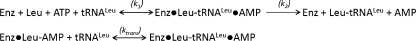Background: Mycoplasma mobile leucyl-tRNA synthetase has lost its CP1 domain via reductive genome evolution resulting in impaired editing.
Results: Fusion of cognate and noncognate bacterial CP1 domains to the aminoacylation canonical core enhances fidelity.
Conclusion: CP1 domain insertions influence amino acid discrimination in the synthetic site.
Significance: Evolutionary addition of the CP1 domain confers multiple mechanisms to achieve fidelity.
Keywords: Amino Acid, Aminoacyl-tRNA Synthetase, Protein Domains, Protein Synthesis, Transfer RNA (tRNA), Translation
Abstract
Statistical proteomes that are naturally occurring can result from mechanisms involving aminoacyl-tRNA synthetases (aaRSs) with inactivated hydrolytic editing active sites. In one case, Mycoplasma mobile leucyl-tRNA synthetase (LeuRS) is uniquely missing its entire amino acid editing domain, called CP1, which is otherwise present in all known LeuRSs and also isoleucyl- and valyl-tRNA synthetases. This hydrolytic CP1 domain was fused to a synthetic core composed of a Rossmann ATP-binding fold. The fusion event splits the primary structure of the Rossmann fold into two halves. Hybrid LeuRS chimeras using M. mobile LeuRS as a scaffold were constructed to investigate the evolutionary protein:protein fusion of the CP1 editing domain to the Rossmann fold domain that is ubiquitously found in kinases and dehydrogenases, in addition to class I aaRSs. Significantly, these results determined that the modular construction of aaRSs and their adaptation to accommodate more stringent amino acid specificities included CP1-dependent distal effects on amino acid discrimination in the synthetic core. As increasingly sophisticated protein synthesis machinery evolved, the addition of the CP1 domain increased specificity in the synthetic site, as well as provided a hydrolytic editing site.
Introduction
The aminoacyl-tRNA synthetases (aaRS)2 discriminate between pools of structurally related amino acids to covalently link their cognate substrates via an aminoacyl bond to the correct tRNA isoacceptor (1, 2). Translation of the genetic code is set in this first step of protein synthesis when the tRNA is aminoacylated for delivery to the ribosome for messenger RNA decoding. Half of the aaRSs cannot fully distinguish between isosteric or structurally overlapping amino acid substrates and have a hydrolytic editing domain that has been fused during evolution to the ancient synthetic core of the synthetase. In these cases, the editing domain clears mistakes to maintain the high fidelity that is required for protein synthesis.
Leucyl- (LeuRS), isoleucyl- (IleRS), and valyl- (ValRS) tRNA synthetases share homologous CP1 domains that are responsible for editing by hydrolytically cleaving one or more noncognate amino acids that have been mischarged to the tRNA (2). Each of these CP1 domains is attached to the catalytic core of the enzyme via two β-strand tethers at a site that splits the primary amino acid sequence of a common Rossmann ATP-binding fold (3, 4) into two halves (5). X-ray crystal structures of LeuRS (Protein Data Bank code 1H3N), ValRS (code 1GAX), and IleRS (code 1ILE) show that the tertiary structure of the “split” Rossmann fold is intact and that the inserted CP1 editing module folds as a discrete domain (6–8).
The crystal structures of the CP1 domains of LeuRS (code 1OBC), IleRS (code 1UDZ), and ValRS (code 1WK9) with amino acid analogues have been reported (9–11). Each of the CP1 domain active sites has diverged to accommodate different amino acid specificities. The IleRS and ValRS CP1 domains hydrolyze mischarged Val-tRNAIle and Thr-tRNAVal, respectively (12–15). The LeuRS CP1 domain requires a broader specificity to accommodate a number of potential synthetic active site mistakes that include methionine, isoleucine, and valine, as well as the nonstandard amino acids norvaline and norleucine (16–18).
We identified the only known example of a LeuRS, IleRS, or ValRS that is completely missing its CP1 domain (19). The gene for Mycoplasma mobile LeuRS (MmLeuRS) has shed the DNA encoding the editing domain of the enzyme, resulting in statistical mutations at leucine sites in the proteome. This contrasts with an artificial LeuRSΔCP1 that we constructed from Escherichia coli LeuRS, where the CP1 domain was deleted and maintained fidelity (20). In this latter case, the unnatural deletion mutant recovered a “pre-transfer” editing activity associated with the canonical Rossmann fold and clears mistakes by hydrolyzing misactivated aminoacyl-adenylate intermediates.
