Background: UDP-galactopyranose mutase (UGM) is a critical enzyme for the proper formation of the cell wall of pathogenic microbes.
Results: The structure of UGM in complex with substrate reveals novel features of substrate binding.
Conclusion: Oxidation and reduction of the flavin cofactor causes rearrangements in the active site that affect substrate binding.
Significance: These are the first structures of UGM from a eukaryotic pathogen.
Keywords: Carbohydrate-binding Protein, Cell Wall, Enzyme Mutation, Enzyme Structure, Membrane Biogenesis
Abstract
UDP-galactopyranose mutase (UGM) is a flavin-containing enzyme that catalyzes the reversible conversion of UDP-galactopyranose (UDP-Galp) to UDP-galactofuranose (UDP-Galf). As in prokaryotic UGMs, the flavin needs to be reduced for the enzyme to be active. Here we present the first eukaryotic UGM structures from Aspergillus fumigatus (AfUGM). The structures are of UGM alone, with the substrate UDP-Galp and with the inhibitor UDP. Additionally, we report the structures of AfUGM bound to substrate with oxidized and reduced flavin. These structures provide insight into substrate recognition and structural changes observed upon substrate binding involving the mobile loops and the critical arginine residues Arg-182 and Arg-327. Comparison with prokaryotic UGM reveals that despite low sequence identity with known prokaryotic UGMs the overall fold is largely conserved. Structural differences between prokaryotic UGM and AfUGM result from inserts in AfUGM. A notable difference from prokaryotic UGMs is that AfUGM contains a third flexible loop (loop III) above the si-face of the isoalloxazine ring that changes position depending on the redox state of the flavin cofactor. This loop flipping has not been observed in prokaryotic UGMs. In addition we have determined the crystals structures and steady-state kinetic constants of the reaction catalyzed by mutants R182K, R327K, R182A, and R327A. These results support our hypothesis that Arg-182 and Arg-327 play important roles in stabilizing the position of the diphosphates of the nucleotide sugar and help to facilitate the positioning of the galactose moiety for catalysis.
Introduction
Galactofuranose (Galf)3 residues are important building blocks for cell wall synthesis within many pathogenic microorganisms, including Mycobacterium tuberculosis and Aspergillus fumigatus. UDP-galactofuranose (UDP-Galf) is the active precursor for production of Galf residues found in Galf-containing glycoconjugates. In eukaryotic species, Galf is found in a number of different cell coat components. In Aspergillus spp. Galf is found in the galactomannan layer, glycoprotein oligosaccharides, and glycolipids. Although Galf is found in glycoinositol phospholipids within both Leishmania major and Trypanosoma cruzi, it is also found in the lipophosphoglycan of Leishmania spp. and in glycoprotein oligosaccharides of T. cruzi (1).
UDP-galactopyranose mutase (UGM) is the enzyme responsible for interconversion of UDP-galactopyranose (UDP-Galp) and UDP-Galf. The critical role Galf plays in the pathogenicity of many microorganisms combined with the fact that UGM and Galf are not found in humans makes UGM an attractive target for drug discovery (2–4).
Although eukaryotic UGMs are less well characterized than the prokaryotic homologues and the sequence conservation between prokaryotic and eukaryotic UGMs is low (15–20%), a number of features are conserved, including the presence of FAD (5, 6). UGM has been identified in the following eukaryotic species: L. major, T. cruzi, Caenorhabaditis elegans, A. fumigatus, Aspergillus nidulans, and Aspergillus niger (6–12). Deletion of UGM from L. major, A. fumigatus, A. nidulans, and A. niger revealed the corresponding knock-out strains were all completely lacking in Galf and exhibited marked alterations to their cell coats (11, 13–15). Additionally, infection of mice by UGM deletion strains of either L. major or A. fumigatus showed significant attenuation to virulence (14, 16).
UGM has been well characterized from a number of bacteria, including Escherichia coli, Klebsiella pneumonia, M. tuberculosis, and Deinococcus radiodurans (17–20). The bacterial enzyme is a flavoprotein found as a dimer, which requires the co-factor to be in the reduced state for activity (21). The structures of prokaryotic UGMs with and without bound substrate have also been reported (20–24). Examination of these structures confirmed that the active site of UGM closes around the substrate as it binds and is controlled by an invariant arginine residue that interacts with the diphosphate of the substrate (5, 20, 24).
Here we present the first eukaryotic structures of UDP-galactopyranose mutase, from A. fumigatus (AfUGM). Despite low sequence identity with known prokaryotic UGMs, the overall fold is largely conserved. The observed structural differences between prokaryotic UGMs and AfUGM are attributed to additional inserts present in AfUGM. Additionally, we have determined the structures of AfUGM complexed with the substrate UDP-Galp and with UDP. These structures allowed for the rationalization of the previously observed differences between eukaryotic and prokaryotic UGMs.
EXPERIMENTAL PROCEDURES
Mutagenesis of AfUGM Active Site Residues
A pET22b plasmid containing the AfUGM gene with a C-terminal His tag (E. coli clone 2212) was used for overexpression of AfUGM (14). Site-directed mutagenesis of the AfUGM mutants (R182K, R182A, R327K, and R327A) was performed using the QuikChangeTM site-directed mutagenesis kit (Stratagene, Inc.) according to the manufacturer's protocol. Sense and antisense primers for all mutants were designed according to the manufacturer's protocol. Overexpression vector pET22b harboring the AfUGM gene was used as the template DNA. The PCR mixture contained 50 ng of template DNA and 15 pmol of each primer. PCR amplifications were carried out in a GeneAmp PCR PTC100 System. The original methylated plasmid was digested with DpnI, and 2 μl of each reaction was used to transform E. coli DH5α competent cells (Novagen). Ampicillin-resistant colonies were selected from the LB plates, and the specific mutations were verified by DNA sequencing (NRC-PBI). The plasmid DNA was isolated from DH5α cells and transformed into BL21-Gold cells (Novagen) for protein overexpression.
Crystallization of AfUGM
AfUGM was overexpressed, purified, and crystallized as previously described (25). All crystallization experiments were carried out using the microbatch method (26). For detailed description of the crystallization experiments, including actual crystallization buffers, see supplemental Text. Briefly drops were prepared by mixing equal volumes of protein solution (with or without ligand) and crystallization solution and overlaid with oil. Plate-like crystals were obtained within 1 week. Crystals were flash-cooled in liquid nitrogen after cryoprotection with cryoprotectant.
Crystallization of Unliganded AfUGM
Bright yellow plate-like crystals were obtained within 2 weeks using the microbatch method (26) at 4 °C.
Crystallization of Non-reduced AfUGM·UDP-Galp Complex
Crystals of non-reduced AfUGM·UDP-Galp complex were obtained using the microbatch method at 4 °C. Before crystallization UDP-Galp (final concentration 17 mm) was added to the protein solution (25 mm Tris malonate, pH 8.0) and incubated for 30 min at room temperature.
