Background: Calsequestrin is a calcium storage/buffer protein within the sarcoplasmic reticulum and binds large amounts of Ca2+ in a unique manner.
Results: The specific coordination, geometry, and cooperative effects of Ca2+ binding were determined.
Conclusion: The oligomeric state of calsequestrin is directly related to high-capacity Ca2+ binding.
Significance: This is the first report of specific Ca2+ coordination sites in calsequestrin and provides a pathological link to related disorders.
Keywords: Calcium-binding Proteins, Protein-Metal Ion Interaction, Sarcoplasmic Reticulum (SR), Skeletal Muscle, X-ray Crystallography
Abstract
Calsequestrin, the major calcium storage protein in both cardiac and skeletal muscle, binds large amounts of Ca2+ in the sarcoplasmic reticulum and releases them during muscle contraction. For the first time, the crystal structures of Ca2+ complexes for both human (hCASQ1) and rabbit (rCASQ1) skeletal calsequestrin were determined, clearly defining their Ca2+ sequestration capabilities through resolution of high- and low-affinity Ca2+-binding sites. rCASQ1 crystallized in low CaCl2 buffer reveals three high-affinity Ca2+ sites with trigonal bipyramidal, octahedral, and pentagonal bipyramidal coordination geometries, along with three low-affinity Ca2+ sites. hCASQ1 crystallized in high CaCl2 shows 15 Ca2+ ions, including the six Ca2+ ions in rCASQ1. Most of the low-affinity sites, some of which are μ-carboxylate-bridged, are established by the rotation of dimer interfaces, indicating cooperative Ca2+ binding that is consistent with our atomic absorption spectroscopic data. On the basis of these findings, we propose a mechanism for the observed in vitro and in vivo dynamic high-capacity and low-affinity Ca2+-binding activity of calsequestrin.
Introduction
Ca2+ is stored in the endoplasmic reticulum and sarcoplasmic reticulum (SR),3 from where it is released in transient Ca2+ surges for various signaling purposes (1, 2). In the case of the SR, release of Ca2+ stores stimulates muscle contraction, with Ca2+ reuptake returning the actin-myosin fibrils to a relaxed state.
The most abundant protein among the acidic SR resident proteins, present at concentrations of up to 100 mg/ml, is the ∼44-kDa Ca2+-binding protein calsequestrin (CASQ). CASQ was suggested to act as a Ca2+ buffer inside the SR by lowering free luminal Ca2+ concentrations to facilitate further uptake of Ca2+ by the Ca2+-ATPase (sarco/endoplasmic reticulum Ca2+-ATPase), probably in concert with other acidic SR luminal proteins (1–3). CASQ binds Ca2+ with high capacity (40–50 mol of Ca2+/mol of CASQ) but with moderate affinity (Kd ∼ 103 m−1) over a Ca2+ concentration range of 0.01–1 m, which stands in marked contrast from the lower 0.1–2 μm cytosolic Ca2+ concentration range handled by the EF-hand proteins (4). This unique Ca2+-buffering range allows CASQ to act as the underlying Ca2+-binding/storage mechanism responsible for the large storage capacity and rapid release of Ca2+ from the SR. In addition, CASQ was reported to actively participate in the Ca2+ release process by localizing Ca2+ at the release channel, sensing the SR Ca2+ concentration, and regulating the amount of Ca2+ released (5). A network of proteins in the junctional face of the SR, such as junctin and triadin, is implicated in regulating the interaction between CASQ and the ryanodine receptor, although details of this association remain uncertain (6).
The two major isoforms of CASQ, skeletal muscle calsequestrin (CASQ1) and cardiac muscle calsequestrin (CASQ2), are encoded by the CASQ1 and CASQ2 genes located on chromosomes 1q21 and 1p13.3-p11, respectively, in humans. CASQ isoforms are also present in the endoplasmic reticulum vacuolar domains of some neurons and smooth muscles (1, 7, 8). In general, CASQ isoforms exhibit 60–70% sequence identity and, consistent with their high Ca2+-binding capacity, contain a large proportion of aspartate and glutamate, with 28.5% of the primary sequence of human CASQ1 (hCASQ1), for example, being composed of aspartate and glutamate, the majority of which are located in the C-terminal region (Fig. 1). The high-capacity Ca2+ binding by CASQ that accompanies its polymerization likely occurs through cross-bridging of clustered acidic residues by Ca2+, establishing on-rates close to the diffusion limit (kon ∼ 109 m−1 s−1) and high off-rates (koff ∼ 106 s−1), resulting in a moderate-affinity binding constant of ∼103 m−1. However, to date, the exact mechanism of Ca2+ coordination by CASQ and its structural impacts have not been elucidated. In this work, we identified, for the first time, bound Ca2+ ions and their exact coordination geometries in both hCASQ1 and rabbit CASQ1 (rCASQ1). We also present structural evidence that cooperative coordination of Ca2+ between CASQ monomers mediates their oligomerization.
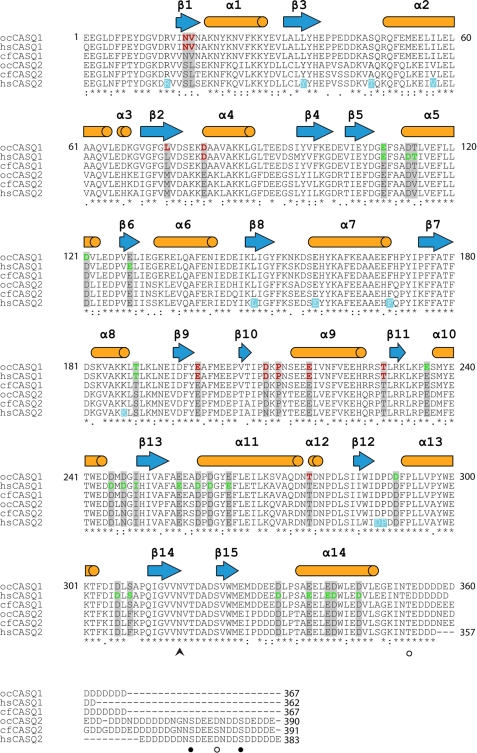
FIGURE 1.
