Background: Degradative amino acid decarboxylation pathways in bacteria have diverse physiological functions.
Results: A histidine decarboxylation pathway introduced in L. lactis improves acid stress survival, and synergy with the glycolytic pathway is demonstrated.
Conclusion: The physiological benefit of the new pathway is strongly dependent on the properties of the host organism.
Significance: Acquisition of the histidine decarboxylation pathway mimics successful horizontal gene transfer.
Keywords: Amine Transport, Amino Acids Transport, Bacteria, Energy Metabolism, Gene Transfer, Acid Stress Resistance, Acid Stress Survival, Histidine Decarboxylation, Horizontal Gene Transfer, Proton Motive Force
Abstract
Degradative amino acid decarboxylation pathways in bacteria generate secondary metabolic energy and provide resistance against acid stress. The histidine decarboxylation pathway of Streptococcus thermophilus CHCC1524 was functionally expressed in the heterologous host Lactococcus lactis NZ9000, and the benefits of the newly acquired pathway for the host were analyzed. During growth in M17 medium in the pH range of 5–6.5, a small positive effect was observed on the biomass yield in batch culture, whereas no growth rate enhancement was evident. In contrast, a strong benefit for the engineered L. lactis strain was observed in acid stress survival. In the presence of histidine, the pathway enabled cells to survive at pH values as low as 3 for at least 2 h, conditions under which the host cells were rapidly dying. The flux through the histidine decarboxylation pathway in cells grown at physiological pH was under strict control of the electrochemical proton gradient (pmf) across the membrane. Ionophores that dissipated the membrane potential (ΔΨ) and/or the pH gradient (ΔpH) strongly increased the flux, whereas the presence of glucose almost completely inhibited the flux. Control of the pmf over the flux was exerted by both ΔΨ and ΔpH and was distributed over the transporter HdcP and the decarboxylase HdcA. The control allowed for a synergistic effect between the histidine decarboxylation and glycolytic pathways in acid stress survival. In a narrow pH range around 2.5 the synergism resulted in a 10-fold higher survival rate.
Introduction
In bacteria, decarboxylation of amino acids is part of simple catabolic pathways in which a precursor amino acid is taken up from the medium, decarboxylated in the cytoplasm, and excreted into the medium as its amine derivative. In the decarboxylation reaction, a proton is consumed, and CO2 is released. The precursor uptake and excretion of the product across the membrane is mediated via an exchange mode of transport that is electrogenic. The excreted product carries one more positive charge than the precursor (1–6). Thus, the coupled reactions of decarboxylation and electrogenic exchange in the pathways generate a proton motive force consisting of both a transmembrane pH gradient (inside alkaline) and electrical potential (inside negative) and provide the cell with metabolic energy (16). In addition to pmf3 generation, the pathways may function in pH regulation and acid stress resistance. In Escherichia coli, four amino acid decarboxylation systems involved in acid stress resistance have been described, termed AR2, AR3, AR4, and AR5, constituting glutamate/γ-amino butyric acid, arginine/agmatine, lysine/cadaverine, and ornithine/putrescine pathways, respectively (7). Although AR2 and AR3 enable survival at extreme low pH, e.g. while traveling through the gastrointestinal tract, AR4 and AR5 help in surviving moderately acidic environments (e.g. pH 4.5). Homologous systems were found in other Gram-negative bacteria (8–10). In Gram-positive bacteria, much of the research on amino acid decarboxylation pathways is focused on the undesirable formation of biogenic amines in fermented foods, especially by lactic acid bacteria (11–13). Pathways for histidine (1, 14–16), tyrosine (3, 17–19), ornithine (4, 20, 22), and glutamate (23, 24) decarboxylation have been identified. The glutamate decarboxylation system of Listeria monocytogenes was shown to confer acid stress resistance, enabling it to survive the acidity of the gastric fluid (23), and tyrosine decarboxylation in Enterococcus faecium resulted in higher survival rates at pH 2.5 (17). Generation of proton motive force was demonstrated for the histidine decarboxylation pathway of Lactobacillus buchneri (1), the tyrosine decarboxylation pathway of E. faecium (17) and Lactobacillus brevis (3), and the aspartate decarboxylation pathway of Tetragenococcus halophilus (25).
Typically, in Gram-positives, the presence of gene clusters encoding amino acid decarboxylation pathways is strain-specific rather than species-specific, indicating that the clusters have been spread through horizontal gene transfer (14, 16, 18, 26). The physiological benefit of the newly acquired pathway for the receiving host, essential for maintenance of the gene cluster, will depend on the metabolic capacity and robustness of the host and the habitat in which the host must survive. Once a function is established, further evolution of the genes and gene cluster will optimize the integration of the pathway in the host organism. Here, we mimic the horizontal gene transfer event, the first step in the successful acquisition of new functionality in this evolutionary process, by introducing the histidine decarboxylation pathway of the yoghurt bacterium Streptococcus thermophilus CHCC1524 (16, 26) in the cheese bacterium Lactococcus lactis and analyzing the beneficial effects for the receiving organism. The gene cluster encoding the pathway consists of the histidine decarboxylase gene hdcA, the histidine/histamine exchanger gene hdcP, and the HdcA maturation factor gene hdcB and is transcribed as one mRNA, thus forming one operon (16, 26). Recently, the complete hdcA-hdcP-hdcB operon was functionally expressed in L. lactis as part of a study demonstrating the function of HdcB in the cleavage of the HdcA proenzyme, an essential step in the formation of the pyruvoyl group cofactor (27, 28). Transcriptional regulation studies in S. thermophilus suggested that the physiological role of the pathway is generation of secondary metabolic energy rather than acid stress resistance (16). This study reveals a major benefit of the pathway for L. lactis in survival at low pH and uncovers a synergistic effect of the histidine decarboxylation pathway and the glycolytic pathway in acid stress survival.
EXPERIMENTAL PROCEDURES
Strains, Media, and Growth Conditions
L. lactis strain NZ9000 (29) was used as a host for the expression of the hdcAPB operon from plasmid pNZ-hdcAPB, in which the hdcAPB operon is located directly behind the nisin-inducible promoter PnisA, or pNZ-PhdcAPB, which contains the hdcAPB operon, including 600 bp of its upstream promoter region (28). Plasmid pNZ-hdcAS82A-PB was derived from pNZ-hdcAPB by inverted PCR with primers hdcA-SA-f and hdcA-SA-r, leading to a serine to alanine mutation at position 82 in HdcA (28). Cells were grown at 30 °C in M17 broth supplied with 25 mm glucose and 5 μg/ml chloramphenicol unless stated otherwise. For expression of hdcAPB from pNZ-hdcAPB, cells grown to mid-exponential phase (A600 = 0.6) were induced by addition of 5 ng/ml nisin and left to grow for another hour.