Crystallization of Reduced AfUGM·UDP-Galp Complex
Crystals of reduced AfUGM·UDP-Galp complex were obtained using the microbatch method at room temperature. UDP-Galp (final concentration of 10 mm) was added to the protein solution (10 mg/ml in 50 mm Tris, pH 8.0, 5 mm DTT) and prior to crystallization was reduced by adding sodium dithionite (final concentration 10 mm). The cofactor was re-reduced by chemical reduction of the crystals with sodium dithionite.
Crystallization of AfUGM·UDP Complex
Crystals of the AfUGM·UDP complex were obtained using the microbatch method at room temperature. UDP (final concentration of 10 mm) was added to the protein solution (10 mg/ml of protein in 25 mm Tris malonate, pH 8.0) and prior to crystallization was reduced by adding sodium dithionite (final concentration 10 mm).
Crystallization of AfUGM Mutants R327K, R327A, and R182K with UDP-Galp
Crystals of R182K-, R327A-, and R327K-AfUGM complexed with UDP-Galp were obtained using the microbatch at room temperature. UDP-Galp (final concentration of 15 mm) was added to the protein solution (10 mg/ml in 25 mm Tris malonate, pH 8.0) and prior to crystallization was reduced by adding sodium dithionite (final concentration 10 mm).
Data Collection and Processing
Diffraction data were collected at 100 K from single crystals at the Canadian Light Source. Datasets of unliganded AfUGM, reduced AfUGM·UDP-Galp complex, and the non-reduced AfUGM·UDP-Galp complex were collected at beamline 08ID-1 on a MAR CCD225 detector. Datasets for AfUGM in complex with UDP and the three AfUGM mutants R182K, R327K, and R327A in complex with UDP-Galp were collected at beamline 08B1-1 on a Rayonix MX300HE x-ray detector. The datasets were processed and scaled using autoprocess (27) and d*TREK (28). In the case of the reduced AfUGM·UDP-Galp complex the data could only be processed and scaled in space group P1. The data collection statistics are shown in Table 1A.
TABLE 1.
Crystallographic data
Numbers in parentheses represent values for the highest resolution shell.
Structure Determination and Refinement
The structure of the non-reduced AfUGM·UDP-Galp complex was determined by molecular replacement using MrBUMP (29) within the CCP4 package (30) using MOLREP (31). The structure solution was found using a partially refined model of UGM from L. major as the search model (25). The reduced AfUGM·UDP-Galp structure was determined by molecular replacement using the non-reduced AfUGM·UDP-Galp complex structure as the search model. For all other complex structures, the reduced AfUGM·UDP-Galp complex structure was used as the search model. First all solvent molecules, cofactor, ligands, and ions were removed from the search model. MOLREP (31) was then used to find molecular replacement solutions. All structures were refined using PHENIX (32). Initially, rigid body refinement was done, followed by simulated annealing using Cartesian dynamics at 2000 K to remove model bias. Clear positive density was present in the electron difference maps contoured at 3σ level for FAD and bound ligand molecules (UDP or UDP-Galp). The models were then further refined using restrained refinement. NCS restraints were used throughout the refinement for all models. The refinement of the R327A structure in space group P212121 was hampered by twinning, which could not initially be detected due to NCS and psuedo-translational symmetry. The data were reprocessed in P21 and refined using the twin operator (h, -k, -l). Rebuilding of the models was carried out using COOT (33). Placement of cofactor and ligands was carried out using ligandFit in PHENIX (32). Libraries for FAD and UDP-Galp were generated with ELBOW in PHENIX (32). The refinement progress was monitored by following Rfree and inspecting the electron density maps. When Rfree dropped below 30%, water molecules were added using water update refinement in PHENIX, and their positions were manually checked using COOT. The final round of refinement was done with optimized refinement target weights for best geometry. Final refinement statistics are shown in Table 1B.
Structural Analysis
The stereochemistry of all models was validated with MOLPROBITY (34) as part of PHENIX (32) and the ADIT validation server at RCBS-Rutgers. Superpositions were calculated with DALI-lite (35) and SUPERPOSE within the CCP4 package (30). Superpositions of the different AfUGM structures were done with SUPERPOSE by superimposing specific residue selections (residues 2–175 and residues 210–510) and excluding the two mobile loops. Structure-based sequence alignments were generated with SEQUOIA (36). Figures were prepared with PYMOL and ESPript (37).
Enzyme Kinetics
Kinetic constants for wild type and mutant AfUGMs were determined following a kinetic assay modified from Partha et al. (20). The conversion of UDP-Galp to UDP-Galf was monitored at 262 nm using HPLC (Agilent Technologies, 1100 Infinity). A fixed concentration of wild type AfUGM and AfUGM mutants was used to have less than 40% conversion to the product UDP-Galp. Reactions were carried out with varying amounts of UDP-Galf (0–200 μm) in a final volume of 100 μl, 50 mm phosphate buffer, pH 7.0, containing 20 mm freshly prepared sodium dithionite. The incubations were carried out for 1 min at 37 °C and quenched with 100 μl of 1-butanol. The R182A and R327A mutants were incubated for 30 min at 37 °C. After centrifugation the aqueous phase was injected on a Carbopac PA1 column. The nucleotide sugars were eluted isocratically with 0.2 m ammonium acetate, pH 7.0. The amount of conversion was determined by integration of the UDP-Galp and UDP-Galf peaks. The initial velocity was calculated from the substrate concentration and percentage UDP-Galp conversion. Kinetic parameters were determined with GraphPad Prism software (GraphPad Software, San Diego, CA) using nonlinear regression analysis. All experiments were performed in duplicate.
Protein Data Bank Accession Numbers
Coordinates have been deposited in the PDB with accession codes: 3UKA (unliganded AfUGM), 3UKF (reduced AfUGM:UDPgalp), 3UKH (non-reduced AfUGM:UDPgalp), 3UKL (AfUGM:UDP), 3UKK (R182K-AfUGM:UDPgalp), 3UKP (R327A-AfUGM:UDPgalp), and 3UKQ (R327K-AfUGM:UDPgalp).
