Background: Rho GTPases are involved in STAT-dependent transcription. However, the connecting signaling pathways are unknown.
Results: Activated RhoA induces STAT3 phosphorylation via ROCK, JNK, CCL1 synthesis, and secretion.
Conclusion: Auto/paracrine cytokine signaling is a link between RhoA and STAT3.
Significance: The knowledge about the connection between RhoA and STAT signaling is crucial for understanding several diseases, especially cancer.
Keywords: Cancer Biology, G Proteins, Rho GTPases, Signal Transduction, Toxins
Abstract
RhoA is reportedly involved in signal transducers and activators of transcription (STAT)-dependent transcription. However, the pathway connecting the GTPase and STAT signaling has not been characterized. Here, we made use of bacterial toxins, which directly activate Rho GTPases to analyze this pathway. Cytotoxic necrotizing factors (CNFs) are produced by pathogenic Escherichia coli strains and by Yersinia pseudotuberculosis. They activate small GTPases of the Rho family by deamidation of a glutamine, which is crucial for GTP hydrolysis. We show that RhoA activation leads to phosphorylation and activation of STAT3 and identify signal proteins involved in this pathway. RhoA-dependent STAT3 stimulation requires ROCK and Jun kinase activation as well as AP1-induced protein synthesis. The secretion of one or more factors activates the JAK-STAT pathway in an auto/paracrine manner. We identify CCL1/I-309 as an essential cytokine, which is produced and secreted upon RhoA activation and which is able to activate STAT3-dependent signaling pathways.
Introduction
Rho GTPases are essential regulators of a wide variety of cellular functions, such as actin remodeling, endocytosis, secretion, integrin signaling, cell motility, transformation, and apoptosis (for review, see Ref. 1). Like most members of the Ras superfamily of small GTPases, Rho proteins cycle between a GDP-bound inactive and GTP-bound active form. The exchange of GDP for GTP is catalyzed by guanosine nucleotide exchange factors, whereas inactivation of Rho proteins results from hydrolysis of the bound GTP, a process that is stimulated by GTPase-activating proteins. A post-translationally attached isoprenyl anchor mediates membrane binding. Rho proteins can also be transported in the cytosol when complexed with the guanosine nucleotide dissociation inhibitor (for review see Ref. 2). Several studies show a connection between these molecular switches and signal transducers and activators of transcription (STAT)2 proteins (3, 4).
STAT proteins are transcription factors, which play a main role in cell growth and survival (5). They are activated by phosphorylation as a response to extracellular stimuli or by intracellular non-receptor tyrosine kinases. In several tumor types STAT3 is found to be permanently activated (6). Whereas STAT1 seems to be required for growth inhibition, STAT3 and STAT5 stimulate proliferation (5). In human hepatoma cells active Cdc42 induces STAT1 phosphorylation by the effector kinase Ack1 (7), whereas Src kinase has been shown to stimulate STAT3 via a Rac-dependent pathway (8). Recently, it has been shown that overexpression of RhoH induces STAT1 but inhibits STAT5 signaling, suggesting an anti-tumorigenic potential of RhoH (9). It is evident that Rho GTPases are involved in STAT-dependent transcription.
However, the signaling pathway between Rho GTPases and STAT proteins remains elusive. Here, we used bacterial toxins as pharmacological tools to directly activate Rho GTPases and studied the connection between RhoA and STAT. The cytotoxic necrotizing factors CNF1, CNF2, and CNF3 from pathogenic Escherichia coli strains and CNFY from Yersinia pseudotuberculosis belong to a family of protein toxins that activate small GTPases of the Rho family by deamidation of a specific glutamine (Gln63 in RhoA), which is crucial for GTP hydrolysis. Thus inactivation of the Rho proteins is blocked (10, 11). The toxins differ in substrate specificity. CNFY specifically modifies RhoA, RhoB, and RhoC (12). CNF1, CNF2, and CNF3 deamidate RhoA, Rac, and Cdc42. However, CNF3 activates RhoA much stronger as compared with CNF1 and CNF2. CNF2 is not used in this study, because it acts like CNF1 (85% identity with CNF1 (13–15)). We treated HEK293 cells with CNFY and CNF3, respectively, and showed that toxin-induced RhoA activation stimulates STAT signaling. The induction is indirect and requires AP1-dependent protein synthesis as well as secretion of the chemokine CCL1/I-309 to activate the JAK-STAT pathway in an auto/paracrine manner.