Internal pH (ΔpH) and Membrane Potential (ΔΨ) Measurements
Cells expressing the hdcAPB operon were washed and loaded with 2,7-bis-(2-carboxyethyl)-5(6)-carboxyfluorescein as described (30). Fluorescence was monitored at 30 °C under continuous stirring in a 1-cm cuvette at 525 nm emission (bandwidth 4 nm) with excitation at 502 nm (bandwidth 16 nm) in an SLM Aminco Bowman series 2 spectrometer. The cytoplasmic pH was calculated as described in Ref. 30.
Membrane potential was measured qualitatively using the fluorescent probe 3,3′-dipropylthiodicarbocyanine (diSC3(5)) (31). 3,3′-dipropylthiodicarbocyanine was added to the cell suspensions at a concentration of 2 μm, after which the suspension was incubated for 5 min at 30 °C under continuous stirring. Fluorescence was monitored in an SLM Aminco Bowman series 2 spectrometer with excitation and emission wavelengths of 625 nm (bandwidth 8) and 690 nm (bandwidth 8), respectively.
Histamine Production by Resting Cells of L. lactis
Cells expressing hdcAPB from pNZ-hdcAPB were washed in 100 mm KPi buffer (pH 5) and resuspended to an A600 of 1 in 100 mm KPi (pH 5.0) unless stated otherwise. At t = 0, l-histidine was added to a final concentration of 5 mm. Glucose was added to 10 mm final concentration unless stated otherwise. Valinomycin, nigericin, and CCCP were added to 1 μm, 1 μm, and 25 μm final concentrations, respectively. Cells were permeabilized by treatment with 0.2% Triton X-100 for 5 min prior to the start of the experiment. For experiments at pH values below 5, 100 mm KPi/citrate buffer was used instead of 100 mm KPi. Samples were taken at the indicated time points. Cells were spun down immediately, and the supernatants were incubated at 95 °C to stop any remaining histidine decarboxylation activity.
Histidine-Histamine Exchange Assay
HdcP was expressed from pNZ-hdcP (28) in an L. lactis strain derived from NZ9000, in which the lysQ gene, encoding a histidine transporter, was deleted (unpublished data). Cells were washed twice in 100 mm KPi (pH 6.0) and resuspended to an A600 of 2.0 in 100 mm KPi (pH 6.0). The cells were loaded with histamine by incubation in the presence of 10 mm glucose and 5 mm histamine for 20 min at room temperature, after which the cells were put on ice until use. External histamine was removed by washing the cells three times in 200 mm KPi (pH 6.0). The cells were resuspended to an A600 of 2.0, and 100-μl aliquots were incubated for 5 min at 30 °C with constant stirring. At time zero, L-[U-14C]histidine (PerkinElmer) was added to a final concentration of 1.5 μm. Uptake was stopped at t = 10, 30, or 60 s by addition of 2 ml of ice-cold 0.1 m LiCl, followed by filtration through a 0.45-μm-pore size nitrocellulose filter ((BA 85, Whatman GmbH, Dassel, Germany). The filter was washed once with 2 ml of 0.1 m LiCl and submerged in Emulsifier Scintillator Plus scintillation fluid (Packard Bioscience, Meriden, CT), and radioactivity was measured in a Tri-Carb 2000CA liquid scintillation analyzer (Packard Instruments).
Determination of Histamine, Glucose, and Lactate Concentrations
Histamine concentrations were determined in a two enzyme-based fluorimetric assay as described (32) with minor modifications. The assay mixture contained 100 μl of 0.5 m KPi (pH 7.0), 100 μl of diamine oxidase (0.7 units/ml, from porcine kidney, Sigma), 10 μl of horseradish peroxidase (100 units/ml), 100 μl of 3-(4-hydroxyphenyl)propionic acid (1 mg/ml) (Sigma-Aldrich), and 190 μl H2O. Aliquots of 100 μl of the supernatant diluted to a concentration within the linear range of the assay (0–75 μm histamine) was added to the assay mixture. After 3 h of incubation at 37 °C, 2.5 ml of 0.1 m NaOH was added. Fluorescence of the oxidation product 2,2′-dihydroxybiphenyl-5,5′-dipropionic acid was measured as emission at 410 nm with excitation at 320 nm (bandwidths 5 nm) using an SLM Aminco Bowman series 2 luminescence spectrometer. Histamine concentrations were calculated using a standard curve derived from histamine solutions of known concentration.
Glucose consumption was measured either directly by measuring the glucose concentration or indirectly by measuring L-lactate produced in the supernatants. Glucose concentrations were measured in a similar assay as described above using glucose oxidase (from Aspergillus niger, type VII, Sigma) instead of diamine oxidase, added to 0.025 units/ml final concentration.
Lactate concentrations were determined using an enzymatic assay in which lactate was oxidized by lactate dehydrogenase (Sigma-Aldrich) in the presence of NAD+ to form pyruvate and NADH. A high pH buffer (pH 9) and hydrazine, which reacts with pyruvate to give hydrazone, were used to allow the reaction to run to complete oxidation of lactate. A reaction mix was prepared consisting of 0.5 m glycine and 0.4 m hydrazine sulfate and adjusted to pH 9 with NAOH, to which 2.5 mm NAD+ and 2 units/ml lactate dehydrogenase were added. 20 μl of sample was added to 200 μl of the reaction mix in 96-wells microtiter plates. After 1 h of incubation at 25 °C, NADH was measured at 340 nm. Lactate concentrations were calculated using a standard curve derived from lactate solutions of known concentration (0–2 mm).
Acid Stress Survival Assay
Cells were grown to stationary phase (pNZ-PhdcAPB) or to A600 = 0.6 and induced for 1 h with nisin (pNZ-hdcAPB and pNZ-hdcAS82-PB). Following harvesting and washing, cells were resuspended in M17 medium containing 10 mm glucose and 10 mm histidine as indicated and adjusted to the desired pH using HCl as indicated. The suspensions were incubated for the indicated time, after which cell survival was determined by plating 10 μl droplets of serial dilutions of samples on GM17 medium and counting colonies after overnight incubation at 30 °C.