RESULTS AND DISCUSSION
Overall Crystal Structure of AfUGM Shows Structural Differences Compared with Prokaryotic UGMs
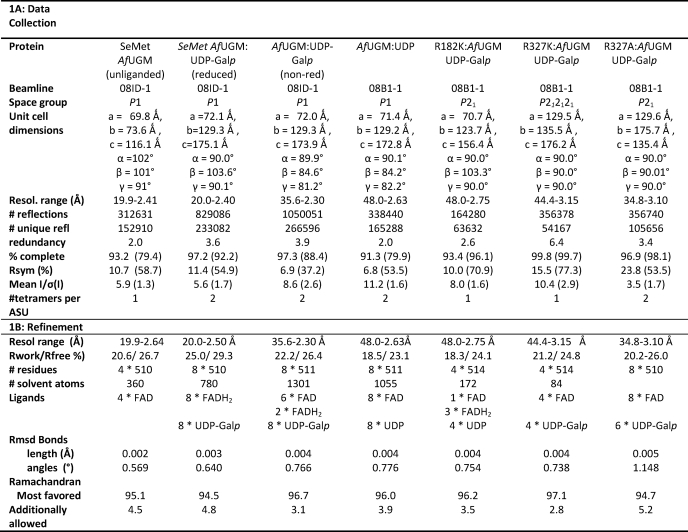
Crystal structures of AfUGM have been determined without substrate UDP-Galp (unliganded AfUGM) and in complex with UDP-Galp (both reduced and oxidized flavin) and UDP (oxidized flavin). All structures are highly similar, except for two mobile loops (loop I (residues 179–187) and loop II (residues 198–208)) (Fig. 1). AfUGM is active as a homotetramer (8) and also crystallizes as a homotetramer (Fig. 1B). Each monomer in the tetramer is composed of 3 domains (FAD binding domain, α-helical domain, and β-sheet domain; Fig. 1C), similar to that of the known prokaryotic UGMs (Fig. 1D) (20–22). A structure-based sequence alignment of AfUGM with known prokaryotic UGMs is shown in supplemental Fig. S1. Superposition of monomer A from the reduced AfUGM·UDP-Galp and D. radiodurans UGM (DrUGM)·UDP-Galp complex structures reveals that 351 residues overlap with a root mean square deviation of 3.0 Å. Despite low sequence identity with known prokaryotic UGMs, the overall fold is largely conserved. The structural differences, due to inserts in AfUGM, will be explained below (Fig. 1D and supplemental Fig. S1).
FIGURE 1.
A, stereodiagram of the monomer from AfUGM, with numbering of the helices and sheets. The numbers correspond to the labels in supplemental Fig. S1, a structure-based sequence alignment. Domain 1 is colored blue, domain 2 is colored green, and domain 3 is colored black. The mobile loops are colored yellow and the additional inserts in AfUGM are colored red. The FAD and UDP-Galp are shown as ball-and-stick representations. B, ribbon representation of reduced AfUGM·UDP-Galp tetramer. Individual subunits are colored red, green, yellow, and blue. FADH2 and UDP-Galp are shown in stick representation. C, superposition of unliganded AfUGM (blue) and reduced AfUGM·UDP-Galp complex (green). Open conformation of mobile loops I and II are shown in yellow. Closed conformation of mobile loops I and II shown in red. The two conformations of loop III are colored the same as for the mobile loops. Arg-182, FADH2, and UDP-Galp are shown in stick representation. D, overall monomer structure of reduced AfUGM·UDP-Galp shown as a ribbon representation (left) and overall monomer structure of reduced DrUGM·UDP-Galp (right). Coloring scheme is the same as for A.
Domain 1 is the FAD binding domain and consists of residues 2–90, 209–291, and 421–510. This domain contains the characteristic βαβ Rossmann-fold but compared with prokaryotic UGMs contains extra secondary structure elements. Residues 242–261 together with residues 5–9 form a small four-stranded mainly anti-parallel β-sheet consisting of strands β1 and β11–β13. Domain 1 has a very long C-terminal extension (residues 474–510), which contains three additional α-helices (α14–α16) located above helix α13 and folds around this helix in a large U-shape. This extension is involved in the formation of the tetramer. In addition helix α15 sits above the entrance of the active site, limiting access to the active site when compared with prokaryotic UGMs.
Domain 2, the α-helical domain, consists of five α-helices (α3–α7) and two 310 helices (η3 and η4). The mobile loops are part of this domain and form the entrance to the active site. This domain shows significant structural differences compared with prokaryotic UGMs (Fig. 1D). Prokaryotic UGMs also have two mobile loops (20–22), however, these mobile loops are different in AfUGM (Fig. 1D, loops highlighted in yellow). Mobile loop 1 in DrUGM is a long flexible loop, whereas in AfUGM this region contains one long α-helix (α3) (residues 113–134) and a short loop (residues 135–141), which is not mobile in AfUGM. Mobile loop 2 in prokaryotic UGMs is much longer in AfUGM and contains an additional inserted α-helix α7 (residues 188–197) thus generating the two separated mobile loops I and II (Fig. 1, A and D, inserts highlighted in red). In AfUGM both α-helix 3 and α-helix 7 are involved in oligomerization.
Domain 3, the β-sheet domain, is built up by residues 91–103 and 292–420. This domain has two additional inserts (Fig. 1, A and D, inserts highlighted in red). Insert 1 (residues 340–366) forms an additional long hairpin-like extension between 310 helix η7 and β-strand β19. Insert 2 (residues 381–420) contains an additional α-helix (α11) and additional β-strands β20 and β21 thus forming a seven-stranded mainly anti-parallel β-sheet instead of the six-stranded anti-parallel β-sheet observed in prokaryotic UGMs (20–22).
AfUGM Undergoes Significant Structural Changes upon Ligand Binding
To study the conformational changes induced in AfUGM upon substrate binding, we have determined the structure of unliganded AfUGM and both AfUGM complexes with UDP and UDP-Galp. The structures were all determined under similar conditions, eliminating the possibility that the changes are due to crystal artifacts. The overall structures for liganded and unliganded AfUGM are all highly similar with only movements in the active site mobile loops (I, II, and III) significantly different (Fig. 1C). The root mean square deviation for all other Cα positions between the structures is less than 0.6 Å.
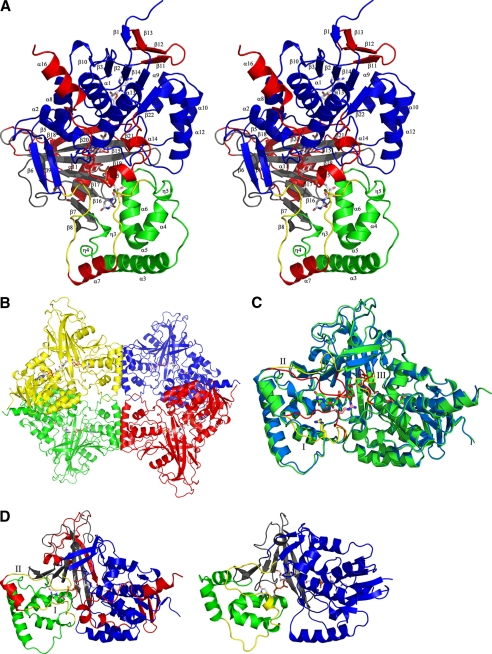
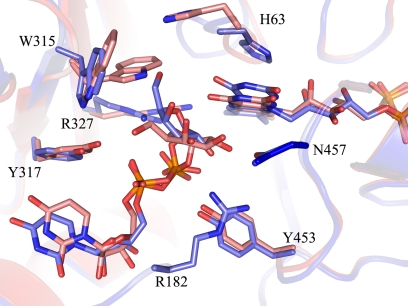
In the unliganded AfUGM crystal structure, the isoalloxazine ring of the cofactor has a flat conformation, indicating that the cofactor is in its oxidized form (FAD) (Fig. 2A). The two mobile loops (I and II) are in the open conformation as seen in unliganded prokaryotic UGMs (21, 22). The reduced AfUGM·UDP-Galp complex was obtained by chemically reducing the FAD cofactor prior to crystallization and was re-reduced prior to data collection resulting in colorless crystals. The isoalloxazine ring of the cofactor has a “butterfly” bent conformation, confirming that the cofactor is reduced (Fig. 2A). The two mobile loops (I and II) are in the closed conformation. The substrate is located in a similar pocket as found in prokaryotic UGMs (14). Most of the residues involved in substrate binding are highly conserved among all UGMs, except for residues in the sugar binding site near the entrance of the active site (Fig. 2B and Table 2). The galactose moiety is bound in the same orientation as observed in the DrUGM·UDP-Galp complex (Fig. 2B). The C1 atom is positioned below N5 of the isoalloxazine ring at a distance of 3.6 Å, lending support to the proposed FAD-galactose adduct (Fig. 2A). Thus this structure represents the productive binding mode.
