hpi, the protein was predominantly localized at the PM, as shown by co-expression with a PM marker (Fig. 1A). Observations at longer times after infiltration revealed fluorescence mainly in the vacuole (supplemental Fig. S3B). Because fluorescence was homogeneous in the vacuole, it probably corresponds to free RFP released following cleavage of the fusion protein in the vacuole. In the case of stably transformed BY2 cells, fluorescence was observed both at the PM and in the vacuole (supplemental Fig. S3C).
FIGURE 1.
NFP localizes to the PM and NFP-S67F localizes in the ER in N. benthamiana. A, NFP localizes to the PM in N. benthamiana. NFP-RFP was co-expressed in N. benthamiana leaves with a PM marker (PMA4-GFP), and the leaves were analyzed by confocal microscopy at 3 dpi. Superposition of the fluorescence images shows clear co-localization of NFP-RFP with the PM marker at the cell boundary. B, NFP localizes to the PM in M. truncatula roots. Roots of M. truncatula nfp-1 were transformed with the Pro35S:NFP-RFP construct or a control vector. Immunocytolabeling was performed using anti-RFP as primary antibodies and the tyramide signal amplification method (green) on root longitudinal sections corresponding to cortical cells. Proteins are stained with Evans blue (red), and the nucleus is stained with DAPI (blue). C, NFP-S67F is retained in the ER in N. benthamiana. The protein encoded by the nfp-2 allele (NFP-S67F) was fused to RFP and co-expressed with the PM (PMA4-GFP) or ER (HDEL-GFP) markers, and the leaves were analyzed by confocal microscopy at 3 dpi. Co-localization with the ER marker in a reticulated network in the cell and around the nucleus suggests that the protein is retained in the ER. Bars, 20 μm.
To study the localization of NFP in M. truncatula roots, we used immunocytolabeling with anti-RFP antibodies on A. rhizogenes transformed roots expressing NFP-RFP from Pro35S. Using a classical procedure, we were again unable to detect NFP. However, using a highly sensitive amplification method, we were finally able to detect a signal corresponding to NFP-RFP at the cell periphery in root cortical cells, whereas little or no signal was detected in vector-transformed control roots (Fig. 1B). This method also detected a PM localization of the NFP-RFP and NFP-YFP proteins in N. benthamiana leaves similarly to the localization of the fluorescence of these proteins (data not shown). Together, these results show that NFP localizes at the PM in both M. truncatula roots and in heterologous plant expression systems.
Number of N-Glycosylation Sites Is Variable between LysM-RLKs, and Individual N-Glycans Are Not Necessary for NFP Activity
Previous work had shown that NFP is highly N-glycosylated and that all of the 10-predicted N-glycosylation sites (NX(S/T)) are most likely occupied (15). The nfp-2 allele contains a point mutation leading to replacement of Ser-67 located in a putative N-glycosylation site in the first LysM domain (LysM1) by Phe and, like nfp-1, is a strong allele showing no NF responses (3). In some receptors, N-glycosylation can affect receptor function by affecting ligand binding and signal transduction (36). In NFP, eight of the N-glycosylation sites occur in the LysM domains (Fig. 2A and supplemental Fig. S4), and four of them, including the one affected in the nfp-2 allele, are conserved in NFP orthologous proteins of both pea (SYM10) and L. japonicus (NFR5) (Fig. 2B). However, the NFP N-glycosylation sites are not well conserved within the LYR and LYK proteins of the LysM-RLK family from M. truncatula, although in some cases, other sites are found in close vicinity to those of NFP (Fig. 2B and supplemental Fig. S4). The number of N-glycosylation sites in the LysM domains of these proteins is variable and can be as few as two (in LYR2, LYK3, and LYK4), whereas NFP, with eight, contains the most (supplemental Fig. S4).
FIGURE 2.
N-Glycosylation sites are not conserved between NFP and various LysM-RLKs, and trafficking of NFP to the PM is not blocked by inhibition of N-glycosylation. A, scheme of the extracellular region of NFP showing positions of the Asn (N) and Cys (C) residues (putatively being involved in post-translational modifications) in relation to the LysM domains. SP, signal peptide; TM, transmembrane domain; numbering is from the start of the signal peptide. B, conservation of the NFP Cys and N-glycosylation site residues in the orthologues from L. japonicus (NFR5) and P. sativum (SYM10) and the LYR and LYK proteins from M. truncatula. +, a site found at the same position; +/−, a site found at a close position (see alignment in supplemental Fig. S4). C, tunicamycin effectively inhibits N-glycosylation of NFP. N. benthamiana leaves expressing NFP-YFP at 2 and 3 dpi were treated with 10 μm tunicamycin or solvent only (diluted DMSO) for 20 h prior to analysis by immunoblotting. Tunicamycin treatment led to NFP-YFP detected at the size predicted from the unmodified AA sequence (93 kDa), whereas the protein in the untreated leaves contains about 20 kDa of N-glycans. D, NFP can reach the PM in the absence of N-glycosylation. The same material described in C (leaves at 3 dpi treated with 10 μm tunicamycin for 20 h) was observed by confocal microscopy; two images are shown. The arrow points to labeling of perinuclear ER, whereas most of the NFP protein localizes to the PM. WB, Western blot. Bars, 20 μm.
In order to test whether N-glycosylation is required for NFP function, we first treated N. benthamiana leaves expressing NFP-YFP with tunicamycin, a drug inhibiting an early step of N-glycosylation. Immunoblot analysis showed that NFP from the tunicamycin-treated sample migrated at the size predicted from its unmodified AA sequence (Fig. 2C), suggesting that the tunicamycin treatment had substantially inhibited NFP N-glycosylation. Microscopy analysis revealed that the protein was located predominantly at the cell periphery at 72 hpi, probably at the PM (Fig. 2D, a), although in some cells NFP fluorescence was observed in perinuclear membranes (arrow in Fig. 2D, b), as also seen in control leaves treated only with diluted DMSO.
We then individually replaced the Asn of each potential N-glycosylation site present in the LysM domains with Gln and took advantage of our nfp-1 complementation system using Pro35S:NFP-RFP tagged proteins to study the effect of the mutations. All mutated proteins were able to complement the absence of nodulation in the nfp-1 mutant (Table 1), showing that each N-glycosylation site, individually, is not essential for NFP function. We combined some of these mutations in order to remove all N-glycosylation sites from LysM1, LysM2, or LysM3. Again, expression of the mutated proteins restored the nodulation in the M. truncatula nfp-1 mutant (Table 1), showing that N-glycosylation of each individual LysM domain is not essential for the biological role of the protein and that the loss of function of nfp-2 is thus not due to the absence of an N-glycan.