RESULTS
Proton Motive Force Inhibits the Flux through the Histidine Decarboxylation Pathway
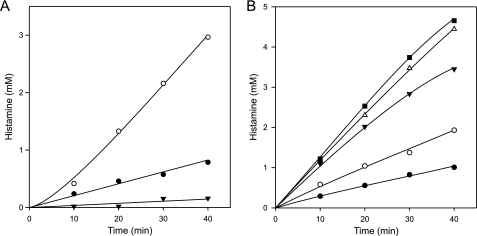
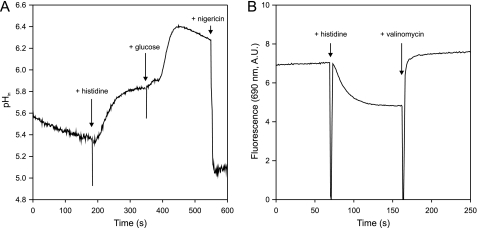
The histidine decarboxylation gene cluster of S. thermophilus CHCC1524, encoding histidine decarboxylase HdcA, the histidine/histamine exchanger HdcP, and the HdcA maturation enzyme HdcB, was functionally expressed in L. lactis NZ9000. Resting cells expressing the pathway were resuspended at an A600 of 1.0 in 100 mm potassium phosphate (KPi) (pH 5.0) buffer. In the presence of 5 mm histidine, the cells produced histamine at a constant rate of ∼18 μm/min for at least 40 min (Fig. 1A). The histidine decarboxylation pathway generates pmf by electrogenic histidine/histamine exchange (membrane potential, ΔΨ), catalyzed by HdcP and proton consumption in the decarboxylation of histidine in the cytoplasm (pH gradient, ΔpH) by HdcA. The observed flux through the pathway generated a pH gradient of 0.5 units as measured by the increase of the internal pH monitored by the fluorescent probe 2,7-bis-(2-carboxyethyl)-5(6)-carboxyfluorescein (Fig. 2A) and a significant membrane potential reported by the fluorescent probe 3,3′-dipropylthiodicarbocyanine (Fig. 2B). In the presence of 25 μm of the protonophore carbonyl cyanide m-chlorophenylhydrazone (CCCP) that completely dissipated both components of the pmf (not shown), a 3- to 4-fold increase in histamine production rate was observed (Fig. 1A). In contrast, in the presence of 10 mm glucose, which resulted in a significantly higher pH gradient of 1 unit (Fig. 2A), the histamine production rate decreased 4-fold (Fig. 1A). The opposite effects of CCCP and glucose on the histamine production rates demonstrate that under the conditions of the experiment, the flux through the histidine decarboxylation pathway is under control of the electrochemical proton gradient. The two components of the pmf may affect different parts of the pathway. The membrane potential ΔΨ may affect the exchanger HdcP, and the pH gradient ΔpH may affect both HdcP and the decarboxylase HdcA, the latter indirectly via changes in the cytoplasmic pH. Valinomycin is a K+-ionophore that selectively dissipates ΔΨ in high K+-buffers (Fig. 2B). In the presence of 1 μm valinomycin, the rate of histamine production by resting cells of L. lactis expressing the HDC pathway in 200 mm KPi (pH 5) buffer containing 5 mm histidine increased almost 2-fold (Fig. 1B). When instead of valinomycin 1 μm of nigericin, a K+/H+ exchanging ionophore that selectively dissipates ΔpH (Fig. 2A), was included, the histamine production rate increased ∼3.5-fold. Adding both ionophores at the same time, which dissipates the complete pmf, resulted in an even higher rate of histamine production that was comparable with the rate in the presence of the protonophore CCCP (Fig. 1B). Therefore, the effects of valinomycin and nigericin were additive. The results show that dissipation of both components of the pmf, i.e. membrane potential and ΔpH, leads to higher histamine production rates. It follows that the control of the pmf over the flux through the HDC pathway is a combined effect of the membrane potential and pH gradient exerted on both HdcP and HdcA.
FIGURE 1.
The histidine decarboxylation pathway is controlled by the pmf. Histamine production by resting cells expressing the HDC pathway. Cells were incubated at an A600 of 1 in 100 mm KPi (A) and 200 mm KPi (B) (pH 5.0) containing 5 mm histidine without further additions (A and B, ●) in the presence of 25 μm CCCP (A, ○; B, ■), 10 mm glucose (A, ▾), and in the presence of 1 μm valinomycin (B, ○), 1 μm nigericin (B, ▾), 1 μm valinomycin, and 1 μm nigericin (B, △).
FIGURE 2.
The histidine decarboxylation pathway generates pmf. A, internal pH. 10 μl of 2,7-bis-(2-carboxyethyl)-5(6)-carboxyfluorescein-loaded cells of L. lactis NZ9000 expressing the HDC pathway were resuspended in 3 ml of 200 mm KPi (pH 5.0) buffer and incubated for 5 min at 30 °C. Histidine, glucose, and nigericin were added at final concentrations of 5 mm, 5 mm, and 1 μm, respectively, at the indicated times. B, membrane potential. 10 μl of cells expressing hdcAPB resuspended at ∼20 mg protein/ml were added to 3 ml of 200 mm KPi (pH 5.0). Histidine and valinomycin were added at final concentrations of 5 mm and 1 μm, respectively.
Effect of pmf on Histidine/Histamine Exchange Catalyzed by HdcP
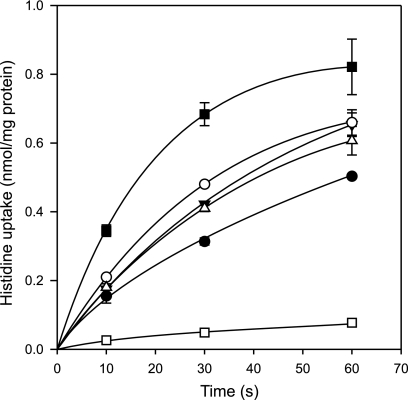
HdcP was expressed from plasmid pNZ-hdcP (28) in a strain of L. lactis with low histidine uptake activity because of deletion of the lysQ gene encoding a histidine transporter (unpublished data). The cells were loaded with 5 mm histamine (see “Experimental Procedures”), the counter substrate for histidine uptake by HdcP. At a concentration 1.5 μm, histidine was taken up at an initial rate of 3 nmol/min/mg protein (Fig. 3). Control cells not loaded with histamine showed an initial uptake rate of less than 0.1 nmol/min/mg protein (not shown), demonstrating that the observed uptake was in exchange with histamine. Dissipation of the pmf by the addition of 25 μm CCCP increased the uptake rate by more than a factor of 2, whereas imposing pmf in the presence of glucose inhibited the initial rate by more than a factor of 6. When ΔΨ was dissipated selectively by valinomycin, an increase in uptake rate was observed, but to a lesser extent than observed with CCCP. A similar increase was observed when ΔpH was selectively dissipated by nigericin. In the presence of both ionophores, a slight additive effect was observed but not to the extent observed with CCCP. The results show that the pmf controls the histidine/histamine exchange rate at both the levels of membrane potential and pH gradient.