FIGURE 2.
A, close-up of active sites from unliganded (left) and liganded AfUGM (right). Unliganded AfUGM, mobile loop III (residues 59–66) above the isoalloxazine ring with His-63 pointing into the highly conserved hydrophobic pocket. Arg-327 is at hydrogen bonding distance from N5 and O4 of FAD. Mobile loop III in the reduced AfUGM·UDP-Galp complex has flipped. His-63 sits above N5 of FADH2. Arg-327 is at hydrogen bonding distance of UDP-Galp. The Fo − Fc-electron density of the ligand (contoured at 3σ) is shown as a green wireframe in the right panel. B, stereodiagram comparison of the active sites from AfUGM·UDP-Galp (green) and DrUGM·UDP-Galp (blue). Labeling of active site residues is according to AfUGM sequence. C, stereodiagram of AfUGM active site with bound UDP-Galp. Residues shown in thick lines are within 4 Å from UDP-Galp. Residues in red are from closed mobile loops I and II. Residues in green (90–114) help to stabilize loop II and in positioning the uracil portion of UDP-Galp. D, overlay of reduced AfUGM·UDP-Galp (blue) on AfUGM·UDP (red). The binding mode of UDP is the same as the binding mode of the UDP moiety in the reduced UDP-Galp crystal structure. The only notable difference with the reduced UDP-Galp complex structure is that upon oxidation of the flavin the Phe-66 side chain moved outwards the active site and loop III at N3 side of the isoalloxazine ring moved ∼1 Å down.
TABLE 2.
Conserved active site residues in AfUGM compared to prokaryotic UGMs
The OH group of Tyr-317 makes the same hydrogen bonding interaction with the α-phosphate of the nucleotide as OH group of Tyr-209 in DrUGM and Tyr-185 in KpUGM (single letter amino acid abbreviations are indicated).
| AfUGM | DrUGM | KpUGM | EcUGM | MtUGM |
|---|---|---|---|---|
| H63 | H85 | H60 | H56 | H65 |
| F66 | H88 | H63 | H59 | H68 |
| R91 | H109 | N84 | N80 | H89 |
| F158 | F175 | F151 | L147 | F157 |
| M159 | F176 | F152 | I148 | V158 |
| Y162 | Y179 | Y155 | Y151 | Y161 |
| N163 | T180 | T156 | T152 | T162 |
| W167 | W184 | W160 | W156 | W166 |
| R182 | R198 | R174 | R170 | R180 |
| P206 | Y209 | Y185 | Y181 | Y191 |
| N207 | F210 | F186 | F182 | F192 |
| W315 | T294 | V268 | V266 | T162 |
| Y317 | N296 | N270 | N268 | N282 |
| R327 | R305 | R280 | R278 | R292 |
| E373 | E325 | E301 | E298 | E315 |
| Y419 | Y335 | Y314 | Y311 | Y328 |
| R447 | R364 | R343 | R340 | R360 |
| Y453 | Y370 | Y349 | Y346 | Y366 |
| N457 | N372 | D351 | D348 | D368 |
| Q458 | M373 | M352 | M349 | M369 |
In prokaryotic UGMs, the positions of the majority of the active site residues remain unchanged when the substrate binds, but in AfUGM, several active site residues change position in order for the substrate to bind in a productive mode (Fig. 2A). The strictly conserved active site arginine, Arg-327, has a different conformation than in prokaryotic UGMs (Arg-305 in DrUGM). In unliganded AfUGM, the guanidinium moiety of Arg-327 is positioned 3.7 Å from O4 and N5 of the FAD and is located 3.8 Å from the planar face of a conserved Trp-315, forming a cation-π interaction. In prokaryotic UGMs, this arginine (Arg-305 in DrUGM) is too far away from the FAD to form hydrogen bonds. Furthermore, the observed cation-π interaction between Arg-327 and Trp-315 does not exist in prokaryotic UGMs because Trp-315 is not conserved in prokaryotes.
Upon ligand binding to AfUGM the loop (residues 306–315) connecting β13 and β14 has moved and the Trp-315 side chain has rotated so that the nitrogen of the indole ring is now forming a hydrogen bond with Tyr-317 (Fig. 2C). Furthermore, its planar face is now within 4.5 Å of the C6 sugar atom. In addition Ile-92 has moved 3.5 Å away from the active site and the conserved Arg-91 side chain has moved toward loop II forming several hydrogen bonds with the main chain to stabilize the closed conformation of loop II. The conserved loop II residues Trp-204, Gly-205, and Pro-206 have moved ∼13.8 Å (Cα position of Pro-206) to close the entrance of the active site and Asn-207 (which is a strictly conserved phenylalanine in prokaryotic UGMs) is positioned within 3.5 Å of C4-OH of the galactose moiety (Fig. 2C). In addition, residues 104–113 (containing helix η3) have moved toward the active site so that the side chain of Tyr-104 stacks with the planar face of the uracil moiety and the side chain of Gln-107 forms hydrogen bonds with the uracil moiety. In addition the main chain amine of Phe-106 forms a hydrogen bond with the uracil moiety (Fig. 2C). This rearrangement is necessary to allow the Trp-315 side chain to rotate and for correct positioning the uracil-ribose moiety of the nucleotide sugar upon loop closure.
Conserved Arginines Play Critical Roles in Activity and Substrate Orientation
In prokaryotic UGMs, the enzyme undergoes significant loop movements upon substrate binding. The re-orientation of this loop results in a 11 Å shift in the Cα position of Arg-174 in KpUGM. This residue is essential for substrate binding in prokaryotic UGMs (5).
