Background: The Sox2-protein interactome in ESC has not been identified.
Results: ESC that exogenously express Oct4, Sox2, Klf4, and c-Myc self-renew. This permitted the identification of the Sox2-interactome in ESC.
Conclusion: Sox2 associates with >70 proteins, and the knockdown of the Sox2-associated protein Smarcd1 induces the differentiation of ESC.
Significance: This is the first description of the Sox2-interactome in undifferentiated ESC.
Keywords: Cell Differentiation, Embryonic Stem Cell, Proteomics, Stem Cells, Transcription Factors, Klf4, MudPIT, Oct4, Sox2, c-Myc
Abstract
Unbiased proteomic screens provide a powerful tool for defining protein-protein interaction networks. Previous studies employed multidimensional protein identification technology to identify the Sox2-interactome in embryonic stem cells (ESC) undergoing differentiation in response to a small increase in the expression of epitope-tagged Sox2. Thus far the Sox2-interactome in ESC has not been determined. To identify the Sox2-interactome in ESC, we engineered ESC for inducible expression of different combinations of epitope-tagged Sox2 along with Oct4, Klf4, and c-Myc. Epitope-tagged Sox2 was used to circumvent the lack of suitable Sox2 antibodies needed to perform an unbiased proteomic screen of Sox2-associated proteins. Although i-OS-ESC differentiate when both Oct4 and Sox2 are elevated, i-OSKM-ESC do not differentiate even when the levels of the four transcription factors are coordinately elevated ∼2–3-fold. Our findings with i-OS-ESC and i-OSKM-ESC provide new insights into the reasons why ESC undergo differentiation when Sox2 and Oct4 are elevated in ESC. Importantly, the use of i-OSKM-ESC enabled us to identify the Sox2-interactome in undifferentiated ESC. Using multidimensional protein identification technology, we identified >70 proteins that associate with Sox2 in ESC. We extended these findings by testing the function of the Sox2-assoicated protein Smarcd1 and demonstrate that knockdown of Smarcd1 disrupts the self-renewal of ESC and induces their differentiation. Together, our work provides the first description of the Sox2-interactome in ESC and indicates that Sox2 along with other master regulators is part of a highly integrated protein-protein interaction landscape in ESC.
Introduction
Interest in pluripotent stem cells and their potential value in regenerative medicine has grown continuously due in large measure to the isolation of mouse and human embryonic stem cells (ESC)6 and, more recently, to the production of induced pluripotent stem cells from both mouse and human somatic cells. An in-depth understanding of the molecular mechanisms responsible for the self-renewal of pluripotent stem cells will be required to realize the full potential of these cells. Numerous genes, including genes that code for the core pluripotency transcription factors Sox2, Oct4, and Nanog, have been identified that are essential for the maintenance of ESC. However, the mechanisms by which these transcription factors and other essential gene products act are only partially understood.
Sox2, Oct4, and Nanog, which function as master regulators in ESC, not only regulate the expression of many other essential genes, they also regulate their own expression by both positive- and negative-feedback loops (1, 2). Importantly, the levels of these transcription factors must be precisely regulated in pluripotent stem cells. In the cases of Oct4 and Sox2, knockdown of either factor induces the differentiation of ESC into trophectoderm-like cells (3, 4). Moreover, small increases of either Oct4 or Sox2 promote the differentiation of ESC, albeit to different cell types (3, 5). Increases in Nanog enable mouse ESC to self-renew without LIF (6), whereas knockdown of Nanog increases the propensity of ESC to differentiate (7). Similar to Nanog, ectopic expression of Klf4 or c-Myc sustains the self-renewal of ESC in the absence of LIF, and the knockdown of either Klf4 or c-Myc promotes the differentiation of ESC (8, 9).
Considerable effort has been devoted to understanding how Sox2, Oct4, and Nanog control the fate of ESC. Genome-wide binding studies determined that there is substantial overlap in the target genes of Sox2, Oct4, and Nanog (10, 11), and in the cases of Sox2 and Oct4 they often bind to adjacent cis-regulatory elements of their target genes. Recently, unbiased proteomic screens have identified proteins that associate with Nanog and Oct4 in ESC (12–16) and Sox2 in ESC undergoing differentiation (17). A proteomic screen using an epitope-tagged form of Nanog in ESC identified 17 Nanog-associated proteins, many of which are required for the self-renewal of ESC, including Oct4, Sall4, and Nac1 (12). A subsequent study identified additional Nanog-associated proteins in ESC, including subunits of the SWI/SNF chromatin-remodeling complex and the repressive NuRD complex (13). More recently, three groups used epitope-tagged forms of Oct4 expressed at levels close to or only slightly higher (∼30%) than Oct4 expressed endogenously by ESC and identified a large network of Oct4-associated proteins (14–16). Thus far, the proteins that associate with Sox2 in ESC have not been reported. However, our laboratory recently described an unbiased Sox2 proteomic screen in ESC undergoing differentiation and identified >60 Sox2-associated proteins (17). Interestingly, several Sox2-associated proteins also associate with Oct4, Nanog, and other pluripotency-associated factors (17).
Identification of the Sox2-protein interactome in self-renewing ESC has been hindered by the lack of highly specific and sensitive Sox2 antibodies that are necessary for conducting an unbiased proteomic screen. To circumvent this shortcoming, we postulated that elevating master regulators in various combinations would identify conditions under which epitope-tagged Sox2 (FLAG-Strep tagged Sox2, (fs)Sox2) could be expressed in ESC without inducing differentiation. To test this possibility, we initially examined whether elevating (fs)Sox2 together with Oct4 could sustain the self-renewal of ESC. We determined that elevating (fs)Sox2 and Oct4 together disrupts the self-renewal of ESC and induces their differentiation. In contrast, elevating (fs)Sox2 along with Oct4, Klf4, and c-Myc in ESC does not disrupt their self-renewal or induce differentiation. Remarkably, ESC are able to maintain their morphology, overall gene expression profile, and self-renewal capacity despite an ∼2-fold elevation of total cellular Sox2 levels when the four transcription factors Oct4, (fs)Sox2, Klf4, and c-Myc are expressed simultaneously. Using ESC engineered to express these four transcription factors from an inducible promoter, we conducted an unbiased proteomic screen of Sox2 and its associated proteins. For this purpose, we isolated Sox2-protein complexes using antibodies that recognize the FLAG epitope and used multidimensional protein identification technology (MudPIT) to analyze the composition of the Sox2-proteins complexes. We identified >70 proteins that associate with Sox2 in ESC and demonstrate that knockdown of the Sox2-associated protein Smarcd1 disrupts the self-renewal of ESC and induces their differentiation.
EXPERIMENTAL PROCEDURES
Cell Culture
KH2 (Open Biosystems, Huntsville, AL), i-OSKM-ESC, and i-OS-ESC were maintained as described previously (5). For experiments with transgene induction, cells were cultured in 4 μg/ml doxycycline (Dox) unless otherwise indicated. Embryoid bodies were produced by culturing i-OSKM-ESC, which had been grown in the presence of Dox for three passages under non-adherent conditions in DME supplemented with 15% fetal bovine serum and 100 μm β-mercaptoethanol. After 4–9 days, as indicated in Fig. 5, the embryoid bodies were transferred to gelatin-coated tissue culture plastic in medium consisting of DME supplemented with 15% fetal bovine serum and 100 μm β-mercaptoethanol. After 5 days in culture, RNA was isolated from the cell population as described below.
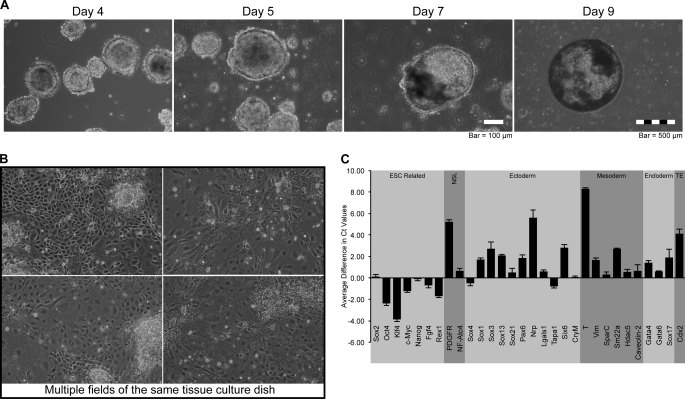
FIGURE 5.
