Background: Survivin is an oncogenic protein that is acetylated by CBP, which restricts its location to the nuclear compartment and blocks its anti-apoptotic effect.
Results: HDAC6 deacetylates survivin to promote its nuclear exit in estrogen receptor-positive breast cancer cells.
Conclusion: Cross-talk between estrogen, CBP, and HDAC6 regulate the amount of nuclear acetylated survivin.
Significance: Understanding how estrogen regulates survivin nuclear export may influence breast cancer treatment.
Keywords: Breast Cancer, CBP, Estrogen, Histone Deacetylase, Survivin, Acetylation
Abstract
Survivin is an oncogenic protein that is highly expressed in breast cancer and has a dual function that is dependent on its subcellular localization. In the cytosol, survivin blocks programmed cell death by inactivating caspase proteins; however, in the nucleus it facilitates cell division by regulating chromosomal movement and cytokinesis. In prior work, we showed that survivin is acetylated by CREB-binding protein (CBP), which restricts its localization to the nuclear compartment and thereby inhibits its anti-apoptotic function. Here, we identify histone deacetylase 6 (HDAC6) as responsible for abrogating CBP-mediated survivin acetylation in the estrogen receptor (ER)-positive breast cancer cell line, MCF-7. HDAC6 directly binds survivin, an interaction that is enhanced by CBP. In quiescent breast cancer cells in culture and in malignant tissue sections from ER+ breast tumors, HDAC6 localizes to a perinuclear region of the cell, undergoing transport to the nucleus following CBP activation where it then deacetylates survivin. Genetically modified mouse embryonic fibroblasts that lack mhdac6 localize survivin predominantly to the nuclear compartment, whereas wild-type mouse embryonic fibroblasts localize survivin to distinct cytoplasmic structures. Together, these data imply that HDAC6 deacetylates survivin to regulate its nuclear export, a feature that may provide a novel target for patients with ER+ breast cancer.
Introduction
Histone deacetylase inhibitors have shown promising early results in the adjuvant treatment of several cancers (1–3). Although there is empiric support for these agents, their mechanism of action in tumor cells is incompletely understood. Histone deacetylase inhibitors block tumor survival pathways at the transcriptional level by inducing expression of tumor suppressor genes such as p53 and p21 as well as by post-translationally modifying non-histone proteins through acetylation of lysine residues (4–6). Global acetylation studies demonstrate that proteins involved in proliferation, cell growth, cell cycle, differentiation, and migration are functionally regulated through acetylation (7). Acetylation/deacetylation alters protein stability, subcellular localization, and subsequent interaction with other proteins. Together, this process can profoundly change protein function, potentially reversing oncogenic potential and increasing tumor susceptibility to treatment.
Our group previously demonstrated that the inhibitor of apoptosis protein survivin is acetylated in a growth factor-dependent manner by the histone acetyltransferase, CREB-binding protein (CBP)2 in breast cancer cells (8). We showed that CBP-dependent acetylation at Lys-129 maintains survivin nuclear localization by promoting its homodimerization and stability in this compartment. By contrast, mutation at this residue to a non-acetylatable amino acid inhibits survivin homodimerization, promotes binding to the nuclear export protein Crm1, and results in export to the cytoplasmic compartment. Because the anti-apoptotic function of survivin is dependent on its cytoplasmic abundance (9, 10), inhibiting survivin nuclear export may be a novel approach to cancer therapy. In support of this concept, studies have demonstrated that increases in nuclear survivin promote apoptosis and increase susceptibility of cancer cells to chemical- and radiation-induced cell death (9, 10). Understanding the mechanism of survivin deacetylation could provide new therapeutic targets to inhibit its nuclear export and thereby regulate its anti-apoptotic function.
Histone deacetylase proteins (HDACs) are classified based on their yeast homologues and include class I (HDACs 1, 2, 3, and 8), class IIa (HDACs 4, 5, 7, and 9), class IIb (HDACs 6 and 10), class III HDACs (sirtuins (silent mating type information regulation 2 homolog) 1–7), and class IV (HDAC 11) (1, 2, 11, 12). Except for the class III NAD+-dependent HDACs, the other zinc-dependent HDACs are the current targets of histone deacetylase inhibitor anti-cancer drugs (13). Class I and II HDACs are expressed at high levels in some primary solid tumors, including breast cancer, and have been demonstrated to be directly involved in cancer development in animal models (13).
Through a series of biochemical assays, we identified HDAC6 as a survivin deacetylase that inhibits CBP-dependent survivin acetylation. HDAC6 primarily localized to the nuclear-cytoplasmic junction in quiescent breast tumor cells in culture and in tumor tissue from breast cancer patients. Once activated by CBP in an estrogen-dependent manner, HDAC6 enters the nucleus, promoting survivin deacetylation and nuclear export. Taken together, our findings reveal a novel mechanism of how HDAC6 and survivin interact at the nuclear membrane to control the acetylation state of survivin.
EXPERIMENTAL PROCEDURES
Cells and Culture
HEK293T, HeLa, and MCF-7 cell lines were cultured as described previously (8). mhdac6-null and wild-type mouse embryonic fibroblasts (MEFs) were a kind gift from the laboratory of Dr. Tso-Pang Yao (Duke University Medical Center, Durham, NC). MEF cells were cultured in DMEM media supplemented with 10% FBS and 1% penicillin/streptomycin. For experiments involving estrogen, MCF-7 cells were cultured for 48 h in phenol red-free Dulbecco's modified Eagle's medium (DMEM) containing 5% charcoal-stripped FBS then serum-starved overnight prior to treatment with estradiol (10 nm) for 6 h. Trichostatin A (TSA) and nicotinamide were purchased from Sigma.