Here, we fused CP1 domains from E. coli LeuRS, IleRS, and ValRS to the M. mobile LeuRS and determined a surprising increase in amino acid discrimination in the first step of the aminoacylation reaction. This suggests that the evolutionary addition of the CP1 domain to the class I enzyme enhanced fidelity of the synthetic aminoacylation reaction in the canonical synthetic core, in addition to conferring a hydrolytic editing activity.
EXPERIMENTAL PROCEDURES
Design and Construction of Hybrid LeuRSs
Using polymerase chain reaction (PCR) mutagenesis, an SpeI restriction endonuclease site (A↓CTAGT) was introduced into the plasmid p14bLiMmLeuRS (19) that encodes the wild type MmLeuRS gene at the sites encoding the CP1 domain fusion position of Glu-229 and Gly-232. The resultant plasmid p14bMBMmLeuRSSpeI was used to clone DNA that encoded the CP1 domain. PCR-amplified gene fragments carrying DNA sequence of the CP1 domain from E. coli LeuRS (p14LiMBMmLeuRS/LeuCP1H9), ValRS (p14LiMBMmLeuRS/ValCP1H1), and IleRS (p14LiMBMmLeuRS/IleCP1H11) were ligated into p14bMBMmLeuRSSpeI plasmid that was linearized with SpeI endonuclease.
Purification of Hybrid MmLeuRS
Wild type MmLeuRS and each of the His6-tagged hybrid proteins were synthesized recombinantly in E. coli and affinity-purified using an FPLC HisTrap HP column (GE Healthcare) (20) followed by purification on a MonoQ 5/50 GL (GE Healthcare) column using low salt and high salt buffers: 20 mm Tris-HCl, pH 8.2, 5 mm NaCl and 20 mm Tris-HCl, pH 8.2, 1 m NaCl. The ion exchange column purification was followed by purification on a size exclusion Superdex 200 column (GE Healthcare) in 10 mm Tris-HCl, pH 8.0, 20 mm NaH2PO4, 100 mm NaCl, 5% glycerol to ensure that residual E. coli LeuRS, IleRS, and ValRS were removed prior to experiments.
Rapid Quench Kinetics
Multiple turnover kinetic reactions containing 5 μm protein, 12.5 μm M. mobile tRNAUAALeu (tRNALeu) transcript, and 50 μm [14C]leucine (318 mCi/mmol) were carried out as described (21, 22). Single turnover kinetic reactions contained 2 μm tRNALeu and 10 μm wild type or hybrid LeuRSs complexed with leucyl-adenylate intermediate and fitted into a single exponential equation to measure ktrans (21, 22).
Transcription and Misaminoacylation of M. mobile tRNALeu
M. mobile tRNALeu was in vitro transcribed (19, 23) and charged with [14C]isoleucine or [35S]methionine using T252Y E. coli LeuRS editing-defective mutant (18).
Pyrophosphate Exchange
In a pyrophosphate exchange assay (20, 24), Km and kcat for activation of leucine and also isoleucine, methionine, and valine were measured using equivalent 1 mm concentrations of amino acid, ATP, and [32P]PPi (900 mCi/mmol) at 30 °C. Pyrophosphate and ATP were separated by thin-layer chromatography and quantitated by phosphorimaging.