TABLE 1.
Extracellular Cys residues and an intracellular kinase residue, but not N-glycosylation sites, are essential for NFP biological activity
Constructs coding for tagged NFP or the indicated mutant variants of NFP were transformed into M. truncatula nfp-1. Transgenic roots were inoculated with S. meliloti, and complementation for nodulation was analyzed 2 weeks later. When indicated, proteins from transgenic roots were analyzed for sensitivity to PNGase F as shown in Fig. 3 (an indicator of ER localization) and localization in N. benthamiana (as shown in Fig. 1 and supplemental Figs. S3 and S5). AA numbering starts from the first Met of the predicted signal peptide. JM, juxtamembrane; A-segment, activation segment; C-tail, C-terminal tail. <, before the indicated LysM domain; ><, between the indicated LysM domains.
| Construct/NFP mutation | Domain mutated | Nodulation | PNGase F sensitivity | Localization in N. benthamiana |
|---|---|---|---|---|
| Empty vector | − | |||
| NFP and NFP-S67F | ||||
| NFP | + | − | PM | |
| S67F | LysM1 | − | + | ER |
| S67T | LysM1 | + | −/+ | |
| S67A | LysM1 | + | − | |
| N-Glycosylation sites | ||||
| N65Q | LysM1 | + | ||
| N71Q | LysM1 | + | ||
| N65Q/N71Q | LysM1 | + | ||
| N112Q | LysM2 | + | ||
| N133Q | LysM2 | + | ||
| N146Q | LysM2 | + | ||
| N112Q/N133Q/N146Q | LysM2 | + | ||
| N189Q | LysM3 | + | ||
| N211Q | LysM3 | + | ||
| N219Q | LysM3 | + | ||
| N189Q/N211Q/N219Q | LysM3 | + | ||
| Cys residues | ||||
| C39A | <LysM1 | + | ||
| C47A | <LysM1 | − | + | ER |
| C39A/C47A | <LysM1 | − | + | ER |
| C102A | LysM1><LysM2 | + | + | |
| C104A | LysM1><LysM2 | + | ||
| C102A/C104A | LysM1><LysM2 | − | + | ER |
| C164A | LysM2><LysM3 | + | + | |
| C166A | LysM2><LysM3 | − | + | |
| C164A/C166A | LysM2><LysM3 | − | + | ER |
| C102A/C164A | + | + | ||
| C102A/C166A | − | + | ||
| C104A/C164A | − | + | ||
| C104A/C166A | − | + | ||
| C102A/C104A/C164A/C166A | − | + | ER | |
| Intracellular region | ||||
| T281A/S282A/S283A | JM | + | ||
| K339A | VAIK | + | ||
| D435A | HRD | + | ||
| T459A/S460A/T461A | A-segment | + | ||
| G474A | α-Helix F | + | −/+ | |
| G474E | α-Helix F | − | −/+ | |
| T482A | α-Helix F | + | ||
| T578A/S579A | C-tail | + | ||
NFP Trafficking to PM Is Regulated in ER
In order to investigate the reason for the lack of function of the nfp-2 allele, a construct coding for NFP-S67F fused to RFP was used to transiently transform N. benthamiana leaves. In contrast to NFP, which localized at the PM at 72 hpi, NFP-S67F localized to the periphery of the nucleus and in a reticulated network throughout the cell and showed co-localization with an ER marker rather than with the PM marker (Fig. 1C).
To study the trafficking of these proteins in M. truncatula, we made use of the information that the N-glycans of NFP are mostly insensitive to PNGase F (15), probably because they are fucosylated on the proximal GlcNAc residue in the Golgi apparatus (19). Therefore, the sensitivity of NFP to PNGase F treatment could be used to test whether the protein has passed through the Golgi and presumably inserted into the PM. We thus studied the effect of PNGase F on different variants of NFP, expressed in M. truncatula roots. NFP showed a small reduction in size following PNGase F treatment (Fig. 3A), probably due to one or two N-glycans not being modified to a complex, fucosylated form, as shown previously (15). In contrast, NFP-S67F was completely sensitive to PNGase F, leading to a protein of the size predicted for unmodified NFP (Fig. 3A). This result suggests that its N-glycans were not modified in the Golgi apparatus, consistent with an ER localization of this protein in M. truncatula.
FIGURE 3.
Trafficking of NFP-S67F to the PM is blocked in M. truncatula, and NFP-S67F protein interacts with the ER-located BIP chaperones in N. benthamiana. A, NFP-S67F lacks Golgi matured N-glycans in M. truncatula. Protein extracts from nfp-1 roots expressing NFP-RFP or the indicated mutated variants were treated with PNGase F and analyzed by immunoblotting. The lower size band after treatment indicates a sensitivity of NFP N-glycans to PNGase F and reflects an absence of maturation of NFP N-glycans in the Golgi apparatus and hence retention of the protein in the ER. The higher band size reflects transport through the Golgi to the PM. B, NFP-S67F interacts with the BIP chaperones in N. benthamiana. NFP-RFP or NFP-S67F-RFP expressed in N. benthamiana leaves were solubilized and purified using anti-RFP antibodies. Immunoblotting reveals that NFP-S67F, but not NFP, interacts strongly with BIP chaperones. NT, nontransformed leaves. C, BIP chaperones are not overexpressed in roots of nfp-2 mutant plants. An immunoblot of protein extracts from roots of WT, nfp-1, and nfp-2 plants reveals similar quantities of BIP chaperones, suggesting that the accumulation of NFP-S67F protein in the ER does not lead to an “unfolded protein response.” The ponceau red staining shows the protein loading. WB, Western blot.
In order to investigate in more detail the effect of the S67F mutation, we replaced Ser-67 with Ala (S67A) or made a conservative substitution by replacing Ser-67 with Thr (S67T), which does not destroy the putative N-glycosylation site. Both mutations allowed complementation of the nfp-1 for nodulation (Table 1), showing that the presence of a Phe at position 67 was responsible for the loss of function of the protein coded by nfp-2. Sensitivity to PNGase F was different for NFP-S67A (similar to NFP, mostly insensitive) compared with NFP-S67T (partially sensitive) (Fig. 3A). In both cases, a fraction of these proteins contained PNGase F-insensitive N-glycans and thus had passed through the Golgi, consistent with this fraction being located at the PM and restoring nodulation. This result shows that only a small amount of correctly localized NFP is enough to perform its function.