MATERIALS AND METHODS
Cell Culture and Reagents
HEK293 cells were grown in DMEM containing 10% fetal bovine serum (FBS), 1% nonessential amino acids, 4 mm penicillin, and 4 mm streptomycin. Starvation of the cells occurred in DMEM containing 1% bovine albumin, 1% nonessential amino acids, 4 mm penicillin, and 4 mm streptomycin. For intoxication of cells, GST-CNF or PMT was directly applied into the medium at a concentration of 100 ng/ml or as indicated. For stimulation of cells with conditioned medium, cells were intoxicated every 24 h with 100 ng/ml of CNF for at least 48 h, these supernatants were centrifuged to pellet detached cells and added onto untreated cells not longer than 20 min. Actinomycin D from Streptomyces sp. and brefeldin A from Penicillium brefeldianum were obtained from Sigma, Y-27632, H1152 dihydrochoride, SP600125, and TCS-JNK5a were from Tocris, JAK Inhibitor I and PP2 were from Calbiochem, YM-254890 was purchased from Astellas (Tokyo, Japan). The recombinant proteins CCL1, CCL3, CCL7, and CCL8 were obtained from Biomol, TGFβ2 was from PeproTech and the blocking peptide BP-CCL1 from Santa Cruz.
Expression and Purification of CNF Proteins
CNF1, CNFY, CNF3, and the respective inactive mutants CNF1-C866S(1-CS), CNFY-C866S(Y-CS), and CNF3-C865S(3-CS) were expressed as N-terminal glutathione S-transferase (GST) fusion proteins and purified as previously described (16). In brief, bacteria expressing the recombinant toxins were lysed by sonication, centrifuged, and GST fusion proteins were purified by affinity chromatography with glutathione-Sepharose beads (GE Healthcare), followed by elution with 10 mm glutathione. Protein content was determined by the Bradford protein assay.
Dual Luciferase Assays and Expression of Recombinant Proteins
Activation of STAT and AP1 proteins were measured in a reporter luciferase assay. HEK293 cells transfected with pGRR5-Luc for general STAT activation (17), pIRF7-Luc (interferon regulatory factor 7) for STAT1 and STAT2 activation (18), pISRE-Luc (interferon stimulated response element) for STAT1 activation (19), pSIE-Luc (c-sis inducible element of c-fos promotor) for STAT3 activation (20), pLHRE-Luc (lactogenic hormone-responsive element) for STAT5 activation (21), and pAP1-Luc obtained from Stratagene for AP1 activation. As an internal control, the pRL-TK vector from Promega was used. Cells were seeded in 96-well plates and transfected with 200 ng/well of reporter plasmids using calcium phosphate transfection. After 5 h the medium was changed, and cells were either treated with inhibitors and/or stimulated with toxin. 24 h after transfection cells were lysed and luciferase assays were performed following the manufacturer's protocol (dual luciferase reporter assay kit, Promega).
For expression of RhoA or STAT3, plasmids encoding wild-type RhoA (pcDNA3-His-RhoA-wt), the constitutive active form of RhoA (pcDNA3-His-RhoA-Q63L), RhoA lacking the isoprenylation motif (pcDNA3-His-RhoA-dCAAX), the constitutive active form of STAT3 (pRc-CMV-STAT3ca) (22), or the respective empty vectors were either transfected in HEK293 cells together with reporter plasmids pRL-TK and pGRR5-Luc or pAP1-Luc by using the calcium phosphate technique. Following an incubation period of 5 h at 37 °C, medium was exchanged and cells were further cultivated. The effects of recombinant protein expression were analyzed by the luciferase reporter assay, whereas the level of expression was determined by immunoblotting.