RESULTS
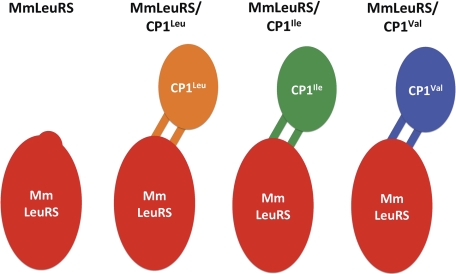
We used M. mobile LeuRS as a scaffold to fuse CP1 domains to the catalytic class I aaRS core and reconstruct an additional hydrolytic editing step that was introduced during evolution to increase fidelity for protein synthesis. We created hybrid M. mobile LeuRS proteins that contained the CP1 domain from E. coli LeuRS (MmLeuRS/CP1Leu) and also the IleRS (MmLeuRS/CP1Ile) and ValRS (MmLeuRS/CP1Val) CP1 domains (Fig. 1). In each case, we retained the β-strand linkers that are native to the CP1 domain that was introduced. The N- and C-terminal β-strands of the CP1 domains were fused to the Rossmann fold of M. mobile LeuRS at Glu-229 and Gly-232 (···224WIGKEEIDG232···), respectively. This corresponds to Glu-228 and Ala-362, the naturally occurring E. coli LeuRS fusion sites that link the flexible β-strands of the CP1 domain to the Rossmann fold (19, 25).
FIGURE 1.
Schematic of M. mobile LeuRS and CP1 hybrid mutants. The canonical synthetic core of MmLeuRS is shown in red. The fused CP1 domains and their respective β-strands from E. coli LeuRS (CP1Leu), IleRS (CP1Ile), and ValRS (CP1Val) are shown in orange, green, and blue, respectively.
SCHEME 1.
Using rapid quench kinetics, we compared the aminoacylation properties of the wild type M. mobile LeuRS that is missing the CP1 domain with each of the hybrid constructs that contained the LeuRS core fused to an E. coli CP1 domain from LeuRS, IleRS, or ValRS. Similar to other class I aaRSs including E. coli LeuRS, the multiple turnover kinetic profile was biphasic (21, 26) for the wild type M. mobile LeuRS and its hybrid constructs (Table 1).
TABLE 1.
Multiple and single turnover kinetic measurements during tRNA aminoacylation with leucine
Methods are described in Hellmann and Martinis (21). Kinetic constants can be defined in Scheme 1.
| k1 | k2 | ktrans | |
|---|---|---|---|
| s−1 | |||
| MmLeuRS | 18.5 ± 3.4 | 2.1 ± 0.11 | 15.2 ± 2.1 |
| MmLeuRS/CP1Leu | 9.7 ± 1 | 1.37 ± 0.12 | 12.6 ± 3.7 |
| MmLeuRS/CP1Ile | 10.6 ± 0.13 | 1.67 ± 0.13 | 14.8 ± 1.1 |
| MmLeuRS/CP1Val | 10.7 ± 0.23 | 0.42 ± 0.05 | 10.5 ± 1.1 |
The rate constant k2 correlates to a rate-limiting product release step for charged tRNA (21, 26). The addition of the CP1 domain from either LeuRS or IleRS only slightly lowered k2. The largest decrease was 5-fold for the LeuRS hybrid that contained CP1Val, suggesting that charged Leu-tRNALeu product release was slowed further by the addition of the CP1 domain. Because the “exit complex” of LeuRS has been proposed to have the 3′-charged end of the tRNA positioned near the hydrolytic active site of the CP1 editing domain (27), it is possible that either the noncognate mismatch between CP1Val and tRNALeu or the charged leucine amino acid affects the mechanism of product release. The faster rate constant, k1, measured during multiple turnover kinetics, was decreased by less than 2-fold (Table 1) for each of the hybrid LeuRSs as compared with the wild type M. mobile LeuRS that is missing its CP1 domain.
The ktrans, measured during single turnover kinetic analysis, represents the kinetic rate constant for the transfer of the amino acid from the adenylate intermediate to tRNALeu and correlated to other class I aaRSs (26). As would be expected as compared with other class I aaRSs (21, 26), ktrans was most similar to k1 for the wild type M. mobile LeuRS and hybrid constructs. Significantly, this supports that the integrity of the synthetic aminoacylation site is maintained despite deep cuts into the Rossmann fold to introduce CP1 domains from different enzymes with distinct specificities.