In order to understand how NFP-S67F is retained in the ER, NFP and NFP-S67F expressed in N. benthamiana leaves were solubilized from membranes, purified using the RFP tag, and submitted to SDS-PAGE. Antibodies raised against the ER lumenal BIP chaperones, which bind and retain unfolded proteins in the ER, were used to test whether BIPs interact with NFP or NFP-S67F. Indeed, BIPs were co-purified with NFP-S67F to a much larger extent than with NFP (Fig. 3B), suggesting that NFP-S67F is not correctly folded. Misfolded proteins occasionally induce an “unfolded protein response,” which leads to overexpression of BIP chaperones (20). We compared the level of BIP in roots from nfp-1 and nfp-2 mutants and wild-type plants grown in aeroponic conditions. No differences in the levels of BIP were detected between the plants (Fig. 3C) despite the accumulation of NFP-S67F·BIP complexes in the ER in nfp-2 plants.
Cysteine Pairs Are Conserved in All LysM-RLKs and Are Essential for NFP Activity
An intriguing feature of all plant LysM-RLKs characterized to date is the presence of CXC motifs between the three LysM domains. In NFP, these occur at Cys-102/Cys-104 between LysM1 and LysM2 and at Cys-162/Cys-164 between LysM2 and LysM3 (Fig. 2, A and B). Further inspection of the extracellular regions revealed two other Cys residues (Cys-39 and Cys-47), separated by seven other residues, present in the N-terminal region of NFP before LysM1, and these residues are clearly conserved in NFR5, SYM10, and LYR1 (supplemental Fig. S4 and Fig. 2B). Two Cys residues in similar positions are also found before LysM1 in all M. truncatula LysM-RLKs, although their exact positions and separation (between three and six residues) are less well conserved. Some of these proteins also contain Cys residues in the predicted signal peptide and thus would not be present in the mature protein. It is noteworthy that five of the proteins (LYR2–LYR6) contain an additional Cys residue in the N-terminal region, after the predicted signal peptide, and also a Cys residue in the LysM2. In addition, LYR5 and LYR6 contain a third CXC motif after LysM3, and these two proteins and LYR2 contain two additional Cys residues in this extracellular juxtamembrane region (supplemental Fig. S4). In all cases, the LysM-RLKs contain an even number of Cys residues in the extracellular regions of the predicted mature proteins, which suggests the presence of S-S bridges.
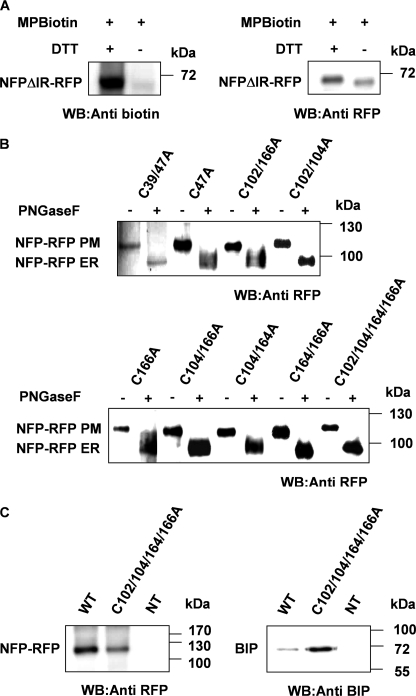
In order to investigate the presence of such putative S-S bridges, we expressed, in N. benthamiana leaves, an RFP fusion of NFP truncated from its intracellular region in order to eliminate unwanted Cys residues in the IR. This truncated form (NFPΔIR-RFP) contains the six Cys residues from the NFP extracellular region, potentially involved in S-S bridges, and one Cys residue in the RFP tag, which faces the cytosol and cannot be involved in an S-S bridge. NFPΔIR-RFP localized at the PM similarly to the full-length NFP (supplemental Fig. S5A). Membrane fractions were prepared from leaves, and free Cys residues were labeled with maleimide-PEG2-biotin before and after treatment with DTT, which reduces the S-S bridges. The truncated NFP proteins were then purified using the RFP tag. An immunoblot using anti-biotin antibodies (Fig. 4A) revealed a substantial increase in Cys residue labeling of NFP after the reduction of the S-S bridges; the increase corresponds to the expected difference (1/7) for all of the six Cys residues of the NFP extracellular region being involved in S-S bridges.
FIGURE 4.
NFP contains three disulfide bridges and mutants in the CXC motifs are retained in the ER. A, NFP bears three S-S bridges. A truncated version of NFP, without its intracellular region, fused to RFP (NFPΔIR-RFP) was expressed in N. benthamiana leaves. The microsomal fraction derived from these leaves was first treated or not with DTT, and then free Cys residues were labeled with maleimide-PEG2-biotin. NFP was then solubilized and purified using anti-RFP antibodies. Proteins were separated by SDS-PAGE and analyzed by immunoblotting; fusion proteins were detected using anti-RFP antibodies, and labeled Cys residues were detected using anti-biotin antibodies. The result suggests that the six Cys residues of NFP extracellular region are involved in S-S bridges, and only the single Cys residue of the RFP is labeled in the absence of DTT. B, NFP mutated in the Cys pairs localizes to the ER in M. truncatula. Protein extracts from nfp-1 roots expressing the indicated NFP-RFP mutated variants were treated as described in Fig. 3A. The sensitivity of the proteins to PNGase F suggests lack of N-glycan modification and hence retention in the ER. C, NFP mutated in the CXC motifs interacts with BIP chaperones. NFP and the quadruple Cys to Ala mutant proteins were expressed and purified from N. benthamiana leaves, and complexes with BIP were analyzed by SDS-PAGE and immunoblotting. The mutant NFP protein co-purifies BIP chaperones, suggesting that it is retained in the ER by binding to BIP chaperones. NT, nontransformed leaves; WB, Western blot.
In order to determine the requirement of these S-S bridges in NFP activity, we replaced each Cys residue, individually or in combination, with Ala and analyzed the proteins for their ability to complement the nfp-1 for nodulation (Table 1). Among the six Cys residues in the extracellular region, Cys-47 and Cys-166 were found to be essential for NFP nodulation activity (Table 1). Analysis of combinations of Cys residue mutations revealed that either Cys-102 or Cys-104 must be present for nodulation activity and that Cys-164 is required if Cys-104 is mutated (Table 1). The mutated proteins that were unable to complement nfp-1 for nodulation were then tested for their sensitivity to PNGase F, and all of them were found to be sensitive (Fig. 4B), suggesting that these proteins contain non-modified N-glycans and that these proteins did not reach the Golgi apparatus. Even the mutated proteins complementing the absence of nodulation in nfp-1 were detected as sensitive to PNGase F (supplemental Fig. S5C), suggesting that these proteins were severely affected in trafficking and that only a small amount of functional protein reached the PM to allow complementation. The ER localization of proteins mutated in the Cys residue is supported by the finding that when the proteins were expressed in N. benthamiana (e.g. C102A/C104A/C164A/C166A), they localized to regions, similar to the ER marker (supplemental Fig. 5B and Table 1). Further analysis of the protein encoded by NFP mutated in the two CXC motifs showed that it co-purified with BIP chaperones from N. benthamiana leaves (Fig. 4C).