In AfUGM, both mobile loops I and II undergo significant rearrangement and are in the closed conformation upon substrate binding, similar to that seen in prokaryotic UGMs (Fig. 1, C and D). The re-orientation of loop I results in a 11 Å shift in the Cα position of Arg-182. The side chain of Arg-182 (corresponding to Arg-198 in DrUGM) points into the active site (Figs. 1C and 2B), but the guanidinium moiety is rotated and points away from the β-phosphate unlike in the prokaryotic UGM complex structures (20). Furthermore, the terminal nitrogen of Arg-182 is within 3.3 Å of the main chain oxygen of Tyr-453 and the side chain oxygen of Asn-457. The other terminal nitrogen of Arg-182 is between 3.3 and 3.6 Å of both C1-OH and C2-OH of the galactose moiety (Fig. 2A). In addition the guanidinium moiety of Arg-182 forms a cation–π interaction with Tyr-453 (Fig. 2D). This suggests that Arg-182 might not be critical for stabilizing the negative charge on the β-phosphate of UDP-Galp as believed for prokaryotic UGMs, but rather it might serve to anchor loop I to the FAD domain and help to stabilize the galactose orientation for catalysis. However, the crystal structure of the AfUGM·UDP complex is also found in the closed conformation, with Arg-182 and UDP located in a similar position to the AfUGM·UDP-Galp complex (Fig. 2D). This suggests that, as in prokaryotic UGMs, the driving force for closing of mobile loop I is likely an ionic interaction between Arg-182 and the diphosphate of UDP.
The second absolutely conserved arginine residue that was identified in prokaryotic UGMs as essential for substrate binding (5) corresponds to Arg-327 in AfUGM. Unlike the prokaryotic homolog, this residue moves when substrate binds. The guanidinium moiety of Arg-327 interacts with both the O5 of the galactose moiety and with the β-phosphate of the UDP moiety (Fig. 2A). Consistent with prokaryotic UGMs, this likely helps both to position the anomeric C1 atom for nucleophilic attack by the flavin N5 and to stabilize the UDP intermediate during turnover (5, 38). In prokaryotic UGMs these two conserved arginines stabilize the negative charge on the β-phosphate of the nucleotide sugar and are essential for UGM activity (5, 38, 39). Mutation of these two residues to alanine in KpUGM (R174A and R280A) resulted in inactive enzyme (5).
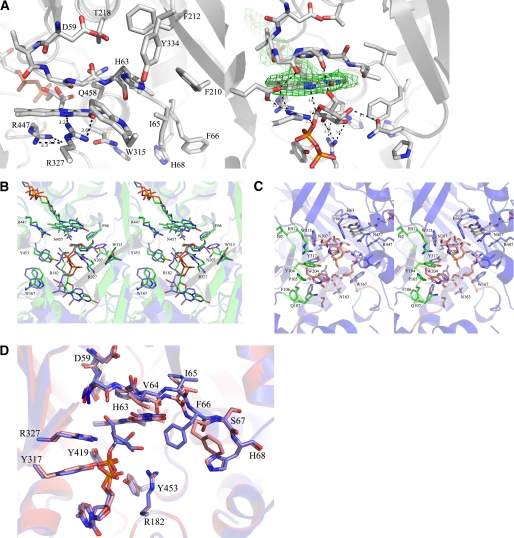
We have mutated both Arg-182 and Arg-327 to alanine and lysine (Table 3). The R327A mutant is inactive. However, the crystal structure of R327A complexed with UDP-Galp contains the substrate with loops I and II in the closed conformation, with Arg-182 in the same position as in wild type AfUGM (Fig. 3A). The diphosphates and galactose moiety have a different orientation compared with the reduced AfUGM·UDP-Galp complex structure. UDP-Galp is bound in a nonproductive binding mode. These results support our hypothesis that Arg-327 plays an important role in stabilizing the position of the diphosphates and helps to facilitate positioning of the galactose moiety for catalysis. The R182A mutant, on mobile loop I, is still active unlike the same mutation in prokaryotic UGM (5). The substrate binds with lower affinity (Km = 607 μm) than wild type AfUGM (Km = 42.5 μm) (Table 3) and shows decreased catalytic efficiency. This suggests that other residues on loop I (e.g. Glu-181) may help facilitate loop closure and substrate binding. We have been unable to crystallize the R182A mutant complexed with UDP-Galp.
TABLE 3.
Kinetic parameters for wild type AfUGM and arginine mutants
| Km | kcat | kcat/Km | |
|---|---|---|---|
| μm | min−1 | μm−1min−1 | |
| Wild type | 42.5 ± 3.7 | 523 ± 30 | 12.3 ± 1.2 |
| R327K | 42.9 ± 5.5 | 7.46 ± 0.08 | 0.18 ± 0.04 |
| R182K | 22.0 ± 1.5 | 26.3 ± 0.4 | 1.20 ± 0.02 |
| R327A | No activity | No activity | No activity |
| R182A | 607 ± 62 | 4.72 ± 0.30 | 0.0078 ± 0.0001 |
FIGURE 3.
Binding mode of UDP-Galp in arginine mutants. A, superposition of R327A AfUGM with bound UDP-Galp (blue) on reduced AfUGM·UDP-Galp (red). UDP-Galp is bound in a non-productive binding mode in R327A AfUGM. Arg-182 is in the same position as in wild type AfUGM. B, the R182K AfUGM mutant with bound UDP-Galp. Only the UDP moiety is visible and no density for Galp moiety. R182K is within 3.6 Å from the Asn-457 side chain but has no interaction with the phosphates or the galactose moiety. Phe-66 points out of the active site. C, R327K AfUGM with bound UDP-Galp. NZ of R327K is in the same position as NH1 of Arg-327 and makes the same interactions with galactose moiety and the β-phosphate as in wild type complex. Phe-66 points into active site.
We have determined the crystal structure of both R182K (Fig. 3B) and R327K (Fig. 3C) mutants with bound UDP-Galp. Both mutants show reduced activity compared with wild type AfUGM (Table 3). The crystal structure of R182K contains one tetramer in the asymmetric unit, with identical subunits (average root mean square deviation of 0.2 Å for all atoms). Three monomers have reduced FADH2 and one monomer has FAD. There is no clear density for the galactose moiety, but the UDP moiety is in the same orientation as seen in the other structures and both loop I and loop II are in the closed conformation. The lysine side chain is in the same orientation as the arginine residue that it replaces. The side chain of Lys-182 is within 3.6 Å of the Asn-457 side chain and has no interaction with the phosphates or the galactose moiety (Fig. 3B). Unexpectedly, the R182K mutation resulted in a 2-fold improvement in binding affinity. This may be due to the shorter lysine side chain improving the orientation of the UDP moiety. Previously, we have shown that the UDP moiety provides the majority of the binding energy (38, 39). The lack of density for the galactose moiety in the R182K mutant would suggest that Arg-182 is indeed important for orienting the sugar moiety for catalysis.