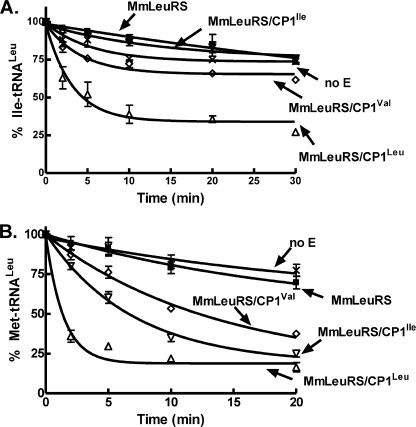
We screened each of the hybrid LeuRS tRNA deacylation activities, which would be reminiscent of a canonical LeuRS post-transfer editing activity that hydrolyzes noncognate amino acids from tRNA. The hybrid composed of the LeuRS CP1 domain fused to the M. mobile LeuRS aminoacylation core (MmLeuRS/CP1Leu) was faithful to its cognate specificities that ensure fidelity in producing Leu-tRNALeu. In particular, it hydrolyzed Ile-tRNALeu and Met-tRNALeu with a kobs of 0.31 and 0.71 min−1, respectively (Fig. 2). In comparison, the wild type E. coli LeuRS respectively deacylated Ile-tRNALeu and Met-tRNALeu with a kobs of 1.43 and 0.69 min−1. This is similar to natural LeuRSs, which have evolved to edit aliphatic standard amino acids that threaten protein synthesis fidelity (16, 18, 23, 28).
FIGURE 2.
Deacylation of charged tRNA by M. mobile LeuRS and hybrid mutants. A and B, deacylation of 2 μm misaminoacylated Ile-tRNALeu (A) or Met-tRNALeu (B) by 1 μm M. mobile LeuRS wild type or hybrid proteins that contain an E. coli CP1 domain. Symbols used are as follows: ×, nonenzymatic tRNA deacylation (no E); ■, MmLeuRS (wild type); △, MmLeuRS/CP1Leu; ▿, MmLeuRS/CP1Ile; ◊, MmLeuRS/CP1Val. Error bars represent the S.D. values based on three separate experiments.
Both hybrid MmLeuRS/CP1Val and MmLeuRS/CP1Ile hydrolyzed Met-tRNALeu (Fig. 2B), with a kobs of 0.07 and 0.14 min−1, respectively. In comparison, the wild type E. coli IleRS hydrolyzed Met-tRNAIle with a kobs rate of 1.32 min−1. The flexibility of the unbranched aliphatic side chain of methionine likely facilitates binding in the hydrolytic editing active site similar to full-length aaRSs. Consistent with the robustness and the specificities of the CP1 hydrolytic activities of the hybrid M. mobile LeuRSs, mischarging did not occur for MmLeuRS/CP1Leu, MmLeuRS/CP1Ile, or MmLeuRS/CP1Val (data not shown). This contrasts with the wild type M. mobile LeuRS, which is prone to mistakes that it fails to efficiently clear in the absence of the CP1 domain editing module (19).
Fusion of the CP1 domain from IleRS or ValRS to M. mobile LeuRS failed to cleave Ile-tRNALeu. This would be expected based on the natural editing specificities of full-length IleRS and ValRS that would block isoleucine from the amino acid-binding pocket of the hydrolytic active site (10, 29, 30). Pyrophosphate exchange assays (19, 24) support that isoleucine is activated by each of the hybrid M. mobile LeuRS proteins, albeit at a higher Km as compared with the cognate leucine substrate (Fig. 3 and supplemental Table S1). However, isoleucine is not mischarged to tRNALeu by any of the hybrid LeuRS proteins that contain a CP1 domain (data not shown). In contrast, the core M. mobile LeuRS that is missing a CP1 domain and serves as a scaffold for these hybrid proteins produces mischarged Ile-tRNALeu (19).
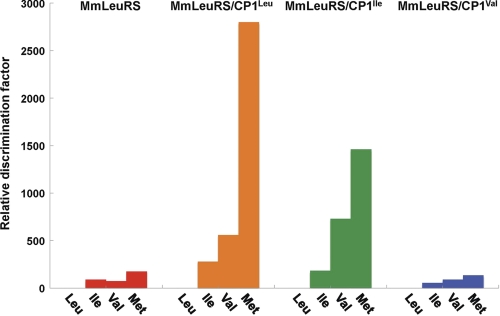
FIGURE 3.
Bar graph representing amino acid discrimination factors relative to leucine. Data for M. mobile LeuRS are shown in red, whereas data for hybrid proteins MmLeuRS/CP1Leu, MmLeuRS/CP1Ile, and MmLeuRS/CP1Val are shown in orange, green, and blue, respectively. This graph is based on the measurements listed in supplemental Table S1.