Finally, we deduced from the mutation analysis the positions of the NFP S-S bridges (see “Discussion” for details) and used this information to build a model of the entire NFP extracellular region (Fig. 5A). Stability of the model supports S-S bridges linking Cys-39 to Cys-104, Cys-47 to Cys-166, and Cys-102 to Cys-164. Such S-S bridges pack together the three LysM domains and the N-terminal region (Fig. 5A). Together, these data suggest that S-S bridges in the NFP extracellular region are essential for correct folding of NFP and trafficking to the PM and that in their absence, the protein is retained in the ER by binding to BIP chaperones.
FIGURE 5.
Molecular modeling of the NFP extracellular region. A, molecular modeling showing the suggested positions of the S-S bridges. LysM domains are colored blue (LysM1), red (LysM2), and green (LysM3) ribbons, whereas the preceding loops are represented in pale blue, pale red, and pale green, respectively. Cys residues involved in S-S bridges are shown as yellow sticks. The S-S bridge positions deduced from mutation and complementation analysis (Table 1) were used for modeling. B, superposed molecular modeling of NFP and NFP S67F. An NFP model is shown before (violet ribbons) and after mutation (pink ribbons); local structural changes are highlighted at the LysM1 level. The arrow points to the position of residue 67. C and D, solvent-accessible surfaces colored according to the lipophilic potential, from brown (hydrophobic) to blue (polar), calculated for the NFP model before (C) and after the S67F mutation (D). The mutation zone is circled.
NFP Intracellular Region Is Necessary for Nodulation
We recently showed that homology with the kinase domain of human interleukin-1 receptor-associated kinase 4 (of which the structure is known) can be used to compare the structures of the predicted kinase domains of plant RLKs, due to the common evolutionary origins of these plant and animal kinases (10). Alignment of the NFP IR with the kinase domains of HsIRAK-4, LYK3, and an Arabidopsis thaliana flagellin receptor (FLS2) confirmed that the NFP IR lacks some essential features that are conserved in active kinases (3), including part of the Gly-rich loop and most of the activation segment (supplemental Fig. S6). However, other residues conserved in kinases, such as the VAIK (phosphotransfer) motif and the HRD motif in the catalytic loop, which are involved in ATP hydrolysis, are still present in NFP. Thus, we investigated whether the intracellular region of NFP and the conserved kinase residues are important for its biological function.
We first tested whether the NFP IR is essential for nodulation and found that a construct lacking the IR (NFPΔIR) did not complement nfp-1 for nodulation. We then tested Ala substitutions in conserved residues in the VAIK motif (Lys-339) and in the HRD motif (Asp-435) and found that both mutant proteins complemented nfp-1 for nodulation (Table 1). We then tested Ala substitutions in residues that are predicted to be phosphorylated (NetPhos program) in the juxtamembrane region (Thr-281, Ser-282, and Ser-283), in α-helix F (Thr-482), in the incomplete activation segment (Thr-459, Ser-460, and Thr-461), and near the C terminus of the protein (Thr-578 and Ser-579). All of the constructs coding NFP mutated in these residues complemented nfp-1 for nodulation (Table 1). Finally, we tested the role of Gly-474, which is a highly conserved kinase residue in predicted α-helix F and which has been shown to be essential for the activity of A. thaliana FLS2 (37). An NFP-G474E mutant protein did not allow complementation of nfp-1 for nodulation, whereas a protein with a more conservative substitution (G474A) complemented. A comparison of the sensitivity to PNGase F of the NFP-G474E and NFP-G474A proteins from M. truncatula roots suggests that at least a fraction of both proteins has passed the Golgi apparatus and should be located at the PM (supplemental Fig. 5D). This result suggests that the G474E mutant is affected not only in trafficking but also in biological function.
DISCUSSION
Genetic studies have shown that NFP plays an essential role in lipochitooligosaccharidic NF perception and signaling. In this paper, using a structure function approach, we have identified key elements of the protein that are required for folding, localization to the PM, and biological activity of the protein.
We used fluorescent protein fusions to the C terminus of the protein to follow NFP localization. Studies in three plant heterologous expression systems suggest that NFP follows a typical cycle for a cell surface receptor: synthesis at the ER, transit through the Golgi apparatus, localization at the PM, and later internalization and degradation in the vacuole (Fig. 1 and supplemental Fig. S3). Although the fluorescence of the fusion proteins was well detected by microscopy when expressed in heterologous systems under the control of a strong promoter (Pro35S), it was not detected in M. truncatula roots when expressed either under the control of its own promoter (ProNFP) or under the control of the Pro35S. In contrast, RLKs of the lectin family, expressed under the control of the Pro35S, were detected in M. truncatula roots using similar microscopy (38). Plasma membrane localization was confirmed in M. truncatula roots by immunocytolabeling, but only using a highly sensitive amplification method. Together, these data suggest that the level of NFP is tightly controlled and kept at a relatively low level in M. truncatula by post-transcriptional or post-translational regulation mechanism(s). Nevertheless, NFP fusion proteins expressed from the Pro35S localized to the PM and complemented the nfp-1 mutant for nodulation, suggesting that NFP fusion proteins are biologically active and thus can be used to monitor the activity of NFP. Moreover, the fusion proteins were detected by immunoblotting, and we used the resistance to PNGase F of Golgi-modified NFP N-glycans to follow NFP trafficking in M. truncatula. We thus used a combination of microscopy and protein biochemistry to examine whether particular NFP structures play a role in its biological function.
Concerning the IR of NFP, our data show that it is essential for biological activity because its deletion leads to a protein that is not functional in nodulation but is still capable of locating to the PM and thus is probably defective in signal transduction. Previous studies had shown that the IR of NFP lacks kinase activity (3), and this is reinforced by our observation that mutants in two highly conserved “catalytic” residues (Lys-339 in the VAIK motif and Asp-435 in the HRD motif) are still able to complement for nodulation (Table 1). In the animal epidermal growth factor receptor kinase family, the kinase-dead ErbB3, which binds the ligand, activates signal transduction through oligomerization and phosphorylation by the kinase-active ErbB2, which does not bind the ligand (39). In our studies of M. truncatula NFP, we have not been able to show transphosphorylation by its potential partner LYK3 (10), although the LYK3 kinase domain is capable of transphosphorylating another symbiotic protein, PUB1 (29). In L. japonicus, the kinase domain of the LYK3 ortholog, NFR1, has been shown in vitro to transphosphorylate the intracellular region of the NFP ortholog, NFR5 on Ser-282 in the juxtamembrane region (8). However, this Ser residue is not essential for nodulation activity of NFR5 in L. japonicus or, as shown here, of NFP in M. truncatula. Unlike in L. japonicus, where NFR5 and NFR1 are both implicated in the initial steps of nodulation, in M. truncatula, LYK3 is only required for later stages of nodulation and infection. Thus, for these early steps, NFP may interact with a yet unidentified RLK. Alternatively, its kinase-dead domain could be activated by ligand-dependent conformational changes, leading to activation of a different signal transduction protein.