Plasmids and Transfections
6×Myc survivin 129K or 129E and HA-CBP were generated as described previously (8). FLAG-HDAC plasmids 1, 2, 3, and 6 were kind gifts from Dr. Edward Seto (Lee Moffitt Cancer Institute, Tampa, FL). Truncated HDAC6 constructs Hdase I (1–414), Hdase II (415–903), and ZIF (904–1215) were generated using PCR amplification from human full-length HDAC6 cDNA with primers containing XbaI and BamHI restriction sites (Hdase I) or XbaI and EcoRI restriction sites (Hdase II and ZIF) and cloned into the N-terminal p3XFLAG-CMV-9 expression vector (Sigma). The HDAC6-dead mutant was a kind gift from Dr. Stuart Schreiber (The Broad Institute and Harvard University, Cambridge, MA). Plasmids were transfected into HEK293T and HeLa cells using either Lipofectamine 2000 or Lipofectamine LTX (Invitrogen) for 48 h. CBP and control siRNA (sc-29244) were purchased from Santa Cruz Biotechnology. MCF-7 cells were transfected with control siRNA (300 pmol) or CBP siRNA (300 pmol) using LipoRNAiMax (Invitrogen).
Immunoprecipitation and Immunoblotting
Treated and transfected cells were washed twice with cold PBS, lysed on ice with cell lysis buffer (20 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm Na2 EDTA, 1% Triton X-100, 2.5 mm sodium pyrophosphate) plus protease inhibitors (1 mm β-glycerophosphate, 1 mm Na3VO4, 1 μg/ml leupeptin, 1 μg/ml aprotinin, 1 mm DTT, and 1 mm PMSF), scraped, and collected in microcentrifuge tubes and incubated on ice for 30 min. For immunoprecipitations, lysates were precleared with protein A/G plus agarose beads (Santa Cruz Biotechnology). Supernatants were incubated at 4 °C with anti-Myc (Santa Cruz Biotechnology) overnight and then incubated on a rocking shaker at 4 °C with protein A/G plus agarose beads for 4 h. Samples were washed, and pellets were denatured in SDS loading buffer and boiled prior to separation by SDS-PAGE. Immunoblotting of immunoprecipitation and total protein samples was performed on PVDF membranes with antibodies for acetylated survivin (Novus Biologicals), full-length survivin, CBP, HDAC6, Myc, actin (Santa Cruz Biotechnology), FLAG (Cell Signaling), α-tubulin (Sigma), and HDAC1 (Affinity Bioreagents).
Subcellular Fractionation
For subcellular fractionation, cells were collected in low salt hypotonic buffer (buffer A; 10 mm Tris, pH 7.5, 10 mm KCl, 0.5 mm EGTA, 1% IGEPAL (Nonidet P-40)) plus protease inhibitors (1 mm β-glycerophosphate, 1 mm Na3VO4, 1 μg/ml leupeptin, 1 μg/ml aprotinin, 1 mm DTT, and 1 mm PMSF) and incubated for 15 min at 4 °C on a rotating shaker. Lysates were then centrifuged at 14,000 rpm for 20 min at 4 °C. Supernatants were collected in separate tubes (cytoplasmic fraction), and pellets were washed with buffer A and recentrifuged as above. Pellets were washed with cold PBS and centrifuged. Pellets were resuspended in high salt hypotonic buffer (buffer B; 20 mm Tris pH 7.9, 25% glycerol, 1.5 mm MgCl2, 400 mm NaCl, and 0.5 mm EGTA) plus protease inhibitors and then incubated for 30 min in 4 °C on a rotating shaker. Lysates were then centrifuged, and supernatants were collected in separate tubes (nuclear fraction). Membranes were immunoblotted with antibodies to acetylated survivin (Novus Biologicals), full-length survivin, CBP, HDAC6, Myc, actin (Santa Cruz Biotechnology), FLAG (Cell Signaling), α-tubulin (Sigma), and HDAC1 (Affinity Bioreagents).
Immunofluorescence
Cells were fixed in 3.7% formaldehyde then blocked and permeabilized in 0.1% Triton X-100, 3% BSA in PBS. Primary antibody was rabbit anti-acetylated survivin (Novus Biologicals), mouse anti-survivin (D-8), and rabbit anti-HDAC6 (H-300) (Santa Cruz Biotechnology). Secondary antibody was anti-rabbit IgG conjugated to Dylight 488 and anti-mouse IgG conjugated to Dylight 594 (ThermoFisher Scientific). Slides were mounted with Prolong anti-fade reagent (Invitrogen). Images were captured using a Nikon C1si Confocal microscope.
RESULTS
Survivin Is Deacetylated at Lys-129 by Member of Class I/II HDACs
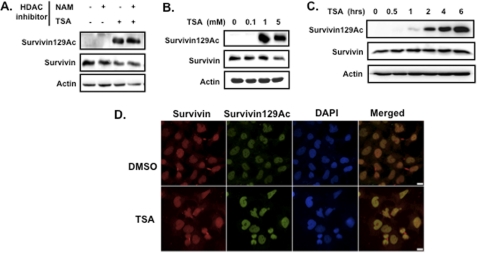
In previous work, we demonstrated that the histone acetyltransferase protein CBP induced survivin acetylation on multiple lysine residues in the estrogen receptor-positive breast cancer cell line, MCF-7 (8). Through mutational analyses, we also showed that loss of CBP-dependent acetylation at Lys-129 facilitates Crm1-mediated nuclear export of deacetylated survivin, suggesting that survivin acetylation dictates whether it functions as a nuclear or cytosolic protein. To determine the histone deacetylase(s) responsible for deacetylating survivin at this and other residues, we treated MCF-7 cells with either the class I/II inhibitor TSA for 6 h or the class III inhibitor nicotinamide for 24 h, or with both, harvested for total cellular protein, and then performed Western blots using an antibody generated to the specific survivin-acetylated Lys-129 residue or an antibody generated to full-length (total) survivin. In TSA-treated MCF-7 cells, robust levels of acetylated survivin protein were observed (Fig. 1A). By contrast, nicotinamide had no effect on survivin acetylation, and no additional increases in acetylation were observed after treatment with both TSA and nicotinamide, suggesting that although class I/II HDACs play a role in deacetylating survivin, class III proteins do not. Total survivin was unchanged in either condition when detected with the antibody against the full-length protein, as expected given that acetylated survivin is a small fraction of the total survivin pool. To determine the dose- and time-dependent effects of TSA on survivin acetylation, we treated MCF-7 cells with increasing concentrations of TSA over a series of time points. Our results demonstrated that 1 μm TSA induced maximal survivin Lys-129 acetylation (Fig. 1B), first detected at 1 h and steadily increasing by 6 h (Fig. 1C), indicating a time- and concentration dependence that should be linked to a functional role for survivin acetylation states. Similar results were also observed in HeLa cells (supplemental Fig. S1).