Verification of i-OSKM-ESC pluripotency after treatment with Dox. i-OSKM-ESC were cultured in medium supplemented with 4 μg/ml Dox continuously for 3 passages. i-OSKM-ESC were then subcultured into non-adherent Petri dishes coated with 0.5% agarose in medium containing 15% DME and 100 μm β-mercaptoethanol but without Dox or LIF. A, photomicrographs were taken at the time points indicated after plating the cells under adherent-free conditions. The day 9 photo is representative of a cystic embryoid body. B, after 4 days in non-adherent culture, embryoid bodies generated from i-OSKM-ESC were transferred to gelatin-coated tissue culture plastic in medium containing 15% DME and 100 μm β-mercaptoethanol. Medium was changed every other day, and photomicrographs were taken 5 days after subculture onto gelatin. C, quantification of mRNA levels of transcripts associated with differentiation and development from cells described above in B. mRNA was collected after 5 days of culture on gelatin-coated tissue culture plastic, and cDNA was synthesized and used as template in triplicate RT-qPCR (n = 3). Ct values were averaged and normalized to GAPDH (as presented) and to HDAC1 (data not shown). Normalization of Ct values to HDAC1 shifted all bars up ∼0.25 Ct value but did not alter the conclusions drawn in “Results.” Error bars represent S.D. between replicates. Values greater than zero represent increased abundance of mRNA from a particular gene.
Construction of TetO-FUW-O(fs)SKM and TetO-FUW-O(fs)S Lentiviral Vectors
The TetO-FUW-O(fs)SKM lentiviral transfer vector was constructed using TetO-FUW-OSKM (Plasmid 20321, Addgene, Cambridge, MA) as the template in two PCR reactions to add a FLAG and a Strep epitope-tag sequence (fs) to the 5′ end of Sox2. In the first PCR step, primers 1U/1L and 2U/2L were used with the TetO-FUW-OSKM vector as template to amplify two fragments with partially overlapping sequences that contained the FLAG and Strep sequences. In the second PCR step, primers 1U and 2L were used with template consisting of the two overlapping PCR products generated in the first step. This second PCR product was then inserted into the NheI and Rsr II sites downstream of the Oct4 coding region in the TetO-FUW-OSKM vector. The sequence containing the FLAG and Strep tags, inserted between NheI and Rsr II sites, was verified by sequencing. Forward primers 1U (CTTTGGCACCCCAGGCTATGGAAGC) and 2U (ATGGACTACAAGGACGACGATGACAAGGGTCGGCCGCCAACTGGAGCCACCCACAATTCGAGAAGGGCGGAATGTATAACATGATGGAGACGGAGCTG). Reverse primers 1L (TCCGCCCTTCTCGAATTGTGGGTGGCTCCAGTTGGCGGCCGACCCCTTGTCATCGTCGTCCTTGTAGTCCATGCATGCAGGCCCGGGGTTTTCTTCAAC) and 2L (GGCCGGTATTTATAATCCGGGTGCTC). The FLAG tag sequence is underlined and in bold, and Strep tag sequence is underlined.
To generate the TetO-FUW-O(fs) vector, a PCR reaction using TetO-FUW-O(fs)SKM as template was used to isolate the O(fs)S coding region, introduce a stop codon at the end of the Sox2 CDS, and flank the O(fs)S with EcoR1 restriction sites. Upper primer (CCGAATTCGCCATGGCTGGAC) and lower primer (TTTGAATTCACATGTGCGACAGGGGCAGTGT) were used; EcoRI sites are underlined, and the stop codon is in bold. TetO-FUW-O(fs)SKM was cut with EcoR1 and re-ligated to remove the polycistronic element, resulting in a TetO-FUW vector. The O(fs)S PCR fragment was then cut with EcoR1 and ligated into TetO-FUW, also cut with EcoR1, to generate TetO-FUW-O(fs)S.
Generation of i-OSKM-ESC and i-OS-ESC
TetO-FUW-O(fs)S and TetO-FUW-O(fs)SKM were packaged into VSV-G pseudotyped lentiviruses, and KH2 ESC were infected with lentiviruses using a protocol described previously (18) and plated at clonal density. Single clones were isolated and genotyped for positive infection with the TetO-FUW-O(fs)SKM or TetO-FUW-O(fs)S construct using 1U and 2L primers described above.
Nuclear Extract Preparation and Western Blot Analysis
To examine protein expression in i-OSKM-ESC, the cells were cultured in the presence or absence of Dox for 48 h and harvested. For all experiments, cells cultured with Dox were supplemented with 4 μg/ml Dox in their culture medium. Nuclear extracts were prepared using the Pierce NE-PERTM nuclear and cytoplasmic extraction kit (Pierce) and resolved on 4–20% gradient-gels (Pierce) as described previously (17). Primary antibodies used were: α-Sox2 (1:1000, ab5603, Abcam, Cambridge, MA), α-Oct4 (1:500, sc-8628, Santa Cruz Biotechnology, Santa Cruz, CA), α-Klf4 (1:1000, #39745, Active Motif, Carlsbad, CA), α-cMyc (1:500, 06–340, Millipore, Billerica, MA), α-HDAC1 (1:2000, ab7028–50, Abcam), and α-Rpa1 (1:500, sc-48425, Santa Cruz). Secondary antibodies used were: α-goat IgG-AP (A4187, Sigma), α-mouse IgG-AP (A4312, Sigma), and α-rabbit IgG-AP (A3687, Sigma). Proteins were detected using ECF substrate (Amersham Biosciences) and scanned on a Typhoon Variable Mode Imager (GE Healthcare). Quantitation of protein expression was performed using ImageQuant 5.0 analysis software.
RNA Analysis
Total RNA was isolated by Tri-Reagent using the manufacturer's protocol (Molecular Research Center, Inc., Cincinnati, OH). Procedures and primers used for RT-qPCR analysis were described previously (17). The upper primer AGGATCTGATTGCAGAGCC and the lower primer CAGGTCTTCACAAAAGGCAT were used to detect caveolin-2 levels, which have been shown to be a marker of smooth muscle development (19). HDAC1 levels were detected using the upper primer CTGGACTTACGAAACAGCGG and the lower primer CTCCTCATCTTCATCCCCAC. Genome-wide RNA expression was determined by performing microarray analysis using Affymetrix GeneChip Mouse Gene 1.0 ST (Santa Clara, CA). The University of Nebraska Medical Center DNA Microarray Core Facility performed data collection. The results were normalized and analyzed with Expression Console Software package (Affymetrix). Microarray data has been deposited with the GEO Repository under accession number GSE34801 (www.ncbi.nlm.nih.gov).
Alkaline Phosphatase (AP) Staining, Immunocytochemistry, and Photomicrographs
AP and immunofluorescence staining were performed as described previously (18). Primary antibodies used for immunocytochemistry were: α-SSEA-1 (1:10, MC480, Developmental Studies Hybridoma Bank, Iowa City, IA), α-Sox2 (1:50, sc-17320, Santa Cruz), α-Oct4 (1:50, sc-8628, Santa Cruz), and α-Nanog (1:50, AF2729, R&D Systems, Minneapolis, MN). Secondary antibodies used were: α-mouse IgM-FITC (F9252, Sigma) and α-goat IgG-FITC (F2016, Sigma). Nuclei were stained with DAPI (D9542, Sigma). Photomicrographs for Fig. 4 were taken with an Axiovert 200 m microscope and a Hamamatsu Photonics Camera C4742-95-12ER using identical exposure settings with the Slidebook 4.0.2.2 microscope control program. All other photomicrographs were taken using an Olympus IMT-2 microscope and Canon Rebel XTi camera.
FIGURE 4.
Immunocytochemistry of i-OSKM-ESC with and without induction of exogenous Oct4, Sox2, Klf4, and c-Myc. i-OSKM-ESC (clone 3.0) were cultured for 48 h without Dox or with Dox to induce the expression of the four exogenous transcription factors. Cells were fixed and stained for SSEA1, Oct4, Sox2, and Nanog. Nuclei were stained by DAPI.
MudPIT Analysis of Sox2-Protein Complexes
Sox2-protein complex purification and MudPIT analysis were described in detail previously (17). i-OSKM-ESC cultured without and with Dox were harvested, and nuclear extracts were prepared using Dounce homogenization (20). Sox2-protein complexes were immunoprecipitated using EZ viewTM red anti-FLAG M2 affinity beads (Sigma) and eluted with 3× FLAG peptide (Sigma). Elutes were precipitated with TCA and subjected to MudPIT analysis as described previously (17). The MS/MS datasets were examined using SEQUEST and the NCBI Mus musculus protein data base (NCBI, 2007-2006-22 release). To estimate relative protein levels, distributed Normalized Spectral Abundance Factors (dNSAF) were calculated for each detected protein, as described elsewhere (21). Three independent experiments were performed, and statistical analysis was performed as described previously (17). Proteins with a statistically significant p value (<0.05) are presented and grouped into three categories: proteins identified only in all three Dox-treated samples but not control samples, proteins enriched at least 5-fold in Dox-induced samples compared with uninduced (control) samples, and proteins identified in two of three Dox-treated samples. False discovery rate (FDR) was determined as described previously (17, 22).