Our work has also shown an important role of Gly-474 in NFP activity. This residue is important for the activity of another plant RLK (37) and is located in the predicted α-helix F just after the activation segment, which is the region that is most highly conserved between NFP, other plant RLKs, and a related human kinase, IRAK-4 (supplemental Fig. S6). This helix is postulated to relay conformation changes leading to substrate-binding. Flexibility due to the Gly residue could be essential for these conformational changes (40).
Concerning the extracellular region of NFP, through studies on the protein encoded by the strong nfp-2 allele, NFP-S67F, we have shown that NFP is exquisitely sensitive to ER-QC. In N. benthamiana, the NFP-S67F fusion protein accumulated in the ER and associated with the BIP chaperones (Fig. 3), which recognize misfolded proteins (20). In M. truncatula, the PNGase F sensitivity of the N-glycans of NFP-S67F (Fig. 3) suggests that the mutant protein also accumulates in the ER in the homologous system.
Modeling the effect of the S67F mutation suggests that the Phe residue should not perturb the global three-dimensional structure of LysM1 (Fig. 5B) but causes changes in the local hydrophobic properties of LysM1 (Fig. 5, C and D). However, it is more likely that steric hindrance rather than increased hydrophobicity is responsible for the ER localization of proteins mutated at position 67 because an Ala residue is more hydrophobic and less bulky than a Thr residue, and S67A does not affect NFP, whereas the S67T mutation led to most of the protein accumulating in the ER (Fig. 3A). In the case of a mutation by a Phe residue, a change in the surface hydrophobicity (Fig. 5, C and D) could participate, in addition to the changes in the LysM1 structure, in ER-QC detection. Thus it is not surprising to find that Ser-67, which is located in an α-helix of the LysM1 (15), is conserved in most members of the M. truncatula LysM-RLK family or is substituted with Ala (supplemental Fig. S4). These results suggest that the phenotype of the strong nfp-2 allele is due to misfolding of the protein and that NFP trafficking to the PM requires correct folding of its extracellular region.
Although Ser-67 is part of an N-glycosylation site, the lack of N-glycosylation is not the reason for the ER-QC of NFP-S67F because NFP-S67A locates to the PM (Fig. 3). Previous work (15) and this study (Fig. 2) have shown that all of the 10 predicted N-glycosylation sites in NFP are probably occupied and that eight of them occur on the surface of the LysM domains. NFP indeed contains more N-glycosylation sites than other M. truncatula LysM-RLKs (Fig. 2). In N. benthamiana, following inhibition of N-glycosylation by tunicamycin treatment, NFP was still able to reach the PM, suggesting that the N-glycans are not essential for NFP folding and trafficking to the PM, although it does not exclude the possibility that they facilitate it. Moreover, by mutating each of these N-glycosylation sites or combinations of them, we have shown that the proteins are still capable of restoring nodulation of the nfp-1 mutant (Table 1), suggesting that N-glycosylation of the LysM domains is not essential for NFP activity and NF perception. Mutations of the N-glycosylation site in the LysM1, N65Q, and of combinations of N-glycosylation sites in the LysM domains led to partial sensitivity to PNGase F (data not shown). In the case of N65Q, the N-glycan itself is not responsible for this partial sensitivity because the mutation S67A, destroying the same N-glycosylation site, is as insensitive to PNGase F as NFP. Thus, the nature of the residues modifying a predicted N-glycosylation site is important because even conservative substitutions in NFP (N65Q and S67T) affect trafficking to the PM.
Nevertheless, although N-glycans might facilitate NFP folding and trafficking to the PM, they are not essential for its function. In contrast, several other RLKs show an N-glycan requirement for their activity. This property is protein-specific because differences in sensitivity to the ER-QC and in N-glycan requirement for activity have been reported for the closely related LRR-RLKs of A. thaliana FLS2 and EFR (36, 41–44).
In contrast to N-glycosylation, S-S bridge formation appears to be essential for NFP activity. By studying a plant expressed NFP fusion, we have shown that all of the six Cys residues in the NFP extracellular region are likely to be involved in S-S bridges (Fig. 4). NFP variants mutated in these Cys residues have PNGase F-sensitive N-glycans (Fig. 4), suggesting that the proteins are retained in the ER. This localization has been demonstrated for the C102A/C104A/C164A/C166A protein in N. benthamiana, and moreover this protein interacts with BIP chaperones (Fig. 4). Protein mutated in the Cys residues might thus be incorrectly folded and/or might interact with ER-QC proteins through either N-glycans, hydrophobic exposed regions or mixed S-S bridges. These results, suggest that the S-S bridges are necessary for NFP folding and trafficking to the PM. Cys residues are present in the extracellular region of many plant RLKs and receptor-like proteins and have been shown to be important for the function of some of these proteins, including Cf9 (36, 45) and BRI1 (46) but not CLV2 (47). For BRI1, the bri1–5 allele contains a point mutation in Cys-69, which leads to retention of the protein in the ER and interaction with the ER chaperones, calnexin and BIP (46).
LysM-RLKs have a very characteristic arrangement of Cys residues with the presence of a CXC motif in each of the two spacer regions between the three LysM domains (Fig. 2). This arrangement occurs also in related LYM proteins (11). In addition, some LysM-RLKs, including M. truncatula LYR5 and LYR6, contain a third CXC motif before the transmembrane-spanning segment (supplemental Fig. S4). CXC motifs are found in several protein families, including some plant proteins (e.g. prolamins and lipid transfer proteins containing an 8-Cys motif (48)), some scorpion toxins and related natural peptides (49), and the animal CXC-type chemokines (50). In some of these proteins, the CXC motif has been shown to be involved in S-S bridges. In most of these cases, each Cys residue of the CXC motif forms an S-S bridge with another Cys residue in the protein, and in only a few very rare examples does the CXC motif form an intramotif S-S bridge leading to a thermodynamically disfavored 11-member ring structure (49, 51).