Amino acid sequence alignment of mature CASQ isoforms from rabbit (Oryctolagus cuniculus (oc)), human (Homo sapiens (hs)), and canine (Canis familiaris (cf)). The amino acid residues participating in Ca2+ coordination are highlighted in red (high affinity) and green (low affinity). Ten mutation sites in human CASQ2 implicated in causing CPVT are highlighted in light blue. The secondary structural elements are indicated with blue arrows on top of the corresponding sequences. The confirmed post-translational modification sites are shown as circles (phosphorylation) or the arrowhead (glycosylation). Bond distances and specific oxo-ligands are provided in Supplemental Tables 1 and 2.
EXPERIMENTAL PROCEDURES
Preparation of Recombinant hCASQ1 and rCASQ1
The open reading frames for both CASQ1 genes without the N-terminal signal peptide (GenBankTM accession numbers EAW52736.1 (human) and M15747.1 (rabbit)) were commercially synthesized (GenScript) and inserted into pET28a using NcoI and XhoI sites. Each plasmid was then transformed into Escherichia coli DH5α cells. After sequence confirmation, the plasmid was transformed into Rosetta BL21(DE3) E. coli. Purification of both rCASQ1 and hCASQ1 was performed following the same procedures as described previously (9).
Crystallization and Structure Determination of Recombinant hCASQ1 and rCASQ1
Both recombinant hCASQ1 and rCASQ1 at an initial concentration of 10 mg/ml in 20 mm HEPES (pH 7.0) and 0.5 m NaCl were crystallized at 4 °C by the vapor diffusion method using 2-methyl-2,4-pentanediol as the precipitating agent. The final reagent concentrations under the hCASQ1 crystallization conditions were 50 mm BisTris propane (pH 5.5), 50 mm CaCl2, and 23% 2-methyl-2,4-pentanediol. The final reagent concentrations under the rCASQ1 crystallization conditions were 50 mm HEPES (pH 7.0), 0.1 m NaCl, 2 mm CaCl2 or SrCl2, and 14% 2-methyl-2,4-pentanediol. The crystals of hCASQ1 and rCASQ1 belonged to the P1 and C2221 space groups, respectively. The diffraction data were collected to resolutions of 2.0 Å for hCASQ1 and 1.9 Å for rCASQ1.
Intensity data were collected at the Advanced Light Source (beamlines BL8.2.1 and BL8.2.2) for both hCASQ1 and rCASQ1 crystals using 1.0-Å wavelength radiation. All data sets were collected at 93 K. The 180º rotation data were collected at the absorption edge of Sr2+ for the maximum Friedel pairs. All data were reduced and scaled using HKL2000 (10). Iterative model adjustment and refinement were completed using the programs Coot (11) and PHENIX (12) with the coordinates of rCASQ1 (Protein Data Bank code 1A8Y). From the Ramachandran plot generated by PHENIX, the final structures were found to have good stereochemistry, with 97.8% (rCASQ1) and 98.0% (hCASQ1) of the residues in the most favored region and 100% (rCASQ1) and 99.7% (hCASQ1) of the residues in the allowed region. The coordinates and diffraction data (3TRP (rCASQ1) and 3UOM (hCASQ1)) have been deposited in the Protein Data Bank. Bond distances and contributing oxo-ligands for Ca2+ coordination are provided in supplemental Table 1 for hCASQ1 and supplemental Table 2 for rCASQ1.
Equilibrium Dialysis/Atomic Absorption Spectrophotometry
To estimate the fractional occupancy (y = [bound Ca2+]/[total hCASQ1]) for recombinant CASQ1, equilibrium dialysis and atomic absorption spectrophotometry were performed as described previously (13). Data fitting methods are provided under supplemental “Methods”.
Molecular Mass Determination by Multiangle Light Scattering
Chromatography and light scattering experiments were performed as described previously (13).
RESULTS
Structure of Human Skeletal Calsequestrin
We crystallized hCASQ1 and rCASQ1 (Table 1) in buffers containing 50 and 2 mm CaCl2, respectively. Although the structures of several CASQ isoforms have been determined (9, 14–16), this is the first report of its Ca2+ complex form. The previously established rCASQ1 crystallization conditions contained a substantial amount of the Ca2+-chelating citrate ion, which made co-crystallization of CASQ with Ca2+ difficult. In addition, under those reported crystallization conditions (9, 14, 15), crystals shattered upon exposure to Ca2+ ions, which disabled a Ca2+ diffusion-based approach. The Ca2+-complexed hCASQ1 and rCASQ1 structures show a significantly increased resolution (2.0 and 1.9 Å, respectively) and resolve previously unresolved areas encompassing residues 327–332 and additional residues at N and C termini. Nevertheless, the electron density for the C-terminal residues 354EDDDDDDDD362 in hCASQ1 and 354EDDDDDEDDDDDDDD368 in rCASQ1 could not be resolved, indicating their disordered nature even at those Ca2+ concentrations. As expected from the high level of sequence similarity (99%) and identity (96%) between these two isoforms, the structure of monomeric hCASQ1 is very similar to that of rCASQ1, with an average root mean square deviation of 1.01 Å (Fig. 2). However, there are noticeable differences in four areas (Fig. 2), and the cause of these differences is likely due to Ca2+ coordination compacting an overall conformation. In addition, the Cα atoms of the previous apo-rCASQ1 (Protein Data Bank code 1A8Y) (9, 14, 15) and the new Ca2+-complexed rCASQ1 are superimposable, with a root mean square deviation of 0.16 Å.