Cytosolic and Nuclear Fractionation
Cytosolic and nuclear extracts were prepared as described (23). HEK293 cells grown on 10-cm dishes were starved overnight and intoxicated for the indicated time intervals. Cells were washed with cold PBS and harvested with a cell scraper in 300 μl of ice-cold hypotonic lysis buffer (20 mm Hepes (pH 7.9), 10 mm KCl, 1 mm EDTA, 10% glycerol, 0.2% Nonidet P-40, complete EDTA-free protease inhibitor mixture from Roche Applied Science and a phosphatase inhibitor mixture (Sigma)). After incubation on ice to allow swelling of the cells, the cytosolic fractions were separated by centrifugation for 1 min at 10,000 × g. The remaining pellets were resuspended in cold high salt buffer (20 mm Hepes (pH 7.9), 10 mm KCl, 1 mm EDTA, 20% glycerol, 420 mm NaCl, 0.2% Nonidet P-40, protease and phosphatase inhibitors), incubated on ice for 30 min, and centrifuged for 10 min at 20,000 × g. Supernatants containing the nuclear extracts were collected. Cytosolic and nuclear fractions were stored at −80 °C until analysis. Protein content was determined using the BC Assay kit (Uptima), following the manufacturer's protocol. Protein samples from cytosolic or nuclear fractions were adjusted to 50 μg of total protein and applied for Western blotting.
Rhotekin Pulldown Assay
Cells were intoxicated with CNF1, CNFY, or CNF3 (100 ng/ml each) for the indicated time periods. After washing with cold phosphate-buffered saline, cells were detached with a cell scraper in lysis buffer (10% glycerol, 50 mm Tris (pH 7.4), 100 mm NaCl, 1% Nonidet P-40, 2 mm MgCl2, and 1 mm PMSF). Lysates were cleared by centrifugation. Supernatants were subjected to Western blotting for determination of total RhoA content and for pulldown experiments. Therefore, GST-Rhotekin (1–90 amino acids of Rhotekin) immobilized to glutathione-Sepharose beads (GE Healthcare) were incubated with the supernatants for 60 min at 4 °C. Beads were precipitated by centrifugation, washed with lysis buffer, and denatured in SDS sample buffer.
Gene Silencing by RNAi
HEK293 cells were seeded in 12-well plates at 4 × 105 cells/well. RhoA targeting siRNA, control siRNA (MISSION siRNA universal negative control), and shuffled RNAi (CAGAUACCGAUGUUAUACU) was obtained from Sigma. Cells were transfected in serum-containing medium using the N-TER nanoparticle siRNA transfection system from Sigma following the manufacturer's recommendations. After a 24-h incubation cells were intoxicated or left untreated, respectively. In addition, siRNA-treated cells were re-transfected. Following incubation overnight, effects of RhoA gene silencing were assayed in whole cell extracts.
Whole Cell Extracts and Immunoblot Analysis
Cell lysates were prepared by harvesting the cells with a scraper in 500 μl of ice-cold PBS containing complete EDTA-free protease inhibitor mixture from Roche Applied Science and phosphatase inhibitor mixtures 1, 2, or 3 from Sigma. Lysis was performed in a buffer containing 150 mm NaCl, 1% sodium deoxycholate, 1 mm EDTA, 25 mm Tris (pH 7.4), 1% Triton X-100, complete EDTA-free protease inhibitor mixture from Roche Applied Science, and phosphatase inhibitor mixtures (Sigma). After incubation for 10 min on ice, lysates were centrifuged and the supernatant was stored for further analysis at −80 °C. Protein contents of the samples were determined using the BC Assay (Uptima) and adjusted to 50 μg each. Immunoblotting was performed by separation of the samples on 7 or 12.5% SDS-PAGE, transferred to PVDF membrane, and incubated with the respective antibodies, followed by incubation with horseradish peroxidase-coupled anti-rabbit or anti-mouse antibody (Rockland) and detection by enhanced chemiluminescence. STAT activation was determined using the phosphospecific STAT3 (Tyr705) or the general STAT3 antibody, for analysis of JAK2 activation the phosphospecific JAK2 (Tyr1007/Tyr1008) and the general JAK2 antibody was used, and for detection of ATF2 activation the phosphospecific ATF2 (Thr71) and the general ATF2 antibody were used (all from Cell Signaling Technology). The nuclear marker protein p84 was detected by an antibody from Abcam, anti-CCL1 was from Abnova, the anti-α-tubulin was obtained from Sigma, anti-RhoA from Santa Cruz, and the GAPDH and His tag antibodies were from Millipore.