Examination of the extracellular region of NFP and other LysM-RLKs revealed two other Cys residues before LysM1, separated by 3–7 residues (supplemental Fig. S4). Mutation analysis revealed that the only individual Cys residues with an essential role in NFP activity are Cys-47 in the N-terminal region and Cys-166, located between LysM2 and LysM3. Thus, an S-S bridge of primordial importance might occur between these two residues. Two other S-S bridges that are individually less essential might occur between the two CXC motifs and between the first CXC motif and Cys-39 in the N-terminal region. Double mutations of NFP-C102A/C104A and NFP-C104A/C164A that would disrupt these two S-S bridges lead to non-functional NFP proteins and suggest that the S-S bridges link Cys-39 to Cys-104 and Cys-102 to Cys-164. An arrangement involving each of the Cys residues before LysM1 interacting with one of the CXC motifs, although speculative, is supported by the observation that some plant LysM-RLKs (e.g. LYR2-LYR6 of M. truncatula) contain a Cys residue in the middle of LysM2 and an additional one in the N-terminal region. In these proteins, a fourth S-S bridge could form between these residues, forming a link between the N-terminal region and LysM2. Taking into account this prediction, we modeled the entire extracellular region of NFP (Fig. 5A), and the stability of the model supports the S-S bridge arrangement we propose. However, our model is an interpretation of the data, and we cannot exclude other arrangements.
Protein-disulfide isomerase, thioredoxin, and some prokaryotic periplasmic enzymes involved in protein oxidation (e.g. DsbA) contain CXXC motifs that have disulfide isomerase activity. Deletion of one AA to form a CXC motif in E. coli thioredoxin greatly increases the disulfide isomerase activity, and even a synthetic CXC tripeptide possesses such activity (52). In addition, the presence of a CXC motif at the C terminus of two enzymes from hyperthermophilic Archaea was shown to be necessary for folding of these enzymes (53), and in their absence, the addition of synthetic peptides, containing the CXC region, was able to restore the folding. These peptides were also shown to have disulfide isomerase activities (53). Thus, it is possible that the CXC motifs in NFP and other LysM-RLKs have disulfide isomerase activities and that these proteins have the ability to catalyze the formation of their own S-S bridges. Because mutations in the Cys residues inhibit NFP trafficking, it is difficult to assess whether the S-S bridges in the extracellular region are also important for ligand binding. In the LysM domains of the chitinase of the fern Pteris ryukyuensis, four Cys residues are involved in intradomain S-S bridges and stabilization of the LysM domain structure (54). For NFP and other plant LysM-RLKs, it is likely that the S-S bridges are important for stabilizing the three-dimensional geometry of the three LysM domains relative to one another (Fig. 5A). Indeed, biochemical analysis showed that the three LysM domains of CERK1 are required for ligand binding (55). Homology modeling and docking suggest that each LysM domain of NFP may bind an NF ligand (15), and genetic analysis suggests differences in importance of the three domains, with LysM2 of NFP (13) and NFR5 (14) playing the most important role in rhizobial selectivity. The configuration defined by the proposed S-S bridges (Fig. 5A), in which the N and C termini of LysM1 and LysM2 and the N terminus of LysM3 are physically very close, may be important for the perception of complex or multiple ligands and/or for cooperativity of ligand binding. The demonstration in NFP of the importance of the Cys residues and their involvement in S-S bridges will aid strategies to produce functional plant LysM-RLK and LYM proteins for ligand binding studies.
Supplementary Material
Acknowledgments
We thank Sophie Valière for help with NFP purification; Sandra Bensmihen, Jean-Jacques Bono, and Clare Gough (Laboratoire des Interactions Plantes-Microorganismes) for critically reading the manuscript; and Prof. Pierre-Alain Carrupt (School of Pharmaceutical Sciences, University of Geneva, University of Lausanne) for technical support with molecular modeling tools.
This work was supported in part by the European Community Marie Curie Research Training Network Program Contract MRTN-CT-2006-035546 “NODPERCEPTION” and French National Research Agency Contracts “NodBindsLysM,” “LCOinNONLEGUMES,” and “SYMPASIGNAL”. This work was performed in the Laboratoire des Interactions Plantes-Microorganismes, part of the “Laboratoire d'Excellence (LABEX) entitled TULIP (ANR-10-LABX-41).

This article contains supplemental Figs. S1–S6.
- NF
- Nod factor
- RLK
- receptor-like-kinase
- AA
- amino acid(s)
- hpi
- hours postinoculation
- dpi
- days postinoculation
- RFP
- red fluorescent protein
- IR
- intracellular region
- S-S
- disulfide
- LysM
- lysin motif
- PM
- plasma membrane
- ER
- endoplasmic reticulum
- PNGase F
- peptide:N-glycosidase F
- qRT-PCR
- quantitative RT-PCR
- LRR
- leucine-rich repeat.