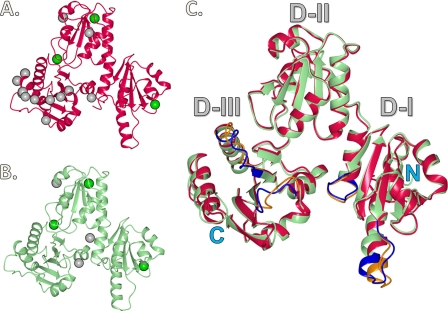
FIGURE 2.
Ribbon diagram representing crystal structures of rCASQ1 and hCASQ1 with identified Ca2+ sites. A, hCASQ1 monomer with high-affinity Ca2+ (green) and low-affinity Ca2+ (gray) sites. B, rCASQ1 monomer with high-affinity Ca2+ (gray) and low-affinity Ca2+ (green) sites. C, superimposed views of hCASQ1 and rCASQ1 with N and C termini indicated. Four major differences are highlighted in blue (rCASQ1) and orange (hCASQ1). D-I, D-II, and D-III, Domains I–III, respectively. These figures were generated using CCP4MG Version 2.5.0.
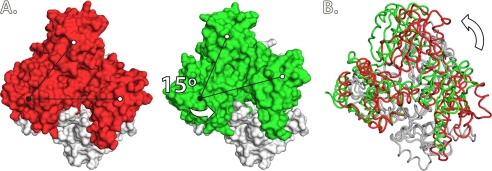
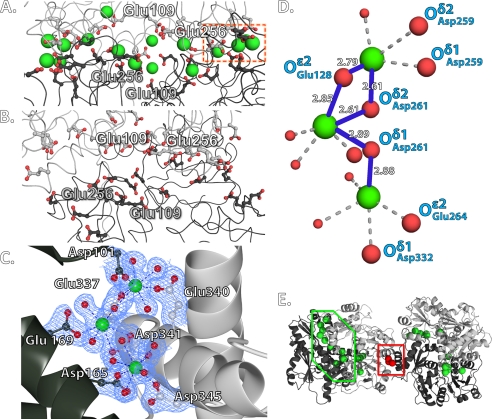
The structures of both hCASQ1 and rCASQ1 are made up of three nearly identical thioredoxin-fold domains (Domains I–III) (Fig. 2). Each of these thioredoxin-like domains is made up of a five-stranded central β-sheet with two α-helices on both sides and has a hydrophobic core with acidic residues on the exterior (Fig. 2). Like other apo-CASQs, the Ca2+-complexed structures of both hCASQ1 and rCASQ1 molecules show a dimer formed by monomers exchanging their extended N-terminal ends. The N-terminal residues of one CASQ1 binds along a groove between two β-strands of Domain II of its dimeric partner. In both apo-rCASQ1 (Protein Data Bank code 1A8Y) and apo-rCASQ1 crystallized in 2 mm CaCl2, the orientation of the two monomers in the front-to-front interface is superimposable. However, the observed orientation of the two monomers in the same interface of hCASQ1 is clearly different from that of both rCASQ1 structures. As shown in Fig. 3, the orientation between two hCASQ1 monomers in the front-to-front dimer interface is rotated ∼15º relative to that of rCASQ1. This conformational change results in bringing multiple acidic residues of each hCASQ1 monomer into close contacts and allowing coordination of multiple Ca2+ ions (Fig. 4, A and B). A hinge of this conformational change is located at the region of hydrophobic interaction among three amphiphilic α-helices: α2 and α5 from one monomer and α13 from the other monomer. The hydrophobic sides of all three helices are clustered together between two exchanged N-terminal arms serving as a pivot (Fig. 3). At the other side of the front-to-front interface, there exists a second type of intermolecular interaction among dimers, forming a back-to-back interface (Figs. 4E and 5A), which is also substantially different from that observed in rCASQ1 (Fig. 5, B and C). The electronegative surfaces of two hCASQ1 dimers are brought together in a back-to-back dimer (Fig. 4, C and E), in which the acidic residues in Domain III of one molecule (Glu-337, Glu-340, and Asp-341) coordinate three Ca2+ ions together with the other acidic residues in both Domain II (Asp-165 and Glu-169) and Domain I (Asp-101) of the neighboring hCASQ1 molecule in this interface. The extension of the arm exchange between monomers is significantly weakened due to this new conformation in the back-to-back interaction that prevents the specific interaction between the tip of the N-terminal peptide and the C-terminal end of the other CASQ molecule (9, 14, 15).
FIGURE 3.
A, relative rotation motion between two monomers in the front-to-front interface of rCASQ1 (left) and hCASQ1 (right). One pivot (black circle) and two reference points (white circles) are indicated for visual aid. B, superimposed views of hCASQ1 (green) and rCASQ1 (red). These figures were generated using Open-Source PyMOLTM Version 1.4.
FIGURE 4.