Purification of (fs)Sox2-Protein Complexes via Strep Epitope
i-OSKM-ESC were cultured with or without Dox for 24 h, nuclear extracts were prepared using the NE-PERTM kit (Pierce), and protein was quantified. (fs)Sox2 and associated proteins were immunoprecipitated using Strep-Tactin spin columns according to manufacturer's protocols (IBA, St. Louis, MO). Next, 900 μg of protein was diluted in Buffer W (100 mm Tris, pH 8.0, 150 mm NaCl, 1 mm EDTA), loaded onto Strep-Tactin spin columns via centrifugation (700 × g, 30 s), washed 4× with Buffer W, and eluted twice with 50 μl of 10 mm d-biotin. The resultant elutes were pooled and analyzed by Western blot analysis.
Computational Analysis
Protein interaction landscapes and gene target analysis were generated using Cytoscape 2.6.3 (23). Gene ontology analysis of Sox2-associated proteins in ESC was performed using DAVID. Mouse Genomics Information and OMIM databases were queried for knock-out phenotypes and associated diseases of Sox2-associated proteins, respectively.
Knockdown of Smarcd1
Smarcd1 was knocked down using lentiviral constructs that express shRNA against Smarcd1 (supplemental Table S8), which were obtained from Open Biosystems (RMM4534). A nonspecific shRNA (Scrambled), described previously (18, 24), was used as a negative control. Infection was conducted in D3 ESC (25), as described above, and infected cells were subcultured into 12-well plates. Forty-eight hours later, AP-staining was conducted, and photomicrographs were taken. For cloning efficiency, infected cells were subcultured at clonal density. Six days later, AP staining was performed, and colonies were scored as ES cells, mixed or differentiated, in 10 random fields by 2 independent observers unaware of sample designation. For Western blot analysis, nuclear proteins were isolated 6 days after infection and blotted as described above.
RESULTS
Identification of the protein interactomes of master regulators in ESC has been facilitated by the expression of epitope-tagged proteins, which elevated their overall levels (12, 14–16). However, in the case of Sox2, elevating Sox2 on its own in ESC induces differentiation. To circumvent the propensity of Sox2 to perturb the self-renewal of ESC, we postulated that elevating Sox2 together with other master regulators would preserve the self-renewal of ESC and enable us to express epitope-tagged Sox2 in undifferentiated ESC.
Generation and Characterization of i-OS-ESC
Initially, we examined whether simultaneously elevating Sox2 with Oct4 would preserve the fate of ESC because Sox2 and Oct4 are known to work together cooperatively (10, 11). For this purpose, KH2 ESC were infected with the Dox-inducible TetO-FUW-O(fs)S lentivirus, as described under “Experimental Procedures.” KH2 ESC were selected for this study because they constitutively express the reverse tet-transactivator (26) and because they were used previously to identify the Sox2-interactome in ESC undergoing differentiation (17). ESC stably infected with the TetO-FUW-O(fs)S lentivirus express Oct4 and epitope-tagged Sox2 from a Dox-inducible polycistronic message that separates the coding sequences for Oct4 and (fs)Sox2 by a self-cleaving peptide (Fig. 1A).
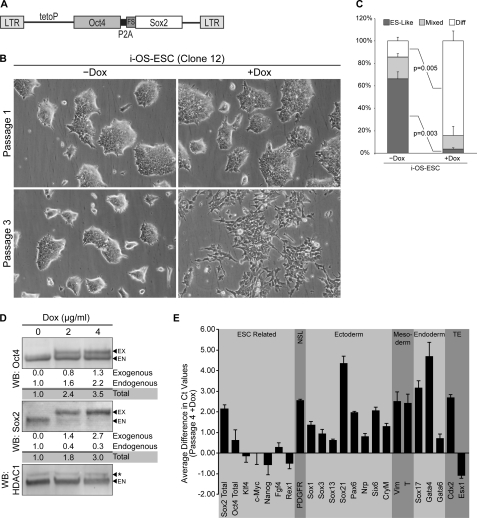
FIGURE 1.
Characterization of i-OS-ESC. A, shown is a schematic of the Dox-inducible polycistronic vector used to engineer i-OS-ESC. The region labeled P2A represents a self-cleaving peptide. Sox2 is dual epitope-tagged at the N terminus with FLAG and Strep epitopes (fs). B, shown are photomicrographs of i-OS-ESC cells (Clone 12) grown in the absence (−Dox) or presence (+Dox) of exogenous expression of Oct4 and Sox2 from a Dox-inducible promoter. Cells labeled as Passage 1 were photographed after ∼48 h of continued Dox exposure; cells labeled as Passage 3 were photographed after ∼6 days of continued Dox exposure. C, shown is a cloning efficiency assay of i-OS-ESC cultured in the absence and presence of Dox for five passages, seeded at clonal density, and scored 48 h later by two observers unaware of sample designation. Colonies were classified according to colony morphology (ES-Like, Mixed, or differentiated (Diff)) in 10 random 10× fields. The results were averaged and graphed as percentages. Error bars represent S.D. between counts of the two scorers. p values were calculated using Student's t test. D, shown are Western blot (WB) analyses of Sox2 and Oct4 isolated from i-OS-ESC after culture in increasing concentrations of Dox for 48 h. Expression levels in parentheses were normalized to HDAC1, and untreated cells were set to one. Exogenous protein is labeled EX, endogenous protein is labeled EN, and nonspecific signal is labeled with an asterisk. E, quantification of mRNA levels of transcripts associated with differentiation and development of ESC is shown. mRNA was collected from i-OS-ESC grown continuously in the absence or presence of Dox for four passages. cDNA was synthesized and used as the template in triplicate RT-qPCR (n = 3). Ct values were averaged and normalized to GAPDH (as presented), a previously validated benchmark for measuring mRNA transcript levels in ESC and ESC undergoing differentiation (40). Ct values were also normalized to HDAC1 (data not shown), which shifted all bars down 1 Ct value but did not alter the conclusions drawn in the text. Error bars represent S.D. between replicates. Values greater than zero represent increased abundance of mRNA from a particular gene. Genes marked with NSL do not represent a specific cell lineage. TE, trophectoderm.
Several stably infected clones were isolated, and two were initially tested for their response to the exogenous expression of Oct4 and Sox2. The addition of Dox to the culture medium for 48 h did not significantly alter the morphology of either clone of ESC (Fig. 1B, supplemental Fig. S1A). However, continued culture of these cells in Dox for 2 additional passages (6 days) caused their morphology to change to a flattened phenotype, characteristic of differentiated cells (Fig. 1B, supplemental Fig. S1A). We also examined the expression levels of Sox2 and Oct4 in these cells by Western blot analysis. For this purpose, nuclear proteins were isolated from the cells cultured without or with Dox during the initial 48-h time period. Exogenous Oct4 and Sox2 migrate more slowly on SDS-PAGE because of a residual self-cleaving peptide and FLAG-Strep epitopes, respectively. In both clones, Sox2 levels were elevated ∼3-fold (Fig. 1D, supplemental Fig. S1B). Comparison of Sox2 and Oct4 levels in clone 12, hereafter referred to as i-OS-ESC, indicated that the total levels of both Sox2 and Oct4 were ∼2–3-fold higher than their endogenous counterparts, and the ratio of total Sox2 to total Oct4 levels was ∼1:1.2 at the highest concentration of Dox (4 μg/ml) tested (Fig. 1D).
To further characterize the responses of ESC to elevating both Sox2 and Oct4, we examined the cloning efficiency of i-OS-ESC because this is a sensitive test of their self-renewal capacity (5). For this purpose, i-OS-ESC were cultured in the absence or presence of Dox for 4 passages and then replated at clonal density (8000 cells per T25 culture flask). Forty-eight hours after subculture at clonal density, cells were scored according to colony morphology (ES-like, differentiated or mixed) by two observers, unaware of sample designation (Fig. 1C). Induction of exogenous Sox2 and Oct4 significantly reduced the proportion of ES-like colonies (70% in −Dox, 3% in +Dox) and significantly enriched the proportion of differentiated colonies (12% in −Dox, 90% in +Dox). To further corroborate our observation that i-OS-ESC differentiate in response to exogenous Sox2 and Oct4, we examined gene markers associated with lineage specification during differentiation. For this purpose, RNA was isolated from i-OS-ESC cultured in the absence or presence of Dox for 4 passages (8 days). RT-qPCR analysis determined that a number of transcripts associated with maintaining ESC fate were decreased, whereas markers for ectoderm, mesoderm, endoderm, and trophectoderm were elevated (Fig. 1E). Thus, simultaneous elevation of Sox2 and Oct4 disrupts the self-renewal of ESC and induces their differentiation.
