Background: Motility is an important environmental response, but flagellar regulation has been studied only in a few model microorganisms.
Results: Essential regulators in the two-component His-Asp phosphorelay system were identified for flagella/motility in Geobacter.
Conclusion: A novel multicomponent His-Asp phosphorelay system controls flagella/motility in Geobacter.
Significance: Elucidating the mechanisms of signal transduction systems for flagella/motility advances the understanding of cellular responses to environments.
Keywords: Gene Regulation, Genomics, Microbiology, Phosphorylation, Signal Transduction, Enhancer-binding Protein, Fe(III) Reduction, FliA, RpoN, Two-component System
Abstract
Geobacter species play an important role in the natural biogeochemical cycles of aquatic sediments and subsurface environments as well as in subsurface bioremediation by oxidizing organic compounds with the reduction of insoluble Fe(III) oxides. Flagellum-based motility is considered to be critical for Geobacter species to locate fresh sources of Fe(III) oxides. Functional and comparative genomic approaches, coupled with genetic and biochemical methods, identified key regulators for flagellar gene expression in Geobacter species. A master transcriptional regulator, designated FgrM, is a member of the enhancer-binding protein family. The fgrM gene in the most studied strain of Geobacter species, Geobacter sulfurreducens strain DL-1, is truncated by a transposase gene, preventing flagellar biosynthesis. Integrating a functional FgrM homolog restored flagellar biosynthesis and motility in G. sulfurreducens DL-1 and enhanced the ability to reduce insoluble Fe(III) oxide. Interrupting the fgrM gene in G. sulfurreducens strain KN400, which is motile, removed the capacity for flagellar production and inhibited Fe(III) oxide reduction. FgrM, which is also a response regulator of the two-component His-Asp phosphorelay system, was phosphorylated by histidine kinase GHK4, which was essential for flagellar production and motility. GHK4, which is a hybrid kinase with a receiver domain at the N terminus, was phosphorylated by another histidine kinase, GHK3. Therefore, the multicomponent His-Asp phosphorelay system appears to control flagellar gene expression in Geobacter species.
Introduction
Motility is considered to be an important attribute that permits Geobacter species to effectively compete for Fe(III) in subsurface environments. The current model for Fe(III) reduction by Geobacter species in the subsurface (1–3) is that Geobacter species are primarily planktonic when electron donors are plentiful. This is because Fe(III) minerals are heterogeneously dispersed in soils and sediments, and thus, once the Fe(III) in one microenvironment is reduced, Geobacter species must search for another source of Fe(III). This contrasts with reduction of soluble electron acceptors, which will continuously diffuse toward a zone in which they are being depleted. It is speculated that Geobacter species are able to continue respiration in the planktonic state because the abundant c-type cytochromes function as capacitors (4, 5), accepting electrons derived from electron donor oxidation as the cells transition between Fe(III) sources. Once a new Fe(III) source is located, the electrons are discharged, presumably via conductive pili (5, 6) and associated cytochromes (7).
Genes for flagellar production are highly conserved in the available genomes of Geobacter species, further suggesting the importance of motility (8, 9). The observation that Geobacter metallireducens produces flagella during growth on Fe(III) oxide, but not soluble chelated Fe(III) (10), suggests that flagellar expression is regulated based on the physiological status of the cell.
Despite the importance of motility to Geobacter species, there is no information on the molecular mechanisms that control the expression of flagella. Until recently, investigations on motility were stymied by the fact that Geobacter sulfurreducens strain DL-1, the only strain of Geobacter species that could be genetically manipulated (11), does not produce flagella (12), despite possessing the full complement of genes for flagellar production found in other Geobacter species (8, 9, 13). However, G. sulfurreducens strain KN400, which was recovered from electrodes poised at low potential (14), does produce flagella. The genome of this strain is available (15), and it can be genetically manipulated with the same procedures that are commonly employed with G. sulfurreducens strain DL-1.
In other bacteria, a highly regulated transcriptional hierarchy controls expression of flagellar genes to ensure that flagella are assembled in a highly ordered manner (16–18). However, the mechanisms for transcriptional control vary. For example, in Enterobacteriaceae with lateral flagella, the master regulator FlhDC is at the top of the regulatory hierarchy (19). FlhDC activates flagellar genes whose expression is dependent on the RNA polymerase σ factor RpoD. In contrast, in Pseudomonadaceae and Vibrionaceae, which have a polar flagellum, the master regulators are FleQ (20) and FlrA (21), respectively. FleQ and FlrA are members of the enhancer-binding protein (EBP)2 family. EBPs typically consist of three domains: an N-terminal sensory (regulatory) domain, a central AAA/σ54 activation domain, and a C-terminal DNA-binding domain (22, 23). The N-terminal domains of FleQ and FlrA show some similarity to the receiver domain of the response regulator of the two-component His-Asp phosphorelay system but lack highly conserved residues such as a phosphorylation site (20, 21). Thus, they are not likely to be response regulators. The two-component His-Asp phosphorelay system is known to be an important molecular signaling device for sensing and responding to environmental changes. The two-component system typically consists of a sensor histidine kinase, which senses environmental signals, and a response regulator, which often exerts gene expression necessary for the adaptation (24). The sensor histidine kinase autophosphorylates its own histidine residue upon signal sensing and subsequently transfers the phosphoryl group to the aspartate residue in the receiver domain of the response regulator. Phosphorylation of the receiver domain results in the activation of the effector (output) domain of the response regulator. FleQ and FlrA activate flagellar genes whose expression is RpoN-dependent, consistent with the fact that EBPs generally promote transcription of RpoN-dependent genes (22, 23). It has previously been suggested that the expression of some flagellar genes in G. sulfurreducens may be RpoN-dependent (25, 26).
