Background: The Smc5/6 complex has six non-SMC elements, including Nse5.
Results: Utilizing two mutants of NSE5, we separated Smc5 sumoylation from Smc5/6 complex function.
Conclusion: Nse5 integrity is important for Smc5/6 complex stability, which in turn is essential for localization of the complex to stalled forks.
Significance: Our results provide the first in vivo characterization of Nse5 for Smc5/6 complex function.
Keywords: Cell Cycle, DNA Repair, DNA Replication, Sumoylation, Yeast Genetics, Smc5/6 Complex, Fork Stability, Replication Stress
Abstract
The Smc5/6 complex belongs to the SMC (structural maintenance of chromosomes) family, which also includes cohesin and condensin. In Saccharomyces cerevisiae, the Smc5/6 complex contains six essential non-Smc elements, Nse1–6. Very little is known about how these additional elements contribute to complex function except for Nse2/Mms21, which is an E3 small ubiquitin-like modifier (SUMO) ligase important for Smc5 sumoylation. Characterization of two temperature-sensitive mutants, nse5-ts1 and nse5-ts2, demonstrated the importance of Nse5 within the Smc5/6 complex for its stability and functionality at forks during hydroxyurea-induced replication stress. Both NSE5 alleles showed a marked reduction in Smc5 sumoylation to levels lower than those observed with mms21-11, a mutant of Mms21 that is deficient in SUMO ligase activity. However, a phenotypic comparison of nse5-ts1 and nse5-ts2 revealed a separation of importance between Smc5 sumoylation and the function of the Smc5/6 complex during replication. Only cells carrying the nse5-ts1 allele exhibited defects such as dissociation of the replisome from stalled forks, formation of fork-associated homologous recombination intermediates, and hydroxyurea sensitivity that is additive with mms21-11. These defects are attributed to a failure in Smc5/6 localization to forks in nse5-ts1 cells. Overall, these data support the premise that Nse5 is important for vital interactions between components within the Smc5/6 complex, and for its functionality during replication stress.
Introduction
The SMC (structural maintenance of chromosomes) family of proteins controls chromosomal organization throughout the cell cycle. The Smc5/6 complex is a member of the SMC complex family, which also includes cohesin and condensin (1–3). The Smc5/6 complex is recruited to DNA damage and is enriched at rDNA repeats (4–6). Upon treatment with the damaging agent methyl methanesulfonate (MMS),2 the complex promotes the resolution of hemicatenane-like recombination DNA intermediates that form at replication forks. These intermediates are visualized as X-shaped structures by two-dimensional gel electrophoresis (7–9). A proposed function of the Smc5/6 complex is to give structural organization to collapsed forks, promoting their processing and preserving genome stability (2, 10).
In Schizosaccharomyces pombe, the Smc5/6 complex is recruited to stalled replication forks and is proposed to play a role during hydroxyurea (HU)-induced replication fork stalling by maintaining fork conformation, allowing the loading of homologous recombination (HR) factors Rad52 and replication protein A (11, 12). In budding yeast, recruitment of the Smc5/6 complex has been detected only at HU-stalled replication forks in checkpoint-deficient cells, where stalling leads to fork collapse (4). Moreover, to date, the visualization of recombination intermediates by two-dimensional gel electrophoresis in Smc5/6-deficient cells has been limited to either cells that have been treated with MMS or cells in which replication forks have collapsed (9, 12). Nonetheless, many Smc5/6 hypomorphic alleles are very sensitive to the drug HU, which suggests that the complex also has a fundamental role when forks stall prior to collapse (11–18).
In addition to the two SMC components Smc5 and Smc6, the complex has six non-Smc elements, Nse1–6. The Mms21 (Nse2) component of the Smc5/6 complex is an E3 small ubiquitin-like modifier (SUMO) ligase (19–21). Deletion of the SUMO ligase domain, as in the mms21-11 mutant allele, results in cellular sensitivity to MMS and HU, suggesting that the complex might regulate targets involved in damage tolerance or replication through sumoylation. Indeed, Mms21 contains an SP-RING-like domain and facilitates the SUMO modification of repair proteins, including Yku70 and Smc5 (19); however, deletion of the SUMO ligase domain dramatically reduces but does not eliminate Smc5 sumoylation (19). This could be due to redundancy with other E3 ligases such as Siz1 and Siz2 (22, 23) or, alternatively, could be due to ligase-independent SUMO conjugation by the E2-conjugating enzyme Ubc9 (24). The essential function of Mms21 is not its E3 ligase activity because mms21-11 mutant cells, lacking ligase activity, grow well in the absence of DNA-damaging agents. By contrast, the full disruption of Mms21 is lethal (19, 20, 25).
The Nse1, Nse3, and Nse4 components form a trimeric subcomplex at the head and adjacent region of Smc5 (26). Little is known about the Nse5 and Nse6 subunits, except, like the other components, they are essential in Saccharomyces cerevisiae. Important to this study, high-throughput two-hybrid screens identified SUMO and Ubc9 as binding partners of Nse5 (27); however, to the best of our knowledge, this has not been verified, and the involvement of Nse5 in sumoylation or replication has never been reported.
In this study, we characterized two hypomorphic Nse5 alleles, nse5-ts1 (28) and nse5-ts2, which were generated by PCR-based mutagenesis and identified by reduced growth at 37 °C. Both alleles have a dramatic reduction in Smc5 sumoylation. The nse5-ts1 allele exhibits replication defects that become additive when combined with the E3 ligase mutant mms21-11. In contrast to nse5-ts1, however, there are no detectable replication defects in nse5-ts2 mutants. Our results support a role for the Smc5/6 complex in preventing fork collapse during stalls in replication and suggest that Smc5/6 complex integrity, which is compromised in nse5-ts1 cells, rather than Smc5 sumoylation, is critical for recovery from HU-induced replication stress.