Generation and Characterization of i-OSKM-ESC
Given the induction of differentiation when Sox2 and Oct4 are both elevated, we decided to examine how ESC would respond to elevating the four transcription factors, Oct4, Sox2, Klf4, and c-Myc, which were first used to reprogram somatic cells. To address this question, we engineered KH2 ESC for the Dox-inducible expression of Oct4, Sox2, Klf4, and c-Myc. For this purpose, lentiviruses were used to introduce a Dox-inducible polycistronic transgene, which expresses a single transcript for the four reprogramming factors separated by self-cleaving peptides (Fig. 2A). As in the case of the TetO-FUW-O(fs)S lentiviral vector, Sox2 was engineered in the TetO-FUW-O(fs)SKM vector to contain FLAG and Strep epitopes at the N terminus of Sox2. After infection and expansion of the cells, three clones were isolated and genotyped, and the induction of the four factors after treatment with Dox was verified by probing for the exogenous expression of Oct4 and Klf4 (Fig. 2D and supplemental Fig. S2B). As noted above, the exogenously expressed Oct4 and Klf4 migrate more slowly on SDS-PAGE than their endogenous counterparts due to retention of self-cleaving peptide sequences at their C termini.
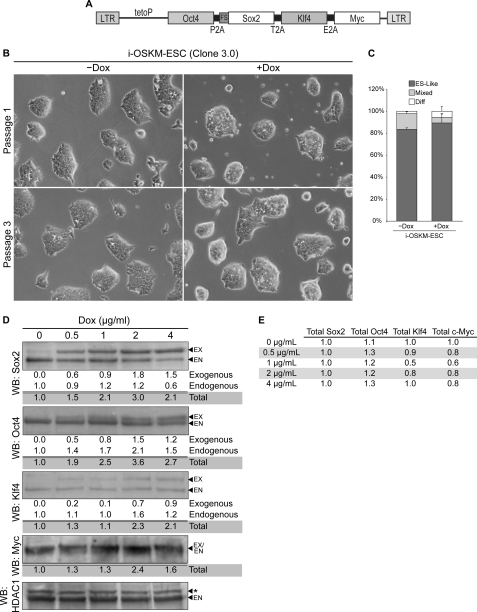
FIGURE 2.
Characterization of i-OSKM-ESC. A, shown is a schematic of Dox-inducible polycistronic lentiviral vector, TetO-FUW-O(fs)SKM, used to engineer i-OSKM-ESC. Regions labeled P2A, T2A, and E2A represent self-cleaving peptides. Sox2 is dual epitope-tagged at the N terminus with FLAG and Strep epitopes (fs). B, shown are photomicrographs of i-OSKM-ESC (clone 3.0) grown in the absence (−Dox) or presence (+Dox) of exogenous expression of Oct4, Sox2, Klf4, and c-Myc from a Dox-inducible promoter. Cells labeled as Passage 1 were photographed after ∼48 h of continued Dox exposure; cells labeled as Passage 3 were photographed after ∼6 days of continued Dox exposure. C, shown is a cloning efficiency assay of i-OSKM-ESC cultured in the absence and presence of Dox for five passages, seeded at clonal density, and scored 48 h later by two observers unaware of sample designation. Colonies were classified according to colony morphology (ES-Like, Mixed, or differentiated (Diff)) in 10 random 10× fields. The results were averaged and graphed as percentages. Error bars represent S.D. between counts of the two scorers. p values were calculated using Student's t test. D, shown are Western blot (WB) analyses of Sox2, Oct4, Klf4, and c-Myc isolated from i-OSKM-ESC after culture in increasing concentrations of Dox for 24 h. Expression levels in parentheses were normalized to HDAC1, and endogenous protein levels in untreated cells were set to one. Exogenous protein is labeled EX, endogenous protein is labeled EN, and nonspecific signal is labeled with an asterisk. E, shown is the relative ratio of total Sox2, Oct4, Klf4, and c-Myc protein expression in i-OSKM-ESC cultured in increasing concentrations of Dox. The protein level of Sox2 measured in subfigure C was set to one, and the relative abundances of other proteins were calculated.
Next, we examined the growth and morphology of the clones after treatment with Dox. Remarkably, elevating the expression of the four transcription factors did not significantly alter the morphology, the expression of the stem cell marker alkaline phosphatase, or the clonal growth of these cells (Fig. 2, supplemental Fig. S2, and data not shown). Moreover, one of the clones (clone 3.0, referred to as i-OSKM-ESC hereafter) was grown continuously for more than 6 passages in the presence of Dox without significant formation of differentiated cells (data not shown). To further substantiate the response of these cells to elevated levels of Oct4, Sox2, Klf4, and c-Myc, i-OSKM-ESC were grown in the absence or presence of Dox for four passages and then seeded at clonal density (8000 cells per T25). Forty-eight hours after passage, cell colonies were scored as ES-like, differentiated, or mixed by two observers, unaware of sample designation. There was no significant change in the ability of i-OSKM-ESC to form colonies at clonal densities in the presence of Dox (Fig. 2C). If anything, there was a small decrease in the number of mixed colonies that consisted of ESC and differentiated cells. Thus, unlike their i-OS-ESC counterparts, which differentiated after 2–3 passages in the presence of Dox (Fig. 1), prolonged elevation of Oct4, Sox2, Klf4, and c-Myc did not induce the differentiation of i-OSKM-ESC.
As part of our analysis of i-OSKM-ESC, we compared the relative expression of Oct4, Sox2, Klf4, and c-Myc after treatment of the cells with different concentrations of Dox. As noted above, exogenously expressed Oct4, Sox2, and Klf4 migrate more slowly than their endogenous counterparts. However, exogenously expressed c-Myc lacks a self-cleaving peptide and is nearly identical in size to endogenous c-Myc. As expected, as the concentration of Dox was increased, the overall protein levels of Oct4, Sox2, Klf4, and c-Myc increased (Fig. 2D). The levels of the four factors peaked at 2 μg/ml Dox. At 4 μg/ml Dox, their total levels were elevated ∼2-fold relative to their endogenous counterparts. In the cases of Oct4 and Sox2, each of their levels rose as much as 3-fold, but the overall ratio of Oct4 to Sox2 did not change substantially (Fig. 2E).
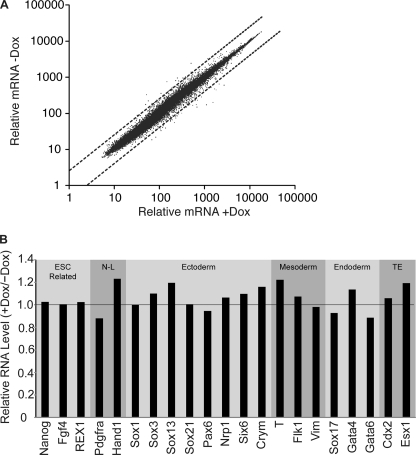
To more broadly examine the effects of overexpressing Oct4, Sox2, Klf4, and c-Myc on the behavior of i-OSKM-ESC, we performed microarray analysis of transcripts expressed by Dox-treated and untreated i-OSKM-ESC to explore their gene expression profiles. Scatter plot analysis indicated that the RNA expression profiles were very similar between the Dox-induced and untreated i-OSKM-ESC (Fig. 3A). Moreover, there was a lack of change in specific lineage markers for ectoderm, endoderm, and mesoderm (Fig. 3B). In addition to these studies, we determined by immunocytochemistry that Sox2, Oct4, and Nanog are localized to the nucleus in the vast majority of the cells both in the presence and absence of Dox (Fig. 4).
FIGURE 3.
Examination of the i-OSKM-ESC transcriptome after induction of exogenous Oct4, (fs)Sox2, Klf4, and c-Myc. A, shown is a scatter plot of transcript expression levels in Dox-treated i-OSKM-ESC (horizontal axis) and untreated i-OSKM-ESC (vertical axis) determined by Affymetrix microarray analysis. Dotted lines denote a 2-fold change in transcript expression levels. B, cell lineage marker expression levels from microarray results are presented as -fold change of Dox-treated i-OSKM-ESC (bars) compared with untreated (horizontal line). Genes marked with N-L do not represent a specific cell lineage, and genes marked TE are representative of trophectoderm.