FIGURE 1.
The class I/II histone deacetylase inhibitors (HDI) TSA induces survivin acetylation in ER+ breast cancer cells. A, acetylated survivin Lys-129 and total survivin levels in response to nicotinamide (NAM), TSA, or both in MCF-7 cells, as determined by Western blotting using a specific antibody to acetylated survivin Lys-129 (Survivin129Ac) or total survivin, respectively. B, dose response of TSA-mediated survivin Lys-129 acetylation in MCF-7 cells. C, time course of survivin Lys-129 acetylation in 1 μm TSA-treated MCF-7 cells. D, subcellular localization of acetylated and total survivin in MCF-7 cells treated with TSA or dimethyl sulfoxide (DMSO; vehicle control) for 6 h and then fixed and co-immunostained with antibodies to acetylated survivin and total survivin (green and red, respectively) and DAPI. Images were taken with a Nikon confocal microscope. Scale bar, 20 μm.
To demonstrate the effects of HDAC inhibition on the subcellular distribution of survivin, we co-immunostained MCF-7 cells with an antibody to total survivin and to acetylated survivin. In the absence of TSA, a small, basal level of acetylated survivin was detected that localized to the nucleus (Fig. 1D, upper panel). Following TSA treatment, the levels of acetylated survivin significantly increased (∼10-fold) in the nuclear compartment (Fig. 1D, lower panel). Total survivin however, was detected in both the cytoplasm and the nucleus of untreated and treated cells. These results were consistent with the Western blot data, suggesting that class I/II HDACs are involved in survivin deacetylation.
HDAC6 Abrogates CBP-dependent Survivin Acetylation
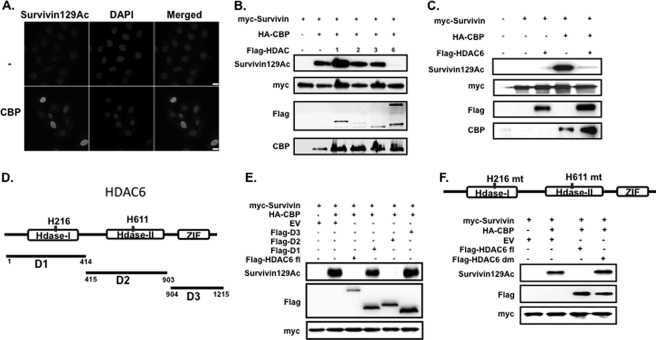
CBP is a central histone acetyltransferase that acetylates multiple cancer-associated proteins and thereby alters their activity (11, 14, 15). To demonstrate the CBP-specific effect on survivin acetylation, we transfected HeLa cells with an expression plasmid encoding HA-tagged CBP then immunostained the cells with the survivin-acetylated antibody. CBP highly induced survivin acetylation, and the acetylated survivin protein displayed a nuclear staining pattern after CBP transfection that was similar to that seen following TSA treatment (Fig. 2A). To identify the specific HDAC(s) that deacetylates survivin, we co-transfected HEK293 cells with a Myc-tagged survivin and CBP construct, along with either empty vector, or one of the class I/II HDACs; HDAC1, HDAC2, HDAC3, or HDAC6 and then performed Western blotting on total cell lysates using the survivin-acetylated antibody. The results showed that CBP-dependent survivin acetylation was unaffected by HDAC1, -2, and -3, but was abolished by HDAC6 (Fig. 2B). These results were recapitulated in HeLa cells (Fig. 2C).
FIGURE 2.
HDAC6 abrogates CBP-mediated survivin acetylation. A, expression of acetylated survivin in HeLa cells transfected with CBP or empty vector (−). Cells were fixed and immunostained with an antibody for acetylated survivin (gray) and DAPI. Images were taken with a Nikon confocal microscope. Scale bar, 20 μm. B, expression of acetylated survivin in HEK293 cells in response to different HDAC proteins. Cells were transfected with Myc-tagged survivin, HA-tagged CBP, and FLAG-tagged HDAC 1, 2, 3, or 6. Total cellular lysates were immunoblotted with the indicated antibodies. C, expression of acetylated survivin in HeLa cells in response to HDAC6. Cells were co-transfected with Myc-tagged survivin, HA-tagged CBP, and FLAG-tagged HDAC6. Total cellular lysates were prepared and immunoblotted with the indicated antibodies. D, schematic diagram of human HDAC6 protein showing the deacetylase domains 1 and 2 and the zinc finger domain III (Hdase I, Hdase II, and ZIF, respectively). His-216 and His-611 are the histidine amino acids mutated to alanine in the HDAC6-dead mutant that inactivates the deacetylase function of Hdase I and Hdase II, respectively. D1 (amino acids 1–414), D2 (amino acids 415–903), and D3 (amino acids 904–1215) illustrate the protein segments for the FLAG-tagged HDAC6 truncated constructs cloned from full-length HDAC6 and used in E. E, HeLa cells were co-transfected with Myc-survivin, HA-CBP, and either FLAG-tagged full-length HDAC6 (fl), FLAG-tagged Hdase I domain 1 (D1), Hdase II domain 2 (D2), ZIF domain 3 (D3), or with empty vector (EV). Total cellular lysates were immunoblotted with the indicated antibodies. F, diagram showing the site of mutations of the HDAC6-dead mutant. HeLa cells were co-transfected with Myc-survivin, HA-CBP, FLAG-HDAC6 full-length (fl), FLAG-HDAC6-dead mutant (dm), or empty vector (EV). Total cellular lysates were immunoblotted with the indicated antibodies.