FIGURE 3.
Histidine/histamine exchange is controlled by the pmf. Cells of L. lactis NZ9000 ΔlysQ expressing HdcP were loaded with 5 mm histamine and resuspended to an A600 of 2 in 200 mm KPi (pH 6.0) buffer. Fifteen seconds before addition of 1.5 μm 14C-histidine, either no further additions were made (●), or 5 μm valinomycin (△), 5 μm nigericin (▾), 5 μm valinomycin, and 5 μm nigericin (○), 25 μm CCCP (■), and 10 mm glucose (□) was added to the cells.
Effect of pH on the Flux through the Histidine Decarboxylation Pathway
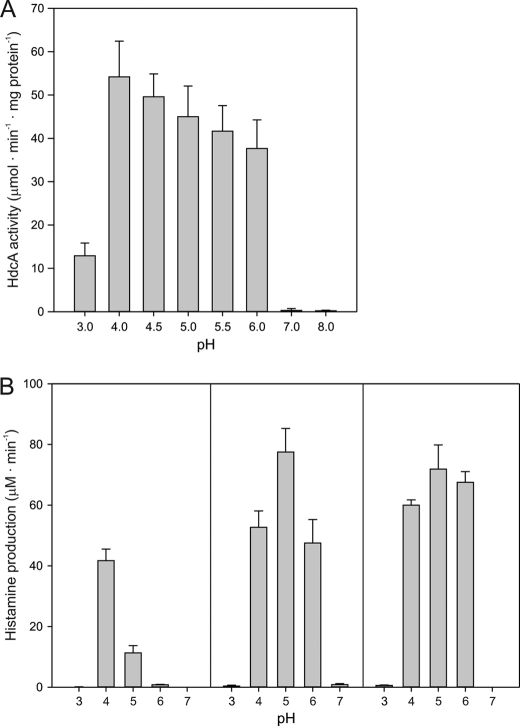
Changes in the proton gradient across the membrane are likely to affect the pH of the cytoplasm. The pH dependence of the purified histidine decarboxylase in the S. thermophilus HDC pathway (HdcA) isolated from E. coli (28) showed an optimum at pH 4 (Fig. 4A). The activity slowly decreased down to 70% of the maximal activity at a pH of 6, resulting in a plateau in the activity in the range pH 4–6. Above pH 6, the activity decreased drastically to no significant activity at pH 7. At the other side of the spectrum, a similar rapid drop in activity was observed at pH values below 4. The results are in line with reported pH profiles of histidine decarboxylases of Gram-positive bacteria that showed pH optima ranging from 4.4 to 5.8 (27).
FIGURE 4.
pH profile of purified HdcA (A) and of cells of L. lactis NZ9000 expressing the HDC pathway (B). A, 0.5 μg of purified HdcA, was added to 500 μl of 200 mm ammonium acetate, adjusted to the indicated pH values, and containing 50 mm L-histidine at 37 °C. Histidine decarboxylation was stopped at 5, 10, 15, 30, 45, and 60 min by incubating 50-μl samples for 5 min at 95 °C. Histamine concentrations were measured as described under “Experimental Procedures.” HdcA of S. thermophilus was purified from E. coli as described before (28). B, cells of L. lactis expressing hdcAPB were resuspended to an A600 of 1 in 100 mm KPi buffer or KPi/citrate buffer, adjusted to the indicated pHs containing 5 mm histidine with no further additions (left panel) in presence of 25 μm CCCP (center panel) and in the presence of 0.2% Triton X-100 (right panel). Error bars represent mean ± S.D. of three independent experiments.
The histamine production rate catalyzed by resting cells of L. lactis expressing the HDC pathway was measured at extracellular pH values in the same range of 3 to 7. The pH profile showed an optimum at pH 4, similar to that observed for the purified decarboxylase, but, in contrast, the rate decreased strongly with increasing pH (Fig. 4B, left panel). At external pH values of 3 and 7, no significant levels of histamine were produced, similar as observed for purified HdcA. It follows that, especially in the external pH range between 4 and 6, the transmembrane pH gradient and/or the membrane potential affect the flux through the HDC pathway. The uncoupler CCCP dissipates the proton gradient and, consequently, the cytoplasmic pH is the same as the extracellular pH. In the presence of 25 μm CCCP, the flux through the pathway increased significantly at pH 4, 5, and 6 but to different extents, and the optimal pH shifted from pH 4 to pH 5 (Fig. 4B, center panel). At neutral pH, no significant activity was detected, which is in line with the lack of activity of purified HdcA at this pH. The sharper pH profile in the presence of CCCP when the pathway is catalyzed by the transporter and decarboxylase in the absence of transmembrane gradients suggests that the activity at pH values flanking the optimum pH is limited by the activity of the transporter. Treatment of the cells with 0.2% Triton X-100, which permeabilizes the membrane and, thereby, bypasses the transport step and leaves the activity depending only on the decarboxylase, resulted in a pH profile that was similar to that of the purified enzyme (Fig. 4B, right panel).
The Histidine Decarboxylation Pathway Does Not Enhance Growth
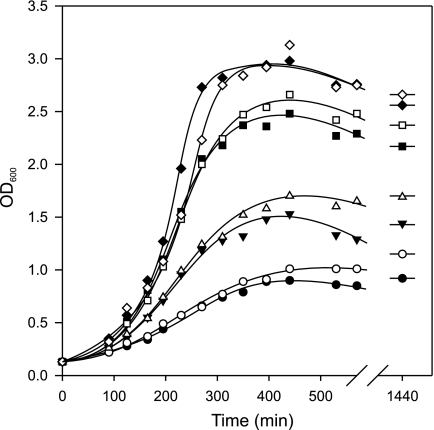
The histidine decarboxylation pathway generates metabolic energy in the form of proton motive force that may improve growth of the host organism. L. lactis expressing the HDC pathway was grown in rich GM17 medium (containing ∼86 μm histidine (33)) with or without addition of 10 mm histidine at four different initial pH values of the medium (Fig. 5). In the absence of added histidine, the growth rates and biomass yields increased in the order pH 5 < 5.5 < 6 < 6.5. The specific growth rate increased from 0.42 h−1 at pH 5 to 0.71 h−1 at pH 6.5. The presence of 10 mm added histidine did not significantly alter the growth rate in the exponential phase. However, upon entering the stationary phase, some increase in biomass was observed relative to the cultures without added histidine. For instance, overnight incubation of the cultures started at pH 5.5 with and without added histidine yielded optical densities of 1.7 and 1.4, respectively. A control growth experiment in medium at pH 5.5 with L. lactis cells producing a HDC pathway with an inactive decarboxylase (HdcA maturation mutant S82A (28), see below) showed that the additional histidine in the growth medium had no influence on growth whatsoever (not shown). The increased biomass production in the presence of histidine is most likely due to the alkalinizing effect of the decarboxylation reaction that counteracts the acidification resulting from glucose metabolism. The pH values of the cultures after overnight incubation were between 0.1 and 0.5 pH units lower in the cultures without added histidine. In conclusion, a functional histidine decarboxylation pathway does not enhance the growth rate of L. lactis in rich medium but results in slightly higher biomass production in batch culture.