Importantly, the exogenous elevation of Oct4, Sox2, Klf4, and c-Myc does not prevent the differentiation of the cells after Dox is removed. For this purpose, i-OSKM-ESC were cultured in the presence of Dox for 1 week. Cells were then cultured in medium without Dox for 2 days and subsequently cultured in medium containing 5 μm retinoic acid for an additional 2 days to induce differentiation (supplemental Fig. S3). i-OSKM-ESC were able to differentiate in response to retinoic acid, suggesting that exogenous Oct4, Sox2, Klf4, and c-Myc do not irrevocably alter the fate of ESC. Additionally, we determined that cultivation of i-OSKM-ESC in the presence of Dox for three passages does not alter their pluripotent properties. When embryoid bodies formed from these cells (Fig. 5A) were transferred to gelatin-coated tissue culture plastic, we observed differentiated cells that exhibited a wide range of cellular morphologies (Fig. 5B) and expressed markers typical of cells derived from each of the three embryonic germ layers (Fig. 5C). Moreover, differentiated cells derived from cystic embryoid bodies formed from these cells also produced rhythmically contacting muscle and cells that exhibited neuronal-like projections (data not shown). Taken together, our findings argue that simultaneous elevation of Oct4, Sox2, Klf4, and c-Myc preserves ESC fate despite the elevation of both Sox2 and Oct4.
Proteomic Identification of Sox2-interacting Proteins in ESC
Recent proteomic studies conducted in ESC identified the interactomes of nine pluripotency-associated proteins: Nanog, Oct4, Sall4, Esrrb, Nac1, Tcfcp2l1, Rex1, Dax1, and Zfp281 (12–16). In most of these studies epitope-tagged forms of these proteins were used to enhance the purification and isolation of their associated proteins (12, 14–16). Missing from this analysis is the Sox2-interactome. Importantly, our observation that simultaneous elevation of Oct4, Sox2, Klf4, and c-Myc does not disrupt the fate of ESC provided an avenue to examine the Sox2-protein interactome in ESC.
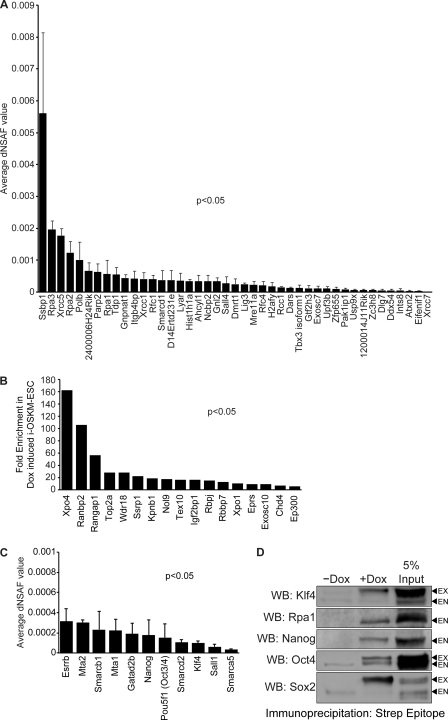
To address this important gap, we used i-OSKM-ESC to perform an unbiased proteomic screen of Sox2-associated proteins. Treatment of i-OSKM-ESC cells with Dox enabled us to take advantage of the fact that Sox2 expressed from the transgene is epitope-tagged, which substantially improved our ability to isolate Sox2-protein complexes. To identify the Sox2-interactome in ESC, we performed three independent isolations of Sox2-protein complexes. As described previously (17), Sox2-protein complexes were isolated using M2-beads (a FLAG antibody conjugated to agarose beads). After extensive washing of the M2 beads to remove non-specifically associated proteins, Sox2-protein complexes were eluted by the addition of 3×-FLAG peptide. MudPIT analysis was performed on TCA-precipitated eluates isolated from Dox-treated or untreated (control) ESC. This analysis identified 43 high-confidence Sox2-associated proteins present in all three Dox-treated ESC, but not in the control ESC, with statistically significant (p < 0.05) dNSAF values (Fig. 6A, supplemental Table S1). We also identified another 17 Sox2-associated proteins enriched in all three MudPIT analyses (>5-fold relative to the untreated control) with statistically significant (p < 0.05) dNSAF values (Fig. 6B, supplemental Table S2). In addition to this group of 60 proteins, we identified 11 proteins not detected in the untreated controls but present in two of the three MudPIT analyses (Fig. 6C, supplemental Table S3). These proteins are included in our discussion below because each was absent in the same Dox-treated sample and because several of them (Klf4, Nanog, and Oct4) were independently confirmed (see below). Furthermore, a fourth protein (Sall1) in this list has been reported recently to associate with Sox2 in ESC (27). Across all Sox2-protein preparations, the spectral false discovery rate was <0.25% (data not shown) (22).
FIGURE 6.
Sox2-associated proteins in ESC identified by MudPIT analysis and downstream validation. A, Sox2-associated proteins with statistically significant dNSAF values (p < 0.05) were identified only in Dox-treated i-OSKM-ESC (clone 3.0) in three MudPIT analyses. B, shown are Sox2-associated proteins present at low levels in uninduced samples but enriched (>5-fold) in Dox-induced i-OSKM-ESC. C, Sox2-associated proteins with statistically significant dNSAF values (p < 0.05) are identified only in Dox-treated i-OSKM-ESC in two of three MudPIT analyses, with known biological functions in ESC. Additional details regarding dNSAF values from each biological replicate are presented in supplemental Tables S1–S3. D, nuclear proteins were isolated from i-OSKM-ESC cultured without and with (4 μg/ml) Dox and quantified. Equal amounts of protein (μg) from −Dox and +Dox conditions were loaded onto Strep-Tactin columns, and (fs)Sox2 along with its associated proteins were immunoprecipitated as described under “Experimental Procedures.” For Western blot (WB) analysis, equal volumes (μl) of immunoprecipitate eluate were run on SDS-PAGE. Nuclear protein from the +Dox condition was included as a control (5% Input). Exogenous protein expressed from the polycistronic transgene is labeled EX, and endogenous protein is labeled EN.
To validate the proteins identified in this proteomic screen, we independently isolated Sox2-protein complexes using a completely different protein isolation protocol. As described under “Experimental Procedures,” Sox2-protein complexes were isolated using a Strep-Tactin column that took advantage of the second epitope tag (Strep tag) included in the FLAG-Strep-tagged Sox2 fusion protein. Complexes isolated using Strep-Tactin columns were probed by Western blot analysis for four proteins, Klf4, Rpa1, Nanog, and Oct4, each of which was present in the Strep-Tactin-isolated Sox2-protein complexes (Fig. 6D). In addition, using a Sox2 antibody, we demonstrated that Sall4 and Rpa1, identified in our MudPIT analysis (Fig. 6A), associate with endogenous Sox2 in untreated i-OSKM-ESC (data not shown).
Role of Sox2-interacting Proteins in Development and Genetic Diseases
Among the 71 Sox2-associated proteins are transcription factors, components of chromatin remodeling complexes (e.g. NuRD and SWI/SNF), DNA repair machinery (Xrcc1, Xrcc5, Xrcc7, Rbpj, Top2a), and DNA replication machinery (Polb, Rpa1, Rpa2, Rpa3) (Fig. 6). Additionally, many of the Sox2-associated proteins are required for the self-renewal of ESC, including: Sall4, Esrrb, Mta1, and Mta2 (12, 13, 28). To understand the roles of Sox2-associated proteins, we searched the Mouse Genome Informatics database for phenotypes of knock-out mice and determined that 46% (33/71) of Sox2-associated proteins have defects during mouse development (supplemental Table S4). We also searched the OMIM data base for Sox2-associated proteins and found 17% (12/71) of Sox2-associated proteins are related to human genetic diseases (supplemental Table S4). Thus, Sox2-associated proteins play important roles during development as well as in human genetic diseases.
Highly Integrated Global Interactome in Embryonic Stem Cells
Previous studies described the interactomes of nine proteins in ESC: Oct4, Nanog, Sall4, Tcfcp2l1, Esrrb, Nac1, Rex1, Dax1, and Zfp281(12–16). Using the Sox2-interactome described in this study and the interactomes of these nine proteins, including the Oct4-interactomes described in three separate reports, we generated a virtual protein-protein interaction landscape in ESC (Fig. 7). This analysis indicated that 28 Sox2-associated proteins associate with two or more of the pluripotency-associated factors examined (supplemental Table S5). Moreover, some of the Sox2-associated proteins, such as Sall4, Sall1, Esrrb, and Smarcd1 associate with at least four of the pluripotency-associated factors. Thus, there appears to be a high degree of integration and interdependence at the protein-protein interaction level between these 10 factors in ESC. As a further indication of the high level of integration among the proteins in this landscape, ChIP-chip and ChIP-seq data indicate that the regulatory regions of 27 of the genes that code for the 71 Sox2-associated proteins are bound by both Sox2 and Oct4, and 18 of their genes are bound by Sox2, Oct4, and Nanog (supplemental Fig. S4 and Table S6). Furthermore, the vast majority of the genes that code for Sox2-associated proteins are bound by a large number of transcription factors that influence the fate of ESC (supplemental Table S6).
FIGURE 7.