Splitting the primary structure of the Rossmann fold to accommodate the addition of a CP1 domain decreased kcat/Km for leucine activation (supplemental Table S1). Remarkably, however, the fusion to either CP1Leu or CP1Ile dramatically enhanced discrimination of the cognate leucine substrate, relative to the noncognate aliphatic amino acids (Fig. 3 and supplemental Table S1). In the case of MmLeuRS/CP1Leu, which reconstructs a canonical LeuRS from the naturally occurring M. mobile LeuRS, a small increase in the Km of 3-fold for leucine contrasts to increases ranging from 24-fold for isoleucine, 35-fold for methionine, and 64-fold for valine. Similarly, the addition of CP1Ile or CP1Val yielded only 2–5-fold increases in Km for leucine in the synthetic active site, as compared with at least 10-fold increases for noncognate amino acids. The addition of CP1Leu and CP1Ile to the canonical core of M. mobile LeuRS increased the discrimination factor for isoleucine 2–3-fold and lowered efficiency of isoleucine activation by almost 100-fold, based on kcat/Km ratios (Fig. 3).
Similar effects were observed for valine activation in the synthetic site of both hybrid LeuRSs. The discrimination factor for valine increased almost 9-fold, with decreasing enzymatic efficiency. Although we did not observe dramatic changes in the discrimination factor for noncognate amino acids when CP1Val was fused to the M. mobile LeuRS Rossmann fold, the efficiency of valine activation was lowered. In addition, the hybrid mutant MmLeuRS/CP1Val did not display mischarging activity, similar to the CP1Leu and CP1Ile hybrids, and maintained similar catalytic efficiency during the aminoacylation reaction (Table 1). Thus, these mechanistic links between the fused CP1 domain and the canonical cores have diverged in an idiosyncratic way.
DISCUSSION
As protein synthesis evolved to greater sophistication, the cellular demand for fidelity of the proteome also increased. The addition of the CP1 editing domain added a hydrolytic sieve to clear aaRS aminoacylation mistakes. This double sieve model to increase aaRS fidelity has been capitalized upon by at least half of the aaRS enzyme family (15, 31).
Reports on IleRS show that its CP1 domain is intimately linked with tRNA-dependent pre-transfer editing activity (32–35). We have shown in LeuRS that removal of the CP1 domain can influence pre-transfer editing that is associated with the synthetic core (20). We hypothesize that the CP1 domain also influences adenylate stability and hydrolysis in the synthetic active site. Interestingly, the insertion editing domain (INS) of the class II bacterial prolyl-tRNA synthetase (ProRS) has been shown to play a role in facilitating or stabilizing adenylate formation (36).
Specificity and fidelity of the aaRSs are initially dependent on the finely tuned synthetic core of the enzymes, which binds and activates amino acids in an ATP-dependent mechanism for aminoacylation to tRNA. The fusion of the CP1 editing domain splits the primary structure of the ancient Rossmann fold that comprises the class I aaRS catalytic core of LeuRS, ValRS, and IleRS. Remarkably, however, rather than disrupting the enzyme core, the insertion of the LeuRS CP1 editing domain distally enhances specificity in the synthetic aminoacylation active site. Thus, fusion of these two protein domains provided a second mechanism to minimize errors that goes beyond the acquisition of the hydrolytic editing site. This finding is consistent with the previously published molecular dynamic simulation that the CP1 domain contributes to the cognate amino acid specificity in tryptophanyl-tRNA synthetase (37). It is also consistent with the editing domains of the class II ProRS, which have been fused or appended to the synthetic core at diverse sites (36, 38).
Our results support that the evolutionary fusion of the CP1 protein domain to the Rossmann fold of the aaRSs not only introduced a hydrolytic active site for editing to improve fidelity, but also enhanced substrate discrimination within the synthetic active site in the ancient catalytic core of the enzyme. This was also found for the class II ProRS editing domain, which is fused to a catalytic core that has a completely different fold (36, 38). Surprisingly, in the case of LeuRS, this increase in discrimination was also conferred by the addition of the noncognate CP1Ile to LeuRS. This suggests that there is inherent overlap between CP1Leu and CP1Ile or in the way these domains are linked to the canonical core. Because the addition of CP1Val did not significantly enhance substrate discrimination in the canonical core per se, it is also possible that these enzymes acquired a common CP1 domain, which subsequently diverged to accommodate varied specificities.