HDAC6 Domain 2 Is Required for Survivin Deacetylation
HDAC6 is a unique member of the HDAC class IIb family, structurally composed of duplicate catalytic domains and a third ubiquitin-binding domain (11, 16). To identify the domain responsible for the survivin deacetylase activity, we constructed three truncated forms of HDAC6 from the full-length HDAC6 cDNA, corresponding to each domain; histone deacetylase domain 1 (amino acids 1–414), histone deacetylase domain 2 (amino acids 415–903), and ubiquitin binding domain 3 (amino acids 904–1215) (Fig. 2D). We co-transfected HeLa cells with Myc-survivin; HA-CBP; FLAG-HDAC6 domain 1, 2, or 3; full-length HDAC6 or empty vector; with the latter two as positive and negative deacetylase controls, respectively. We performed Western blotting on total cell lysates using the survivin-acetylated antibody. Our results demonstrated that similar to full-length HDAC6, domain 2 alone abolished CBP-mediated survivin acetylation, whereas domains 1 and 3 had little to no effect (Fig. 2E). To further validate that HDAC6 domain 2 is responsible for survivin deacetylation, we utilized an HDAC6 construct (HDAC6-dm) containing a double point mutation that lacks deacetylase activity (12). Our results demonstrated that CBP-dependent survivin acetylation is restored when the HDAC6 deacetylase activity is inactivated (Fig. 2F). Taken together, the data suggest that domain 2 of HDAC6 is necessary and sufficient to inhibit CBP-dependent survivin acetylation. This is consistent with published reports demonstrating that domain 2 is responsible for the deacetylase activity of other proteins (17, 18).
HDAC6 Regulates Survivin Deacetylation in Different Subcellular Pools
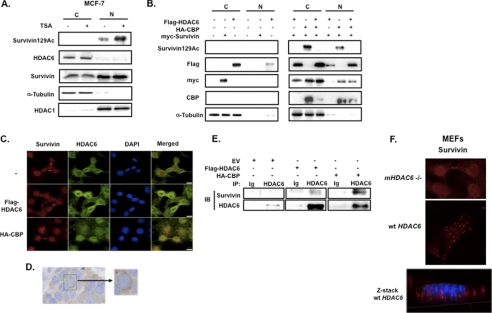
As HDAC6 is mainly cytoplasmic but can be stimulated to enter the nucleus via its nuclear localization signal (19), we sought to determine the subcellular compartment(s) within which HDAC6 deacetylates survivin. To examine the endogenous distribution of HDAC6 and acetylated survivin in MCF-7 cells, we performed subcellular fractionation in the presence and absence of TSA. Untreated MCF-7 cells demonstrated a basal level of nuclear acetylated survivin, which significantly increased following TSA treatment (Fig. 3A), consistent with our previous microscopy results (Fig. 1D). HDAC6 primarily localized to the cytoplasm under both conditions. HDAC6 protein levels were not altered by TSA, consistent with TSA-mediated inhibition of HDAC6 enzymatic activity but not protein steady state.
FIGURE 3.
HDAC6 deacetylates survivin for its nuclear export. A, MCF-7 cells were treated with 1 μm TSA or vehicle control for 6 h. Nuclear (N) and cytoplasmic (C) lysates were prepared by subcellular fractionation then immunoblotted with anti-acetylated survivin, anti-survivin, anti-HDAC6, α-tubulin, and HDAC1, the latter two of which were cytoplasmic and nuclear fraction controls, respectively. B, HEK293 cells were co-transfected with FLAG-HDAC6, HA-CBP, and Myc-survivin. Nuclear (N) and cytoplasmic (C) lysates were prepared and immunoblotted (IB) with anti-acetylated survivin, anti-FLAG, anti-Myc, anti-CBP, and anti-tubulin. C, HeLa cells were co-transfected with FLAG-HDAC6, HA-CBP, or empty vector (−) and then fixed and co-immunostained with anti-survivin (red), anti-HDAC6 (green), and DAPI. Cells were imaged by a Nikon confocal microscope. Scale bars, 20 μm. D, 4 μm paraffin-fixed estrogen-receptor positive breast cancer tissue was immunostained with anti-HDAC6, as described previously (23). E, HeLa cells were transfected with FLAG-HDAC6, HA-CBP or empty vector (EV) then immunoprecipitated (IP) with anti-HDAC6 or immunoglobulin control (Ig) and immunoblotted with anti-survivin and anti-HDAC6. F, MEFs isolated from mHDAC6 null (−/−) or WT mice were immunostained with anti-survivin (red) and imaged by a Nikon confocal microscope. Scale bar, 20 μm. Optical sectioning through the depth of the cell was performed using z-stacking (red, survivin; blue, DAPI).