Network of Ca2+ ions in dimer interfaces. A, observed Ca2+ ions in the front-to-front interface of hCASQ1. The Ca2+ ions are depicted as green spheres, and coordinating carboxylate oxygen atoms are depicted as red spheres. The acidic residues from each hCASQ1 monomer are shown in black and gray. B, the corresponding acidic residues of rCASQ1 at the same front-to-front interface as in A are not in close proximity and there is no Ca2+ ion. C, observed Ca2+ ions in the back-to-back interface of hCASQ1. Dashed blue lines indicate the Ca2+ coordination. The electron density maps are contoured at 1.5σ. Contributing residues from a dimeric partner are shown in dark green. D, blowup view of the area depicted by the dashed orange box in A showing Ca2+ coordination with μ-oxo-like bonds. The oxygen atoms of the acidic side chains and water are shown as large and small red balls, respectively. E, global view of the front-to-front (green box) and back-to-back (red box) interfaces in four hCASQ1 monomers. The same Ca2+ ions in A and C are depicted as green and red spheres, respectively. These figures were generated using CCP4MG Version 2.5.0.
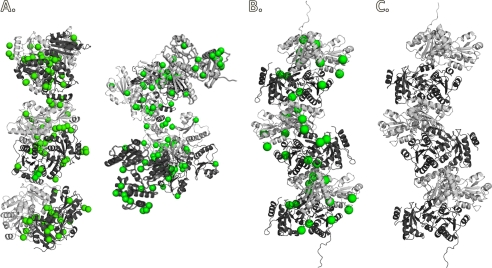
FIGURE 5.
Observed oligomeric interactions in CASQ1. CASQ1 is shown in gray and black ribbon representation, with the observed Ca2+ ions shown as green spheres. A, the full hexamer of the P1 unit cell content (asymmetric unit) of hCASQ1 crystallized in 50 mm CaCl2. Left, choice of linearly arranged hexamer; right, choice of hexamer in a branched arrangement. B, the hexamer of rCASQ1 crystallized in 2 mm CaCl2. C, the hexamer of rCASQ1 crystallized in 0 mm CaCl2. These figures were generated using Open-Source PyMOLTM Version 1.4.
Ca2+ Coordination in Calsequestrin
Six Ca2+ ions are located in rCASQ1 crystallized in the presence of 2 mm CaCl2 (Fig. 2). The corresponding electron densities for all except one are confirmed by the anomalous difference map of the Sr2+-complexed rCASQ1 diffraction data. Approximately 15 Ca2+ ions, including all six Ca2+ ions of rCASQ1, are found per hCASQ1 molecule (Figs. 2 and 5). Most of these Ca2+ ions are located in the front-to-front dimer (Fig. 4A) and back-to-back interfaces (Fig. 4C).
Ca2+-binding Capacity Assay
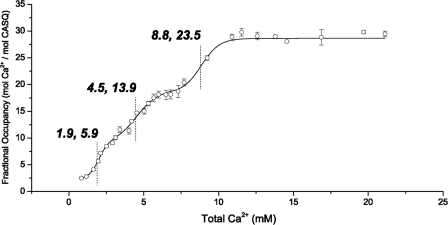
To study the Ca2+-binding capacity of hCASQ1, atomic absorption spectroscopy was performed under the same buffer conditions as used for multiangle light scattering (MALS). As shown in Fig. 6, hCASQ1 shows a multiphasic Ca2+-binding curve, which is indicative of the stepwise formation of higher order polymeric structures of hCASQ1 as Ca2+ concentration increases. Consistent with MALS results (Fig. 7), hCASQ1 begins its initial transition at 1 mm Ca2+ and reaches its half-maximal value at ∼1.9 mm Ca2+. The next transition begins at ∼3.0 mm and reaches its half-maximal point at ∼4.5 mm Ca2+, obtaining a fractional occupancy of ∼14. The final transition begins at 7.5 mm Ca2+ and reaches the transition point at ∼9 mm Ca2+, obtaining an occupancy of 23.5. Beyond this transition range of Ca2+ concentration, the occupancy value reaches the maximum value of ∼27. The initial transition is the most responsive to the changes in [Ca2+], with an approximate slope of 1.32 ± 0.59. The second and third transitions are less responsive to [Ca2+], with slopes of 0.61 ± 0.18 and 0.74 ± 0.17, respectively.
FIGURE 6.
Ca2+-binding capacity of hCASQ1 determined by atomic absorption spectroscopy. The number of Ca2+ ions bound to hCASQ1 was determined by equilibrium dialysis and atomic absorption spectroscopy. Fractional occupancy (y = [bound Ca2+]/[total protein]) was plotted against total [Ca2+]. Half-maximal points are indicated as (total [Ca2+], fractional occupancy).
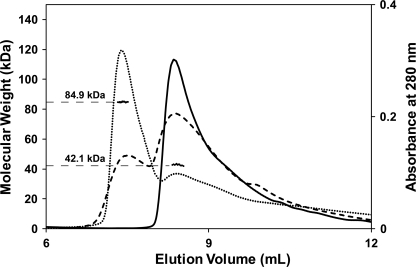
FIGURE 7.
MALS elution profiles of hCASQ1 with and without Ca2+. Elution profiles are shown as molecular mass (kDa) versus elution volume. The solid line indicates the elution profile in the absence of CaCl2, and the dashed line represents the elution profile in the presence of 1 mm CaCl2. The dotted line represents the elution profile in 2 mm CaCl2. The molecular masses as determined by static light scattering are shown as dots, which were extended to the left axis for ease of interpretation. In addition, the average molecular masses for each major peak are indicated.
MALS
Recombinant hCASQ1 was studied by MALS to determine the effect of Ca2+ in forming oligomers. The results clearly indicate the monomeric nature of hCASQ1 in solution containing 300 mm KCl without any Ca2+ (Fig. 7, solid line). In the presence of 1 mm Ca2+, recombinant hCASQ1 was predominantly in a monomeric state, with ∼25% in a dimeric state (Fig. 7, dotted line). However, in 2 mm Ca2+, most of the population became dimeric, consistent with the transitory nature of this range of Ca2+ concentrations.