FIGURE 5.
Growth of L. lactis NZ9000 pNZ-PhdcAPB in GM17 medium adjusted to pH 6.5 (♢ and ♦), 6.0 (□ and ■), 5.5 (△ and ▴), or 5.0 (○ and ●) in the presence (♢, □, △, and ○) or absence (♦, ■, ▴, and ●) of 10 mm histidine.
The Histidine Decarboxylation Pathway Improves Survival under Acid Stress
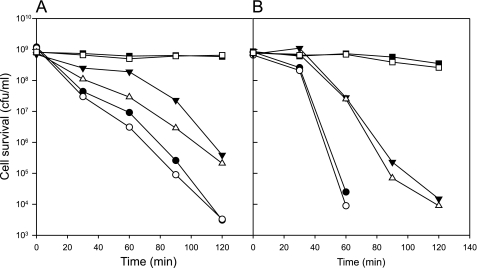
Acid stress survival of L. lactis was determined by incubation of the cells in GM17 medium set to a pH of 3 and at 30 °C. Under these conditions the cells do not grow. L. lactis expressing the HDC pathway from the native S. thermophilus HdcA promoter was grown to stationary phase in GM17 medium where the expression of the pathway was shown to be highest (28). The cells were washed and resuspended in the acidic GM17 medium either with or without addition of 10 mm histidine. After incubation for 2 h at 30 °C, aliquots of serial dilutions were spotted on GM17 agar plates to determine cell survival in the medium at pH 3 (Fig. 6A). Survival of cells expressing hdcAPB and incubated with 10 mm histidine was ∼100%, whereas the same cells incubated without histidine showed a survival of less than 0.5%. Compared with the control cells harboring the empty vector, the addition of histidine to the medium negatively affected the viability of the cells, resulting in 1–2 orders of magnitude less viable cells. A similar reduction of viability in the absence of added histidine was observed with cells containing the HDC pathway. The higher vulnerability of cells expressing the pathway by itself in acidic medium parallels the slightly lower growth rates in medium of normal pH relative to the control cells. Despite these effects, the cells expressing the HDC pathway revealed a remarkable higher viability in the presence of histidine, conditions that allow flux through the pathway. In fact, within the error of the experiment, the survival rate appeared to be 100%.
FIGURE 6.
Acid stress survival by histidine decarboxylation in L. lactis. A, L. lactis NZ9000 harboring pNZ-PhdcAPB (HDC+) or the empty vector pNZ8048 (HDC-) was grown in GM17 to stationary phase and washed and resuspended in GM17 adjusted to pH 3 with or without 25 mm histidine (His+ or His-, respectively). After 2-h incubation, 10 μl of droplets of serial dilutions were put on GM17 agar plates to determine colony-forming units (CFU)/ml. Dilution factors are indicated at the top. B, survival of L. lactis NZ9000 expressing the active (pNZ-hdcAPB, ■) or inactive pathway (pNZ-hdcA-S82S-PB, □) resuspended in GM17 containing 10 mm histidine adjusted to pH 3. At the indicated times, CFU/ml were determined as described above.
Mutation S82A in histidine decarboxylase HdcA of S. thermophilus results in the production of HdcA proenzyme that is not able to mature into active decarboxylase (28). Cells of L. lactis producing the pathways containing wild-type and mutant HdcA show identical growth rates, and the cytoplasmic levels of proenzyme and mature enzyme are comparable (28). Comparison of cells containing the wild-type and mutant pathways allows for a direct comparison of the viability in the presence and absence of flux through the decarboxylation pathway without the negative effects of histidine in the medium and the production of the pathway itself. As observed above, a time series demonstrated that L. lactis cells containing the active HDC pathway were 100% viable at pH 3 in the presence of histidine for at least 2 h (Fig. 6B). Under the same conditions, cells containing the inactive pathway were viable for the first 20 min, after which the viable cell count dropped dramatically by a factor of 105 after 1 h. In the case of the cells containing the active pathway, a similar reduction in viability was obtained after 2 h of incubation in medium at pH 2.5 (not shown). It follows that active turnover of the histidine decarboxylation pathway provides L. lactis with a remarkable higher survival rate at low pH.
Synergy between Glucose Metabolism and Histidine Decarboxylation in Acid Stress Survival
In GM17 medium set at a pH of 2.5 and containing 10 mm of histidine, the viable count of L. lactis containing the HDC pathway reduced by a factor of 10 about every 40 min (Fig. 7A). When, in addition to histidine, 10 mm of glucose was present during the incubation, the survival rate was significantly and reproducibly higher. The largest effect of over a factor of 10 in surviving cells was observed between 1 to 1.5 h of incubation, after which the difference diminished again. The effect of glucose could only be observed in a very narrow pH range. At pH 2.7, no difference was observed simply because the cells all survived, and, already at the slightly lower pH of 2.4, the protective effect of glucose was significantly smaller. Importantly, no significant effect of glucose was observed in the absence of an active decarboxylation pathway. The viability of cells containing the inactive HDC pathway (HdcA-S84A mutant, see above) incubated at pH values resulting in more or less the same death rate as observed with the active pathway was the same in the presence or absence of glucose (Fig. 7B). It follows that the additional protection provided by glucose depends on the activity of the HDC pathway.
FIGURE 7.
Acid stress survival in the presence of histidine and glucose. A, cells of L. lactis NZ9000 harboring pNZ-hdcAPB were incubated in M17 containing 10 mm histidine and adjusted to pH 2.7 (□ and ■), pH 2.5 (△ and ▴), or pH 2.4 (○ or ●) in presence (■, ▴, and ●) and absence (□, △, and ○) of 10 mm glucose. B, survival of L. lactis NZ9000 pNZ-hdcA-S82A-PB in M17 containing 10 mm histidine and adjusted to pH 3.5 (□ and ■), pH 3.25 (△ and ▴), and pH 3.0 (○ or ●) in the presence (■, ▴, and ●) or absence (□, △, and ○) of 10 mm glucose. Survival of the cells was determined at the indicated times as described in the legend for Fig. 4.