To determine the compartment(s) where HDAC6 deacetylates survivin, we first performed subcellular fractionation. We co-transfected HEK293 cells with Myc-survivin and FLAG-HDAC6 constructs in the presence or absence of CBP then analyzed the nuclear and cytoplasmic fractions by Western blot. In the absence of CBP-driven acetylation, total survivin and HDAC6 were localized primarily within the cytoplasm (Fig. 3B, left panel). Following CBP-driven acetylation, both total survivin and HDAC6 dramatically increased within the nuclear compartment (Fig. 3B, right panel), indicating that CBP-dependent acetylation induces nuclear survivin accumulation and promotes HDAC6 nuclear trafficking, potentially as a control mechanism to regulate the nuclear levels of acetylated survivin. Correspondingly, CBP-induced acetylated survivin was detected in both nuclear and cytoplasmic compartments and was abolished in both compartments by wild-type HDAC6.
HDAC6 Enters Nucleus to Bind Survivin and Facilitate Survivin Nuclear Export
To further investigate the subcellular region(s) where HDAC6 deacetylates survivin under different conditions, we transfected HeLa cells with and without HDAC6 or CBP and performed immunofluorescence microscopy. In non-transfected cells, endogenous HDAC6 exhibited a perinuclear staining pattern with minimal colocalization with nuclear survivin (Fig. 3C, top row). This pattern was similarly observed after HDAC6 transfection, with a substantial increase in staining noted around the nuclear membrane (Fig. 3C, middle row). The same perinuclear staining pattern was observed in tissue sections obtained from ER-positive breast carcinomas immunostained with anti-HDAC6 (Fig. 3D), supporting the cell culture results. Following CBP transfection in HeLa cells, HDAC6 localized primarily within the nucleus and colocalized with nuclear survivin (Fig. 3C, bottom row). These results show that CBP can induce HDAC6 nuclear import, while maintaining acetylated survivin sequestered in the nuclear compartment. Although it has been reported that HDAC6 binds nuclear proteins (20) and that its structure includes an N-terminal nuclear import signal (19), factors regulating its entry into the nucleus have not been identified. The demonstration that CBP can stimulate HDAC6 nuclear entry strongly suggests that HDAC6 itself is modified by histone acetyltransferase proteins.
To establish whether survivin is a direct substrate of HDAC6, HDAC6 was immunoprecipitated from non-transfected HeLa cells or after transfection with HDAC6 or CBP. HDAC6-survivin immunoprecipitants were not well visualized in the non-transfected cells; however, binding between the two proteins was observed in HDAC6-transfected cells and was further increased in CBP-transfected cells (Fig. 3E), consistent with a direct association between HDAC6 and survivin in the nucleus (Fig. 3C).
To examine the potential in vivo requirements for HDAC6 regulating survivin function, we immunostained HDAC6 null and wild-type (WT) MEFs (kind gift from the laboratory of Dr. Tso-Pang Yao, Duke University Medical Center, Durham, NC) with anti-survivin. The localization pattern of survivin differed in these two cell types. Although the WT cells displayed a punctate, cytoplasmic pattern that was excluded from the nucleus (Z-stack, Fig. 3F), the HDAC6 null cells exhibited a primarily nuclear pattern (Fig. 3F), suggesting that HDAC6-mediated survivin deacetylation is required for survivin nuclear export. Collectively, these results support a role for HDAC6 as a guardian or gatekeeper at the nuclear-cytoplasmic border, inhibiting acetylated survivin nuclear export under resting conditions. When stimulated by histone acetyltransferase proteins, HDAC6 is imported into the nucleus and binds survivin to deacetylate the protein as a likely control mechanism to regulate increasing levels of nuclear acetylated survivin and perhaps to increase its cytoplasmic concentration for its anti-apoptotic activity (9).
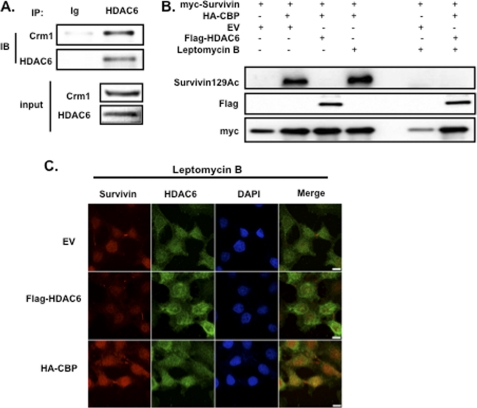
HDAC6-mediated Survivin Deacetylation Is Independent of Crm1
HDAC6, like survivin, is exported from the nucleus by Crm1 (21); therefore, we investigated whether an HDAC6-Crm1 complex is required for survivin deacetylation. Endogenous HDAC6 immunoprecipitants showed strong binding to Crm1 under unstimulated conditions (Fig. 4A). To determine whether Crm1 is required for survivin deacetylation, we treated HeLa cells with the Crm1 inhibitor, leptomycin B (LB), and performed Western blot assays for acetylated survivin. HDAC6 inhibited survivin acetylation both in the presence and absence of LB (Fig. 4B), suggesting that Crm1 is not required for survivin deacetylation by HDAC6. These findings were corroborated by immunofluorescence, demonstrating that HDAC6 localizes to the nucleus and colocalizes with survivin under conditions where Crm1 is inactivated by LB (Fig. 4C).
FIGURE 4.
Survivin deacetylation by HDAC6 is independent of the nuclear export protein Crm1. A, untreated HeLa cells were lysed and immunoprecipitated with anti-HDAC6 or immunoglobulin control (Ig) and immunoblotted with anti-Crm1 and anti-HDAC6. Total Crm1 and total HDAC6 protein are shown as input. B, HeLa cells were co-transfected with FLAG-HDAC6, HA-CBP, and Myc-survivin then treated with or without LB, as described previously (8). Total cellular lysates were immunoblotted (IB) with anti-acetylated survivin, anti-Myc, and anti-FLAG. C, HeLa cells were co-transfected with expression plasmids encoding FLAG-HDAC6, HA-CBP, or an empty vector control (EV) and then treated with LB as described previously (3). Cells were fixed and immunostained with anti-survivin (red), anti-HDAC6 (green), and DAPI and imaged by a Nikon confocal microscope. Scale bar, 20 μm. IP, immunoprecipitation.