Although it maintains its CP1 domain, the human mitochondrial LeuRS is similar to M. mobile LeuRS in that it has acquired mutations in its hydrolytic active site that have abolished its editing activity (39). Significantly, kinetic measurements showed that the synthetic site has increased its discrimination to exceed 1/3000. Likewise, yeast ProRS has an inactive editing domain that has been retained in conjunction with an active site that has similarly increased amino acid discrimination (38). This is the threshold for aaRSs that has been proposed to require an editing domain to maintain fidelity (2, 40).
It is possible that the human LeuRS enzyme retains its CP1 domain because it increases amino acid discrimination in the synthetic site similar to the hybrid M. mobile LeuRSs. In contrast, fidelity is decreased for the wild type M. mobile LeuRS due to a loss of discrimination in the aminoacylation active site, as well as the shedding of its CP1 editing domain (19). Because the M. mobile LeuRS has uniquely lost its CP1 domain during evolution, we hypothesize that this compromise in amino acid fidelity provides an idiosyncratic selective advantage to this host-dependent pathogen.
Supplementary Material
Acknowledgments
We thank Dr. Christopher Francklyn and Dr. Ethan Guth for valuable discussions and training in quench flow kinetics.
This work was supported, in whole or in part, by National Institutes of Health Grant GM063789 (to S. A. M. and M. T. B.). This work was also supported by National Science Foundation Grant MCB0843611.

This article contains supplemental Table 1.
- aaRS
- aminoacyl-tRNA synthetase
- LeuRS
- leucyl-tRNA synthetase
- IleRS
- isoleucyl-tRNA synthetase
- ValRS
- valyl-tRNA synthetase
- ProRS
- prolyl-tRNA synthetase
- MmLeuRS
- M. mobile LeuRS.
REFERENCES
- 1. Ibba M., Söll D. (1999) Quality control mechanisms during translation. Science 286, 1893–1897 [DOI] [PubMed] [Google Scholar]
- 2. Mascarenhas A. P., Martinis S. A., An A. E., Rosen A. E., Musier-Forsyth K. (2008) in Protein Engineering (RajBhandary U. L., Koehrer C., eds) pp. 153–200, Springer-Verlag New York Inc., New York [Google Scholar]
- 3. Adams M. J., McPherson A., Jr., Rossmann M. G., Schevitz R. W., Wonacott A. J. (1970) The structure of the nicotinamide-adenine dinucleotide coenzyme when bound to lactate dehydrogenase. J. Mol. Biol. 51, 31–38 [DOI] [PubMed] [Google Scholar]
- 4. Rao S. T., Rossmann M. G. (1973) Comparison of super-secondary structures in proteins. J. Mol. Biol. 76, 241–256 [DOI] [PubMed] [Google Scholar]
- 5. Zelwer C., Risler J. L., Brunie S. (1982) Crystal structure of Escherichia coli methionyl-tRNA synthetase at 2.5 Å resolution. J. Mol. Biol. 155, 63–81 [DOI] [PubMed] [Google Scholar]
- 6. Cusack S., Yaremchuk A., Tukalo M. (2000) The 2 Å crystal structure of leucyl-tRNA synthetase and its complex with a leucyl-adenylate analogue. EMBO J. 19, 2351–2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nureki O., Vassylyev D. G., Tateno M., Shimada A., Nakama T., Fukai S., Konno M., Hendrickson T. L., Schimmel P., Yokoyama S. (1998) Enzyme structure with two catalytic sites for double-sieve selection of substrate. Science 280, 578–582 [DOI] [PubMed] [Google Scholar]
- 8. Fukai S., Nureki O., Sekine S., Shimada A., Tao J., Vassylyev D. G., Yokoyama S. (2000) Structural basis for double-sieve discrimination of l-valine from l-isoleucine and l-threonine by the complex of tRNAVal and valyl-tRNA synthetase. Cell 103, 793–803 [DOI] [PubMed] [Google Scholar]