Detection of CCL1 Protein in Cell Culture Supernatants
HEK293 cells were grown in a T175 flask to confluence, medium was replaced with DMEM without serum or BSA, and intoxicated every 24 h for at least 48 h. Supernatants were cleared from cell debris by centrifugation (800 × g, 3 min) and precipitated by adding the same volume of ice-cold 10% TCA (trichloroacetic acid). After centrifugation at 20,000 × g, 40 min, at 4 °C the pellets were washed with 10% TCA, 5% TCA, and resuspended in a buffer containing 6 m urea, 200 mm Tris-HCl (pH 8.5), 10 mm EDTA (pH 7.5), 0.5% SDS. Subsequently, Western blotting was performed using Tricine/SDS-PAGE.
RNA Isolation and PCR Array
HEK293 cells were starved and intoxicated with CNFY overnight or left untreated, respectively. RNA was isolated using the RNeasy kit including DNase digestion from Qiagen. 1 μg of RNA each was applied to reverse transcription using the RT2 First Strand Kit and afterward to the human Common Cytokines RT2 Profiler PCR Array or the human Chemokines & Receptors RT2 Profiler PCR Array (SABiosciences/Qiagen) following the manufacturer's recommendations.
RESULTS
Activation of Rho GTPases by CNFs Induces STAT Activation
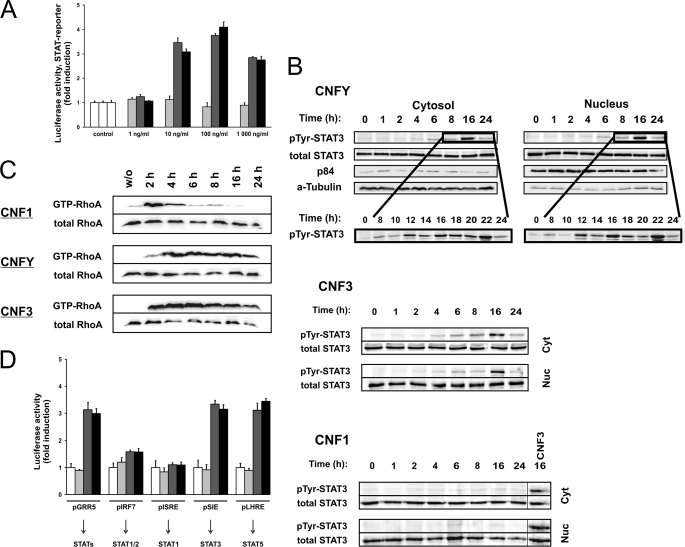
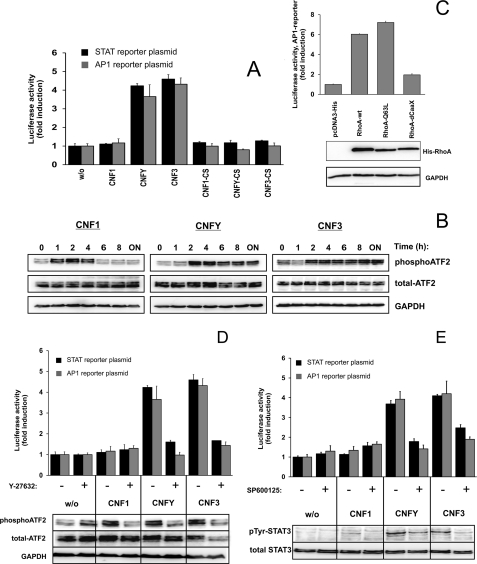
It has been shown that activation of Rho GTPases is involved in the modulation of STAT-dependent transcription (3, 4). We intended to study whether activation of Rho GTPases is sufficient to induce STAT activation. In a first approach, we treated HEK293 cells with CNFs, bacterial toxins that directly activate Rho GTPases by deamidation, and measured the STAT-mediated transcriptional activity in a luciferase assay. Therefore, we transfected cells with the luciferase reporter plasmid pGRR5 that contains a γ-interferon-activated site as promotor, which is recognized by all STAT molecules (17). Transcriptional activity was assessed in a dose-dependent manner with increasing concentrations of CNF1, CNFY, or CNF3, respectively. As shown in Fig. 1A, CNF3 and CNFY induced STAT-dependent transcription about 3–4-fold as compared with untreated cells.