Estrogen Induces Survivin and HDAC6 Levels and Their Nuclear Localization
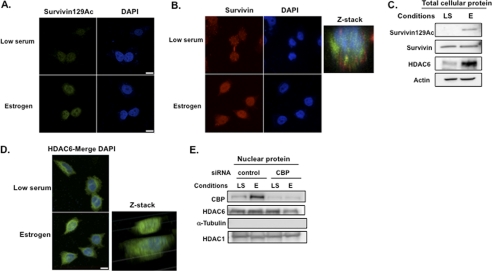
Estrogen was shown recently to increase CBP acetyltransferase activity through coactivator-associated arginine methyltransferase-induced methylation (22). Given our observations that CBP acetylates nuclear survivin, we examined whether estrogen affects survivin expression and localization in MCF-7 cells. Cells were starved for 48 h, treated with 10 nm estradiol or vehicle control for 6 h, and then fixed and co-immunostained with anti-survivin and anti-acetylated survivin. Estrogen treatment mimicked the effects observed previously after CBP transfection (shown in Fig. 2B), promoting an increase in the levels of nuclear acetylated survivin (Fig. 5A). As expected from the observed increase in acetylated survivin, estrogen also enhanced total survivin nuclear localization (Fig. 5B). Interestingly, under conditions of low serum and no estrogen, survivin localized in a punctate, cytoplasmic granular pattern (Fig. 5B), similar to the staining pattern of survivin observed in the HDAC6 WT MEFs, suggesting a potential link between estrogen, survivin, and HDAC6. These effects were also observed when cells were treated with the estrogen receptor antagonist, tamoxifen (supplemental Fig. S2). To determine the effect of estrogen on HDAC6 levels, we performed Western blots on total cell lysates isolated from the estrogen-treated MCF-7 cells. Estrogen treatment was associated with higher levels of HDAC6 as well as acetylated survivin proteins (Fig. 5C), supporting results of clinical studies that show an increase in HDAC6 levels in estrogen receptor-positive breast tumors (23, 24). To examine the effects of estrogen on HDAC6 subcellular localization, we performed immunofluorescence microscopy. In the absence of estrogen, HDAC6 localized to perinuclear regions of the cell. However, after estrogen treatment its localization increased within the nucleus, as demonstrated by z-stack analysis (Fig. 5D). Together, these data show that estrogen treatment leads to an increase in both acetylated survivin and nuclear HDAC6 levels, possibly through increasing CBP activity.
FIGURE 5.
Estrogen induces acetylated survivin and HDAC6 levels and promotes their nuclear localization through CBP. For all the experiments in this figure, MCF-7 cells were cultured in low serum (LS) for 48 h and then treated with 10 nm estradiol (E) for 6 h. A and B, cells were fixed and immunostained with an antibody to acetylated survivin (green, A) or total survivin (red, B), stained with DAPI, and then imaged with a Nikon confocal microscope. Scale bar, 20 μm. Optical sectioning through the depth of the cell was performed using z-stacking (red, survivin; blue, DAPI). C, total cellular lysates were immunoblotted with anti-acetylated survivin, anti-survivin, anti-HDAC6, and anti-actin. D, cells were fixed and immunostained with anti-HDAC6 (green) and DAPI (blue) then imaged with a Nikon confocal microscope. Scale bar, 20 μm. Optical sectioning through the depth of the cell was performed using z-stacking. E, MCF-7 cells were transfected with CBP or control siRNA, cultured in low serum (LS) with or without estrogen (E), followed by isolation of cell nuclei. Nuclear protein was immunoblotted with antibodies to CBP, HDAC6, α-tubulin, and HDAC1.
To determine the requirement for CBP in the nuclear induction of HDAC6 in response to estrogen, we knocked down CBP using siRNA in MCF-7 cells. We then isolated cell nuclei and examined the levels of HDAC6 under conditions of low serum and following estrogen treatment. MCF-7 cells transfected with CBP siRNA resulted in an ∼90% decrease in nuclear CBP protein (Fig. 5E). Interestingly, estrogen led to an increase in endogenous CBP levels in the nuclei of control siRNA-treated cells, consistent with a stabilization of CBP protein in this compartment mediated by estrogen. However, knockdown of CBP in the presence of estrogen resulted in a decrease in HDAC6 protein within cell nuclei (75% of control levels). This finding supports a requirement for CBP in an estrogen-mediated induction of nuclear HDAC6 in MCF-7 cells.
DISCUSSION
HDAC proteins have become pervasive cancer treatment targets due to their involvement in multiple signaling pathways that provide a survival advantage for tumor cells (5). The class IIb deacetylase, HDAC6 increases cancer cell motility and promotes metastatic spread in breast cancer models (24, 25). Inhibition of HDAC6 leads to deregulation of acetylation of multiple proteins, resulting in a disruption of microtubule dynamics and aggresome formation, promoting cancer cell death (26). Here, we identified another target for HDAC6, the anti-apoptotic protein survivin. Principally understood as a cytoplasmic protein (27), we demonstrate a novel nuclear role for HDAC6 in deacetylating survivin. Following estrogen-dependent CBP activation, HDAC6 enters the nucleus and interacts with survivin in a mechanism to regulate its acetylation state and promote its nuclear exit.