Another σ factor, FliA, is involved in the expression of additional flagellar genes in some microorganisms (16–18). The master regulator FlhDC in Enterobacteriaceae or FlrA in Vibrionaceae controls expression of fliA (19). In contrast, in Pseudomonadaceae, fliA expression is independent of the master regulator FleQ (27). Genome sequence analysis revealed a homolog of FliA, but no homolog of FlhDC, FleQ, or FlrA in Geobacter species (8, 9, 13).
Here, we report on studies with G. sulfurreducens that identify the multicomponent His-Asp phosphorelay system regulating flagellar gene expression in Geobacter species. This is an important step in identifying the environmental cues controlling flagellar expression and motility in Geobacter species.
EXPERIMENTAL PROCEDURES
Strains and Growth Conditions
G. sulfurreducens strains DL-1 (12) and KN400 (14) were grown anaerobically in NBAF medium (acetate and fumarate as the electron donor and acceptor, respectively) or FWA medium (acetate as the electron donor) containing Fe(III) citrate or Fe(III) oxide as the electron acceptor (11). The medium was supplemented with the appropriate antibiotics when necessary.
Cell growth was monitored by measuring the absorbance at 600 nm when fumarate served as the electron acceptor. The concentrations of Fe(II) in the cultures containing Fe(III) as the electron acceptor were determined by the FerroZine assay (28). Swimming motility was examined on soft agar (0.3%) plates by spotting 10-μl cultures. The FWA medium containing Fe(III) citrate for soft agar plates was supplemented with 0.0008% (w/v) CaCl2 and 0.002% (w/v) MgSO4. Plate manipulations were conducted at 30 °C in an anaerobic glove box containing a N2/CO2/H2 (83:10:7) atmosphere. Escherichia coli DH5α (29) was used for plasmid preparation and grown in LB medium (30) supplemented with the appropriate antibiotics when necessary.
DNA Binding Assay
The DNA fragment containing the flhA promoter region used as a probe was prepared by PCR with primers flhA-pro1 and flhA-pro2 (supplemental Table S1) and labeled with [γ-32P]ATP by T4 polynucleotide kinase. DNA binding assays with EBPs were carried out as described previously (31). The fgrM gene from the KN400 strain was amplified by PCR with primers fgrM-DB1 and fgrM-DB2 (supplemental Table S1) and cloned in pET24b (Novagen). FgrM protein was prepared with a histidine tag at the C terminus as described previously (32).
Integration of fgrM Gene from KN400 Strain into DL-1 Strain
The fgrM gene from the KN400 strain was amplified by PCR with primers fgrM-int1 and fgrM-int2 (supplemental Table S1) and cloned in pET24b. The resultant plasmid was integrated into the chromosome of the G. sulfurreducens DL-1 strain (supplemental Fig. S1A) by electroporation (11).
RT-PCR
Total RNA was prepared from G. sulfurreducens DL-1 strains grown in NBAF medium. cDNA was prepared using reverse transcriptase with primers fgrM-RTPCR1 (fgrM), flhA-RTPCR1 (flhA), cheW1-RTPCR1 (cheW1), and dcuB-RTPCR1 (dcuB) (supplemental Table S1). cDNA thus prepared was amplified by PCR with primers fgrM-RTPCR1 and fgrM-RTPCR2 (fgrM), flhA-RTPCR1 and flhA-RTPCR2 (flhA), cheW1-RTPCR1 and cheW1-RTPCR2 (cheW1), and dcuB-RTPCR1 and dcuB-RTPCR2 (dcuB) (supplemental Table S1). The amplified DNA fragments were analyzed by agarose gel electrophoresis, followed by staining with EtBr.
Interruption of fgrM Gene in KN400 Strain
The internal region of the fgrM gene from the KN400 strain was amplified by PCR with primers fgrM-int3 and fgrM-int4 (supplemental Table S1) and cloned in pBluescript II KS(−) (Stratagene). The resultant plasmid was integrated into the chromosome of the KN400 strain by electroporation (11), and the fgrM gene was interrupted (supplemental Fig. S1B).
Transmission Electron Microscopy
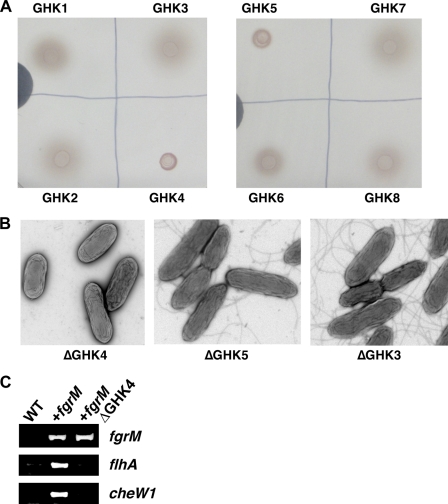
Cells were grown in NBAF liquid medium. The cells were placed on 400-mesh carbon-coated copper grids, incubated for 2 min, and then stained with 2% uranyl acetate. Cells were observed using a JOEL 100 transmission electron microscope at an accelerating voltage of 80 kV. Images were taken digitally using MaxIm DL software and analyzed using ImageJ (rsbweb.nih.giv/ij/index.html).
Construction of Deletion Mutants of Histidine Kinases
Genes for GHK1–8 (Geobacter histidine kinase) were replaced with a spectinomycin resistance gene. Double-crossover homologous recombination was carried out by electroporation with the linear DNA fragment consisting of the spectinomycin resistance gene flanked by 0.7-kilobase pair DNA fragments containing the upstream and downstream regions of the kinase domains. These flanking DNA fragments were amplified by PCR with the primers listed in supplemental Table S1. The DNA fragment of the spectinomycin resistance gene was amplified by PCR with primers Sp-fwd and Sp-rev (supplemental Table S1) using pSJS985 (33) as a template. The replacement was confirmed by PCR amplification.