EXPERIMENTAL PROCEDURES
Yeast Strains, Plasmids, Primers, and Antibodies
Yeast strains are listed in supplemental Table S1. All experiments were performed at 25 °C unless indicated otherwise. For yeast two-hybrid analysis, the NSE5 genes were cloned into a pEG202-derived bait plasmid (29), creating Nse5-LexA fusion proteins under the control of a galactose-inducible promoter. SMT3 was cloned into pJG4-6-derived prey vectors (29), creating a B42-activating domain fusion protein under the control of a galactose-inducible promoter. Inserting a stop codon after amino acid 96 created the Smt3ΔGG mutant. All constructs were confirmed by sequencing, and protein expression was confirmed by Western blot analysis with anti-LexA (2-12) and anti-HA (F7) antibodies (Santa Cruz Biotechnology).
Detection of Sumoylated Proteins
Nickel-nitrilotriacetic acid (Ni-NTA) purification of His8-Smt3 was performed as described by Wohlschlegel et al. (30) with the following changes. Pellets of 2 × 109 cells were treated with N-ethylmaleimide (Sigma) to preserve sumoylation. Proteins were immunoprecipitated with Ni-NTA (Qiagen) overnight at 4 °C, followed by washing three times with 6 m guanidine hydrochloride, 100 mm NaH2PO4 (pH 8), and 0.05% Tween 20 and five times with 8 m urea, 100 mm NaH2PO4 (pH 6.3), and 0.05% Tween 20. Beads were resuspended in SDS loading buffer, boiled, and run on 4–20% gradient gels (Bio-Rad).
Two-dimensional Gel Electrophoresis
Samples were psoralen cross-linked, and the purification of DNA intermediates, the two-dimensional gel procedure, and the quantification of replication intermediates were carried out as described previously (31). A complete description of the quantification is provided in supplemental Text S1. The DNA samples were digested with NcoI and analyzed with probes recognizing ARS305. Each experiment was performed independently at least twice with qualitatively identical results. Representative results are shown in the figures.
Chromatin Immunoprecipitation
ChIP/quantitative real-time PCR (qPCR) was performed as described previously (32–34). DNA quantification by real-time PCR was performed on an ABI 7900 sequence detector system at the Southern Alberta Cancer Research Institute. The -fold enrichment represents recovery at the origin sites relative to a late replicating non-origin site as determined by BrdU incorporation.
BrdU/IP-chip and ChIP-chip
BrdU/IP-chip and ChIP-chip experiments were performed as described previously (4, 35) 60 min after release from the mating pheromone α-factor into 0.2 m HU at 25 °C with the following modifications. For FLAG-Smc6 ChIP, 150 ml of culture (A600 = 0.8–1.0) was used. Cells were lysed using a 6870 Freezer/Mill (SPEX). Anti-FLAG® monoclonal antibody M2 (F1804, Sigma) was used, and the recovered DNA was hybridized to S. cerevisiae tiling arrays from Affymetrix® at the Bioinformatics and Expression Analysis Core Facility of Karolinska Institutet. Analysis and map making were performed as described previously (36). Complete maps are included in supplemental Data Sets S1–S4.
Two-hybrid Analysis
Constructs were transformed into JC1280. For drop assays, strains were grown in the absence of glucose and plated on medium containing 2% galactose and lacking His and Trp (to select for plasmids) and additionally Leu (to measure expression from lexAop6-LEU2). Liquid culture yeast two-hybrid assays were also performed in strain JC1280, which was additionally transformed with the lacZ reporter plasmid pSH18034. Protein-protein interactions were detected by quantitative β-galactosidase activity for permeabilized cells and represent the averages of three independent experiments, with error bars indicating S.D. (37).
Co-immunoprecipitation Assays
Cells containing HA-tagged Nse6 and Myc-tagged Smc5 were grown at 25 °C to log phase before cells were lysed with glass beads in lysis buffer (50 mm HEPES, 140 mm NaCl, 1 mm EDTA, and 1% Triton X-100). Protein extracts were applied to anti-Myc antibody-coupled Dynabeads (Invitrogen) and immunoprecipitated for 2 h at 4 °C. Following immunoprecipitation, samples were split and washed by shaking at 1400 rpm for 5 min once in lysis buffer and twice in wash buffer (100 mm Tris (pH 8), 0.5% Nonidet P-40, 1 mm EDTA, and either 300 mm or 1 m NaCl). Beads were resuspended in SDS loading buffer and run on 8% SDS-polyacrylamide gels, followed by Western blotting with anti-HA (F7) and anti-Myc (9E10) antibodies.
RESULTS
Nse5 Interacts with SUMO and Is Required for Smc5 Sumoylation
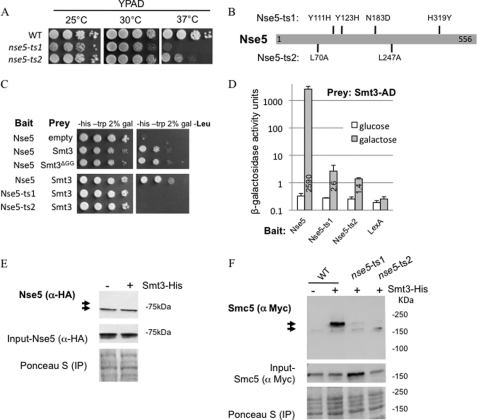
We sought to characterize Nse5 within the Smc5/6 complex by utilizing two temperature-sensitive alleles, nse5-ts1 and nse5-ts2 (Ref. 28 and this study). Both alleles exhibited normal growth at 25 °C; however, nse5-ts1 cells were inviable at 37 °C, and nse5-ts2 cells were extremely slow growing (Fig. 1A). (Unless stated otherwise, all experiments hereafter were performed at 25 °C.) DNA sequencing revealed that nse5-ts1 included four point mutations, Y111H, Y123H, N183D, and H319Y, and that nse5-ts2 had two, L70A and L247A (Fig. 1B).
FIGURE 1.