The multiple functions of survivin are dependent on its highly regulated expression in distinct subcellular pools (28). We showed previously that survivin nuclear export was dependent on its acetylation state (8). Here, we identify that HDAC6-mediated deacetylation promotes its nuclear exit. In wild-type MEFs and in serum-starved breast cancer cells, survivin localizes to cytoplasmic aggregates, whereas in hdac6 null MEFs and estrogen-treated breast cancer cells, survivin localizes diffusely within the nucleus. Although the nature of the survivin cytoplasmic aggregates is yet unclear, HDAC6-dependent aggresome formation exhibits a similar cytoplasmic staining pattern, suggesting they may represent a component of aggresomes or be involved in autophagy (29). Deacetylation of survivin may potentially promote its interaction with α-tubulin, the binding site for which is located within the domain containing amino acids 99–142 (30), which includes the lysine 129 site.
HDAC6 is an estrogen-regulated protein (24) and has been investigated as a potential prognostic factor for ER+ breast cancer. Most ER+ tumors analyzed express HDAC6; however, there is conflicting data on how increased expression may influence patient prognosis. Although Yoshida et al. (31) suggest that high HDAC6 expression correlates with poor prognosis, Zhang et al. (23) report that it correlates with better prognosis. Our results suggest that HDAC6 expression levels may not be as critical as its subcellular localization, providing a potential explanation for previous clinical discrepancies. A distinct perinuclear staining pattern was observed by immunohistochemistry in invasive ductal carcinoma, which correlates with the pattern we observed in untreated breast cancer cells in vitro. By contrast, treatment of breast cancer cells with estrogen stimulated its nuclear localization. Together, our results suggest that estrogen signaling may be a key factor that promotes HDAC6 nuclear localization and may regulate its deacetylation of survivin as well as other yet unidentified nuclear oncoproteins.
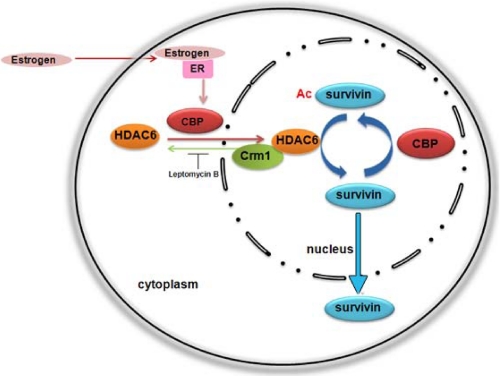
In summary, here we identify HDAC6 as a survivin deacetylase at the nuclear-cytosolic junction of ER+ breast cancer cells. HDAC6 is induced to enter the nucleus by the histone acetyltransferase protein CBP and is required for survivin nuclear export, suggesting a control mechanism that promotes trafficking of deacetylated survivin out of the nucleus, perhaps to enhance its function within the cytosol (Fig. 6). In a similar manner, the growth hormone estrogen induces both survivin and HDAC6 protein levels as well as HDAC6 nuclear entry, suggesting its importance in regulating survivin function in ER+ breast tumors. The dynamic regulation of survivin acetylation/deacetylation may provide an opportunistic target for the treatment of these tumors.
FIGURE 6.
Model of HDAC6-mediated survivin deacetlyation for survivin nuclear export. ER+ breast cancer cells can be stimulated by estrogen that activates the histone acetyltransferase protein CBP. CBP induces HDAC6 expression and promotes its nuclear entry where it deacetylates survivin, independent of Crm1, and stimulates its nuclear export in a mechanism to control nuclear acetylated survivin levels.
Supplementary Material
Acknowledgment
We thank Ginny Horvasian for help with confocal microscopy.
This work was supported, in whole or in part, by National Institutes of Health Grant P20RR017695 (to R. A. A.).

This article contains supplemental Figs. S1 and S2.
- CBP
- CREB binding protein
- CREB
- cAMP-responsive element-binding protein
- HDAC
- histone deacetylase protein
- MEF
- mouse embryonic fibroblast
- TSA
- trichostatin A
- LB
- leptomycin B.
REFERENCES
- 1. Federico M., Bagella L. (2011) Histone deacetylase inhibitors in the treatment of hematological malignancies and solid tumors. J. Biomed. Biotechnol. 2011, 475641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Glozak M. A., Seto E. (2007) Histone deacetylases and cancer. Oncogene 26, 5420–5432 [DOI] [PubMed] [Google Scholar]
- 3. Marks P., Rifkind R. A., Richon V. M., Breslow R., Miller T., Kelly W. K. (2001) Histone deacetylases and cancer: Causes and therapies. Nat. Rev. Cancer 1, 194–202 [DOI] [PubMed] [Google Scholar]
- 4. Dickinson M., Johnstone R. W., Prince H. M. (2010) Histone deacetylase inhibitors: Potential targets responsible for their anti-cancer effect. Invest. New Drugs 28, S3–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Marks P. A. (2010) Histone deacetylase inhibitors: A chemical genetics approach to understanding cellular functions. Biochim. Biophys. Acta 1799, 717–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Marks P. A. (2010) The clinical development of histone deacetylase inhibitors as targeted anticancer drugs. Expert Opin. Investig. Drugs 19, 1049–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Choudhary C., Kumar C., Gnad F., Nielsen M. L., Rehman M., Walther T. C., Olsen J. V., Mann M. (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 [DOI] [PubMed] [Google Scholar]