Protein-protein interaction landscape of ESC. An integrated network of proteins necessary for the self-renewal and pluripotency of ESC and their associated proteins is presented. In total, 334 proteins are represented, 10 of which serve as nodes in the protein-protein interaction network. Yellow circles represent high confidence Sox2-associated proteins, and light yellow circles represent proteins identified in two of three MudPIT analyses. A summary of Sox2-associated proteins that associate with other transcription factors is presented in supplemental Table S5.
The high degree of interdependence between these 10 pluripotency-associated factors suggested that blocking the expression of proteins that associate with three or more pluripotency factors would dramatically change the protein-protein interaction landscape and alter the fate of ESC. This seemed all the more likely for proteins that are part of chromatin-remodeling complexes, such as Smarca5 and Smarcd1, each of which associates with at least 4 of the 10 pluripotency-associated factors (supplemental Table S5). We tested this prediction by knocking down Smarca5 and Smarcd1. Smarca5 associates with Sox2, Oct4, Tcfcp2l1, and Esrrb, and Smarcd1 associates with Sox2, Sall4, Esrrb, and Tcfcp2l1 (supplemental Table S5). Previous studies reported that ESC lacking Smarca5 expression could not be isolated (29), but it was not determined how established ESC would respond to the knockdown of Smarca5. We determined that the knockdown of Smarca5 disrupts the self-renewal of ESC and induces their differentiation (data not shown).
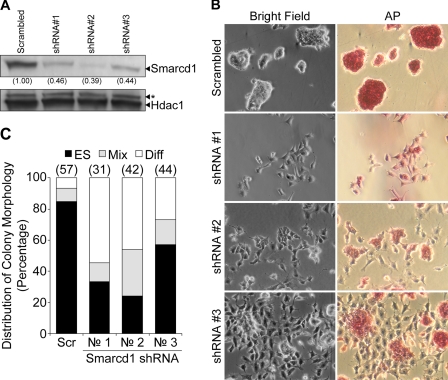
We also determined that the knockdown of Smarcd1 induces the differentiation of ESC. Previous studies reported that Smarcd1 (also referred to as Baf60A) along with Brg1 (Smarca4) and Baf155 (Smarcc1) is present in a chromatin-remodeling complex known as esBAF. Brg1 and Baf 155 have been shown to be required by ESC (30), but the role of Smarcd1 had not been examined. Moreover, the promoter of the Smarcd1 gene is methylated in mouse embryonic fibroblasts but not in ESC (31). We determined that knockdown of Smarcd1 in ESC by three different shRNAs substantially reduced the expression of Smarcd1, especially in the case of shRNA #2 (Fig. 8A). Importantly, unlike a control shRNA, which had no demonstrable effect on the ESC or the expression of Smarcd1, each of the three Smarcd1 shRNAs promoted the differentiation of ESC, as indicated by formation of altered cellular morphology and reduced intensity of AP staining (Fig. 8B). Furthermore, when plated at clonal densities, the percentage of colonies exhibiting the morphology of ESC decreased from >80 to ∼25% in the case of shRNA #2, and there was a large increase in the number and percentage of colonies that exhibited the morphology of mixed or fully differentiated colonies (Fig. 8C). In view of these results, other Sox2-associated proteins that associate with three or more pluripotency-associated factors are also likely to influence the behavior of ESC.
FIGURE 8.
Knockdown of the Sox2-associated protein Smarcd1 in ESC. A, shown is Western blot analysis of Smarcd1 protein levels after knockdown by shRNA constructs. Scrambled shRNA-infected cells served as a control. HDAC1 served as a loading control. Nonspecific signal is labeled with an asterisk. B, photomicrographs of cells infected with lentiviral vectors expressing shRNA constructs against Smarcd1 are presented (shRNA #1, #2, and #3). A non-targeting shRNA construct (Scrambled) served as a negative control. Cells were fixed and stained for AP. shRNA sequences are provided in supplemental Table S8. C, shown are the effects of Smarcd1 knockdown on the self-renewal and morphology of ESC. Scrambled or shRNA infected D3 cells were plated at clonal density 3 days after infection in T25 flasks. Six days later the cells were stained for AP and scored for ES, mixed (Mix), and differentiated (Diff) colonies in 10 random fields by two observers unaware of sample designation. Results are shown as (%) of total colonies. The number above each bar represents the average total number of colonies counted by the two observers.
DISCUSSION
Our studies demonstrate that simultaneously elevating Sox2 and Oct4 together in i-OS-ESC disrupts their self-renewal, whereas simultaneously elevating Oct4, Sox2, Klf4, and c-Myc in i-OSKM-ESC does not disrupt their self-renewal, alter their morphology, or change their overall properties. Using i-OSKM-ESC, we conducted an unbiased proteomic screen of Sox2-associated proteins in undifferentiated ESC. This work identified >70 proteins that associate with Sox2 in ESC. Interestingly, as discussed below, our studies argue that the Sox2-interactome is highly cell type-dependent.
Elevating different combinations of Oct4, Sox2, Klf4, and c-Myc in ESC makes two important points. First, the induction of differentiation of ESC when either Sox2 or Oct4 is elevated is not simply due to changing the ratio between these two master regulators. Previous studies demonstrated that ESC differentiate when Sox2 is elevated ∼2-fold, yet the levels of Oct4 do not change (5). Similarly, when ESC differentiate in response to increases in Oct4, Sox2 levels begin to decrease, at least at the RNA level (3). In contrast, in i-OSKM-ESC, the ratio between Sox2 and Oct4 does not change significantly when the cells are treated with Dox. Thus, changing the relative ratio of Sox2 and Oct4 does not appear to be the primary reason why ESC differentiate when Sox2 or Oct4 are elevated. Second, simply elevating the level of Sox2 and Oct4 together does not automatically induce differentiation. When i-OSKM-ESC are exposed to Dox, cellular levels of Sox2 and Oct4 rise 2–3-fold, yet the cells do not differentiate. Clearly, elevating Klf4 and c-Myc along with Sox2 and Oct4 in these cells has a strong influence over the self-renewal of i-OSKM-ESC. Although the action of Klf4 and c-Myc in this context is far from clear, our studies are consistent with the prominent role played by the combination of Oct4, Sox2, Klf4, and c-Myc during the reprogramming of somatic cells (32). Collectively, our studies suggest that the maintenance of the self-renewal and pluripotency of ESC depends upon a carefully orchestrated balance of multiple master regulators.
i-OSKM-ESC described in this study provided an excellent opportunity to perform an unbiased proteomic screen of proteins that associate with Sox2 in ESC. In this regard, the ability to express epitope-tagged Sox2 enabled us to readily isolate Sox2-protein complexes from ESC. We identified 71 proteins that associate with Sox2, many of which are known to influence the self-renewal and pluripotency of ESC. Moreover, many of the Sox2-associated proteins identified in this study are required for normal mammalian development (supplemental Table S4), and ∼25% of the Sox2-associated proteins have been shown by others to be part of the Oct4-interactome (supplemental Table S5).
When our Sox2-interactome is compared with published proteomic data for other essential pluripotency-associated proteins, it is evident that Sox2 is part of a highly integrated protein-protein interaction landscape (Fig. 7). Equally important, many of the Sox2-associated proteins also associate with several other transcription factors required for the self-renewal and pluripotency of ESC. In this regard, several Sox2-associated proteins (Sall4, Sall1, Esrrb, Smarca5, and Smarcd1) associate with at least 4 of the 10 pluripotency-associated factors. This led us to suggest that proteins found to associate with multiple required transcription factors are also very likely to be required by undifferentiated ESC (33). We tested this possibility by knocking down Smarcd1, which associates with Sox2, Sall4, Esrrb, and Tcfcp2l1 in ESC (supplemental Table S5). As predicted, knockdown of Smarcd1 disrupts the self-renewal of ESC and induces their differentiation (Fig. 8). Interestingly, earlier studies demonstrated that ESC possess a large, multisubunit chromatin remodeling complex known as esBAF (30). This complex contains several apparently interchangeable subunits that contribute to its unique composition. For example, esBAF contains, Brg1 (Smarc4), Baf155 (Smarcc1), and Smarcd1 but not the related subunits Brm (Smarca2), Baf170 (Smarcc2), or Baf60c (Smarcd3), respectively. Although it has been previously reported that the self-renewal of ESC is dependent on Smarca4 and Smarcc1, the role of other subunits in the complex (30), including Smarcd1, had not been examined. Our results indicate that ESC also require Smarcd1. Future studies will be needed to determine whether the knockdown of Smarcd1 prevents the formation of a stable esBAF complex or whether a Smarcd1-depleted esBAF complex is unable to function properly. We also determined that the knockdown of Smarca5, which interacts with four different pluripotency-associated factors (Sox2, Oct4, Tcfcp2l1, and Esrrb - supplemental Table S5), disrupts the self-renewal of ESC and induces their differentiation. Interestingly, previous studies reported that ESC lacking Smarca5 could not be isolated (29); however, it was not determined whether ESC could not be established due to cell death or due to loss of self-renewal capacity coupled with induction of differentiation. Our findings argue for the latter.