Nse5 temperature-sensitive alleles nse5-ts1 and nse5-ts2 are unable to interact with SUMO and are defective in Smc5 sumoylation. A, drop assays on YPAD (yeast extract, peptone, adenine, and dextrose) were performed with exponentially growing cultures for which 1:10 serial dilutions were performed, and plates were incubated at 25, 30, or 37 °C to compare WT (JC470), nse5-ts1 (JC1361), and nse5-ts2 (JC1833) cells. B, sequencing of nse5-ts1 revealed four point mutations, Y111H, Y123H, N183D, and H319Y, whereas sequencing of nse5-ts2 revealed two point mutations, L70A and L247A. C and D, yeast two-hybrid analysis (described under “Experimental Procedures”) demonstrated that Nse5, but not Nse5-ts1 or Nse5-ts2, interacted with both WT Smt3 and Smt3ΔGG, a truncation that cannot be conjugated to target proteins. E, Nse5 does not appear to be a target of SUMO modification. Sumoylated proteins were purified from cells expressing endogenously HA-tagged Nse5 with (JC1960) or without (JC1355) His8-tagged Smt3. Proteins were isolated by Ni-NTA affinity purification of His-Smt3 as described previously (30). Western blotting with anti-HA antibody failed to show a higher mobility shift band corresponding to sumoylated Nse5. F, nse5-ts1 and nse5-ts2 cells are deficient in Smc5 sumoylation. Sumoylated proteins were purified from cells expressing endogenously Myc-tagged Smc5 and His8-tagged Smt3. Proteins were isolated by Ni-NTA affinity purification of His-Smt3 as described previously (30). Western blotting with anti-Myc antibody allowed visualization of sumoylated Smc5 in WT cells (JC1157), but not nse5-ts1 (JC1156) or nse5-ts2 (JC1884) cells or cells lacking the Smt3 tag (JC720). IP, immunoprecipitation.
A high-throughput screen previously reported SUMO to be a binding partner of Nse5 (27). Consistent with this, we confirmed that Nse5 interacted with SUMO (Smt3) in yeast two-hybrid analysis (Fig. 1, C and D). To determine whether this interaction represents the sumoylation of Nse5, we performed a Ni-NTA pulldown assay of His-tagged Smt3 in cells carrying HA-tagged Nse5 (30). Western blot analysis with anti-HA antibody indicated that no high-mobility band shifts representing SUMO-modified Nse5 were present (Fig. 1E, lower arrow indicating the unmodified form), suggesting that Nse5 is not a target of sumoylation. Furthermore, Nse5 interacted with a mutant form of SUMO that cannot be conjugated to target proteins, Smt3ΔGG (Fig. 1C), which indicates that Nse5 associates with SUMO via noncovalent interactions. Strikingly, we observed a significant reduction in the association between Nse5 and SUMO for both Nse5-ts1 and Nse5-ts2 (Fig. 1, C and D) despite being overexpressed at similar levels to the wild type (supplemental Fig. S1).
The localization of Mms21 on the coiled-coil arm of Smc5 is adjacent to the Nse5/Nse6-binding site at the hinge domain of the complex (25, 26), and this led us to question whether Nse5 could play a role in facilitating Mms21-dependent sumoylation of Smc5. To investigate this, we performed Ni-NTA pulldown assays of His-tagged Smt3 in cells carrying Myc-tagged Smc5 in wild-type and mutant cells, followed by Western blot analysis with anti-Myc antibody (30). Similar to previous reports (19), Smc5 sumoylation was readily detected in wild-type cells (Fig. 1F); however, there was a marked decrease in the sumoylation of Smc5 in the nse5-ts1 and nse5-ts2 mutants (Fig. 1F).
During Replication Stress, Defects Are Observed in nse5-ts1 Cells
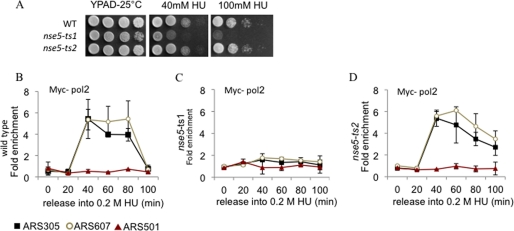
Previous reports have demonstrated that hypomorphic alleles of Smc5/6 components are very sensitive to HU, indicating that the complex likely has a fundamental role when forks stall (11–18). Drop assays showed that nse5-ts1 (but not nse5-ts2) cells are sensitive to HU (Fig. 2A). One measure of replication correctness during replication stress is to monitor replisome association with stalled forks by ChIP. The recovered DNA from the ChIP was quantified by qPCR with primer pairs to two early-firing origins, ARS305 and ARS607, with late-firing ARS501 serving as a negative control (32–34). We determined the association of DNA polymerase ϵ by monitoring Myc-Pol2 recovery with stalled forks when cells were released into S phase in the presence of HU at the indicated time points. Compared with the wild type, we observed a reduction in polymerase association at both early-firing origins in nse5-ts1 cells (Fig. 2, B and C); however, recovery of Myc-Pol2 in nse5-ts2 cells was similar to wild type (Fig. 2D). Furthermore, there was a correlation between DNA polymerase ϵ stability and HU sensitivity (Fig. 2). Taken together with the data in Fig. 1, these results suggest that SUMO modification of Smc5 is not required for Smc5/6 complex functionality in response to HU, nor is Smc5 sumoylation a major contributing factor to survival during replication stress.
FIGURE 2.
nse5-ts1 cells display replisome instability during HU-induced replication stress. A, drop assays (1:10 serial dilutions) with exponentially growing cultures were performed on YPAD ± medium containing the indicated concentrations of HU for WT (JC470), nse5-ts1 (JC1361), and nse5-ts2 (JC1833) cells at 25 °C. B–D, ChIP with anti-Myc antibody 9E10 was performed 25 °C on the following cells released from α-factor into YPAD + 0.2 m HU for Myc-Pol2: WT (JC1805), nse5-ts1 (JC1471), and nse5-ts2 (JC1914) cells. Genomic regions amplified in the ChIP analysis correspond to early-firing origins ARS305 and ARS607 and late-firing origin ARS501 as described previously (32, 34).