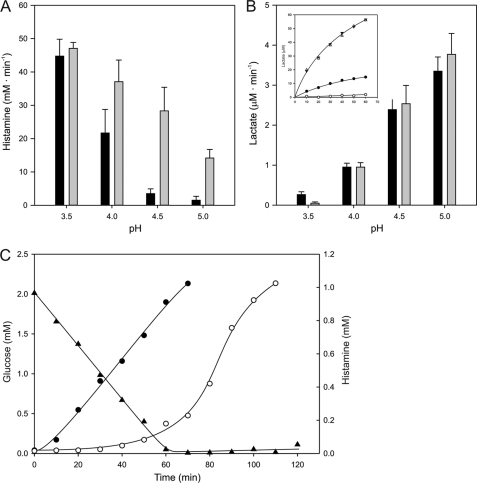
Interaction between the two pathways was demonstrated by measuring the fluxes through the decarboxylation and glycolytic pathways at different pH values in resting cells resuspended in 100 mm KPi/citrate buffers. A progressive inhibition by glucose of the histamine production rate catalyzed by cells expressing the HDC pathway was observed between external pH values of 3.5 and 5 (Fig. 8A). At pH 3.5, only a marginal level of inhibition was observed, whereas strong inhibition resulting in 56, 14, and 6% residual activities was observed at pH 4, 4.5, and 5, respectively (see also Fig. 1). The time course of the histamine production and glucose consumption at pH 5 demonstrated that production of the former was low until glucose was depleted, after which the production rate was similar to what was observed in the absence of glucose from the beginning (Fig. 8C). The complementary experiment showed that the glucose metabolism rate, measured as lactate production, was much less affected by the decarboxylation pathway. In the presence of histidine, the rate of glycolysis catalyzed by the cells containing the inactive HDC pathway dropped rapidly down to the background levels of the assay in the pH range of 5 to 3.5 (Fig. 8B). With cells containing the active HDC pathway, the rates were not significantly different at pH 4, 4.5, and 5. However, at pH 3.5, the rate was significant and at least one order of magnitude higher than observed with the inactive HDC pathway (Fig. 8B, inset). In conclusion, the glycolytic rate is stimulated at low pH by the decarboxylation pathway, but the absolute rate is low.
FIGURE 8.
Interaction between histidine and glucose metabolism. A, histamine production rates by resting cells of L. lactis NZ9000 pNZ-hdcAPB in 100 mm KPi/citrate buffer at the indicated pH and containing 5 mm histidine in the presence (black bars) or absence (gray bars) of 10 mm glucose. B, lactate production rates by resting cells of L. lactis NZ9000 harboring pNZ-hdcAPB (black bars) and pNZ-hdcA-S82A-PB (gray bars) in 100 mm KPi/citrate buffer at the indicated pHs and containing 10 mm glucose and 5 mm histidine. Histamine and lactate production rates were determined from the initial linear part of the time curves. Inset, time curves of lactate production at pH 3.5 (○ or ●) and pH 4.0 (△ and ▴) by cells harboring pNZ-hdcAPB (● and ▴) or pNZ-hdcA-S82A-PB (● and ▴). Error bars represent mean ± S.D. of three different experiments. C, histamine production (○ and ●) and glucose consumption (▴) by L. lactis NZ9000 pNZ-hdcAPB in 100 mm KPi (pH 5.0), 10 mm histidine, in the presence (○) or absence (●) of 2 mm glucose.
DISCUSSION
Degradative amino acid decarboxylation pathways are relatively simple pathways that operate autonomously, independent of the rest of cellular metabolism. The pathways have specific properties that add to the physiology of the host organism in several ways that may contribute to fitness under particular conditions. This explains why, at least in Gram-positives, the presence of the pathways are strain-specific and are acquired through horizontal gene transfer (14, 18). The physiological role of the newly acquired pathway will depend on the host organism and the conditions in the habitat. Possible functions of decarboxylation pathways are 1) production of metabolic energy that will contribute under energy-limiting conditions; 2) counteracting acidification by carbohydrate metabolism, which will postpone inhibition of growth at low pH in batch culture; 3) acid stress resistance, which will improve growth at low pH; and 4) acid stress survival, allowing the organism to survive under extreme acidic conditions. Once a new function is established in a host, the pathway may be optimized in terms of regulation of expression and enzyme activities. For instance, a main function under acid stress conditions may result in pH-controlled expression and optimal activity of the transporter and decarboxylase at acidic pH. Here, the functional expression of the histidine decarboxylation pathway of S. thermophilus CHCC1524, encoded by the hdcAPB operon, in L. lactis NZ9000 was described and the benefits of the pathway added to the physiology of the organism were investigated. The results clearly showed that the strongest benefit for the engineered L. lactis strain was improvement of acid stress survival. Cells expressing the active pathway were able to remain viable during at least 2 h of incubation in acidic medium set to pH 3, whereas less than 0.001% of the control cells survived after 1 h (Fig. 6B). In addition, a small effect was observed on the biomass yield in batch culture, whereas no growth rate enhancement was detected in the pH range 5–6.5, indicating that L. lactis is not energy-limited when growing in M17 under these conditions. The results emphasize that the physiological function of this type of pathway is host-specific. In the endogenous host S. thermophilus, the histidine decarboxylation pathway was suggested to function in pmf generation when conditions are energy-limited (16).
Survival of L. lactis in acidic milieus showed a very steep drop below a threshold pH value. Although incubation in GM17 medium of pH 3.5 hardly affected the viability of the cells, lowering of the pH by half a pH unit to pH 3 resulted in rapidly decreasing numbers of viable cells (Fig. 7B). The increased resistance when an active histidine decarboxylation pathway is operative does not seem to change this pattern but shifts the threshold pH down to more acidic values. Then, the cells can cope easily with an external pH of 3, but, again, run into trouble when the pH drops by half a unit (Fig. 7A). Therefore, the beneficial effect is limited to the pH range 3–3.5. Most likely, the protective effect will depend on the activity of the decarboxylation pathway. The HDC pathway of S. thermophilus is believed to be of relative low activity (16), and it would be interesting to explore the limits of acid survival of L. lactis by expressing more active decarboxylation pathways for histidine or any other amino acid. Strains of L. lactis or lactic acid bacteria in general with improved acid tolerance may be important in the field of probiotics when the cells have to pass the human gastrointestinal tract.