In Vitro Phosphorylation Assays
The DNA fragments encoding GHK3-K (Ala226–Gln499), GHK4-R (Met1–Glu124), GHK4-K (Met424–Arg727), and FgrM-R (Met1–His121), where “K” is kinase domain and “R” is receiver domain, were amplified by PCR with the primers listed in supplemental Table S1. The PCR products were cloned into pET24b. The cloned genes were overexpressed using an autoinduction system (Novagen) as instructed by the manufacturer, and the proteins were prepared with a histidine tag at their C termini. Purification of these proteins was performed as described previously (34). In vitro phosphorylation assays were performed with [γ-32P]ATP as described previously (31). Kinases (10 pmol) were incubated at room temperature for 5 min for the autophosphorylation reaction. Response regulators (10 pmol) were then added and further incubated at room temperature for 5 min for the phosphotransfer reaction.
RESULTS
Master Regulator for Flagellar Gene Expression
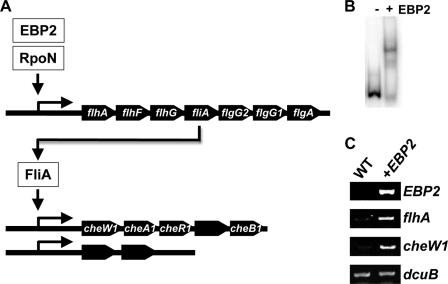
Sequence analysis identified genes involved in flagellar biosynthesis and motility that were predicted to be controlled by RpoN (supplemental Fig. S2A) or FliA (supplemental Fig. S2B). Among them, the flhA operon appears to be a key to flagellar gene regulation in Geobacter species, as it contains the gene encoding FliA (Fig. 1).
FIGURE 1.
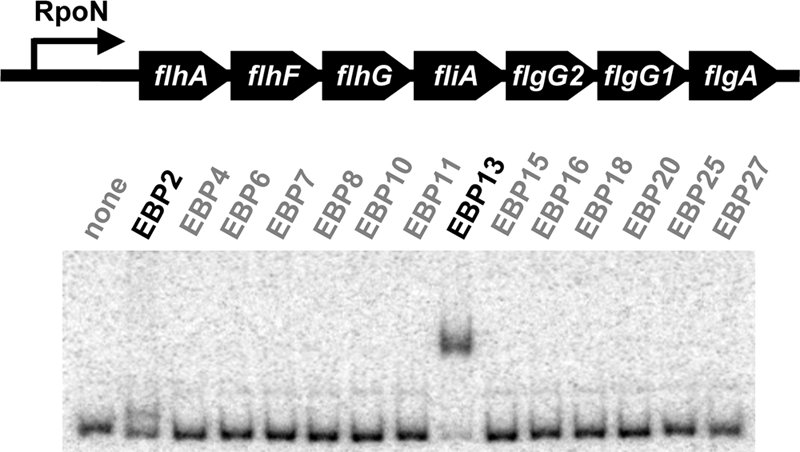
Identification of transcription factor involved in regulation of flagellar gene expression in G. sulfurreducens DL-1. DNA binding assays were conducted with the promoter region of the flhA operon containing flhA, flhF, flhG, fliA, flgG2, flgG1, and flgA (upper). EBPs conserved in Geobacter species were tested for their DNA-binding activity for the flhA promoter (lower).
RpoN is known to require an EBP for transcription initiation (22, 23), suggesting an additional level of regulation of expression of the flagellar genes. Therefore, the 14 previously described EBPs that are conserved in the genomes of G. sulfurreducens and two flagellum-producing species, G. metallireducens and Geobacter uraniireducens (31), were tested for their ability to bind the flhA promoter region of G. sulfurreducens DL-1 (Fig. 1). EBP2 and EBP13 both bound this region (Fig. 1). It should be noted that the EBP2-flhA promoter complex migrated much faster than the EBP13-flhA promoter complex in the DNA binding assay because, as described below, EBP2 is smaller than EBP13. The DNA-binding activity of EBP2 and EBP13 appears to be specific, as EBP2 and EBP13 did not bind other promoters known to be involved in other cellular functions such as nitrogen fixation (31).
EBP2 in G. sulfurreducens DL-1 has a DNA-binding domain but lacks the regulatory domain and most of the AAA/σ54 activation domain, which are typically found in the EBP family and necessary for transcription initiation with RpoN. In contrast, EBP2 homologs in other Geobacter species have all three domains (supplemental Figs. S3 and S4). The EBP2 homologs appear to have a receiver domain of the response regulator in the two-component His-Asp phosphorelay system as the regulatory domain. In G. sulfurreducens DL-1, a gene (GSU0298) encoding a regulatory domain and an AAA/σ54 activation domain that are homologous to those of the EBP2 homologs in other Geobacter species is located upstream of a gene encoding a transposase that is located upstream of the EBP2 gene (supplemental Fig. S3). Thus, it appears that insertion of a transposase might have disrupted the EBP2 gene, which could have the consequence of preventing transcription of key flagellar genes.
In contrast to the EBP2 gene of G. sulfurreducens DL-1, the EBP2 homolog of G. sulfurreducens KN400, which produces flagella, contains all three domains (supplemental Fig. S3). The EBP2 homolog of G. sulfurreducens KN400 bound the flhA promoter region from the DL-1 strain in the DNA binding assay (Fig. 2B).
FIGURE 2.
Induction of flagellum-related genes by KN400 EBP2 in G. sulfurreducens DL-1. A, model for regulatory cascades in flagellar gene expression in Geobacter species. B, DNA-binding activity. EBP2 from the KN400 strain was tested with the DNA fragment containing the flhA promoter region from the DL-1 strain in DNA binding assays. C, expression of flagellum-related genes. Expression of EBP2, flhA, and cheW1 was examined by RT-PCR assays in the wild-type DL-1 strain (WT) and the DL-1 strain containing the complete EBP gene (+EBP2). As a control, the expression of the dcuB gene was examined.