To gain a more global understanding of the function of Sox2-associated proteins, we performed gene ontology analysis. DAVID analysis indicated that the Sox2-associated proteins fall into several major categories: transcription, chromatin assembly/modification, macromolecule metabolism, development, cell cycle progression/cell Division, RNA processing, and DNA repair (supplemental Fig. S5 and Table S7). Proteins that fall into the DNA repair category raise an interesting question. Do DNA repair proteins also participate in gene transcription, or does Sox2 participate in DNA repair? Both may be true. A recent study in yeast reported the RPa complex composed of RPa1, -2, and -3, which we demonstrate associates with Sox2 (Fig. 6), plays an important role during transcriptional elongation (34). Additionally, another report demonstrated that the DNA repair XPC nucleotide excision repair complex, composed of Xpc, Rad23b, and Cetn2, is necessary for Sox2 and Oct4 to activate the expression of the Nanog promoter (35). Interestingly, Sox2 was found to associate in ESC with Rad23 in two of three MudPIT analyses and Cetn2 in one of three analyses (data not shown). In support of the reciprocal possibility, Oct4 has recently been reported to associate with UV-damage chromatin in ESC (36).
Because our proteomic analysis of the Sox2-interactome in ESC was conducted using the same cell system (KH2 ESC), the same level of epitope-tagged Sox2 induction (2-fold) and the same proteomic platform to analyze protein complexes, we also compared the Sox2-interactome in ESC (this report) and in ESC undergoing differentiation (17). Our proteomic analysis indicates that the Sox2-interactome in ESC (Fig. 6) differs significantly from the Sox2-interactome soon after (<24 h) ESC begin to differentiate in response to a 2-fold increase in the level of Sox2 (17). In fact, less than a third of the Sox2-associated proteins are present in the Sox2-interactomes of both ESC and ESC undergoing differentiation (supplemental Fig. S6).
Currently, it is unclear why the Sox2-interactome in ESC differs significantly from the Sox2-interactome in ESC undergoing differentiation. However, other studies that we have conducted indicate that the Sox2-interactome is highly cell type-dependent. In a single MudPIT analysis of Sox2-associated proteins isolated from differentiated cells derived from i-OSKM-ESC after treatment with retinoic acid, only two proteins were found to associate with FLAG-Strep-tagged Sox2 in both datasets, Mta2 and Xpo4 (data not shown). Moreover, our analysis of the Sox-interactome in DAOY cells, a medulloblastoma tumor cell line, identified only seven proteins in common with ESC even though three separate MudPIT analyses identified >200 proteins that associate with FLAG-Strep-tagged Sox2 in DAOY cells (data not shown). Importantly, each of these studies involved the isolation of Sox2-protein complexes using the same protein purification protocol and analysis by the same proteomic platform.
One contributing factor that could help explain the difference between the Sox2-interactome in ESC and ESC undergoing differentiation when Sox2 is elevated on its own (33) is differential expression of the Sox2-associated proteins when ESC begin to differentiate. In this regard, at least four Sox2-associated proteins present in the Sox2-interactome of ESC (Sall1, Rangap2, Rbpj, and Wdr18) eventually decrease when ESC differentiate, whereas the expression of five of the Sox2-associated proteins (Sox21, Msi2, Foxp4, Tial1, and Cstf2) have been reported to increase when ESC differentiate. However, many of these proteins do not change significantly under the conditions used to assess the Sox2-interactome in ESC undergoing differentiation. In this system Oct4 levels do not decrease (5), yet Oct4 is only identified as a Sox2-associated protein in undifferentiated ESC (Fig. 6). Additionally, although the criteria used to classify a protein as being Sox2-associated were stringent, this does not appear to be a major factor for the limited overlap between the Sox2-associated proteins in ESC and ESC undergoing differentiation. For our Sox2-interactome in ESC, included proteins were either present in all three MudPIT analyses or were enriched >6-fold in all 3 MudPIT analyses when i-OSKM-ESC were induced with Dox. Importantly, the majority of proteins identified only in ESC undergoing differentiation (supplemental Fig. S6) were not enriched or identified in any of our proteomic screens of Sox2-associated proteins in i-OSKM-ESC.
Another contributing factor that may explain the change in the Sox2-interactome when ESC begin to differentiate is changes in the post-translational modifications of Sox2-associated proteins. In this regard, changes in the phosphorylation status of several proteins are known to control their interaction with Oct4 and Nanog (37, 38). Remarkably, the human ESC phosphoproteome changes ∼50% 1 h after human ESC are induced to differentiate (39). Thus far, changes in the phosphoproteome of mouse ESC undergoing differentiation have not been reported. However, at least 11 Sox2-associated proteins that are exclusively in the Sox2-interactome in ESC and at least 15 Sox2-associated proteins that are exclusively in the Sox2-interactome in ESC undergoing differentiation have been reported to change their phosphorylation status after 1 h when human ESC begin to differentiate (supplemental Fig. S7).
In conclusion, the studies described in this report demonstrate that simultaneous elevation of four transcription factors, Oct4, Sox2, Klf4, and c-Myc, supports the self-renewal of ESC and provides a means to identify the Sox2 protein interactome in undifferentiated ESC. Importantly, this fills a significant gap in the global proteomic analysis of proteins crucial for maintaining the self-renewal of ESC. Our studies argue that Sox2-interactome in ESC is part of a highly integrated protein-protein interaction landscape that changes significantly shortly after ESC begin to differentiate. Additionally, genome-wide transcription factor binding studies conduced in ESC (10, 11) indicate that many of the genes that code for Sox2-associated proteins are bound by Sox2 and Oct4. Thus, the action of master regulators, such as Sox2 and Oct4, is integrated together at more than one level in ESC.
Supplementary Material
Acknowledgments
We thank Michelle Desler and Phillip Wilder for technical assistance. Core facilities of the UNMC Eppley Cancer Center were supported in part by NCI, National Institutes of Health Cancer Center Support Grant fCA 36727.
This work was supported, in whole or in part, by National Institutes of Health Grant GM 080571. This work was also supported by the Nebraska Department of Health (Stem Cell 2009-01).

This article contains supplemental Tables S1–S8 and Figs. S1–S7.
Microarray data have been deposited with the GEO Repository under accession number GSE34801.
- ESC
- embryonic stem (ES) cells
- i-OS-ESC
- inducible Oct4/(fs)Sox2 embryonic stem cells
- i-OSKM-ESC
- inducible Oct4/(fs)Sox2/Klf4/c-Myc embryonic stem cells
- MudPIT
- multidimensional protein identification technology
- Dox
- doxycycline
- fs
- FLAG-Strep
- dNSAF
- distributed normalized spectral abundance factor
- AP
- alkaline phosphatase
- qPCR
- quantitative PCR.