DISCUSSION
Ca2+ ions found in rCASQ1 and hCASQ1 can be divided into two classes: high and low affinity. Three high-affinity and three low-affinity Ca2+ ions are in rCASQ1, and three high-affinity and 12 low-affinity Ca2+ ions are in each hCASQ1 monomer (Fig. 2). Most of the coordinating residues of both the high-affinity (red) and low-affinity (green) sites are conserved in CASQ1 and CASQ2 among humans, canines, and rabbits (Figs. 1 and 2), making it plausible that these Ca2+-binding sites exist in all of those CASQs, as well. As shown in Fig. 6, the fractional Ca2+ occupancy of hCASQ1 at 1.9 mm is 5.9, which matches the number of Ca2+ ions found in rCASQ1 crystallized in the presence of 2 mm CaCl2. These three high-affinity sites are already filled at the beginning of the curve (Fig. 6). Although no Ca2+ was added to the crystallization buffer for the previous structure of apo-rCASQ1 (Protein Data Bank code 1A8Y) (9, 14, 15), it probably contained high-affinity Ca2+ ions, as they appear to be critical for proper protein folding and thus should have been co-purified bound to the protein (4). The density for these Ca2+ ions might have been overlooked simply due to their marginal resolution.
Low-affinity Ca2+
The low-affinity sites are designated as such based on coordination typically involving acidic residues on the surface of CASQ1 molecules together with water molecules or side chain hydroxyl groups (Fig. 8, G–I). In addition, there are acidic residues forming μ-carboxylate-like bridges between Ca2+ ions (Fig. 4D) as observed in Ca2+-complexed prothrombin fragment-1 (18), however with a significant variation in the mono-μ-oxo bond distance.
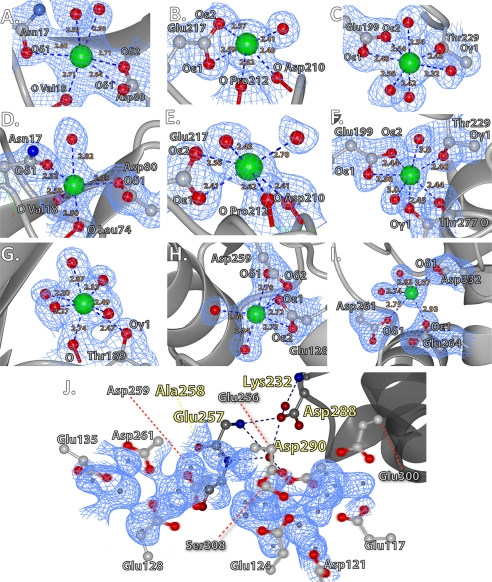
FIGURE 8.
Ca2+ coordination observed in hCASQ1 and rCASQ1. A–C, high-affinity sites from hCASQ1. D–F, the same high-affinity sites found in rCASQ1. G–I, low-affinity sites. J, diffused electron density surrounded by the acidic residues in the front-to-front interface. The hydrogen-bonding network involving Asp-288 is indicated by dashed blue lines, with helix α13 shown in dark gray. These figures were generated using CCP4MG Version 2.5.0. The individual electron density maps were contoured at 1.5σ.
Among the ∼70 low-affinity Ca2+ ions found in the P1 unit cell of hCASQ1 (Fig. 5A), most are located in the front-to-front (Fig. 4A) and back-to-back (Fig. 4B) interfaces, coordinated by interdigitating carboxylate groups from two monomers. The corresponding acidic residues of rCASQ1 and canine CASQ2 (Protein Data Bank code 1SJI) have higher temperature factors or adopt dual conformations of their side chain dihedral angles, reflecting the intrinsic flexibility when in the absence of Ca2+. As mentioned previously, coordination of these interfacial Ca2+ ions requires a rotation between the two monomers to bring the acidic residues of each into close proximity (Fig. 4, A and B). The same rotation could have been the cause of cracking during our unsuccessful attempts at Ca2+ soaking. In addition, the multiphasic nature of the fractional Ca2+ occupancy of hCASQ1 (Fig. 6) reflects the Ca2+-binding cooperativity concurrent with stepwise oligomerization as Ca2+ concentrations increase (2, 9, 17). However, considering the maxima of the fractional occupancy of ∼28 (Fig. 6), the ∼15 Ca2+ ions placed per monomer in our crystal structure is lower than to be expected of a crystal grown in the presence of 50 mm Ca2+. One possible location of the remaining expected Ca2+ ions is in the observed tube of electron density surrounded by flexible carboxyl side chains in each front-to-front interface of hCASQ1 (Fig. 8J). The unresolved densities inside the electronegative cavity could provide a substantial number of nonspecific sites for monovalent and divalent cations without much entropic cost (4, 14). The other possible location is the highly acidic and disordered C terminus, a critical component for Ca2+-dependent tetramer formation (17), where there was a marked difference in the conformations of traced C-terminal residues among the six hCASQ1 monomers. It is likely that a combination of dynamic disorder within a molecule and the static lattice disorder could diminish the overall contour levels of the corresponding electron densities, thereby preventing us from resolving further Ca2+ ions and their associated coordination environment.
A low-affinity site of particular note is the Ca2+ ion coordinated in a monocapped trigonal prismatic configuration by the backbone carbonyl and side chain of Thr-189 and five water molecules that are hydrogen-bonded to a secondary solvation shell (Fig. 8G). Two nearby acidic residues, Glu-194 and Asp-196, appear to promote localization of two of the water ligands through hydrogen bonding or ion-dipole interactions, which boost the negative charge on the water oxygen. The hydroxyl side chain of Thr-189 was previously identified as a phosphorylation site (19, 20). Considering these long-range interactions of Thr-189, the effect of its phosphorylation would be substantial.