- 8. Wang H., Holloway M. P., Ma L., Cooper Z. A., Riolo M., Samkari A., Elenitoba-Johnson K. S., Chin Y. E., Altura R. A. (2010) Acetylation directs survivin nuclear localization to repress STAT3 oncogenic activity. J. Biol. Chem. 285, 36129–36137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chan K. S., Wong C. H., Huang Y. F., Li H. Y. (2010) Survivin withdrawal by nuclear export failure as a physiological switch to commit cells to apoptosis. Cell Death Dis. 1, e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pavlyukov M. S., Antipova N. V., Balashova M. V., Vinogradova T. V., Kopantzev E. P., Shakhparonov M. I. (2011) Survivin monomer plays an essential role in apoptosis regulation. J. Biol. Chem. 286, 23296–23307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yang X. J., Seto E. (2007) HATs and HDACs: From structure, function, and regulation to novel strategies for therapy and prevention. Oncogene 26, 5310–5318 [DOI] [PubMed] [Google Scholar]
- 12. Grozinger C. M., Hassig C. A., Schreiber S. L. (1999) Three proteins define a class of human histone deacetylases related to yeast Hda1p. Proc. Natl. Acad. Sci. U.S.A. 96, 4868–4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Witt O., Deubzer H. E., Milde T., Oehme I. (2009) HDAC family: What are the cancer relevant targets? Cancer Lett. 277, 8–21 [DOI] [PubMed] [Google Scholar]
- 14. Iyer N. G., Ozdag H., Caldas C. (2004) p300/CBP and cancer. Oncogene 23, 4225–4231 [DOI] [PubMed] [Google Scholar]
- 15. Lee K. K., Workman J. L. (2007) Histone acetyltransferase complexes: One size does not fit all. Nat. Rev. Mol. Cell Biol. 8, 284–295 [DOI] [PubMed] [Google Scholar]
- 16. Aldana-Masangkay G. I., Sakamoto K. M. (2011) The role of HDAC6 in cancer. J. Biomed. Biotechnol. 2011, 875824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zou H., Wu Y., Navre M., Sang B. C. (2006) Characterization of the two catalytic domains in histone deacetylase 6. Biochem. Biophys. Res. Commun. 341, 45–50 [DOI] [PubMed] [Google Scholar]
- 18. Zhang Y., Gilquin B., Khochbin S., Matthias P. (2006) Two catalytic domains are required for protein deacetylation. J. Biol. Chem. 281, 2401–2404 [DOI] [PubMed] [Google Scholar]
- 19. Bertos N. R., Gilquin B., Chan G. K., Yen T. J., Khochbin S., Yang X. J. (2004) Role of the tetradecapeptide repeat domain of human histone deacetylase 6 in cytoplasmic retention. J. Biol. Chem. 279, 48246–48254 [DOI] [PubMed] [Google Scholar]
- 20. Palijan A., Fernandes I., Verway M., Kourelis M., Bastien Y., Tavera-Mendoza L. E., Sacheli A., Bourdeau V., Mader S., White J. H. (2009) Ligand-dependent corepressor LCoR is an attenuator of progesterone-regulated gene expression. J. Biol. Chem. 284, 30275–30287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Verdel A., Curtet S., Brocard M. P., Rousseaux S., Lemercier C., Yoshida M., Khochbin S. (2000) Active maintenance of mHDA2/mHDAC6 histone-deacetylase in the cytoplasm. Curr. Biol. 10, 747–749 [DOI] [PubMed] [Google Scholar]
- 22. Ceschin D. G., Walia M., Wenk S. S., Duboé C., Gaudon C., Xiao Y., Fauquier L., Sankar M., Vandel L., Gronemeyer H. (2011) Methylation specifies distinct estrogen-induced binding site repertoires of CBP to chromatin. Genes Dev. 25, 1132–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang Z., Yamashita H., Toyama T., Sugiura H., Omoto Y., Ando Y., Mita K., Hamaguchi M., Hayashi S., Iwase H. (2004) HDAC6 expression is correlated with better survival in breast cancer. Clin. Cancer Res. 10, 6962–6968 [DOI] [PubMed] [Google Scholar]
- 24. Saji S., Kawakami M., Hayashi S., Yoshida N., Hirose M., Horiguchi S., Itoh A., Funata N., Schreiber S. L., Yoshida M., Toi M. (2005) Significance of HDAC6 regulation via estrogen signaling for cell motility and prognosis in estrogen receptor-positive breast cancer. Oncogene 24, 4531–4539 [DOI] [PubMed] [Google Scholar]
- 25. Rey M., Irondelle M., Waharte F., Lizarraga F., Chavrier P. (2011) HDAC6 is required for invadopodia activity and invasion by breast tumor cells. Eur. J. Cell Biol. 90, 128–135 [DOI] [PubMed] [Google Scholar]
- 26. Kawaguchi Y., Kovacs J. J., McLaurin A., Vance J. M., Ito A., Yao T. P. (2003) The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell 115, 727–738 [DOI] [PubMed] [Google Scholar]
- 27. Boyault C., Sadoul K., Pabion M., Khochbin S. (2007) HDAC6, at the cross-roads between cytoskeleton and cell signaling by acetylation and ubiquitination. Oncogene 26, 5468–5476 [DOI] [PubMed] [Google Scholar]
- 28. Altieri D. C. (2008) New wirings in the survivin networks. Oncogene 27, 6276–6284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Iwata A., Riley B. E., Johnston J. A., Kopito R. R. (2005) HDAC6 and microtubules are required for autophagic degradation of aggregated huntingtin. J. Biol. Chem. 280, 40282–40292 [DOI] [PubMed] [Google Scholar]
- 30. Altieri D. C. (2008) Survivin, cancer networks, and pathway-directed drug discovery. Nat. Rev. Cancer 8, 61–70 [DOI] [PubMed] [Google Scholar]
- 31. Yoshida N., Omoto Y., Inoue A., Eguchi H., Kobayashi Y., Kurosumi M., Saji S., Suemasu K., Okazaki T., Nakachi K., Fujita T., Hayashi S. (2004) Prediction of prognosis of estrogen receptor-positive breast cancer with combination of selected estrogen-regulated genes. Cancer Sci. 95, 496–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.