To assess the effects of the EBP2 gene from the KN400 strain on flagellar gene expression in the DL-1 strain, it was integrated into the chromosome of the DL-1 strain. The expression of the KN400 EBP2 gene in the DL-1 strain was confirmed with a RT-PCR assay (Fig. 2C). The integration of the KN400 EBP2 gene induced the expression of the flhA gene and other flagellum-related genes (Fig. 2C).
Furthermore, the expression of genes such as the cheW operon containing chemotaxis genes, which appears to be controlled by FliA (Fig. 2A and supplemental Fig. S2B), was also activated by the integration of the KN400 EBP2 gene in G. sulfurreducens DL-1 (Fig. 2C). In contrast, the expression of the dicarboxylic acid transporter gene dcuB, which has RpoN-dependent promoter elements (25), was not affected (Fig. 2C). These results suggest that the EBP2 gene is malfunctioning in the wild-type G. sulfurreducens DL-1 strain and that EBP2 serves as a master regulator for flagellar gene expression because EBP2 not only directly regulated the expression of flagellar genes such as flhA and fliA but also influenced other flagellum-related genes, including cheW1, that are likely to be regulated by FliA (Fig. 2A). Therefore, EBP2 has been termed FgrM (flagellar gene regulator, master).
Restoration of Flagellar Biosynthesis
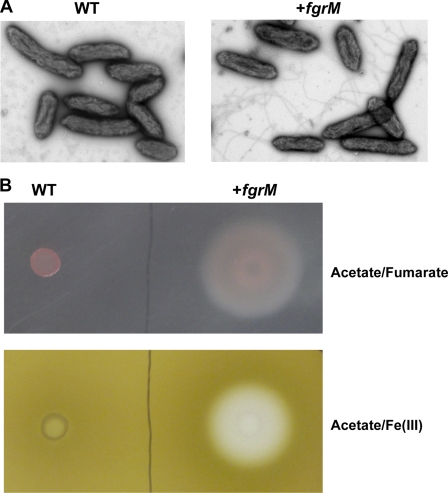
Induced expression of the flagellar genes following the introduction of a complete fgrM gene was accompanied by production of flagella (Fig. 3A) and motility that could be observed on soft agar plates (Fig. 3B). When fumarate was used as the electron acceptor, periodic concentric rings were formed on the soft agar plate by the flagellum-proficient DL-1 strain (Fig. 3B). There was a larger zone of Fe(III) reduction on the soft agar plates containing Fe(III) citrate as the electron acceptor with the flagellum-proficient DL-1 strain than with the wild-type DL-1 strain (Fig. 3B). These results indicate that the repaired flagellar biosynthesis restored motility in the DL-1 strain.
FIGURE 3.
Restoration of flagellar biosynthesis and motility by complete fgrM gene in G. sulfurreducens DL-1. A, flagellar biosynthesis. The wild-type DL-1 cells (WT) and the DL-1 cells containing the complete fgrM gene (+fgrM) were grown in NBAF liquid medium and observed under a transmission electron microscope. B, motility. The wild-type DL-1 cells and the DL-1 cells containing the complete fgrM gene were spotted on agar (0.3%) plates containing NBAF or FWA medium supplemented with Fe(III) citrate.
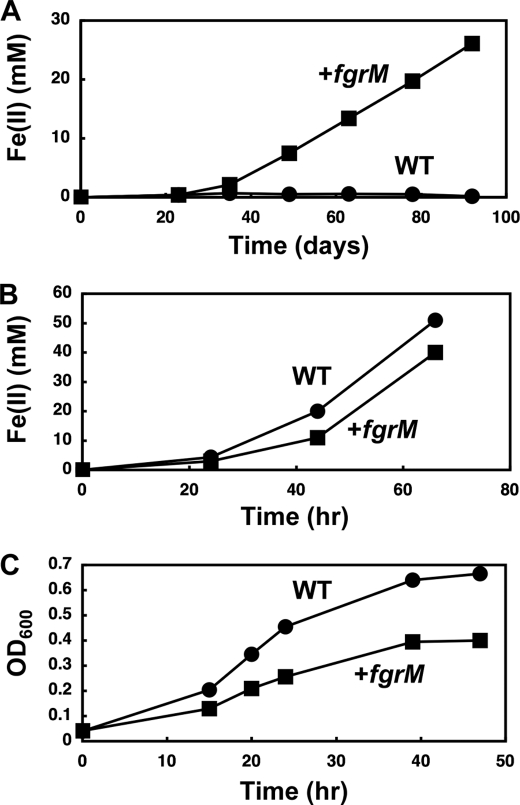
Introduction of the uninterrupted fgrM gene impacted on growth with various electron acceptors. For example, the initial transfer of G. sulfurreducens DL-1 from medium with fumarate as the electron acceptor to medium with Fe(III) oxide as the electron acceptor typically resulted in a long lag period, which was significantly diminished in the strain with the uninterrupted fgrM gene (Fig. 4A). In contrast, the DL-1 strain with the uninterrupted fgrM gene reduced Fe(III) more slowly than the wild-type DL-1 strain in medium with soluble Fe(III) citrate as the electron acceptor (Fig. 4B) and grew slightly slower and reached a lower final culture density than the wild-type DL-1 strain with the soluble fumarate electron acceptor (Fig. 4C).
FIGURE 4.