The crystal structure of the R327K AfUGM·UDP-Galp complex contains two independent dimers per asymmetric unit that together with symmetry related molecules form two AfUGM tetramers. All four molecules are identical (root mean square deviation = 0.2 Å) and have FAD and substrate bound. The UDP-Galp is in a similar orientation to wild type AfUGM and loop I and loop II are in the closed conformation, however, loop II shows more flexibility. The side chain of the lysine residue is in the same position and point in the same direction as the arginine in wild type AfUGM. The terminal nitrogen of Lys-327 is in the same position as that of the terminal nitrogen of Arg-327 and makes the same interactions with galactose moiety and the β-phosphate (Fig. 3C).
Non-reduced AfUGM·UDP-Galp Complex Offers Insight into Conformational Changes
The non-reduced AfUGM·UDP-Galp complex was obtained by incubating the enzyme for 30 min with UDP-Galp prior to crystallization, with no addition of reducing agent. Although the cofactor was not reduced before crystallization all monomers contain bound substrate. Although the non-reduced and reduced AfUGM·UDP-Galp structures are largely similar, there are a number of significant structural differences. Importantly, the eight subunits in the asymmetric unit of the non-reduced AfUGM complex show different loop conformations, enabling us to identify the relationship between FAD oxidation state and structural conformations.
Mobile loops I and II show multiple conformations in the non-reduced AfUGM·UDP-Galp complex structure, with a mixture of open, closed, and semi-open (loop I closed and loop II open) conformations. Two monomers contain reduced flavin (FADH2) as indicated by the bent conformation of the isoalloxazine ring. These monomers are in the closed conformation and make identical interactions with the UDP moiety as in the reduced AfUGM·UDP-Galp complex. All other monomers have FAD bound. The subunits with FAD show a mixture of open and semi-open conformations. These monomers have fewer interactions with the substrate UDP-Galp. The main interactions are between the diphosphate and conserved tyrosines (Tyr-162, -317, -419, and -453) and with O2 and O3 of the ribose ring and the side chain of Asn-163. In addition, helix η3 is in the position observed in the unliganded AfUGM structure and has no interaction with the uracil moiety. The uracil-ribose moiety has moved 1.5 Å toward the entrance of the active site compared with the reduced AfUGM·UDP-Galp structure. Although the cofactor was not reduced before crystallization all monomers contain UDP-Galp in the active site, demonstrating that non-reduced AfUGM is able to bind substrate, in contrast to what was previously reported by Oppenheimer et al. (8).
Alternative Binding of Sugar Moiety in Non-reduced AfUGM·UDP-Galp Complex
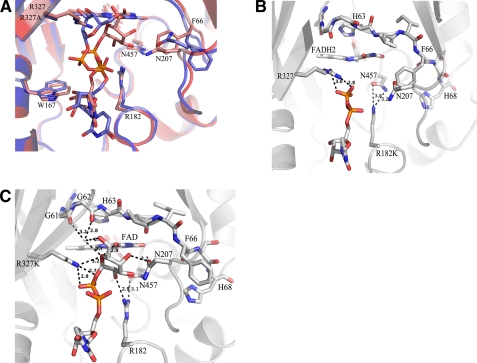
The alteration in flavin oxidation state results in a significant difference in the position of the sugar moiety in the active site of AfUGM (Fig. 4). In the non-reduced AfUGM·UDP-Galp structure, the β-phosphate and galactose moiety are rotated ∼60° compared with the reduced AfUGM·UDP-Galp structure. The galactose C1 atom is ∼3.7 Å away from the flavin N5 in the reduced monomers. The corresponding distance in the non-reduced monomers is ∼5.2 Å, too far away for nucleophilic attack. The alteration in galactose position is likely caused by movement of Trp-315, which is found in two orientations. In one orientation, Trp-315 forms a cation–π interaction with Arg-327 and this conformation would form a steric clash with the galactose moiety if it was bound in the same orientation found in the reduced AfUGM·UDP-Galp structure (Fig. 4). The interactions in the sugar binding site are significantly different compared with the reduced AfUGM·UDP-Galp structure (Figs. 2C and 4). The guanidinium moiety of Arg-327 is positioned to form hydrogen bonds with the O4 and N5 of FAD and with the O5 of the galactose. The side chain of Asn-457 is hydrogen bonded to C3 and C4 hydroxyls of galactose. Arg-182 is within 3.3 Å of the carbonyl oxygen of Tyr-453 and the side chain oxygen of Asn-457 but does not make any contacts with the galactose moiety. Thus, although non-reduced AfUGM·UDP-Galp is able to bind UDP-Galp, the cofactor needs to be in the reduced state for productive binding of the substrate.
FIGURE 4.
Overlay of reduced AFUGM·UDP-Galp (blue) and non-reduced AfUGM·UDP-Galp (red). The interactions in the sugar binding site are significantly different in the non-reduced UDP-Galp complex structure.
Loop III Undergoes Redox-dependent Conformation Changes
A conserved loop (residues 60–66) above the si-face of the isoalloxazine ring of the cofactor (hereafter named loop III) has an altered conformation in unliganded AfUGM compared with prokaryotic UGMs (20–22) (Fig. 2A). The side chain of His-63 is oriented toward the β-sheet of domain 3 and is located in a conserved hydrophobic pocket formed by residues Ile-65, Phe-210, Phe-212, Tyr-334, and Trp-315. Gly-62 sits above the middle ring of the isoalloxazine ring with its main chain amine ∼3.5 Å from N5 of FAD. The carbonyl oxygens of Gly-61 and Gly-62 are within hydrogen bonding distance from the side chain nitrogen of Gln-458 (methionine in prokaryotic UGMs). Phe-66 is situated on the end of the loop and points out of the active site. Ile-65 points into the active site, preventing loop II closure through a steric clash with Asn-207.
When the cofactor is reduced and substrate binds, loop III undergoes a conformation shift (Fig. 2A) and the peptide chain (residues 61–66) rotates almost 180°. His-63 moves to a position above the N5 of the isoalloxazine ring, similar to that seen in prokaryotic UGMs (20–22). The reduction of FAD results in the N3 of the isoalloxazine ring bending away from the active site, Ile-65 moving out of the active site and Phe-66 rotating to point into the active site. Phe-66 is then situated in a highly conserved pocket formed by His-68, Pro-206, Asn-207, Asn-457, and the isoalloxazine ring (Fig. 2A). This type of loop flipping has not been observed in prokaryotic UGMs. In prokaryotic UGMs there is a conserved histidine (His-88 in DrUGM) at the Phe-66 location, pointing into the active site and held in place by a conserved phenylalanine and aspartate (Phe-210 and Asp-212 in DrUGM, respectively). Thus, Phe-66 may act as a switch to control loop III flipping and may be regulated by the redox state of the cofactor.