High-affinity Ca2+
Consistent with the previous prediction that the first few Ca2+ ions at low Ca2+ concentrations may bind CASQ with some specificity (4), three high-affinity sites are observed per rCASQ1 (Figs. 2B and Fig. 8, D–F) and hCASQ1 (Figs. 2A and Fig. 8, A–C). These Ca2+-binding sites are designated as high-affinity based on the observation that they are partially buried in the protein by an inner coordination sphere composed mainly of residue side chains and the polypeptide backbone, making them appear to be essential for proper folding into a functional protein, as seen in the Ca2+-binding site of concanavalin A or the EF-hand proteins such as calmodulin (21, 22).
The first high-affinity site is the Ca2+-binding site in Domain I (Fig. 8, A and D). In rCASQ1, Ca2+ at this site is coordinated in a trigonal bipyramidal arrangement by the side chains of Asn-17 and Asp-80 (monodentate); the backbone carbonyls of Val-18 and Leu-74; and a water molecule, with no apparent secondary solvation shell. A nearby conserved His-38 appears to promote the water ligand at this site through hydrogen bonding, which can boost negative charge on the water or generate a coordinating hydroxide ion through an acid-base mechanism. In hCASQ1, however, His-38 does not assist in coordination, and Leu-74 plays an indirect role by hydrogen bonding to a coordinating water molecule through its backbone carbonyl (Fig. 8A).
The second high-affinity site, located in the acidic crevice between Domains II and III, appears to be important for maintaining the tertiary structure of both hCASQ1 and rCASQ1 (Fig. 8, B and E). In rCASQ1, Ca2+ at this site is coordinated in a pentagonal bipyramidal arrangement by the side chains of Glu-199 (bidentate), Thr-229, and Thr-277; the backbone carbonyl of Thr-277; and two water molecules that are interacting with a secondary solvation shell (Fig. 8E). In hCASQ1, two water molecules replace Thr-277, but the coordination geometry and positions of the remaining ligands are otherwise conserved (Fig. 8B).
The third high-affinity site in rCASQ1 coordinates Ca2+ in an octahedral arrangement through the carboxylate of Glu-217 (bidentate), the backbone carbonyls of Asp-210 and Pro-212, and two water molecules that are interacting with a secondary solvation shell. In hCASQ1, additional coordination by the Asp-210 carboxylate in a monodentate fashion results in a monocapped octahedral arrangement (Fig. 8, C and F).
Intermolecular Interface and Oligomerization
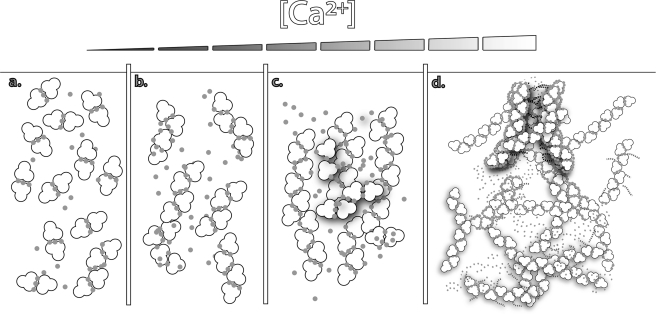
In summary with the crystal structures, MALS, and atomic absorption data accumulated herein, CASQ1 oligomerizes starting from a monomer containing three high-affinity Ca2+ ions (Fig. 9a). The Ca2+-binding curve of hCASQ1 shows transitions at 1.9, 4.5, and 8.9 mm Ca2+, with a substantial increase in Ca2+ occupancy at each step (Fig. 6). Based on MALS data (Fig. 7), the first and second transitions in Fig. 6 are monomer/dimer and dimer/tetramer transitions, respectively. Previously, small-angle x-ray scattering experiments indicated that CASQ exists as octameric oligomers (80 × 175 × 374 Å) at 10 mm Ca2+ (13). This suggests that the transition observed at 8.9 mm is a tetramer/octamer transition (Fig. 9b), after which the Ca2+ occupancy increases by ∼40%. Although the initial populations of monomers and dimers only persist over a range of 2 mm Ca2+, the higher order polymer has, at a minimum, a 5-fold broader range and S.D. from point to point (±0.50) compared with the monomer/dimer (±0.11) (Fig. 6). This indicates that transitions at those high Ca2+ concentrations could be much more dynamic and complex, probably involving previously mentioned intermolecular rotation among monomers (Fig. 3) with concomitant coordination of large numbers of low-affinity Ca2+ ions in the front-to-front interface. In addition, the altered arrangement of the back-to-back interaction of hCASQ1 brings substantial numbers of acidic residues into proximity with each other (Fig. 4C). Unlike the mostly linearly packed lattices of CASQ crystallized in buffers in which the Ca2+ concentrations were low (Fig. 5B) or absent (Fig. 5C), the observed interaction in the new type of back-to-back interface of hCASQ1 allows oligomerization in both linear and branched manners, which could establish a matrix-type network (Fig. 9, c and d). Likewise, the electron microscopic features of skeletal SR from several species indicate that CASQ1 forms an intricate network that is predominantly one-dimensional but frequently ramifies (2, 23), with the presence of connecting tendrils linking the tangle of linear and ramified segments. Therefore, upon exposure to physiological SR Ca2+ concentrations, CASQ likely forms a mesh-like matrix (Fig. 9, c and d), similar to those observed in EM images (2, 23), by employing the back-to-back interaction observed in the hCASQ1 lattice (Fig. 5A). As shown in Fig. 6, it is unlikely that the further developed precipitous matrices would increase the number of bound Ca2+ ions. Significantly, the clusters of disordered C termini for all of the CASQ1 molecules in the P1 lattice are pointing toward the narrow openings between the linear polymers in the matrix, and it is tempting to speculate that this channel-like crevice (Fig. 9d) can form in between CASQ polymers at high concentrations of both Ca2+ and CASQ in junctional SR. This long and narrow port can be assembled/disassembled dynamically in a Ca2+ concentration-dependent manner and effectively conduct Ca2+ ions to the Ca2+ release channel at diffusion-limited rates.