Effects of flagellar biosynthesis on growth with soluble or insoluble electron acceptors. A, insoluble Fe(III) oxide. The wild-type DL-1 strain (WT) and the DL-1 strain containing the complete fgrM gene (+fgrM) were grown in FWA liquid medium containing Fe(III) oxide. Reduction of Fe(III) was monitored by measuring the concentration of Fe(II). B, soluble Fe(III) citrate. The wild-type DL-1 strain and the DL-1 strain containing the complete fgrM gene were grown in FWA liquid medium containing Fe(III) citrate. Reduction of Fe(III) was monitored by measuring the concentration of Fe(II). C, fumarate. The wild-type DL-1 strain and the DL-1 strain containing the complete fgrM gene were grown in NBAF liquid medium. Growth was monitored as A600.
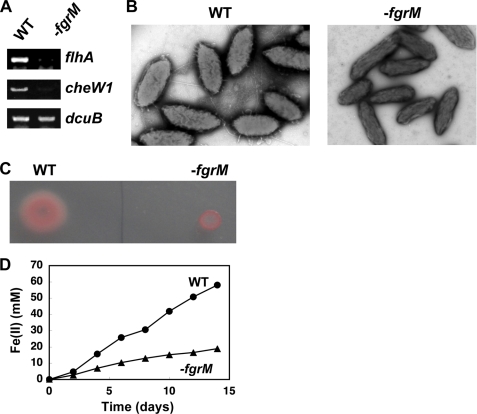
To further evaluate the FgrM function, the uninterrupted fgrM gene in G. sulfurreducens KN400 was interrupted. Interruption of the fgrM gene resulted in a strain that did not express flhA or cheW1 (Fig. 5A), no longer produced flagella (Fig. 5B), was nonmotile (Fig. 5C), and was impaired in its capacity for Fe(III) oxide reduction (Fig. 5D). These results further support the conclusion that FgrM is the master regulator for flagellar gene expression in Geobacter species.
FIGURE 5.
Effects of interruption of fgrM gene. A, expression of flagellum-related genes. Expression of flhA and cheW1 was examined by RT-PCR assays in the wild-type KN400 strain (WT) and the KN400 strain containing the interrupted fgrM gene (−fgrM). As a control, the expression of the dcuB gene was examined. B, flagellar biosynthesis. The wild-type KN400 cells and the KN400 cells containing the interrupted fgrM gene were grown in NBAF liquid medium and observed under a transmission electron microscope. C, Motility. The wild-type KN400 cells and the KN400 cells containing the interrupted fgrM gene were spotted on agar (0.3%) plates containing NBAF medium. D, reduction of insoluble Fe(III) oxide. The wild-type KN400 strain and the KN400 strain containing the interrupted fgrM gene were grown in FWA liquid medium containing Fe(III) oxide. Reduction of Fe(III) was monitored by measuring the concentration of Fe(II).
Identification of Histidine Kinase Involved in Motility and Flagellar Biosynthesis
FgrM appears to belong to a response regulator of the two-component His-Asp phosphorelay system (supplemental Fig. S4), and thus, it is likely that a sensor histidine kinase regulates the activity of FgrM by phosphorylation. Genes for a two-component system are often co-localized in the same operon in other bacteria, but this is often not the case in Geobacter species (8, 9, 13). No genes for a putative sensor histidine kinase are found in the region near the fgrM gene in the genomes of Geobacter species. The genomes of Geobacter species encode an unusually large number of genes for two-component signaling proteins (8, 9, 13). A histidine kinase for FgrM is likely conserved in Geobacter species, as for FgrM. To identify such a histidine kinase, eight histidine kinases (GHK1–8) highly conserved in Geobacter species were selected from 95 putative histidine kinases encoded in the genome of G. sulfurreducens DL-1 by analyzing amino acid sequences of all of the putative histidine kinases (supplemental Table S2). These histidine kinases were inactivated in the DL-1 strain containing the uninterrupted fgrM gene, and the motility of these mutants was examined on a soft agar plate. The GHK4 and GHK5 mutants did not exhibit motility, whereas other mutants did (Fig. 6A). The GHK5 mutant became motile when incubated for a longer period on the soft agar plate, whereas the GHK4 mutant still did not (supplemental Fig. S5). The lack of motility of the GHK4 mutant was likely due to the absence of flagella (Fig. 6B). In contrast, the GHK5 mutant produced flagella (Fig. 6B). It is likely that GHK4 was involved in transcription of genes for flagella and motility, as transcripts for flhA and cheW1 were not detected in the GHK4 mutant (Fig. 6C). These results suggest that GHK4 is involved in the transcriptional activation of genes for flagella and motility.
FIGURE 6.
Identification of histidine kinase involved in flagellar biosynthesis. A, motility. Mutants were spotted on agar (0.3%) plates containing NBAF medium. B, flagellar biosynthesis. Mutants were grown in NBAF liquid medium and observed under a transmission electron microscope. C, expression of flagellum-related genes. Expression of fgrM, flhA, and cheW1 was examined by RT-PCR assays in the wild-type DL-1 strain (WT), the DL-1 strain containing the complete fgrM gene (+fgrM), and the DL-1 strain containing the complete fgrM gene but lacking the GHK4 gene (+fgrMΔGHK4).