Phenotypic Analysis of nse5-ts and mms21-11 Cells
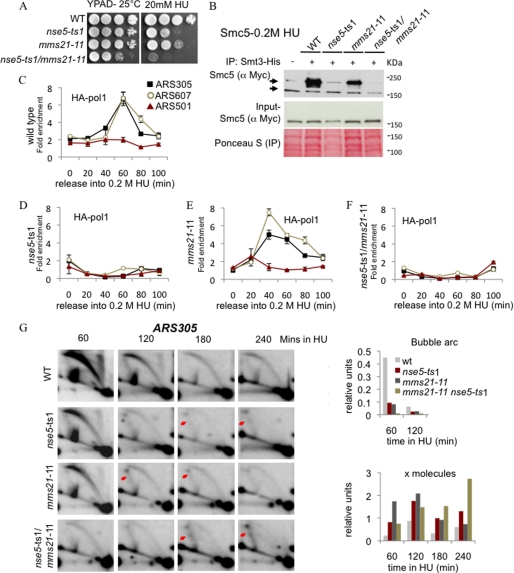
The loss of viability of nse5-ts1 cells after HU treatment and the notable decrease in Smc5 sumoylation for both alleles prompted us to analyze the genetic interactions between NSE5 and the E3 SUMO ligase MMS21. The nse5-ts2/mms21-11 double mutants were not more sensitive to HU than the single mutants alone (supplemental Fig. S2A). However, the nse5-ts1/mms21-11 double mutants grew slowly on rich medium and showed synergistic sensitivity to HU (Fig. 3A and supplemental Fig. S2A). For this reason, we continued our analysis using the nse5-ts1 allele. First, we monitored Smc5 sumoylation, and in a side-by-side comparison, all cells carrying the nse5-ts1 allele were more deficient in Smc5 sumoylation than SUMO ligase-deficient mms21-11 mutants (Fig. 3B).
FIGURE 3.
nse5-ts1 and mms21-11 cells accumulate X-shaped DNA structures during HU treatment. A, drop assays (1:10 serial dilutions) with exponentially growing cultures were performed on YPAD ± medium containing the indicated concentrations of HU for WT (JC470), nse5-ts1 (JC1361), mms21-11 (JC1908), and nse5-ts1/mms21-11 (JC1320) cells at 25 °C. B, Smc5 sumoylation is further reduced in nse5-ts1 than in mms21-11 mutant cells. Smc5 sumoylation was analyzed as described for Fig. 1F in WT (JC1157), nse5-ts1 (JC1156), mms21-11 (JC1155), and nse5-ts1/mms21-11 (JC1124) cells. IP, immunoprecipitation. C–F, ChIP with anti-HA antibody (F7) was performed at 25 °C on the following cells released from α-factor into YPAD + 0.2 m HU for HA-Pol1: WT (JC1805), nse5-ts1 (JC1471), mms21-11 (JC1718), and nse5-ts1/mms21-11 (JC1804) cells. G, two-dimensional gel analysis comparing WT (JC470), nse5-ts1 (JC1361), mms21-11 (JC1908), and nse5-ts1/mms21-11 (JC1320) cells that were arrested in G1 with α-factor and released into YPAD + 0.2 m HU at 25 °C. The replication and recombination intermediates at the ARS305 locus at 60, 120, 180, and 240 min after release into HU were visualized by two-dimensional gel electrophoresis, followed by Southern blot analysis. Arrows indicate the accumulated X-shaped DNA molecules.
Characterization of nse5-ts1/mms21-11 cells involved monitoring the association of replisome components with forks by ChIP. DNA polymerases α and ϵ, as well as replication protein A, were measured when cells were released from α-factor into S phase in the presence of HU. For Myc-Rfa1, the 70-kDa subcomponent of replication protein A, we observed very little difference between the wild type and any of the mutants (supplemental Fig. S3, A–D). Fork-associated DNA polymerases α and ϵ were determined using HA-Pol1 and Myc-Pol2, respectively. We observed a reduction in polymerase association at ARS305 and ARS607 that was most pronounced in cells carrying the nse5-ts1 allele; however, mms21-11 cells looked very similar to the wild type (Fig. 2, C–F, and supplemental Fig. S3, E–G). Compared with mms21-11, nse5-ts1 appears to be a more penetrant allele when forks initially stall, as there was little correlation between Mms21 ligase activity and replisome association.
To understand further the molecular basis of the HU sensitivity, we examined the kinetics and pattern of replication intermediates in nse5-ts1 and mms21-11 cells by two-dimensional gel analysis. Psoralen cross-linking has allowed for enhanced detection of low-abundance X-shaped structures. Using a probe to ARS305 (supplemental Fig. S4), we observed that bubble intermediates derived from origin firing and sister forks moving apart were significantly reduced in both nse5-ts1 and mms21-11 mutants and were almost absent in nse5-ts1/mms21-11 cells in the presence of HU (Fig. 3G). This could be due to problems in origin firing or an increased propensity for fork collapse. In line with the latter hypothesis, we observed that at late time points, X-shaped DNA molecules accumulated in all mutants. This phenotype was most pronounced in nse5-ts1/mms21-11 double-mutant cells at 240 min following HU treatment and suggests that replication forks had potentially regressed or had been remodeled into recombination-like structures in an attempt to repair (Fig. 3G). We noted that in two-dimensional gel analysis, nse5-ts2 cells did not accumulate X-shaped DNA structures and exhibited wild-type levels of initiation (supplemental Fig. S2B).