- 9. Fukunaga R., Fukai S., Ishitani R., Nureki O., Yokoyama S. (2004) Crystal structures of the CP1 domain from Thermus thermophilus isoleucyl-tRNA synthetase and its complex with l-valine. J. Biol. Chem. 279, 8396–8402 [DOI] [PubMed] [Google Scholar]
- 10. Fukunaga R., Yokoyama S. (2005) Structural basis for noncognate amino acid discrimination by the valyl-tRNA synthetase editing domain. J. Biol. Chem. 280, 29937–29945 [DOI] [PubMed] [Google Scholar]
- 11. Lincecum T. L., Jr., Tukalo M., Yaremchuk A., Mursinna R. S., Williams A. M., Sproat B. S., Van Den Eynde W., Link A., Van Calenbergh S., Grøtli M., Martinis S. A., Cusack S. (2003) Structural and mechanistic basis of pre- and post-transfer editing by leucyl-tRNA synthetase. Mol. Cell 11, 951–963 [DOI] [PubMed] [Google Scholar]
- 12. Eldred E. W., Schimmel P. R. (1972) Rapid deacylation by isoleucyl transfer ribonucleic acid synthetase of isoleucine-specific transfer ribonucleic acid aminoacylated with valine. J. Biol. Chem. 247, 2961–2964 [PubMed] [Google Scholar]
- 13. Baldwin A. N., Berg P. (1966) Transfer ribonucleic acid-induced hydrolysis of valyladenylate bound to isoleucyl ribonucleic acid synthetase. J. Biol. Chem. 241, 839–845 [PubMed] [Google Scholar]
- 14. Lin L., Hale S. P., Schimmel P. (1996) Aminoacylation error correction. Nature 384, 33–34 [DOI] [PubMed] [Google Scholar]
- 15. Fersht A. R. (1977) Editing mechanisms in protein synthesis: rejection of valine by the isoleucyl-tRNA synthetase. Biochemistry 16, 1025–1030 [DOI] [PubMed] [Google Scholar]
- 16. Karkhanis V. A., Mascarenhas A. P., Martinis S. A. (2007) Amino acid toxicities of Escherichia coli that are prevented by leucyl-tRNA synthetase amino acid editing. J. Bacteriol. 189, 8765–8768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Englisch S., Englisch U., von der Haar F., Cramer F. (1986) The proofreading of hydroxy analogues of leucine and isoleucine by leucyl-tRNA synthetases from E. coli and yeast. Nucleic Acids Res. 14, 7529–7539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mursinna R. S., Martinis S. A. (2002) Rational design to block amino acid editing of a tRNA synthetase. J. Am. Chem. Soc. 124, 7286–7287 [DOI] [PubMed] [Google Scholar]
- 19. Li L., Boniecki M. T., Jaffe J. D., Imai B. S., Yau P. M., Luthey-Schulten Z. A., Martinis S. A. (2011) Naturally occurring aminoacyl-tRNA synthetases editing-domain mutations that cause mistranslation in Mycoplasma parasites. Proc. Natl. Acad. Sci. U.S.A. 108, 9378–9383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Boniecki M. T., Vu M. T., Betha A. K., Martinis S. A. (2008) CP1-dependent partitioning of pre-transfer and post-transfer editing in leucyl-tRNA synthetase. Proc. Natl. Acad. Sci. U.S.A. 105, 19223–19228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hellmann R. A., Martinis S. A. (2009) Defects in transient tRNA translocation bypass tRNA synthetase quality control mechanisms. J. Biol. Chem. 284, 11478–11484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Francklyn C. S., First E. A., Perona J. J., Hou Y. M. (2008) Methods for kinetic and thermodynamic analysis of aminoacyl-tRNA synthetases. Methods 44, 100–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Karkhanis V. A., Boniecki M. T., Poruri K., Martinis S. A. (2006) A viable amino acid editing activity in the leucyl-tRNA synthetase CP1-splicing domain is not required in the yeast mitochondria. J. Biol. Chem. 281, 33217–33225 [DOI] [PubMed] [Google Scholar]