REFERENCES
- 1. Boer B., Kopp J., Mallanna S., Desler M., Chakravarthy H., Wilder P. J., Bernadt C., Rizzino A. (2007) Elevating the levels of Sox2 in embryonal carcinoma cells and embryonic stem cells inhibits the expression of Sox2:Oct-3/4 target genes. Nucleic Acids Res. 35, 1773–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rizzino A. (2009) Sox2 and Oct-3/4. A versatile pair of master regulators that orchestrate the self-renewal and pluripotency of embryonic stem cells. Wiley Interdiscip. Rev. Syst. Biol. Med. 1, 228–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Niwa H., Miyazaki J., Smith A. G. (2000) Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation, or self-renewal of ES cells. Nat. Genet. 24, 372–376 [DOI] [PubMed] [Google Scholar]
- 4. Chew J. L., Loh Y. H., Zhang W., Chen X., Tam W. L., Yeap L. S., Li P., Ang Y. S., Lim B., Robson P., Ng H. H. (2005) Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol. Cell. Biol. 25, 6031–6046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kopp J. L., Ormsbee B. D., Desler M., Rizzino A. (2008) Small increases in the level of Sox2 trigger the differentiation of mouse embryonic stem cells. Stem Cells 26, 903–911 [DOI] [PubMed] [Google Scholar]
- 6. Mitsui K., Tokuzawa Y., Itoh H., Segawa K., Murakami M., Takahashi K., Maruyama M., Maeda M., Yamanaka S. (2003) The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 113, 631–642 [DOI] [PubMed] [Google Scholar]
- 7. Chambers I., Silva J., Colby D., Nichols J., Nijmeijer B., Robertson M., Vrana J., Jones K., Grotewold L., Smith A. (2007) Nanog safeguards pluripotency and mediates germ line development. Nature 450, 1230–1234 [DOI] [PubMed] [Google Scholar]
- 8. Zhang P., Andrianakos R., Yang Y., Liu C., Lu W. (2010) Kruppel-like factor 4 (Klf4) prevents embryonic stem (ES) cell differentiation by regulating Nanog gene expression. J. Biol. Chem. 285, 9180–9189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cartwright P., McLean C., Sheppard A., Rivett D., Jones K., Dalton S. (2005) LIF/STAT3 controls ES cell self-renewal and pluripotency by a Myc-dependent mechanism. Development 132, 885–896 [DOI] [PubMed] [Google Scholar]
- 10. Boyer L. A., Lee T. I., Cole M. F., Johnstone S. E., Levine S. S., Zucker J. P., Guenther M. G., Kumar R. M., Murray H. L., Jenner R. G., Gifford D. K., Melton D. A., Jaenisch R., Young R. A. (2005) Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen X., Xu H., Yuan P., Fang F., Huss M., Vega V. B., Wong E., Orlov Y. L., Zhang W., Jiang J., Loh Y. H., Yeo H. C., Yeo Z. X., Narang V., Govindarajan K. R., Leong B., Shahab A., Ruan Y., Bourque G., Sung W. K., Clarke N. D., Wei C. L., Ng H. H. (2008) Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133, 1106–1117 [DOI] [PubMed] [Google Scholar]
- 12. Wang J., Rao S., Chu J., Shen X., Levasseur D. N., Theunissen T. W., Orkin S. H. (2006) A protein interaction network for pluripotency of embryonic stem cells. Nature 444, 364–368 [DOI] [PubMed] [Google Scholar]
- 13. Liang J., Wan M., Zhang Y., Gu P., Xin H., Jung S. Y., Qin J., Wong J., Cooney A. J., Liu D., Songyang Z. (2008) Nanog and Oct4 associate with unique transcriptional repression complexes in embryonic stem cells. Nat. Cell Biol. 10, 731–739 [DOI] [PubMed] [Google Scholar]
- 14. van den Berg D. L., Snoek T., Mullin N. P., Yates A., Bezstarosti K., Demmers J., Chambers I., Poot R. A. (2010) An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell 6, 369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pardo M., Lang B., Yu L., Prosser H., Bradley A., Babu M. M., Choudhary J. (2010) An expanded Oct4 interaction network. Implications for stem cell biology, development, and disease. Cell Stem Cell 6, 382–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ding J., Xu H., Faiola F., Ma'ayan A., Wang J. (2012) Oct4 links multiple epigenetic pathways to the pluripotency network. Cell Res. 22, 155–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mallanna S. K., Ormsbee B. D., Iacovino M., Gilmore J. M., Cox J. L., Kyba M., Washburn M. P., Rizzino A. (2010) Proteomic analysis of Sox2-associated proteins during early stages of mouse embryonic stem cell differentiation identifies Sox21 as a novel regulator of stem cell fate. Stem Cells 28, 1715–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cox J. L., Mallanna S. K., Ormsbee B. D., Desler M., Wiebe M. S., Rizzino A. (2011) Banf1 is required to maintain the self-renewal of both mouse and human embryonic stem cells. J. Cell Sci. 124, 2654–2665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Scherer P. E., Lewis R. Y., Volonte D., Engelman J. A., Galbiati F., Couet J., Kohtz D. S., van Donselaar E., Peters P., Lisanti M. P. (1997) Cell-type and tissue-specific expression of caveolin-2. Caveolins 1 and 2 co-localize and form a stable hetero-oligomeric complex in vivo. J. Biol. Chem. 272, 29337–29346 [DOI] [PubMed] [Google Scholar]
- 20. Kelly D., Scholtz B., Orten D. J., Hinrichs S. H., Rizzino A. (1995) Regulation of the transforming growth factor-β 2 gene promoter in embryonal carcinoma cells and their differentiated cells. Differential utilization of transcription factors. Mol. Reprod. Dev. 40, 135–145 [DOI] [PubMed] [Google Scholar]
- 21. Zhang Y., Wen Z., Washburn M. P., Florens L. (2010) Refinements to label free proteome quantitation: how to deal with peptides shared by multiple proteins. Anal. Chem. 82, 2272–2281 [DOI] [PubMed] [Google Scholar]
- 22. Elias J. E., Haas W., Faherty B. K., Gygi S. P. (2005) Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nat. Methods 2, 667–675 [DOI] [PubMed] [Google Scholar]
- 23. Shannon P., Markiel A., Ozier O., Baliga N. S., Wang J. T., Ramage D., Amin N., Schwikowski B., Ideker T. (2003) Cytoscape. A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wiebe M. S., Traktman P. (2007) Poxviral B1 kinase overcomes barrier to autointegration factor, a host defense against virus replication. Cell Host Microbe 1, 187–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Doetschman T. C., Eistetter H., Katz M., Schmidt W., Kemler R. (1985) The in vitro development of blastocyst-derived embryonic stem cell lines. Formation of visceral yolk sac, blood islands, and myocardium. J. Embryol. Exp. Morphol. 87, 27–45 [PubMed] [Google Scholar]
- 26. Beard C., Hochedlinger K., Plath K., Wutz A., Jaenisch R. (2006) Efficient method to generate single-copy transgenic mice by site-specific integration in embryonic stem cells. Genesis 44, 23–28 [DOI] [PubMed] [Google Scholar]
- 27. Karantzali E., Lekakis V., Ioannou M., Hadjimichael C., Papamatheakis J., Kretsovali A. (2011) Sall1 regulates embryonic stem cell differentiation in association with nanog. J. Biol. Chem. 286, 1037–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ivanova N., Dobrin R., Lu R., Kotenko I., Levorse J., DeCoste C., Schafer X., Lun Y., Lemischka I. R. (2006) Dissecting self-renewal in stem cells with RNA interference. Nature 442, 533–538 [DOI] [PubMed] [Google Scholar]
- 29. Stopka T., Skoultchi A. I. (2003) The ISWI ATPase Snf2h is required for early mouse development. Proc. Natl. Acad. Sci. U.S.A. 100, 14097–14102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ho L., Ronan J. L., Wu J., Staahl B. T., Chen L., Kuo A., Lessard J., Nesvizhskii A. I., Ranish J., Crabtree G. R. (2009) An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc. Natl. Acad. Sci. U.S.A. 106, 5181–5186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Farthing C. R., Ficz G., Ng R. K., Chan C. F., Andrews S., Dean W., Hemberger M., Reik W. (2008) Global mapping of DNA methylation in mouse promoters reveals epigenetic reprogramming of pluripotency genes. PLoS Genet. 4, e1000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Takahashi K., Yamanaka S. (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 33. Mallanna S. K., Rizzino A. (2012) Systems biology provides new insights into the molecular mechanisms that control the fate of embryonic stem cells. J. Cell. Physiol. 227, 27–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sikorski T. W., Ficarro S. B., Holik J., Kim T., Rando O. J., Marto J. A., Buratowski S. (2011) Sub1 and RPA associate with RNA polymerase II at different stages of transcription. Mol. Cell 44, 397–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fong Y. W., Inouye C., Yamaguchi T., Cattoglio C., Grubisic I., Tjian R. (2011) A DNA repair complex functions as an Oct4/Sox2 coactivator in embryonic stem cells. Cell 147, 120–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bártová E., Šustáčková G., Stixová L., Kozubek S., Legartová S., Foltánková V. (2011) Recruitment of Oct4 protein to UV-damaged chromatin in embryonic stem cells. PLoS One 6, e27281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Seki Y., Kurisaki A., Watanabe-Susaki K., Nakajima Y., Nakanishi M., Arai Y., Shiota K., Sugino H., Asashima M. (2010) TIF1β regulates the pluripotency of embryonic stem cells in a phosphorylation-dependent manner. Proc. Natl. Acad. Sci. U.S.A. 107, 10926–10931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Moretto-Zita M., Jin H., Shen Z., Zhao T., Briggs S. P., Xu Y. (2010) Phosphorylation stabilizes Nanog by promoting its interaction with Pin1. Proc. Natl. Acad. Sci. U.S.A. 107, 13312–13317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Van Hoof D., Muñoz J., Braam S. R., Pinkse M. W., Linding R., Heck A. J., Mummery C. L., Krijgsveld J. (2009) Phosphorylation dynamics during early differentiation of human embryonic stem cells. Cell Stem Cell 5, 214–226 [DOI] [PubMed] [Google Scholar]
- 40. Murphy C. L., Polak J. M. (2002) Differentiating embryonic stem cells. GAPDH, but neither HPRT nor β-tubulin, is suitable as an internal standard for measuring RNA levels. Tissue Eng. 8, 551–559 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.