FIGURE 9.
Schematic diagram of conformations of CASQ1 at different Ca2+ concentrations as suggested by crystal structures, MALS, small-angle x-ray scattering, EM, and atomic absorption. a, front-to-front dimer formation occurs through an N-terminal arm exchange and coordination of a few Ca2+ ions. b, back-to-back interaction forming a tetramer. c, rotation happens between two monomers in the front-to-front interface, enabling both a large number of Ca2+ ions to be coordinated and new dimer contacts that allow growing linear polymers to branch. d, at higher Ca2+ concentrations, a network of ramified linear polymers could be further developed. A hypothetical tube is established by these CASQ polymers in which their disordered C termini (dotted lines) are arranged inward, thus serving as the basis for channel-like crevices for Ca2+.
Pathological Manifestations of CASQ
The residues involved in both front-to-front and back-to-back interfaces, many of which constitute the Ca2+-binding sites, are the most highly conserved residues in the entire sequence of CASQ (Fig. 1). This situation is reminiscent of the higher conservation of active site residues of other proteins and thus strongly indicates that these two interfaces are the functional contacts involved in coupling polymerization and low-affinity Ca2+ binding (9, 17). Therefore, any disruption or interference of this critical intermolecular interaction by either mutations or binding of small organic molecules could disrupt the functional integrity of CASQ (1, 7, 8, 24). Mutation of residues participating in these high-affinity Ca2+ sites can certainly contribute to the pathology of diseases such as catecholaminergic polymorphic ventricular tachycardia type 2 (CPVT2). CPVT2 is a familial arrhythmogenic cardiac disorder characterized by syncopal events, seizures, or sudden cardiac death at a young age (8). In a state in which sarco/endoplasmic reticulum Ca2+-ATPase is fully activated by adrenergic stimulation, the reduced Ca2+-buffering capacity of mutated CASQ2 would permit luminal free Ca2+ concentrations to overshoot the normal threshold of ryanodine receptor-2 for a store overload-induced Ca2+ release and result in arrhythmia (1). So far, in vitro testing shows that most CASQ2 mutations implicated in causing CPVT2 disrupt either protein folding or Ca2+-induced polymerization (1, 15). For example, the mutation of Arg-33 in human CASQ2 (Arg-14 in rCASQ1), which is highly conserved in CASQ1 and CASQ2 across species, is associated with CPVT2-R33Q (25). Using the high-resolution structure of CASQ1 as a model, the pathological manifestation of CPVT2-R33Q can be attributed to destabilization of a particular high-affinity Ca2+ (Figs. 1 and 6A) that is in close proximity to the mutated Arg-33, which in turn can disrupt both front-to-front and back-to-back interactions.
The other CPVT-causing CASQ2 mutation (CPVT2-D288H) is the point mutation of the completely conserved Asp-288 (Fig. 1). Asp-288 in both hCASQ1 and rCASQ1 takes part in an extended hydrogen-bonding network involving Lys-232, Glu-257, Ser-283, Asp-290, and Asp-291 (Fig. 8J), apparently serving a critical role in stabilizing Domain III. In addition, these residues are located proximal to the front-to-front interface near helix α13 (residues 290–302), which is a pivot point of the intermolecular rotation responding to rising Ca2+ concentrations (Fig. 3). Therefore, the CPVT2-D288H mutation not only could affect the integrity of Domain III but also could disrupt high-capacity Ca2+ binding. As a corollary of CPVT2, ablation of CASQ1 causes malignant hyperthermia (1, 26, 27). The same pathophysiology in connection with the high-affinity Ca2+ sites could be ascribed to an episode of malignant hyperthermia in the case of mutated CASQ1, although such mutations have not yet been reported (1).
Supplementary Material
This work was supported by American Heart Association Grant 850084Z, National Science Foundations Grants MCB 1021148 and DBI 0959778, and the M. J. Murdock Charitable Trust.

This article contains supplemental “Methods” and Tables S1 and S2.
The atomic coordinates and structure factors (codes 3TRP and 3UOM) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- SR
- sarcoplasmic reticulum
- CASQ
- calsequestrin
- hCASQ1
- human CASQ1
- rCASQ1
- rabbit CASQ1
- BisTris propane
- 1,3-bis(tris(hydroxymethyl)methylamino)propane
- MALS
- multiangle light scattering
- CPVT2
- catecholaminergic polymorphic ventricular tachycardia type 2.