His-Asp Phosphorelay
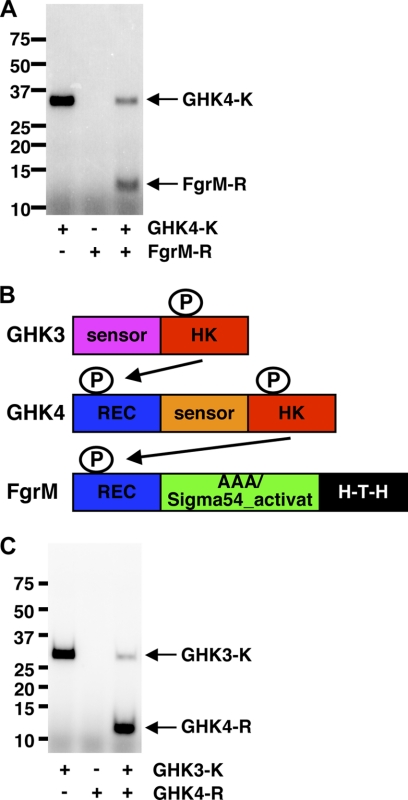
To elucidate the relationship between GHK4 and FgrM in the regulation of flagellar gene expression, the possibility of GHK4 and FgrM functioning as a two-component system was assessed because mutants of GHK4 or FgrM exhibited similar phenotypes, no expression of genes for flagella and motility, no flagellar biosynthesis, and lack of motility, as shown above. The kinase domain of GHK4 (GHK4-K) and the receiver domain of FgrM (FgrM-R) were purified and examined by in vitro phosphorylation assays. GHK4-K showed autophosphorylation activity and phosphorylated FgrM-R (Fig. 7A). These results suggest that GHK4 and FgrM function as a two-component system.
FIGURE 7.
His-Asp phosphorelay. A, in vitro phosphorylation assay with GHK4-K and FgrM-R. GHK4-K and FgrM-R were tested for autophosphorylation and phosphotransfer activity in vitro. The molecular mass standards are shown in kilodaltons. B, model for phosphorelay. HK, histidine kinase; REC, receiver domain; AAA/Sigma54_activat, AAA/σ54 activation domain. C, in vitro phosphorylation assay with GHK3-K and GHK4-R. GHK3-K and GHK4-R were tested for autophosphorylation and phosphotransfer activity in vitro. The molecular mass standards are shown in kilodaltons.
GHK4 has a domain homologous to the receiver domain at the N terminus (Fig. 7B). The gene for GHK3 is located immediately upstream of the gene for GHK4 on the genome, and GHK3 is also conserved in Geobacter species (supplemental Table S2). Thus, it seemed possible that GHK3 might be the kinase for GHK4. To evaluate this possibility, GHK3-K and GHK4-R were purified and examined by in vitro phosphorylation assays. GHK3-K exhibited autophosphorylation activity as well as phosphotransfer activity to GHK4-R (Fig. 7C), suggesting that GHK3 is involved in the regulatory network containing GHK4 and FgrM. Therefore, it is likely that the His-Asp phosphorelay system mediated by GHK3, GHK4, and FgrM controls flagellar gene expression in Geobacter species.
DISCUSSION
In this study, we identified important regulators for the synthesis of flagella in Geobacter species. The FgrM master transcriptional regulator for the synthesis of flagella in Geobacter species is an RpoN-dependent EBP, as found in other bacteria such as Pseudomonadaceae and Vibrionaceae (16–18). However, unlike these other bacterial EBPs, FgrM appears to be a response regulator of the two-component His-Asp phosphorelay system and to be activated via phosphorylation by the histidine kinase GHK4, which is also essential for the expression of genes for flagella and motility. GHK4 has a receiver domain at the N terminus and was phosphorylated in vitro by GHK3, another histidine kinase conserved in Geobacter species. This suggests that GHK4 is regulated by GHK3. However, the GHK3 mutant was able to produce flagella and was motile (Fig. 6, A and B), unlike the GHK4 and FgrM mutants. The role GHK3 plays in the regulation of flagellar gene expression remains to be elucidated. A putative sensor domain of GHK3 shows similarity to the PAS domain, whereas that of GHK4 shows similarity to the PAS and GAF domains (supplemental Table S2). However, they are not highly homologous to known proteins. The PAS domain is a signaling module known to sense light, oxygen, redox, and small molecules (35, 36). The GAF domain is a ubiquitous signaling module known to bind small molecules such as cyclic nucleotide monophosphates, tetrapyrroles, and formate (37, 38). Characterization of these domains should provide critical information for an environmental cue that regulates flagellar biosynthesis. It appears likely that GHK5 is involved in motility, but not in flagellar biosynthesis (Fig. 6, A and B). Elucidation of the function of GHK5 would further advance the understanding of the regulatory network in motility of Geobacter species.
The transcriptional regulatory system for the synthesis of flagella in Geobacter species also appears to employ the σ factor FliA, as found in other bacterial systems (16–18). It is likely that expression of the fliA gene is under the control of FgrM in Geobacter species, as it is in other bacteria such as Enterobacteriaceae and Vibrionaceae. Predicted target genes of FliA include genes for the final steps of flagellar assembly, as well as the chemotaxis signaling system, including chemoreceptors (methyl-accepting chemotaxis proteins). The presence of four cheA genes in the G. sulfurreducens genome suggests that G. sulfurreducens possesses multiple chemotaxis systems or homologs (26). The Che1 system appears to be involved in motility in G. sulfurreducens, as it is likely to be under the control of the transcriptional regulatory system mediated by FgrM and FliA. These findings will make it possible to study chemotaxis in Geobacter species in a more systematic manner. In addition to FgrM, RpoN, and FliA, there may be additional transcription factors involved in the regulation of flagellar biosynthesis in Geobacter species. EBP13 also bound the flhA promoter region in the DNA binding assay, but its function remains to be investigated.
The interruption of the fgrM gene in G. sulfurreducens DL-1 by insertion of a transposase must be a relatively recent phenomenon, as the KN400 strain of the same species contains an uninterrupted gene, as does the closely related G. metallireducens. A similar interruption of the fgrM homolog by a transposase is found in Pelobacter propionicus (supplemental Fig. S6), which is phylogenetically intertwined with Geobacter species (39, 40).
G. sulfurreducens DL-1 was enriched and isolated with soluble chelated Fe(III) (12), whereas G. metallireducens was enriched and purified by dilution in medium with insoluble Fe(III) oxide (41). G. metallireducens cells specifically produce flagella when grown on insoluble Fe(III), but not soluble Fe(III) citrate (10). As shown here, expressing the fgrM gene decreased the rate of growth on soluble electron acceptors, suggesting that the original method for enriching and isolating G. sulfurreducens may have selected for a strain that did not express fgrM. Further investigation into the mechanisms by which environmental signals and regulatory proteins control flagellar gene expression and other potential phenotypic impacts of flagellar expression in Geobacter species is warranted.