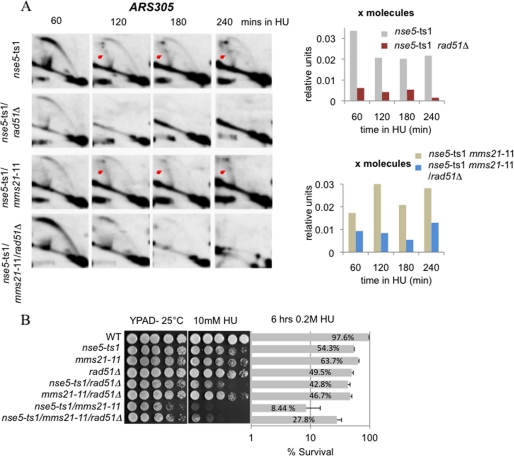
Given the involvement of the Smc5/6 complex in HR, we determined if the accumulation of X-shaped intermediates in HU-treated nse5-ts1 mutants is dependent on Rad51 (8, 38). Similar to previous reports of MMS exposure (8), the accumulation of X-shaped molecules after HU treatment in mms21-11 cells was Rad51-dependent (supplemental Fig. S5). Strikingly, in both nse5-ts1/rad51Δ and mms21-11/nse5-ts1/rad51Δ mutants, the X-shaped structures no longer persisted (Fig. 4A), indicating that their accumulation depends on Rad51-mediated recombination at HU-stalled forks rather than reversed forks that form upon replisome dissociation. Finally, we observed a partial suppression of HU sensitivity in nse5-ts1/mms21-11 mutants when deleting RAD51 (Fig. 4B), suggesting that aberrant HR structures, which persist at HU-stalled forks, contribute to the loss of cell viability.
FIGURE 4.
Formation of X-shaped molecules in nse5-ts1 mutants is Rad51-dependent. A, two-dimensional gel analysis comparing nse5-ts1 (JC1361), nse5-ts1/rad51Δ (JC2031), nse5-ts1/mms21-11 (JC1320), and nse5-ts1/mms21-11/rad51Δ (JC2030) cells that were arrested in G1 with α-factor and released into YPAD + 0.2 m HU at 25 °C. The replication and recombination intermediates at the ARS305 locus at 60, 120, 180, and 240 min after release into HU were visualized by two-dimensional gel electrophoresis, followed by Southern blot analysis. Arrows indicate the accumulated X-shaped DNA molecules. B, drop assays (1:5 serial dilutions) with exponentially growing cultures were performed on YPAD ± medium containing 10 mm HU, and cell viability was monitored as colony outgrowth from asynchronous cultures after transient exposure to 0.2 m HU for 6 h at 25 °C, with values normalized to survival at time point 0 for WT (JC470), nse5-ts1 (JC1361), mms21-11 (JC1908), rad51Δ (JC1362), nse5-ts1/rad51Δ (JC2031), mms21-11/rad51Δ (JC2032), nse5-ts1/mms21-11 (JC1320), and nse5-ts1/mms21-11/rad51Δ (JC2030) cells.
The Smc5/6 Complex Shows Reduced Recruitment to Stalled Forks in nse5-ts1 Mutants
We wanted to understand the source of replication defects in nse5-ts1 cells. First, we monitored replication kinetics in wild-type and nse5-ts1 cells synchronized in G1 and released into S phase in the presence of HU and BrdU. Newly synthesized DNA was labeled with BrdU for 60 min before immunoprecipitation and hybridization onto yeast genome tiling arrays (Fig. 5A). Consistent with both the two-dimensional gel analysis and ChIP for the replisome components (Fig. 3), cells harboring the nse5-ts1 allele exhibited a noticeable decrease in BrdU incorporation; however, sites of replication initiation remained identical to the wild type. Importantly, the lengths of the BrdU tracks were similar between wild-type and nse5-ts1 cells (Fig. 5A), suggesting similar fork rates. This is important because faster progression could have accounted for the reduction in replisome components detected at ARS305 and ARS607 in nse5-ts1 mutants and lower bubble and Y-arc signals by two-dimensional gel analysis (Fig. 3). Taken together, the data strongly support the interpretation that the loss of polymerase recovery in nse5-ts1 cells represents a decrease in replisome association with stalled replication forks. Finally, no enrichment was observed with BrdU at late-firing origins in nse5-ts1 cells (Fig. 5A), indicating that the S phase checkpoint remained intact.
FIGURE 5.
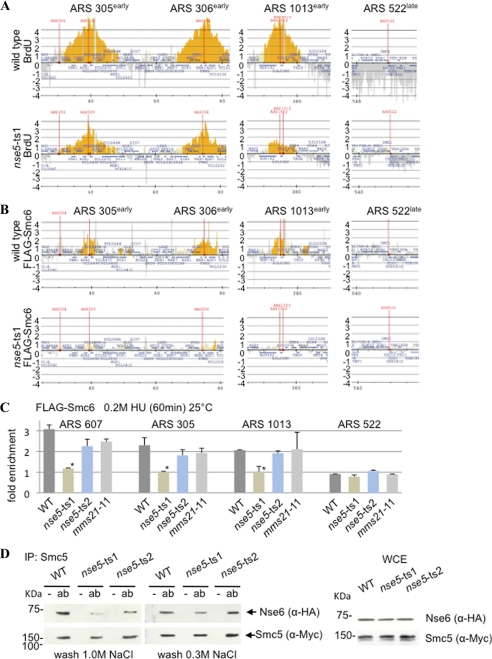
Smc6 recruitment to stalled forks is reduced in nse5-ts1 mutants. BrdU/IP-chip (A) or ChIP-chip (B) with anti-FLAG antibody M2 was performed for FLAG-Smc6 in WT (CB207) and nse5-ts1 (CB1795) cells released from α-factor into 0.2 m HU for 60 min at 25 °C. The signal intensity ratio on a log 2 scale is shown on the y axis, and the chromosome coordinates are shown on the x axis for chromosomes III and X. All coordinates in the yeast genome are available in supplemental Data Sets S1–S4. C, ChIP-qPCR with anti-FLAG antibody M2 was performed for FLAG-Smc6 in WT (JC1594), nse5-ts1 (JC1665), nse5-ts2 (JC1913), and mms21-11 (JC2075) cells that were arrested in G1 with α-factor and released into YPAD + 0.2 m HU at 25 °C as described in the legend to Fig. 2 but with more stringent wash conditions, including 300 mm NaCl and 250 mm LiCl. A probability of p = 0.001 when comparing t values of nse5-ts1 and wild type (*) indicates that the differences are statistically significant. D, shown are the results from co-immunoprecipitation of Myc-tagged Smc5 and HA-tagged Nse6 (as described under “Experimental Procedures”) with washes performed under conditions of 0.3 m NaCl or 1.0 m NaCl in WT (JC2229), nse5-ts1 (JC2230), and nse5-ts2 (JC2231) cells. IP, immunoprecipitation; ab, antibody; WCE, whole cell extract.