Stabilization of the Flavin Redox State
Oppenheimer et al. (8) noted that AfUGM is capable of keeping part of its cofactor reduced while under aerobic conditions. Varying degrees of activity have been reported for prokaryotic UGMs as well (21, 40), making it likely that oxidation of the enzyme is a result of exposure to oxygen during purification, not as a natural state of the enzyme. The increase in stability of the reduced state of the flavin noted for AfUGM compared with prokaryotic UGMs may be a result of the structural changes found in loop III of AfUGM compared with prokaryotic UGMs. Loop III is highly conserved among eukaryotic UGMs but is different from prokaryotic UGMs (supplemental Fig. S1). In prokaryotic UGMs this loop does not alter conformation upon oxidation/reduction of FAD. The large movement of loop III observed in AfUGM may be due to the redox state of the cofactor rather than due to binding of the substrate. The loop III movements seen in the non-reduced AfUGM·UDP-Galp complex are consistent with our observations of the loop movements in the presence of non-reduced and reduced FAD. The monomers containing reduced FADH2 show the conformation of loop III found in the reduced AfUGM·UDP-Galp structure, whereas the monomers with oxidized FAD show the conformation found in unliganded AfUGM.
Oppenheimer et al. (8) found that addition of UDP to reduced AfUGM causes fast oxidation of the cofactor, whereas addition of the substrate UDP-Galp caused only very slow oxidation. The structures of AfUGM·UDP and reduced AfUGM·UDP-Galp are identical except for the galactose moiety. Although there is slow oxidation in the presence of UDP-Galp it seems that the galactose moiety reduces the oxidation speed of the cofactor, possibly by helping to maintain the conformations of the active site residues in positions that favor the reduced FAD cofactor or by limiting access of oxygen to the active site.
Our results provide a detailed examination of the binding of substrate to eukaryotic UGM. Although these structures show similarities to previously reported prokaryotic UGMs, there are a number of structural features that are not found in prokaryotic UGMs. In particular, the structural changes that occur upon reduction/oxidation of the flavin cofactor are not seen in prokaryotic UGM and help to explain the previously reported observations (8). These structures offer new insights into the function of this class of enzymes.
Supplementary Material
Acknowledgments
We thank the Saskatchewan Health Research Foundation for funding the Molecular Design Research Group of the University of Saskatchewan, Dr. David Palmer (University of Saskatchewan) for many helpful discussions and shared equipment, and Dr. Sean Dalrymple for helpful discussions during manuscript preparation. We thank the staff of macromolecular beam line 08ID-1, Canadian Light Source (CLS), Saskatoon, Saskatchewan, Canada, for their technical assistance during data collection. The structural studies described in this paper were performed at the CLS, which is supported by NSERC, the National Research Council of Canada, the Canadian Institutes of Health Research, the Province of Saskatchewan, Western Economic Diversification Canada, and the University of Saskatchewan.
This work was supported in part by grants from Natural Science and Engineering Research Council of Canada (NSERC) and Canadian Institutes of Health Research-Regional Partnership Program (to D. A. R. S.).

This article contains supplemental text and Fig. S1.
- Galf
- galactofuranose
- AfUGM
- A. fumigatus UGM
- UGM
- UDP-galactopyranose mutase
- UDP-Galp
- uridine diphosphate-galactopyranose
- UDP-Galf
- uridine diphosphate-galactofuranose.
REFERENCES
- 1. Tefsen B., Ram A. F., Van Die I., Routier F. H. (2012) Galactofuranose in eukaryotes. Aspects of biosynthesis and functional impact. Glycobiology 22, 456–469 [DOI] [PubMed] [Google Scholar]
- 2. Pan F., Jackson M., Ma Y., McNeil M. (2001) Cell wall core galactofuran synthesis is essential for growth of mycobacteria. J. Bacteriol. 183, 3991–3998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Richards M. R., Lowary T. L. (2009) Chemistry and biology of galactofuranose-containing polysaccharides. ChemBioChem 10, 1920–1938 [DOI] [PubMed] [Google Scholar]
- 4. Pedersen L. L., Turco S. J. (2003) Galactofuranose metabolism. A potential target for antimicrobial chemotherapy. Cell. Mol. Life Sci. 60, 259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chad J. M., Sarathy K. P., Gruber T. D., Addala E., Kiessling L. L., Sanders D. A. (2007) Site-directed mutagenesis of UDP-galactopyranose mutase reveals a critical role for the active-site, conserved arginine residues. Biochemistry 46, 6723–6732 [DOI] [PubMed] [Google Scholar]
- 6. Beverley S. M., Owens K. L., Showalter M., Griffith C. L., Doering T. L., Jones V. C., McNeil M. R. (2005) Eukaryotic UDP-galactopyranose mutase (GLF gene) in microbial and metazoal pathogens. Eukaryot. Cell 4, 1147–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bakker H., Kleczka B., Gerardy-Schahn R., Routier F. H. (2005) Identification and partial characterization of two eukaryotic UDP-galactopyranose mutases. Biol. Chem. 386, 657–661 [DOI] [PubMed] [Google Scholar]
- 8. Oppenheimer M., Poulin M. B., Lowary T. L., Helm R. F., Sobrado P. (2010) Characterization of recombinant UDP-galactopyranose mutase from Aspergillus fumigatus. Arch. Biochem. Biophys. 502, 31–38 [DOI] [PubMed] [Google Scholar]
- 9. Oppenheimer M., Valenciano A. L., Sobrado P. (2011) Isolation and characterization of functional Leishmania major virulence factor UDP-galactopyranose mutase. Biochem. Biophys. Res. Commun. 407, 552–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kleczka B., Lamerz A. C., Bakker H., Wiese M., Gerardy-Schahn R., Routier F. H. (2004) Leishmania major UDP-galactopyranose mutase. Characterization and validation of a potential drug target. Glycobiology 14, 1119–1120 [Google Scholar]
- 11. El-Ganiny A. M., Sanders D. A., Kaminskyj S. G. (2008) Aspergillus nidulans UDP-galactopyranose mutase, encoded by ugmA plays key roles in colony growth, hyphal morphogenesis, and conidiation. Fungal Genet. Biol. 45, 1533–1542 [DOI] [PubMed] [Google Scholar]
- 12. Novelli J. F., Chaudhary K., Canovas J., Benner J. S., Madinger C. L., Kelly P., Hodgkin J., Carlow C. K. (2009) Characterization of the Caenorhabditis elegans UDP-galactopyranose mutase homolog glf-1 reveals an essential role for galactofuranose metabolism in nematode surface coat synthesis. Dev. Biol. 335, 340–355 [DOI] [PubMed] [Google Scholar]