The histidine decarboxylation pathway generates proton motive force, and the experiments revealed a remarkable strong control over the flux through the pathway by the pmf. The highest rate was observed when the pmf was quenched by the presence of a protonophore, and the pathway runs without sensing a counter force (Fig. 1A). A 4-fold lower rate was observed when the pmf generated by the pathway itself was allowed to develop. In the presence of a second, more powerful pmf generating system (glucose), the pathway was almost completely inhibited. Apparently, the pmf generated by the decarboxylation pathway itself is of average magnitude, suggesting that the pathway will only be functional at low pmf, i.e. energy-limited conditions. The mechanism by which the pmf exerts its control over the pathway is complex, and both membrane potential and pH gradient play a role (Fig. 1B). Control by the membrane potential is limited to the histidine/histamine exchanger and will slow down an electrogenic step in the transporter cycle (Fig. 3). In a buffer of defined pH, the pH gradient will affect the cytoplasmic pH, which may affect the activity of both the transporter (Fig. 3) and the decarboxylase. The pH profile of the decarboxylase activity shows that the enzyme is strongly inhibited at pH values below 3.5 and above 6 (Figs. 4 and 8A). L. lactis has been reported to maintain an internal pH of 7 at an external pH of 5 in the presence of glucose (21) which largely explains the low flux through the pathway in the presence of glucose (Fig. 1A) and the stimulation of the rate at pH values of 5 and 6 when the pH gradient was dissipated in the presence of CCCP (Fig. 4B). The control of the transporter on the flux appears to be smaller as a function of cytoplasmic pH but is still apparent at pH 4 and 6. It follows that as may be expected from an evolutionary point of view, control of the pmf on the flux is distributed over the complete pathway, i.e. over both the transporter and the decarboxylase.
The protective effect of the HDC pathway at acidic conditions is due to an increase of the cytoplasmic pH, which prevents denaturing of cytoplasmic enzymes. At the same time, the alkalinizing effect of the pathway may relieve the acidic condition, but this depends a lot on cell density. The same protective effect may be obtained from glycolytic activity when the cytoplasmic pH is raised through proton pumping by F1F0-ATPase driven by ATP generated in glycolysis. The downside would be the acidifying effect of the pathway, which will make things in the environment only worse. Glycolysis is the main metabolic pathway during normal growth in fermentative bacteria, and it is likely that glycolytic activity will drop rapidly with decreasing pH. Nevertheless, the properties of both pathways allow for a synergistic effect in survival at low pH that would result in survival during extended periods of time. At low pH and in the presence of both histidine and glucose, histidine decarboxylation activity would increase the cytoplasmic pH to values where glycolysis becomes active. As a consequence the pmf generated by ATP hydrolysis would slow down the flux through the decarboxylation pathway. The feedback loop results in the preferential use of glucose over histidine in the initial stages of survival. Once the system runs out of glucose, histidine decarboxylation would take over again. The experiments support the feasibility of such a survival mechanism. In the absence of an active HDC pathway, the presence of glucose did not result in a higher survival rate in medium at low pH, most likely because of the lack of activity through the glycolytic pathway under these conditions (Fig. 7B). With the active HDC pathway, the synergistic effect of histidine and glucose was observed but only in a narrow pH range. Between pH 2.4 and 3, an improved survival rate of up to 1 order of magnitude was observed in the presence of glucose (Fig. 7A). Experiments in buffer clearly showed the inhibitory effect of glycolysis on the decarboxylation pathway and the stimulating effect of HDC activity on glycolysis, but the latter effect was only observed at low pH (Fig. 8, A and B). The ordered consumption of glucose and histidine was apparent (Fig. 8C). It is not clear if the observed synergistic mechanism is of physiological importance in wild-type strains. Clearly, optimal performance of the system would require a fine-tuning of the pH profiles of the two pathways by evolution. Such optimization has not taken place in the engineered L. lactis strain used in these studies.
This work was supported by European Community Seventh Framework Programme Grant Agreement No. 211441-BIAMFOOD and by The Netherlands Organization for Scientific Research (NWO-ALW).
- pmf
- proton motive force
- HDC
- histidine decarboxylation
- CCCP
- carbonyl cyanide m-chlorophenylhydrazone.
REFERENCES
- 1. Molenaar D., Bosscher J. S., ten Brink B., Driessen A. J., Konings W. N. (1993) Generation of a proton motive force by histidine decarboxylation and electrogenic histidine/histamine antiport in Lactobacillus buchneri. J. Bacteriol. 175, 2864–2870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abe K., Hayashi H., Maloney P. C. (1996) Exchange of aspartate and alanine. Mechanism for development of a proton-motive force in bacteria. J. Biol. Chem. 271, 3079–3084 [DOI] [PubMed] [Google Scholar]
- 3. Wolken W. A., Lucas P. M., Lonvaud-Funel A., Lolkema J. S. (2006) The mechanism of the tyrosine transporter TyrP supports a proton motive tyrosine decarboxylation pathway in Lactobacillus brevis. J. Bacteriol. 188, 2198–2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pereira C. I., San Romão M. V., Lolkema J. S., Crespo M. T. (2009) Weissella halotolerans W22 combines arginine deiminase and ornithine decarboxylation pathways and converts arginine to putrescine. J. Appl. Microbiol. 107, 1894–1902 [DOI] [PubMed] [Google Scholar]
- 5. Soksawatmaekhin W., Kuraishi A., Sakata K., Kashiwagi K., Igarashi K. (2004) Excretion and uptake of cadaverine by CadB and its physiological functions in Escherichia coli. Mol. Microbiol. 51, 1401–1412 [DOI] [PubMed] [Google Scholar]
- 6. Gong S., Richard H., Foster J. W. (2003) YjdE (AdiC) is the arginine:agmatine antiporter essential for arginine-dependent acid resistance in Escherichia coli. J. Bacteriol. 185, 4402–4409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhao B., Houry W. A. (2010) Acid stress response in enteropathogenic γ proteobacteria. An aptitude for survival. Biochem. Cell Biol. 88, 301–314 [DOI] [PubMed] [Google Scholar]