- 24. Splan K. E., Musier-Forsyth K., Boniecki M. T., Martinis S. A. (2008) In vitro assays for the determination of aminoacyl-tRNA synthetase editing activity. Methods 44, 119–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Betha A. K., Williams A. M., Martinis S. A. (2007) Isolated CP1 domain of Escherichia coli leucyl-tRNA synthetase is dependent on flanking hinge motifs for amino acid editing activity. Biochemistry 46, 6258–6267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhang C. M., Perona J. J., Ryu K., Francklyn C., Hou Y. M. (2006) Distinct kinetic mechanisms of the two classes of aminoacyl-tRNA synthetases. J. Mol. Biol. 361, 300–311 [DOI] [PubMed] [Google Scholar]
- 27. Tukalo M., Yaremchuk A., Fukunaga R., Yokoyama S., Cusack S. (2005) The crystal structure of leucyl-tRNA synthetase complexed with tRNALeu in the post-transfer-editing conformation. Nat. Struct. Mol. Biol. 12, 923–930 [DOI] [PubMed] [Google Scholar]
- 28. Mursinna R. S., Lincecum T. L., Jr., Martinis S. A. (2001) A conserved threonine within Escherichia coli leucyl-tRNA synthetase prevents hydrolytic editing of leucyl-tRNALeu. Biochemistry 40, 5376–5381 [DOI] [PubMed] [Google Scholar]
- 29. Fukunaga R., Yokoyama S. (2006) Structural basis for substrate recognition by the editing domain of isoleucyl-tRNA synthetase. J. Mol. Biol. 359, 901–912 [DOI] [PubMed] [Google Scholar]
- 30. Schmidt E., Schimmel P. (1994) Mutational isolation of a sieve for editing in a transfer RNA synthetase. Science 264, 265–267 [DOI] [PubMed] [Google Scholar]
- 31. Fersht A. R. (1998) Sieves in sequence. Science 280, 541. [DOI] [PubMed] [Google Scholar]
- 32. Nomanbhoy T. K., Hendrickson T. L., Schimmel P. (1999) Transfer RNA-dependent translocation of misactivated amino acids to prevent errors in protein synthesis. Mol. Cell 4, 519–528 [DOI] [PubMed] [Google Scholar]
- 33. Nordin B. E., Schimmel P. (2003) Transiently misacylated tRNA is a primer for editing of misactivated adenylates by class I aminoacyl-tRNA synthetases. Biochemistry 42, 12989–12997 [DOI] [PubMed] [Google Scholar]
- 34. Bishop A. C., Nomanbhoy T. K., Schimmel P. (2002) Blocking site-to-site translocation of a misactivated amino acid by mutation of a class I tRNA synthetase. Proc. Natl. Acad. Sci. U.S.A. 99, 585–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hale S. P., Auld D. S., Schmidt E., Schimmel P. (1997) Discrete determinants in transfer RNA for editing and aminoacylation. Science 276, 1250–1252 [DOI] [PubMed] [Google Scholar]
- 36. Hati S., Ziervogel B., Sternjohn J., Wong F. C., Nagan M. C., Rosen A. E., Siliciano P. G., Chihade J. W., Musier-Forsyth K. (2006) Pre-transfer editing by class II prolyl-tRNA synthetase: role of aminoacylation active site in “selective release” of noncognate amino acids. J. Biol. Chem. 281, 27862–27872 [DOI] [PubMed] [Google Scholar]
- 37. Pham Y., Kuhlman B., Butterfoss G. L., Hu H., Weinreb V., Carter C. W., Jr. (2010) Tryptophanyl-tRNA synthetase Urzyme: a model to recapitulate molecular evolution and investigate intramolecular complementation. J. Biol. Chem. 285, 38590–38601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. SternJohn J., Hati S., Siliciano P. G., Musier-Forsyth K. (2007) Restoring species-specific post-transfer editing activity to a synthetase with a defunct editing domain. Proc. Natl. Acad. Sci. U.S.A. 104, 2127–2132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lue S. W., Kelley S. O. (2005) An aminoacyl-tRNA synthetase with a defunct editing site. Biochemistry 44, 3010–3016 [DOI] [PubMed] [Google Scholar]
- 40. Loftfield R. B. (1963) The frequency of errors in protein biosynthesis. Biochem. J. 89, 82–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.