REFERENCES
- 1. Gough C., Cullimore J. (2011) Lipo-chitooligosaccharide signaling in endosymbiotic plant-microbe interactions. Mol. Plant Microbe Interact. 24, 867–878 [DOI] [PubMed] [Google Scholar]
- 2. Oldroyd G. E., Downie J. A. (2008) Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu. Rev. Plant Biol. 59, 519–546 [DOI] [PubMed] [Google Scholar]
- 3. Arrighi J. F., Barre A., Ben Amor B., Bersoult A., Soriano L. C., Mirabella R., de Carvalho-Niebel F., Journet E. P., Ghérardi M., Huguet T., Geurts R., Dénarié J., Rougé P., Gough C. (2006) The Medicago truncatula lysin [corrected] motif receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol. 142, 265–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Madsen E. B., Madsen L. H., Radutoiu S., Olbryt M., Rakwalska M., Szczyglowski K., Sato S., Kaneko T., Tabata S., Sandal N., Stougaard J. (2003) A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425, 637–640 [DOI] [PubMed] [Google Scholar]
- 5. Radutoiu S., Madsen L. H., Madsen E. B., Felle H. H., Umehara Y., Grønlund M., Sato S., Nakamura Y., Tabata S., Sandal N., Stougaard J. (2003) Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425, 585–592 [DOI] [PubMed] [Google Scholar]
- 6. Smit P., Limpens E., Geurts R., Fedorova E., Dolgikh E., Gough C., Bisseling T. (2007) Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol. 145, 183–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shiu S. H., Bleecker A. B. (2001) Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. U.S.A. 98, 10763–10768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Madsen E. B., Antolín-Llovera M., Grossmann C., Ye J., Vieweg S., Broghammer A., Krusell L., Radutoiu S., Jensen O. N., Stougaard J., Parniske M. (2011) Autophosphorylation is essential for the in vivo function of the Lotus japonicus Nod factor receptor 1 and receptor-mediated signaling in cooperation with Nod factor receptor 5. Plant J. 65, 404–417 [DOI] [PubMed] [Google Scholar]
- 9. Castells E., Casacuberta J. M. (2007) Signaling through kinase-defective domains. The prevalence of atypical receptor-like kinases in plants. J. Exp. Bot. 58, 3503–3511 [DOI] [PubMed] [Google Scholar]
- 10. Klaus-Heisen D., Nurisso A., Pietraszewska-Bogiel A., Mbengue M., Camut S., Timmers T., Pichereaux C., Rossignol M., Gadella T. W., Imberty A., Lefebvre B., Cullimore J. V. (2011) Structure-function similarities between a plant receptor-like kinase and the human interleukin-1 receptor-associated kinase-4. J. Biol. Chem. 286, 11202–11210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fliegmann J., Uhlenbroich S., Shinya T., Martinez Y., Lefebvre B., Shibuya N., Bono J. J. (2011) Biochemical and phylogenetic analysis of CEBiP-like LysM domain-containing extracellular proteins in higher plants. Plant Physiol. Biochem. 49, 709–720 [DOI] [PubMed] [Google Scholar]
- 12. Buist G., Steen A., Kok J., Kuipers O. P. (2008) LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol. Microbiol. 68, 838–847 [DOI] [PubMed] [Google Scholar]
- 13. Bensmihen S., de Billy F., Gough C. (2011) Contribution of NFP LysM domains to the recognition of Nod factors during the Medicago truncatula/Sinorhizobium meliloti symbiosis. PLoS One 6, e26114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Radutoiu S., Madsen L. H., Madsen E. B., Jurkiewicz A., Fukai E., Quistgaard E. M., Albrektsen A. S., James E. K., Thirup S., Stougaard J. (2007) LysM domains mediate lipochitin-oligosaccharide recognition and Nfr genes extend the symbiotic host range. EMBO J. 26, 3923–3935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mulder L., Lefebvre B., Cullimore J., Imberty A. (2006) LysM domains of Medicago truncatula NFP protein involved in Nod factor perception. Glycosylation state, molecular modeling, and docking of chitooligosaccharides and Nod factors. Glycobiology 16, 801–809 [DOI] [PubMed] [Google Scholar]
- 16. Lerouge P., Cabanes-Macheteau M., Rayon C., Fischette-Lainé A. C., Gomord V., Faye L. (1998) N-Glycoprotein biosynthesis in plants. Recent developments and future trends. Plant Mol. Biol. 38, 31–48 [PubMed] [Google Scholar]
- 17. Saijo Y. (2010) ER quality control of immune receptors and regulators in plants. Cell Microbiol. 12, 716–724 [DOI] [PubMed] [Google Scholar]
- 18. Pattison R. J., Amtmann A. (2009) N-glycan production in the endoplasmic reticulum of plants. Trends Plant Sci. 14, 92–99 [DOI] [PubMed] [Google Scholar]
- 19. Gomord V., Fitchette A. C., Menu-Bouaouiche L., Saint-Jore-Dupas C., Plasson C., Michaud D., Faye L. (2010) Plant-specific glycosylation patterns in the context of therapeutic protein production. Plant Biotechnol. J. 8, 564–587 [DOI] [PubMed] [Google Scholar]
- 20. Vitale A., Boston R. S. (2008) Endoplasmic reticulum quality control and the unfolded protein response. Insights from plants. Traffic 9, 1581–1588 [DOI] [PubMed] [Google Scholar]
- 21. Kremers G. J., Goedhart J., van Munster E. B., Gadella T. W., Jr. (2006) Cyan and yellow super fluorescent proteins with improved brightness, protein folding, and FRET Förster radius. Biochemistry 45, 6570–6580 [DOI] [PubMed] [Google Scholar]
- 22. Shaner N. C., Campbell R. E., Steinbach P. A., Giepmans B. N., Palmer A. E., Tsien R. Y. (2004) Improved monomeric red, orange, and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22, 1567–1572 [DOI] [PubMed] [Google Scholar]
- 23. Lefebvre B., Batoko H., Duby G., Boutry M. (2004) Targeting of a Nicotiana plumbaginifolia H+-ATPase to the plasma membrane is not by default and requires cytosolic structural determinants. Plant Cell 16, 1772–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Haseloff J., Siemering K. R., Prasher D. C., Hodge S. (1997) Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. U.S.A. 94, 2122–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ivanchenko M., Vejlupkova Z., Quatrano R. S., Fowler J. E. (2000) Maize ROP7 GTPase contains a unique, CaaX box-independent plasma membrane targeting signal. Plant J. 24, 79–90 [DOI] [PubMed] [Google Scholar]
- 26. Klarenbeek J. B., Goedhart J., Hink M. A., Gadella T. W., Jalink K. (2011) A mTurquoise-based cAMP sensor for both FLIM and ratiometric read-out has improved dynamic range. PLoS One 6, e19170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Boisson-Dernier A., Chabaud M., Garcia F., Bécard G., Rosenberg C., Barker D. G. (2001) Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol. Plant Microbe Interact. 14, 695–700 [DOI] [PubMed] [Google Scholar]
- 28. van Bokhoven H., Verver J., Wellink J., van Kammen A. (1993) Protoplasts transiently expressing the 200K coding sequence of cowpea mosaic virus B-RNA support replication of M-RNA. J. Gen. Virol. 74, 2233–2241 [DOI] [PubMed] [Google Scholar]