The prominent replication phenotypes seen in nse5-ts1 cells prompted us to investigate whether the mutant allele influences Smc5/6 complex localization during replication stress. Although there was evidence for recruitment of the complex to HU-stalled forks in S. pombe by ChIP-qPCR (12), the presence of the complex at forks in S. cerevisiae has not been definitively established (4). We took a genome-wide approach and performed ChIP with FLAG-Smc6, as a marker for the Smc5/6 complex during stalls in replication, followed by hybridization onto tiling arrays. Cell lysis was carried out in liquid nitrogen, and an alternative antibody (anti-FLAG® monoclonal antibody M2) allowed for more sensitive detection of FLAG-Smc6 at HU-arrested forks. Using this methodology, Smc6 was consistently recovered at HU-stalled replication forks in wild-type cells (Fig. 5B), suggesting that its role in fork maintenance is direct in nature. There was a striking reduction in the recovery of Smc6 at sites of replication by both ChIP-chip and ChIP-qPCR in nse5-ts1 mutants (Fig. 5, B and C, and supplemental Fig. S6A). In contrast to nse5-ts1, localization of the Smc5/6 complex to stalled forks in nse5-ts2 and mms21-11 mutants was not statistically different from the wild type (Fig. 5C). These data indicate that in nse5-ts1 cells, defects during replication stress arise because the Smc5/6 complex fails to be recruited to HU-stalled forks.
There are a least two conceivable explanations for why the Smc5/6 complex is not recruited to stalled forks in nse5-ts1 mutants. First, Nse5 could serve as the “recruiting factor” for complex localization, and important interactions are lost in nse5-ts1 cells. An alternative explanation is that the overall stability of the Smc5/6 complex is compromised in cells carrying the nse5-ts1 allele; therefore, it does not properly localize. To assess this, we monitored complex stability by measuring the association of Nse6 (the binding partner of Nse5) with Smc5 by co-immunoprecipitation. Nse6 was recovered with Smc5 in wild-type cells after washing in a high concentration of salt (1.0 m NaCl) (Fig. 5D). In nse5-ts1 mutants, there was a visible decrease in the recovery of Nse6 with Smc5; however, the complex did not completely dissociate because interactions were detected under low-salt conditions (0.3 m NaCl) (Fig. 5D). The loss of complex stability in nse5-ts1 mutants was not a result of lowered Nse5 protein levels, which were equivalent to the wild type (supplemental Fig. S6B). For nse5-ts2 cells, the recovery of Nse6 with Smc5 was slightly reduced, but not to the level of nse5-ts1 mutants (Fig. 5D), and Nse6-Smc5 association was indistinguishable from the wild type in 0.3 m NaCl (Fig. 5D). Taken together, our data suggest that Nse5 is essential for stable Smc5/6 complex association, and this is paramount for complex functionality at stalled forks.
DISCUSSION
One distinguishing feature of the Smc5/6 complex is the presence of additional components within the complex, namely Nse1–6. We have investigated the importance of Nse5 for Smc5/6 complex function by utilizing two mutant alleles, nse5-ts1 and nse5-ts2. We have demonstrated by three independent methods (two-dimensional gel analysis, ChIP-qPCR of replisome components, and BrdU/IP-chip) that cells carrying the nse5-ts1 allele have fork-associated defects under conditions of nucleotide depletion-associated stress. The data presented here are the first to show that Nse5 is essential for the complex integrity of Smc5/6, which in turn is necessary for its recruitment and functionality at stalled forks.
Nse5 interacts with SUMO, and our data suggest that this is through noncovalent interactions because Nse5 itself does not appear to be sumoylated. Although it is unclear what drives these interactions, both mutants show a dramatic decrease in interacting with SUMO when overexpressed in yeast two-hybrid analysis and a clear reduction of Smc5 sumoylation in vivo. The characterization of nse5-ts1 and nse5-ts2 mutants was instrumental in determining the significance of Smc5 sumoylation during replication stress. In many ways, the nse5-ts2 mutant we generated can be viewed as a separation of function allele. We observed wild-type patterns of replication intermediates and survival after HU treatment of nse5-ts2 mutants, which uncouples Smc5 sumoylation from Smc5/6 complex function at stalled forks. Smc5 sumoylation could be a “bystander effect” because of its proximity to the E3 ligase Mms21. The identification and mutation of the target lysine in Smc5 will be a necessary tool to address conclusively the functional outcome of Smc5 sumoylation.
The Smc5/6 complex is integrally involved in the resolution of DNA repair intermediates that form at collapsed replication forks. Indeed, several groups have demonstrated that recombination structures persist at collapsed forks in smc5/6 mutants (8, 15, 39). What we have shown new here is that in nse5-ts1 and mms21-11 cells, X-shaped structures arise when forks are stalled by HU. This suggests that the Smc5/6 complex also has a role during stalling to prevent fork collapse. Moreover, we have shown that the formation and resolution of these intermediates do not depend on Smc5 sumoylation, as X-shaped structures do not accumulate in nse5-ts2 cells, which are clearly deficient in Smc5 sumoylation. These data suggest that Smc5 sumoylation is not a prerequisite for complex functionality in response to HU or a major contributing factor to survival during replication stress. Nonetheless, these findings do not detract from a clear role for the Smc5/6 complex when forks stall. We have demonstrated that the Smc5/6 complex helps prevent replisome dissociation when forks stall, an event that would precede HR resolution and that is disrupted in nse5-ts1 mutants.