Supplementary Material
Acknowledgments
We thank J. Ward and M. Sharma for technical support.
This work was supported by Grants DE-FC02-02ER63446 and DE-SC0004080 from the Office of Science (Biological and Environmental Research), United States Department of Energy.

This article contains supplemental Figs. S1–S6, Tables S1 and S2, and additional references.
- EBP
- enhancer-binding protein.
REFERENCES
- 1. Lovley D. R. (2008) Extracellular electron transfer: wires, capacitors, iron lungs, and more. Geobiology 6, 225–231 [DOI] [PubMed] [Google Scholar]
- 2. Mahadevan R., Palsson B. Ø., Lovley D. R. (2011) In situ to in silico and back: elucidating the physiology and ecology of Geobacter spp. using genome-scale modeling. Nat. Rev. Microbiol. 9, 39–50 [DOI] [PubMed] [Google Scholar]
- 3. Lovley D. R., Ueki T., Zhang T., Malvankar N. S., Shrestha P. M., Flanagan K. A., Aklujkar M., Butler J. E., Giloteaux L., Rotaru A. E., Holmes D. E., Franks A. E., Orellana R., Risso C., Nevin K. P. (2011) Geobacter: the microbe electric's physiology, ecology, and practical applications. Adv. Microb. Physiol. 59, 1–100 [DOI] [PubMed] [Google Scholar]
- 4. Esteve-Núñez A., Sosnik J., Visconti P., Lovley D. R. (2008) Fluorescent properties of c-type cytochromes reveal their potential role as an extracytoplasmic electron sink in Geobacter sulfurreducens. Environ. Microbiol. 10, 497–505 [DOI] [PubMed] [Google Scholar]
- 5. Malvankar N. S., Vargas M., Nevin K. P., Franks A. E., Leang C., Kim B. C., Inoue K., Mester T., Covalla S. F., Johnson J. P., Rotello V. M., Tuominen M. T., Lovley D. R. (2011) Tunable metallic-like conductivity in microbial nanowire networks. Nat. Nanotechnol. 6, 573–579 [DOI] [PubMed] [Google Scholar]
- 6. Reguera G., Nevin K. P., Nicoll J. S., Covalla S. F., Woodard T. L., Lovley D. R. (2006) Biofilm and nanowire production leads to increased current in Geobacter sulfurreducens fuel cells. Appl. Environ. Microbiol. 72, 7345–7348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Leang C., Qian X., Mester T., Lovley D. R. (2010) Alignment of the c-type cytochrome OmcS along pili of Geobacter sulfurreducens. Appl. Environ. Microbiol. 76, 4080–4084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Aklujkar M., Young N. D., Holmes D., Chavan M., Risso C., Kiss H. E., Han C. S., Land M. L., Lovley D. R. (2010) The genome of Geobacter bemidjiensis, exemplar for the subsurface clade of Geobacter species that predominate in Fe(III)-reducing subsurface environments. BMC Genomics 11, 490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Aklujkar M., Krushkal J., DiBartolo G., Lapidus A., Land M. L., Lovley D. R. (2009) The genome sequence of Geobacter metallireducens: features of metabolism, physiology, and regulation common and dissimilar to Geobacter sulfurreducens. BMC Microbiol. 9, 109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Childers S. E., Ciufo S., Lovley D. R. (2002) Geobacter metallireducens accesses insoluble Fe(III) oxide by chemotaxis. Nature 416, 767–769 [DOI] [PubMed] [Google Scholar]
- 11. Coppi M. V., Leang C., Sandler S. J., Lovley D. R. (2001) Development of a genetic system for Geobacter sulfurreducens. Appl. Environ. Microbiol. 67, 3180–3187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Caccavo F., Jr., Lonergan D. J., Lovley D. R., Davis M., Stolz J. F., McInerney M. J. (1994) Geobacter sulfurreducens sp. nov., a hydrogen- and acetate-oxidizing dissimilatory metal-reducing microorganism. Appl. Environ. Microbiol. 60, 3752–3759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Methé B. A., Nelson K. E., Eisen J. A., Paulsen I. T., Nelson W., Heidelberg J. F., Wu D., Wu M., Ward N., Beanan M. J., Dodson R. J., Madupu R., Brinkac L. M., Daugherty S. C., DeBoy R. T., Durkin A. S., Gwinn M., Kolonay J. F., Sullivan S. A., Haft D. H., Selengut J., Davidsen T. M., Zafar N., White O., Tran B., Romero C., Forberger H. A., Weidman J., Khouri H., Feldblyum T. V., Utterback T. R., Van Aken S. E., Lovley D. R., Fraser C. M. (2003) Genome of Geobacter sulfurreducens: metal reduction in subsurface environments. Science 302, 1967–1969 [DOI] [PubMed] [Google Scholar]
- 14. Yi H., Nevin K. P., Kim B. C., Franks A. E., Klimes A., Tender L. M., Lovley D. R. (2009) Selection of a variant of Geobacter sulfurreducens with enhanced capacity for current production in microbial fuel cells. Biosens. Bioelectron. 24, 3498–3503 [DOI] [PubMed] [Google Scholar]
- 15. Nagarajan H., Butler J. E., Klimes A., Qiu Y., Zengler K., Ward J., Young N. D., Methé B. A., Palsson B. Ø., Lovley D. R., Barrett C. L. (2010) De novo assembly of the complete genome of an enhanced electricity-producing variant of Geobacter sulfurreducens using only short reads. PLoS ONE 5, e10922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McCarter L. L. (2006) Regulation of flagella. Curr. Opin. Microbiol. 9, 180–186 [DOI] [PubMed] [Google Scholar]
- 17. Smith T. G., Hoover T. R. (2009) Deciphering bacterial flagellar gene regulatory networks in the genomic era. Adv. Appl. Microbiol. 67, 257–295 [DOI] [PubMed] [Google Scholar]