- 13. Damveld R. A., Franken A., Arentshorst M., Punt P. J., Klis F. M., van den Hondel C. A., Ram A. F. (2008) A novel screening method for cell wall mutants in Aspergillus niger identifies UDP-galactopyranose mutase as an important protein in fungal cell wall biosynthesis. Genetics 178, 873–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schmalhorst P. S., Krappmann S., Vervecken W., Rohde M., Müeller M., Braus G. H., Contreras R., Braun A., Bakker H., Routier F. H. (2008) Contribution of galactofuranose to the virulence of the opportunistic pathogen Aspergillus fumigatus. Eukaryot. Cell 7, 1268–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lamarre C., Beau R., Balloy V., Fontaine T., Wong Sak Hoi J., Guadagnini S., Berkova N., Chignard M., Beauvais A., Latge J. P. (2009) Galactofuranose attenuates cellular adhesion of Aspergillus fumigatus. Cell. Microbiol. 11, 1612–1623 [DOI] [PubMed] [Google Scholar]
- 16. Kleczka B., Lamerz A. C., van Zandbergen G., Wenzel A., Gerardy-Schahn R., Wiese M., Routier F. H. (2007) Targeted gene deletion of Leishmania major UDP-galactopyranose mutase leads to attenuated virulence. J. Biol. Chem. 282, 10498–10505 [DOI] [PubMed] [Google Scholar]
- 17. Nassau P. M., Martin S. L., Brown R. E., Weston A., Monsey D., McNeil M. R., Duncan K. (1996) Galactofuranose biosynthesis in Escherichia coli K12. Identification and cloning of UDP-galactopyranose mutase. J. Bacteriol. 178, 1047–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Köplin R., Brisson J. R., Whitfield C. (1997) UDP-galactofuranose precursor required for formation of the lipopolysaccharide O antigen of Klebsiella pneumoniae serotype O1 is synthesized by the product of the rfbDKPO1 gene. J. Biol. Chem. 272, 4121–4128 [DOI] [PubMed] [Google Scholar]
- 19. Weston A., Stern R. J., Lee R. E., Nassau P. M., Monsey D., Martin S. L., Scherman M. S., Besra G. S., Duncan K., McNeil M. R. (1997) Biosynthetic origin of mycobacterial cell wall galactofuranosyl residues. Tuber. Lung Dis. 78, 123–131 [DOI] [PubMed] [Google Scholar]
- 20. Partha S. K., van Straaten K. E., Sanders D. A. (2009) Structural basis of substrate binding to UDP-galactopyranose mutase. Crystal structures in the reduced and oxidized state complexed with UDP-galactopyranose and UDP. J. Mol. Biol. 394, 864–877 [DOI] [PubMed] [Google Scholar]
- 21. Sanders D. A., Staines A. G., McMahon S. A., McNeil M. R., Whitfield C., Naismith J. H. (2001) UDP-galactopyranose mutase has a novel structure and mechanism. Nat. Struct. Biol. 8, 858–863 [DOI] [PubMed] [Google Scholar]
- 22. Beis K., Srikannathasan V., Liu H., Fullerton S. W., Bamford V. A., Sanders D. A., Whitfield C., McNeil M. R., Naismith J. H. (2005) Crystal structures of Mycobacteria tuberculosis and Klebsiella pneumoniae UDP-galactopyranose mutase in the oxidized state and Klebsiella pneumoniae UDP-galactopyranose mutase in the (active) reduced state. J. Mol. Biol. 348, 971–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gruber T. D., Borrok M. J., Westler W. M., Forest K. T., Kiessling L. L. (2009) Ligand binding and substrate discrimination by UDP-galactopyranose mutase. J. Mol. Biol. 391, 327–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gruber T. D., Westler W. M., Kiessling L. L., Forest K. T. (2009) X-ray crystallography reveals a reduced substrate complex of UDP-galactopyranose mutase poised for covalent catalysis by flavin. Biochemistry 48, 9171–9173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Van Straaten K. E., Sanders D. A. (2012) Towards the crystal structure elucidation of eukaryotic UDP-galactopyranose mutase. Acta Crystallographica Sect. F Struct. Biol. Cryst., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. D'Arcy A., Sweeney A. M., Haber A. (2004) Practical aspects of using the microbatch method in screening conditions for protein crystallization. Methods 34, 323–328 [DOI] [PubMed] [Google Scholar]
- 27. Fodje M. N., Berg R., Black G., Grochulski P., Janzen K. (2010) Proceedings of PCaPAC 2010, October 5–8, 2010, SK Canada Saskatoon, Saskatchewan [Google Scholar]
- 28. Pflugrath J. W. (1999) The finer things in X-ray diffraction data collection. Acta Crystallogr. D Biol. Crystallogr. 55, 1718–1725 [DOI] [PubMed] [Google Scholar]
- 29. Keegan R. M., Winn M. D. (2007) Automated search-model discovery and preparation for structure solution by molecular replacement. Acta Crystallogr. D Biol. Crystallogr. 63, 447–457 [DOI] [PubMed] [Google Scholar]
- 30. Collaborative Computational Project, Number 4 (1994) The CCP4 suite. Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 31. Vagin A., Teplyakov A. (1997) MOLREP, an automated program for molecular replacement. J. Appl. Crystallogr. 30, 1022–1025 [Google Scholar]
- 32. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX, a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Emsley P., Cowtan K. (2004) Coot, model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 34. Davis I. W., Murray L. W., Richardson J. S., Richardson D. C. (2004) MOLPROBITY, structure validation and all-atom contact analysis for nucleic acids and their complexes. Nucleic Acids Res. 32, W615-W619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Holm L., Sander C. (1995) Dali, a network tool for protein structure comparison. Trends Biochem. Sci. 20, 478–480 [DOI] [PubMed] [Google Scholar]
- 36. Bruns C. M., Hubatsch I., Ridderström M., Mannervik B., Tainer J. A. (1999) Human glutathione transferase A4-4 crystal structures and mutagenesis reveal the basis of high catalytic efficiency with toxic lipid peroxidation products. J. Mol. Biol. 288, 427–439 [DOI] [PubMed] [Google Scholar]
- 37. Gouet P., Courcelle E., Stuart D. I., Métoz F. (1999) ESPript, analysis of multiple sequence alignments in PostScript. Bioinformatics 15, 305–308 [DOI] [PubMed] [Google Scholar]
- 38. Yuan Y., Bleile D. W., Wen X., Sanders D. A., Itoh K., Liu H. W., Pinto B. M. (2008) Investigation of binding of UDP-Galf and UDP-[3-F]Galf to UDP-galactopyranose mutase by STD-NMR spectroscopy, molecular dynamics, and CORCEMA-ST calculations. J. Am. Chem. Soc. 130, 3157–3168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yao X. H., Bleile D. W., Yuan Y., Chao J., Sarathy K. P., Sanders D. A. R., Pinto B. M., O'Neill M. A. (2009) Substrate directs enzyme dynamics by bridging distal sites. UDP-galactopyranose mutase. Proteins Struct. Funct. Bioinformat. 74, 972–979 [DOI] [PubMed] [Google Scholar]
- 40. Zhang Q. B., Liu H. W. (2000) Studies of UDP-galactopyranose mutase from Escherichia coli. An unusual role of reduced FAD in its catalysis. J. Am. Chem. Soc. 122, 9065–9070 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.