The accumulation of Rad51-dependent X-shaped structures at stalled forks in mms21-11 and nse5-ts1 cells is consistent with the model that the Smc5/6 complex 1) prevents fork collapse and subsequent recombination-mediated fork restart and 2) promotes the resolution of such intermediates if/when they form. Our data are in line with those of Irmisch et al. (39) and the concept that the Smc5/6 complex has an “early” and “late” function during HU-induced fork stalling. The data shown here suggest that the early function involves complex localization, which helps stabilize the replisome through a mechanism in which Mms21 ligase activity is not that critical. Indeed, in contrast to nse5-ts1 cells, in which the complex fails to properly localize, replisome stability at stalled forks in mms21-11 mutants remains largely intact. X-shaped molecules form during prolonged HU stalling in nse5-ts1 and mms21-11 mutants, suggesting that forks either eventually collapse or try to restart via HR. The activity of Mms21 is important for the late function of resolving HR intermediates. Our data also suggest that in mms21-11 mutants, the defects in HR resolution will ultimately be attributed to a misregulation in the sumoylation of yet-to-be determined targets, as Smc5 sumoylation appears dispensable. The identification of these targets will be critical for understanding the role of the Smc5/6 complex at stalled forks in its entirety.
Supplementary Material
Acknowledgments
We are grateful to E. Johnson for strains and very helpful discussions. We thank Damien D'Amours for instructive co-immunoprecipitation suggestions; X. Zhao for sharing anti-SUMO antibody; and S. Gasser, P. Hieter, and S. Ben-Aroya for strains. We thank all members of the Cobb laboratory for helpful discussions. The ABI 7900 sequence detector system at the Southern Alberta Cancer Research Institute was provided by the Alberta Cancer Foundation.
This work was supported by grants from the Alberta Heritage Foundation for Medical Research (AHFMR), Alberta Cancer Board Grant 23575, and Canadian Institutes of Health Research Grant MOP-82736 (to J. A. C.); Associazione Italiana per la Ricerca sul Cancro (AIRC) Grant IG 10637 and European Research Council (ERC) Grant REPSUBREP242928 (to D. B.); and the European Research Council, the Swedish Research Council, the Swedish Cancer Society, Vinnova, and the Swedish Foundation for Strategic Research (SSF) (to C. S.).

This article contains supplemental Figs. S1–S6, Tables S1 and S2, Text S1, and Datasets S1–S4.
- MMS
- methyl methanesulfonate
- HU
- hydroxyurea
- HR
- homologous recombination
- SUMO
- small ubiquitin-like modifier
- Ni-NTA
- nickel-nitrilotriacetic acid
- qPCR
- quantitative real-time PCR.
REFERENCES
- 1. Murray J. M., Carr A. M. (2008) Smc5/6: a link between DNA repair and unidirectional replication? Nat. Rev. Mol. Cell Biol. 9, 177–182 [DOI] [PubMed] [Google Scholar]
- 2. De Piccoli G., Torres-Rosell J., Aragón L. (2009) The unnamed complex: what do we know about Smc5/6? Chromosome Res. 17, 251–263 [DOI] [PubMed] [Google Scholar]
- 3. Potts P. R. (2009) The Yin and Yang of the MMS21-SMC5/6 SUMO ligase complex in homologous recombination. DNA Repair 8, 499–506 [DOI] [PubMed] [Google Scholar]
- 4. Lindroos H. B., Ström L., Itoh T., Katou Y., Shirahige K., Sjögren C. (2006) Chromosomal association of the Smc5/6 complex reveals that it functions in differently regulated pathways. Mol. Cell 22, 755–767 [DOI] [PubMed] [Google Scholar]
- 5. Torres-Rosell J., Machín F., Farmer S., Jarmuz A., Eydmann T., Dalgaard J. Z., Aragón L. (2005) SMC5 and SMC6 genes are required for the segregation of repetitive chromosome regions. Nat. Cell Biol. 7, 412–419 [DOI] [PubMed] [Google Scholar]
- 6. Pebernard S., Schaffer L., Campbell D., Head S. R., Boddy M. N. (2008) Localization of Smc5/6 to centromeres and telomeres requires heterochromatin and SUMO, respectively. EMBO J. 27, 3011–3023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Choi K., Szakal B., Chen Y. H., Branzei D., Zhao X. (2010) The Smc5/6 complex and Esc2 influence multiple replication-associated recombination processes in Saccharomyces cerevisiae. Mol. Biol. Cell 21, 2306–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Branzei D., Sollier J., Liberi G., Zhao X., Maeda D., Seki M., Enomoto T., Ohta K., Foiani M. (2006) Ubc9- and mms21-mediated sumoylation counteracts recombinogenic events at damaged replication forks. Cell 127, 509–522 [DOI] [PubMed] [Google Scholar]
- 9. Sollier J., Driscoll R., Castellucci F., Foiani M., Jackson S. P., Branzei D. (2009) The Saccharomyces cerevisiae Esc2 and Smc5/6 proteins promote sister chromatid junction-mediated intra-S repair. Mol. Biol. Cell 20, 1671–1682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hwang J. Y., Smith S., Ceschia A., Torres-Rosell J., Aragón L., Myung K. (2008) Smc5/6 complex suppresses gross chromosomal rearrangements mediated by break-induced replications. DNA Repair 7, 1426–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Miyabe I., Morishita T., Hishida T., Yonei S., Shinagawa H. (2006) Rhp51-dependent recombination intermediates that do not generate checkpoint signal are accumulated in Schizosaccharomyces pombe rad60 and smc5/6 mutants after release from replication arrest. Mol. Cell. Biol. 26, 343–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ampatzidou E., Irmisch A., O'Connell M. J., Murray J. M. (2006) Smc5/6 is required for repair at collapsed replication forks. Mol. Cell. Biol. 26, 9387–9401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pebernard S., McDonald W. H., Pavlova Y., Yates J. R., 3rd, Boddy M. N. (2004) Nse1, Nse2, and a novel subunit of the Smc5/6 complex, Nse3, play a crucial role in meiosis. Mol. Biol. Cell 15, 4866–4876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pebernard S., Wohlschlegel J., McDonald W. H., Yates J. R., 3rd, Boddy M. N. (2006) The Nse5-Nse6 dimer mediates DNA repair roles of the Smc5/6 complex. Mol. Cell. Biol. 26, 