FIGURE 1.
CNFY and CNF3 induce STAT transcriptional activity and STAT3 phosphorylation at Tyr705. A, induction of STAT activity. HEK293 cells were transfected with pGRR5-Luc and pRL-TK followed by intoxication overnight with increasing concentrations of CNF1 (light gray bars), CNFY (dark gray bars), CNF3 (black bars), or left untreated (open bars). Relative light unit values of firefly luciferase in the lysates were normalized to those of the internal control pRL-TK. Results are given as fold-induction compared with the untreated control. Shown is one representative experiment out of at least three performed in triplicates ± S.D. B, time course of STAT3 phosphorylation at Tyr705. HEK293 cells were serum-starved, treated with CNF1, CNFY, or CNF3, or left untreated for the indicated time periods. Tyr705 phosphorylation (pTyr) of STAT3 and total STAT3 was determined in cytosolic and nuclear fractions. Additionally, immunoblotting with antibodies specific for the nuclear protein p84 and α-tubulin (α-Tubulin) as a cytosolic marker was performed. Images are representatives of at least 3 independent experiments. C, long-term RhoA activation by CNFY and CNF3. HEK293 cells were intoxicated with CNF1, CNFY, or CNF3 for the indicated time intervals and lysed. The amount of total RhoA in the lysates was analyzed by immunoblotting with a specific antibody. To analyze GTP-loaded RhoA, binding to the effector domain of Rhotekin coupled to Sepharose beads was analyzed. Shown are examples of at least three independent experiments. D, selective STAT activity. Cells were transfected with pRL-TK to allow normalization and in addition with pGRR5-Luc, pIRF7-Luc, pISRE-Luc, pSIE-Luc, or pLHRE-Luc, respectively. Cells were treated overnight with CNF1 (light gray bars), CNFY (dark gray bars), CNF3 (black bars), or left untreated (open bars). Results were obtained from one representative experiment out of three performed in triplicates ± S.D.
Accordingly, using a phosphospecific antibody against tyrosine 705 for Western blotting, we detected increased phosphorylation of STAT3 in the cytosolic and nuclear fractions of CNF3- or CNFY-treated cells, which occurred 6–8 h after toxin addition (Fig. 1B). To ensure equal loading of protein, Western blotting with a STAT3 antibody was performed. Antibodies against the cytosolic marker protein α-tubulin and the nuclear protein p84 were used as controls for efficient fractionation. CNFY- or CNF3-catalyzed RhoA activation was evident 2 h following toxin addition and was stable for more than 24 h (Fig. 1C). Surprisingly, CNF1 neither induced STAT phosphorylation nor STAT-mediated luciferase transcription, although the toxin entered the cells and activated RhoA as monitored by pulldown experiments. CNF1 treatment does not result in long lasting activation of RhoA, whereas CNFY-induced or CNF3-catalyzed activation was stable for more than 24 h (Fig. 1C). Presumably due to the transient activation of RhoA, CNF1 shows no STAT transcriptional activity, even at high concentrations of 1 μg/ml (about 7 nm). The data show that picomolar concentrations of CNF3 and CNFY are sufficient to stimulate STAT3 activation. In contrast, there was no response to a 100-fold increased concentration of CNF1. As expected, the catalytically inactive mutants of all toxins had no effect (supplemental Fig. S1).
STAT3 and STAT5 but not STAT1 are activated
To distinguish which of the STAT proteins was activated in response to CNFs, we used previously described reporter plasmids with different STAT binding sites for luciferase assays. Cells were transfected with pGRR5-Luc (all STATs), pIRF7-Luc (STAT1/2), pISRE-Luc (STAT1), pSIE-Luc (STAT3), or pLHRE-Luc (STAT5), respectively. Intoxication with CNFY or CNF3 exclusively induced activity of STAT3 and STAT5 (Fig. 1D).