- 8. Tanaka Y., Kimura B., Takahashi H., Watanabe T., Obata H., Kai A., Morozumi S., Fujii T. (2008) Lysine decarboxylase of Vibrio parahaemolyticus. Kinetics of transcription and role in acid resistance. J. Appl. Microbiol. 104, 1283–1293 [DOI] [PubMed] [Google Scholar]
- 9. Kieboom J., Abee T. (2006) Arginine-dependent acid resistance in Salmonella enterica serovar Typhimurium. J. Bacteriol. 188, 5650–5653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Giles T. N., Graham D. E. (2007) Characterization of an acid-dependent arginine decarboxylase enzyme from Chlamydophila pneumoniae. J. Bacteriol. 189, 7376–7383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Spano G., Russo P., Lonvaud-Funel A., Lucas P., Alexandre H., Grandvalet C., Coton E., Coton M., Barnavon L., Bach B., Rattray F., Bunte A., Magni C., Ladero V., Alvarez M., Fernández M., Lopez P., de Palencia P. F., Corbi A., Trip H., Lolkema J. S. (2010) Biogenic amines in fermented foods. Eur. J. Clin. Nutr. 64, S95–100 [DOI] [PubMed] [Google Scholar]
- 12. Silla Santos M. H. (1996) Biogenic amines. Their importance in foods. Int. J. Food Microbiol. 29, 213–231T [DOI] [PubMed] [Google Scholar]
- 13. Fernández M., Zúñiga M. (2006) Amino acid catabolic pathways of lactic acid bacteria. Crit Rev. Microbiol. 32, 155–183 [DOI] [PubMed] [Google Scholar]
- 14. Lucas P. M., Wolken W. A., Claisse O., Lolkema J. S., Lonvaud-Funel A. (2005) Histamine-producing pathway encoded on an unstable plasmid in Lactobacillus hilgardii 0006. Appl. Environ. Microbiol. 71, 1417–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Martín M. C., Fernández M., Linares D. M., Alvarez M. A. (2005) Sequencing, characterization and transcriptional analysis of the histidine decarboxylase operon of Lactobacillus buchneri. Microbiology 151, 1219–1228 [DOI] [PubMed] [Google Scholar]
- 16. Calles-Enríquez M., Eriksen B. H., Andersen P. S., Rattray F. P., Johansen A. H., Fernández M., Ladero V., Alvarez M. A. (2010) Sequencing and transcriptional analysis of the Streptococcus thermophilus histamine biosynthesis gene cluster. Factors that affect differential hdcA expression. Appl. Environ. Microbiol. 76, 6231–6238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pereira C. I., Matos D., San Romão M. V., Crespo M. T. (2009) Dual role for the tyrosine decarboxylation pathway in Enterococcus faecium E17. Response to an acid challenge and generation of a proton motive force. Appl. Environ. Microbiol. 75, 345–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lucas P. M., Blancato V. S., Claisse O., Magni C., Lolkema J. S., Lonvaud-Funel A. (2007) Agmatine deiminase pathway genes in Lactobacillus brevis are linked to the tyrosine decarboxylation operon in a putative acid resistance locus. Microbiology 153, 2221–2230 [DOI] [PubMed] [Google Scholar]
- 19. Coton M., Fernández M., Trip H., Ladero V., Mulder N. L., Lolkema J. S., Alvarez M. A., Coton E. (2011) Characterization of the tyramine-producing pathway in Sporolactobacillus sp. P3J. Microbiology 157, 1841–1849 [DOI] [PubMed] [Google Scholar]
- 20. Coton E., Mulder N., Coton M., Pochet S., Trip H., Lolkema J. S. (2010) Origin of the putrescine-producing ability of the coagulase-negative bacterium Staphylococcus epidermidis 2015B. Appl. Environ. Microbiol. 76, 5570–5576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Poolman B., Driessen A. J., Konings W. N. (1987) Regulation of solute transport in streptococci by external and internal pH values. Microbiol. Rev. 51, 498–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guirard B. M., Snell E. E. (1980) Purification and properties of ornithine decarboxylase from Lactobacillus sp. 30a. J. Biol. Chem. 255, 5960–5964 [PubMed] [Google Scholar]
- 23. Cotter P. D., Gahan C. G., Hill C. (2001) A glutamate decarboxylase system protects Listeria monocytogenes in gastric fluid. Mol. Microbiol. 40, 465–475 [DOI] [PubMed] [Google Scholar]
- 24. Sanders J. W., Leenhouts K., Burghoorn J., Brands J. R., Venema G., Kok J. (1998) A chloride-inducible acid resistance mechanism in Lactococcus lactis and its regulation. Mol. Microbiol. 27, 299–310 [DOI] [PubMed] [Google Scholar]
- 25. Abe K., Ohnishi F., Yagi K., Nakajima T., Higuchi T., Sano M., Machida M., Sarker R. I., Maloney P. C. (2002) Plasmid-encoded asp operon confers a proton motive metabolic cycle catalyzed by an aspartate-alanine exchange reaction. J. Bacteriol. 184, 2906–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rossi F., Gardini F., Rizzotti L., La, Gioia F., Tabanelli G., Torriani S. (2011) Quantitative analysis of histidine decarboxylase gene (hdcA) transcription and histamine production by Streptococcus thermophilus PRI60 under conditions relevant to cheese making. Appl. Environ. Microbiol. 77, 2817–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. van Poelje P. D., Snell E. E. (1990) Pyruvoyl-dependent enzymes. Annu. Rev. Biochem. 59, 29–59 [DOI] [PubMed] [Google Scholar]
- 28. Trip H., Mulder N. L., Rattray F. P., Lolkema J. S. (2011) HdcB, a novel enzyme catalysing maturation of pyruvoyl-dependent histidine decarboxylase. Mol. Microbiol. 79, 861–871 [DOI] [PubMed] [Google Scholar]
- 29. de Ruyter P. G., Kuipers O. P., de Vos W. M. (1996) Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Environ. Microbiol. 62, 3662–3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Molenaar D., Abee T., Konings W. N. (1991) Continuous measurement of the cytoplasmic pH in Lactococcus lactis with a fluorescent pH indicator. Biochim. Biophys. Acta 1115, 75–83 [DOI] [PubMed] [Google Scholar]
- 31. Síp M., Herman P., Plásek J., Hrouda V. (1990) Transmembrane potential measurement with carbocyanine dye diS-C3-(5). Fast fluorescence decay studies. J. Photochem. Photobiol. B Biol. 4, 321–328 [DOI] [PubMed] [Google Scholar]
- 32. Matsumoto T., Furuta T., Nimura Y., Suzuki O. (1984) 3-(p-hydroxyphenyl)propionic acid as a new fluorogenic reagent for amine oxidase assays. Anal. Biochem. 138, 133–136 [DOI] [PubMed] [Google Scholar]
- 33. Guimont C. (2002) Change of free amino acids in M17 medium after growth of Streptococcus thermophilus and identification of a glutamine transport ATP-binding protein. Int. Dairy J. 12, 729–736 [Google Scholar]