REFERENCES
- 1. MacLennan D. H., Chen S. R. (2009) Store overload-induced Ca2+ release as a triggering mechanism for CPVT and MH episodes caused by mutations in RYR and CASQ genes. J. Physiol. 587, 3113–3115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Royer L., Ríos E. (2009) Deconstructing calsequestrin. Complex buffering in the calcium store of skeletal muscle. J. Physiol. 587, 3101–3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Meissner G., Wang Y., Xu L., Eu J. (2009) Silencing genes of sarcoplasmic reticulum proteins clarifies their roles in excitation-contraction coupling. J. Physiol. 587, 3089–3090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. MacLennan D. H., Reithmeier R. A. (1998) Ion tamers. Nat. Struct. Biol. 5, 409–411 [DOI] [PubMed] [Google Scholar]
- 5. Györke S., Stevens S. C., Terentyev D. (2009) Cardiac calsequestrin: quest inside the SR. J. Physiol. 587, 3091–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lee K. W., Maeng J. S., Choi J. Y., Lee Y. R., Hwang C. Y., Park S. S., Park H. K., Chung B. H., Lee S. G., Kim Y. S., Jeon H., Eom S. H., Kang C., Kim D. H., Kwon K. S. (2012) Role of junctin protein interactions in cellular dynamics of calsequestrin polymer upon calcium perturbation. J. Biol. Chem. 287, 1679–1687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dulhunty A., Wei L., Beard N. (2009) Junctin: the quiet achiever. J. Physiol. 587, 3135–3137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Knollmann B. (2009) New roles of calsequestrin and triadin in cardiac muscle. J. Physiol. 587, 3081–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Park H., Park I. Y., Kim E., Youn B., Fields K., Dunker A. K., Kang C. (2004) Comparing skeletal and cardiac calsequestrin structures and their calcium binding: a proposed mechanism for coupled calcium binding and protein polymerization. J. Biol. Chem. 279, 18026–18033 [DOI] [PubMed] [Google Scholar]
- 10. Otwinowski Z., Minor W. (1997) Methods in Enzymology 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 11. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 12. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Sanchez E. J., Munske G. R., Criswell A., Milting H., Dunker A. K., Kang C. (2011) Phosphorylation of human calsequestrin: implications for calcium regulation. Mol. Cell. Biochem. 353, 195–204 [DOI] [PubMed] [Google Scholar]
- 14. Wang S., Trumble W. R., Liao H., Wesson C. R., Dunker A. K., Kang C. H. (1998) Crystal structure of calsequestrin from rabbit skeletal muscle sarcoplasmic reticulum. Nat. Struct. Biol. 5, 476–483 [DOI] [PubMed] [Google Scholar]
- 15. Kim E., Youn B., Kemper L., Campbell C., Milting H., Varsanyi M., Kang C. (2007) Characterization of human cardiac calsequestrin and its deleterious mutants. J. Mol. Biol. 373, 1047–1057 [DOI] [PubMed] [Google Scholar]
- 16. Sanchez E. J., Lewis K. M., Munske G. R., Nissen M. S., Kang C. (2012) Glycosylation of skeletal calsequestrin: implications for its function. J. Biol. Chem. 287, 3042–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Park H., Wu S., Dunker A. K., Kang C. (2003) Polymerization of calsequestrin. Implications for Ca2+ regulation. J. Biol. Chem. 278, 16176–16182 [DOI] [PubMed] [Google Scholar]
- 18. Soriano-Garcia M., Padmanabhan K., de Vos A. M., Tulinsky A. (1992) The Ca2+ ion- and membrane-binding structure of the Gla domain of Ca-prothrombin fragment 1. Biochemistry 31, 2554–2566 [DOI] [PubMed] [Google Scholar]
- 19. Nori A., Furlan S., Patiri F., Cantini M., Volpe P. (2000) Site-directed mutagenesis and deletion of three phosphorylation sites of calsequestrin of skeletal muscle sarcoplasmic reticulum. Effects on intracellular targeting. Exp. Cell Res. 260, 40–49 [DOI] [PubMed] [Google Scholar]
- 20. Beard N. A., Laver D. R., Dulhunty A. F. (2004) Calsequestrin and the calcium release channel of skeletal and cardiac muscle. Prog. Biophys. Mol. Biol. 85, 33–69 [DOI] [PubMed] [Google Scholar]
- 21. Hardman K. D., Agarwal R. C., Freiser M. J. (1982) Manganese- and calcium-binding sites of concanavalin A. J. Mol. Biol. 157, 69–86 [DOI] [PubMed] [Google Scholar]
- 22. Finn B. E., Evenäs J., Drakenberg T., Waltho J. P., Thulin E., Forsén S. (1995) Calcium-induced structural changes and domain autonomy in calmodulin. Nat. Struct. Biol. 2, 777–783 [DOI] [PubMed] [Google Scholar]
- 23. Franzini-Armstrong C., Kenney L. J., Varriano-Marston E. (1987) The structure of calsequestrin in triads of vertebrate skeletal muscle: a deep-etch study. J. Cell Biol. 105, 49–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kang C., Nissen M. S., Sanchez E. J., Lam K. S., Milting H. (2010) Potential adverse interaction of human cardiac calsequestrin. Eur. J. Pharmacol. 646, 12–21 [DOI] [PubMed] [Google Scholar]
- 25. Terentyev D., Nori A., Santoro M., Viatchenko-Karpinski S., Kubalova Z., Gyorke I., Terentyeva R., Vedamoorthyrao S., Blom N. A., Valle G., Napolitano C., Williams S. C., Volpe P., Priori S. G., Gyorke S. (2006) Abnormal interactions of calsequestrin with the ryanodine receptor calcium release channel complex linked to exercise-induced sudden cardiac death. Circ. Res. 98, 1151–1158 [DOI] [PubMed] [Google Scholar]
- 26. Dainese M., Quarta M., Lyfenko A. D., Paolini C., Canato M., Reggiani C., Dirksen R. T., Protasi F. (2009) Anesthetic- and heat-induced sudden death in calsequestrin-1-knockout mice. FASEB J. 23, 1710–1720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Protasi F., Paolini C., Dainese M. (2009) Calsequestrin-1: a new candidate gene for malignant hyperthermia and exertional/environmental heat stroke. J. Physiol. 587, 3095–3100 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.