1617–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen Y. H., Choi K., Szakal B., Arenz J., Duan X., Ye H., Branzei D., Zhao X. (2009) Interplay between the Smc5/6 complex and the Mph1 helicase in recombinational repair. Proc. Natl. Acad. Sci. U.S.A. 106, 21252–21257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chavez A., Agrawal V., Johnson F. B. (2011) Homologous recombination-dependent rescue of deficiency in the structural maintenance of chromosomes (Smc) 5/6 complex. J. Biol. Chem. 286, 5119–5125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cost G. J., Cozzarelli N. R. (2006) Smc5p promotes faithful chromosome transmission and DNA repair in Saccharomyces cerevisiae. Genetics 172, 2185–2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hu B., Liao C., Millson S. H., Mollapour M., Prodromou C., Pearl L. H., Piper P. W., Panaretou B. (2005) Qri2/Nse4, a component of the essential Smc5/6 DNA repair complex. Mol. Microbiol. 55, 1735–1750 [DOI] [PubMed] [Google Scholar]
- 19. Zhao X., Blobel G. (2005) A SUMO ligase is part of a nuclear multiprotein complex that affects DNA repair and chromosomal organization. Proc. Natl. Acad. Sci. U.S.A. 102, 4777–4782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Andrews E. A., Palecek J., Sergeant J., Taylor E., Lehmann A. R., Watts F. Z. (2005) Nse2, a component of the Smc5/6 complex, is a SUMO ligase required for the response to DNA damage. Mol. Cell. Biol. 25, 185–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Potts P. R., Yu H. (2005) Human MMS21/NSE2 is a SUMO ligase required for DNA repair. Mol. Cell. Biol. 25, 7021–7032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Johnson E. S., Gupta A. A. (2001) An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 106, 735–744 [DOI] [PubMed] [Google Scholar]
- 23. Takahashi Y., Toh-e A., Kikuchi Y. (2001) A novel factor required for the SUMO1/Smt3 conjugation of yeast septins. Gene 275, 223–231 [DOI] [PubMed] [Google Scholar]
- 24. Johnson E. S., Blobel G. (1997) Ubc9p is the conjugating enzyme for the ubiquitin-like protein Smt3p. J. Biol. Chem. 272, 26799–26802 [DOI] [PubMed] [Google Scholar]
- 25. Duan X., Sarangi P., Liu X., Rangi G. K., Zhao X., Ye H. (2009) Structural and functional insights into the roles of the Mms21 subunit of the Smc5/6 complex. Mol. Cell 35, 657–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Duan X., Yang Y., Chen Y. H., Arenz J., Rangi G. K., Zhao X., Ye H. (2009) Architecture of the Smc5/6 complex of Saccharomyces cerevisiae reveals a unique interaction between the Nse5/6 Subcomplex and the Hinge Regions of Smc5 and Smc6. J. Biol. Chem. 284, 8507–8515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hazbun T. R., Malmström L., Anderson S., Graczyk B. J., Fox B., Riffle M., Sundin B. A., Aranda J. D., McDonald W. H., Chiu C. H., Snydsman B. E., Bradley P., Muller E. G., Fields S., Baker D., Yates J. R., 3rd, Davis T. N. (2003) Assigning function to yeast proteins by integration of technologies. Mol. Cell 12, 1353–1365 [DOI] [PubMed] [Google Scholar]
- 28. Ben-Aroya S., Coombes C., Kwok T., O'Donnell K. A., Boeke J. D., Hieter P. (2008) Toward a comprehensive temperature-sensitive mutant repository of the essential genes of Saccharomyces cerevisiae. Mol. Cell 30, 248–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Aushubel F. M., Brent R., Kinston R., Moore D., Seidman J. J., Smith J., Struhl K. (1994) Current Protocols in Molecular Biology, Unit 13.14.1–13.14.17, John Wiley & Sons, New York [Google Scholar]
- 30. Wohlschlegel J. A., Johnson E. S., Reed S. I., Yates J. R., 3rd (2006) Improved identification of SUMO attachment sites using C-terminal SUMO mutants and tailored protease digestion strategies. J. Proteome Res. 5, 761–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vanoli F., Fumasoni M., Szakal B., Maloisel L., Branzei D. (2010) Replication and recombination factors contributing to recombination-dependent bypass of DNA lesions by template switch. PLoS Genet. 6, 1–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tittel-Elmer M., Alabert C., Pasero P., Cobb J. A. (2009) The MRX complex stabilizes the replisome independently of the S phase checkpoint during replication stress. EMBO J. 28, 1142–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cobb J. A., Schleker T., Rojas V., Bjergbaek L., Tercero J. A., Gasser S. M. (2005) Replisome instability, fork collapse, and gross chromosomal rearrangements arise synergistically from Mec1 kinase and RecQ helicase mutations. Genes Dev. 19, 3055–3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Cobb J. A., Bjergbaek L., Shimada K., Frei C., Gasser S. M. (2003) DNA polymerase stabilization at stalled replication forks requires Mec1 and the RecQ helicase Sgs1. EMBO J. 22, 4325–4336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Katou Y., Kaneshiro K., Aburatani H., Shirahige K. (2006) Genomic approach for the understanding of dynamic aspect of chromosome behavior. Methods Enzymol. 409, 389–410 [DOI] [PubMed] [Google Scholar]
- 36. Kegel A., Betts-Lindroos H., Kanno T., Jeppsson K., Ström L., Katou Y., Itoh T., Shirahige K., Sjögren C. (2011) Chromosome length influences replication-induced topological stress. Nature 471, 392–396 [DOI] [PubMed] [Google Scholar]
- 37. Bjergbaek L., Cobb J. A., Tsai-Pflugfelder M., Gasser S. M. (2005) Mechanistically distinct roles for Sgs1p in checkpoint activation and replication fork maintenance. EMBO J. 24, 405–417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liberi G., Maffioletti G., Lucca C., Chiolo I., Baryshnikova A., Cotta-Ramusino C., Lopes M., Pellicioli A., Haber J. E., Foiani M. (2005) Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev. 19, 339–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Irmisch A., Ampatzidou E., Mizuno K., O'Connell M. J., Murray J. M. (2009) Smc5/6 maintains stalled replication forks in a recombination-competent conformation. EMBO J. 28, 144–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.