- 18. Soutourina O. A., Bertin P. N. (2003) Regulation cascade of flagellar expression in Gram-negative bacteria. FEMS Microbiol. Rev. 27, 505–523 [DOI] [PubMed] [Google Scholar]
- 19. Chilcott G. S., Hughes K. T. (2000) Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 64, 694–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Arora S. K., Ritchings B. W., Almira E. C., Lory S., Ramphal R. (1997) A transcriptional activator, FleQ, regulates mucin adhesion and flagellar gene expression in Pseudomonas aeruginosa in a cascade manner. J. Bacteriol. 179, 5574–5581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Prouty M. G., Correa N. E., Klose K. E. (2001) The novel σ54- and σ28-dependent flagellar gene transcription hierarchy of Vibrio cholerae. Mol. Microbiol. 39, 1595–1609 [DOI] [PubMed] [Google Scholar]
- 22. Studholme D. J., Dixon R. (2003) Domain architectures of σ54-dependent transcriptional activators. J. Bacteriol. 185, 1757–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wigneshweraraj S., Bose D., Burrows P. C., Joly N., Schumacher J., Rappas M., Pape T., Zhang X., Stockley P., Severinov K., Buck M. (2008) Modus operandi of the bacterial RNA polymerase containing the α54 promoter specificity factor. Mol. Microbiol. 68, 538–546 [DOI] [PubMed] [Google Scholar]
- 24. Hoch J. A., Silhavy T. J. (eds) (1995) Two-component Signal Transduction, ASM Press, Washington, D.C [Google Scholar]
- 25. Leang C., Krushkal J., Ueki T., Puljic M., Sun J., Juárez K., Núñez C., Reguera G., DiDonato R., Postier B., Adkins R. M., Lovley D. R. (2009) Genome-wide analysis of the RpoN regulon in Geobacter sulfurreducens. BMC Genomics 10, 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tran H. T., Krushkal J., Antommattei F. M., Lovley D. R., Weis R. M. (2008) Comparative genomics of Geobacter chemotaxis genes reveals diverse signaling function. BMC Genomics 9, 471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dasgupta N., Wolfgang M. C., Goodman A. L., Arora S. K., Jyot J., Lory S., Ramphal R. (2003) A four-tiered transcriptional regulatory circuit controls flagellar biogenesis in Pseudomonas aeruginosa. Mol. Microbiol. 50, 809–824 [DOI] [PubMed] [Google Scholar]
- 28. Lovley D. R., Phillips E. J. (1986) Organic matter mineralization with reduction of ferric iron in anaerobic sediments. Appl. Environ. Microbiol. 51, 683–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hanahan D. (1983) Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166, 557–580 [DOI] [PubMed] [Google Scholar]
- 30. Miller J. H. (1972) Experiments in Molecular Genetics, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 31. Ueki T., Lovley D. R. (2010) Novel regulatory cascades controlling expression of nitrogen-fixation genes in Geobacter sulfurreducens. Nucleic Acids Res. 38, 7485–7499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ueki T., Lovley D. R. (2010) Genome-wide gene regulation of biosynthesis and energy generation by a novel transcriptional repressor in Geobacter species. Nucleic Acids Res. 38, 810–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sandler S. J., Clark A. J. (1994) RecOR suppression of recF mutant phenotypes in Escherichia coli K-12. J. Bacteriol. 176, 3661–3672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ueki T., Inouye S. (2002) Transcriptional activation of a heat-shock gene, lonD, of Myxococcus xanthus by a two-component histidine-aspartate phosphorelay system. J. Biol. Chem. 277, 6170–6177 [DOI] [PubMed] [Google Scholar]
- 35. Taylor B. L., Zhulin I. B. (1999) PAS domains: internal sensors of oxygen, redox potential, and light. Microbiol. Mol. Biol. Rev. 63, 479–506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Taylor B. L. (2007) Aer on the inside looking out: paradigm for a PAS-HAMP role in sensing oxygen, redox, and energy. Mol. Microbiol. 65, 1415–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Anantharaman V., Koonin E. V., Aravind L. (2001) Regulatory potential, phyletic distribution, and evolution of ancient, intracellular small molecule-binding domains. J. Mol. Biol. 307, 1271–1292 [DOI] [PubMed] [Google Scholar]
- 38. Aravind L., Ponting C. P. (1997) The GAF domain: an evolutionary link between diverse phototransducing proteins. Trends Biochem. Sci. 22, 458–459 [DOI] [PubMed] [Google Scholar]
- 39. Holmes D. E., Nevin K. P., Lovley D. R. (2004) Comparison of 16 S rRNA, nifD, recA, gyrB, rpoB, and fusA genes within the family Geobacteraceae fam. nov. Int. J. Syst. Evol. Microbiol. 54, 1591. [DOI] [PubMed] [Google Scholar]
- 40. Lonergan D. J., Jenter H. L., Coates J. D., Phillips E. J., Schmidt T. M., Lovley D. R. (1996) Phylogenetic analysis of dissimilatory Fe(III)-reducing bacteria. J. Bacteriol. 178, 2402–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lovley D. R., Giovannoni S. J., White D. C., Champine J. E., Phillips E. J., Gorby Y. A., Goodwin S. (1993) Geobacter metallireducens gen. nov. sp. nov., a microorganism capable of coupling the complete oxidation of organic compounds to the reduction of iron and other metals. Arch. Microbiol. 159, 336–344 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.