- 29. Mbengue M., Camut S., de Carvalho-Niebel F., Deslandes L., Froidure S., Klaus-Heisen D., Moreau S., Rivas S., Timmers T., Hervé C., Cullimore J., Lefebvre B. (2010) The Medicago truncatula E3 ubiquitin ligase PUB1 interacts with the LYK3 symbiotic receptor and negatively regulates infection and nodulation. Plant Cell 22, 3474–3488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vitha S., Baluska F., Mews M., Volkmann D. (1997) Immunofluorescence detection of F-actin on low melting point wax sections from plant tissues. J. Histochem. Cytochem. 45, 89–95 [DOI] [PubMed] [Google Scholar]
- 31. Timmers A. C., Auriac M. C., Truchet G. (1999) Refined analysis of early symbiotic steps of the Rhizobium-Medicago interaction in relationship with microtubular cytoskeleton rearrangements. Development 126, 3617–3628 [DOI] [PubMed] [Google Scholar]
- 32. Moreau S., Verdenaud M., Ott T., Letort S., de Billy F., Niebel A., Gouzy J., de Carvalho-Niebel F., Gamas P. (2011) Transcription reprogramming during root nodule development in Medicago truncatula. PLoS One 6, e16463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pearlman D. A., Case D. A., Caldwell J. W., Ross W. S., Cheatham T. E. (1995) Comp. Phys. Commun. 91, 1–41 [Google Scholar]
- 34. Powell M. J. D. (1964) An efficient method for finding the minimum of a function of saved variables without calculating derivatives. Comput. J. 7, 55–162 [Google Scholar]
- 35. Waldherr-Teschner M., Goetze T., Heiden W., Knoblauch M., Vollhardt H., Brickmann J. (1992) Advances in Scientific Visualization, pp. 58–67, Springer, Heidelberg [Google Scholar]
- 36. Häweker H., Rips S., Koiwa H., Salomon S., Saijo Y., Chinchilla D., Robatzek S., von Schaewen A. (2010) Pattern recognition receptors require N-glycosylation to mediate plant immunity. J. Biol. Chem. 285, 4629–4636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gómez-Gómez L., Bauer Z., Boller T. (2001) Both the extracellular leucine-rich repeat domain and the kinase activity of FSL2 are required for flagellin binding and signaling in Arabidopsis. Plant Cell 13, 1155–1163 [PMC free article] [PubMed] [Google Scholar]
- 38. Navarro-Gochicoa M. T., Camut S., Timmers A. C., Niebel A., Herve C., Boutet E., Bono J. J., Imberty A., Cullimore J. V. (2003) Characterization of four lectin-like receptor kinases expressed in roots of Medicago truncatula. Structure, location, regulation of expression, and potential role in the symbiosis with Sinorhizobium meliloti. Plant Physiol. 133, 1893–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Boudeau J., Miranda-Saavedra D., Barton G. J., Alessi D. R. (2006) Emerging roles of pseudokinases. Trends Cell Biol. 16, 443–452 [DOI] [PubMed] [Google Scholar]
- 40. Kannan N., Neuwald A. F. (2005) Did protein kinase regulatory mechanisms evolve through elaboration of a simple structural component? J. Mol. Biol. 351, 956–972 [DOI] [PubMed] [Google Scholar]
- 41. Li J., Zhao-Hui C., Batoux M., Nekrasov V., Roux M., Chinchilla D., Zipfel C., Jones J. D. (2009) Specific ER quality control components required for biogenesis of the plant innate immune receptor EFR. Proc. Natl. Acad. Sci. U.S.A. 106, 15973–15978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lu X., Tintor N., Mentzel T., Kombrink E., Boller T., Robatzek S., Schulze-Lefert P., Saijo Y. (2009) Uncoupling of sustained MAMP receptor signaling from early outputs in an Arabidopsis endoplasmic reticulum glucosidase II allele. Proc. Natl. Acad. Sci. U.S.A. 106, 22522–22527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nekrasov V., Li J., Batoux M., Roux M., Chu Z. H., Lacombe S., Rougon A., Bittel P., Kiss-Papp M., Chinchilla D., van Esse H. P., Jorda L., Schwessinger B., Nicaise V., Thomma B. P., Molina A., Jones J. D., Zipfel C. (2009) Control of the pattern-recognition receptor EFR by an ER protein complex in plant immunity. EMBO J. 28, 3428–3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Saijo Y., Tintor N., Lu X., Rauf P., Pajerowska-Mukhtar K., Häweker H., Dong X., Robatzek S., Schulze-Lefert P. (2009) Receptor quality control in the endoplasmic reticulum for plant innate immunity. EMBO J. 28, 3439–3449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. van der Hoorn R. A., Wulff B. B., Rivas S., Durrant M. C., van der Ploeg A., de Wit P. J., Jones J. D. (2005) Structure-function analysis of cf-9, a receptor-like protein with extracytoplasmic leucine-rich repeats. Plant Cell 17, 1000–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hong Z., Jin H., Tzfira T., Li J. (2008) Multiple mechanism-mediated retention of a defective brassinosteroid receptor in the endoplasmic reticulum of Arabidopsis. Plant Cell 20, 3418–3429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Song X., Guo P., Li C., Liu C. M. (2010) The cysteine pairs in CLV2 are not necessary for sensing the CLV3 peptide in shoot and root meristems. J. Integr. Plant Biol. 52, 774–781 [DOI] [PubMed] [Google Scholar]
- 48. José-Estanyol M., Gomis-Rüth F. X., Puigdomènech P. (2004) The eight-cysteine motif, a versatile structure in plant proteins. Plant Physiol. Biochem. 42, 355–365 [DOI] [PubMed] [Google Scholar]
- 49. Zhu Q., Liang S., Martin L., Gasparini S., Ménez A., Vita C. (2002) Role of disulfide bonds in folding and activity of leiurotoxin I. Just two disulfides suffice. Biochemistry 41, 11488–11494 [DOI] [PubMed] [Google Scholar]
- 50. Joseph P. R., Sarmiento J. M., Mishra A. K., Das S. T., Garofalo R. P., Navarro J., Rajarathnam K. (2010) Probing the role of CXC motif in chemokine CXCL8 for high affinity binding and activation of CXCR1 and CXCR2 receptors. J. Biol. Chem. 285, 29262–29269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Derewenda U., Boczek T., Gorres K. L., Yu M., Hung L. W., Cooper D., Joachimiak A., Raines R. T., Derewenda Z. S. (2009) Structure and function of Bacillus subtilis YphP, a prokaryotic disulfide isomerase with a CXC catalytic motif. Biochemistry 48, 8664–8671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Woycechowsky K. J., Raines R. T. (2003) The CXC motif. A functional mimic of protein-disulfide isomerase. Biochemistry 42, 5387–5394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cacciapuoti G., Peluso I., Fuccio F., Porcelli M. (2009) Purine nucleoside phosphorylases from hyperthermophilic Archaea require a CXC motif for stability and folding. FEBS J. 276, 5799–5805 [DOI] [PubMed] [Google Scholar]
- 54. Ohnuma T., Onaga S., Murata K., Taira T., Katoh E. (2008) LysM domains from Pteris ryukyuensis chitinase-A. A stability study and characterization of the chitin-binding site. J. Biol. Chem. 283, 5178–5187 [DOI] [PubMed] [Google Scholar]
- 55. Petutschnig E. K., Jones A. M., Serazetdinova L., Lipka U., Lipka V. (2010) The lysin motif receptor-like kinase (LysM-RLK) CERK1 is a major chitin-binding protein in Arabidopsis thaliana and subject to chitin-induced phosphorylation. J. Biol. Chem. 285, 28902–28911 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.