In line with the results shown above, CNF1 did not induce STAT-dependent transcription of any of the reporter plasmids used. For the following experiments we used solely the reporter plasmid containing the γ-interferon-activated site, which is recognized by all STAT molecules (pGRR5-Luc).
RhoA Is Necessary and Sufficient to Induce STAT Activation
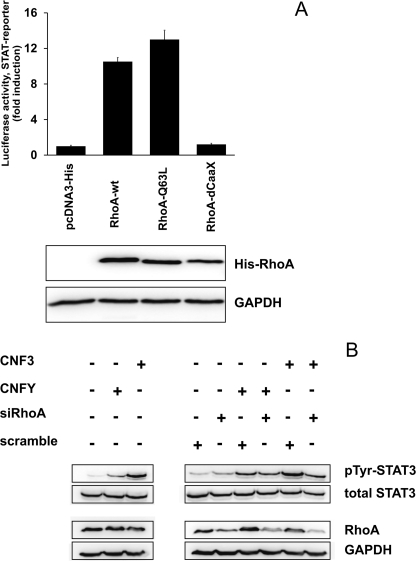
CNFY selectively deamidates RhoA, but not Rac and Cdc42 (12). CNF3 is also a strong RhoA activator (14). Therefore, our data indicate that activation of RhoA is sufficient to induce STAT-dependent luciferase expression. To verify that RhoA is involved in STAT activation, we co-transfected either a plasmid encoding wild-type RhoA, dominant-active RhoA(Q63L), or an inactive mutant of RhoA without an isoprenylation site (RhoA-dCAAX), each together with the STAT reporter plasmid into HEK293 cells and performed luciferase assays. As expected, expression of wild-type or dominant-active RhoA caused increased STAT transcriptional activity. In contrast, expression of the truncated version of RhoA (RhoA-dCAAX), which is not post-translationally modified and thus remains inactive (24), did not induce STAT transcriptional activity (Fig. 2A).
FIGURE 2.
CNF-mediated activation of STAT3 is dependent on RhoA. A, coexpression of RhoA constructs with luciferase reporter plasmids. HEK293 cells were cotransfected with pRL-TK and pGRR5-Luc with pcDNA3.1-plasmids encoding His- tagged RhoA-wild type (RhoA-wt), the constitutive active mutant RhoA-Q63L, RhoA-dCAAX lacking the isoprenylation motif or an empty control vector. 24 h after transfection, luciferase activity in the lysates was determined. Results were shown as fold-induction compared with mock transfected cells. Levels of protein expression were analyzed by immunoblotting with a RhoA specific antibody. Shown is one representative of five experiments. B, RhoA knockdown with siRNA. HEK293 cells were left untransfected (first part) or transfected (second part) with negative control siRNA (scramble) or siRNA targeting RhoA, using the N-TER nanoparticle siRNA transfection system. After 24 h, cells were intoxicated with the toxins as indicated. Lysates were immunoblotted with phosphospecific STAT antibody. Membranes were stripped and re-probed for total STAT3. Efficiency of gene silencing was controlled by detection of RhoA in the cell lysates using a RhoA-specific antibody. Detection of GAPDH was used as loading control. pTyr, phosphotyrosine. Shown is a representative example of three independent experiments.
In a reciprocal experiment, we asked whether toxin-induced STAT phosphorylation requires signaling through RhoA. Therefore, we diminished the amount of endogenous RhoA using RNAi and treated the cells with either CNFY or CNF3. The amount of RhoA in cells transfected with siRNA targeting RhoA (siRhoA) was clearly reduced compared with control siRNA (scramble)-transfected cells (Fig. 2B, right) or to cells transfected with a shuffled RNAi containing two mismatches (supplemental Fig. S2). In contrast, there were no differences of RhoA content following intoxication of cells with CNFs (Fig. 2B, left), STAT3 phosphorylation was detected by Western blotting. As shown in Fig. 2B, right, toxin-induced phosphorylation of STAT3 was less in cells with diminished RhoA content. An antibody against GAPDH was used to control identical protein load. Intoxication with CNFY and CNF3 of untransfected cells (Fig. 2B, left) controlled the induction of STAT3 phosphorylation. The data indicate that RhoA mediates the toxin-induced STAT3 activation.
ROCK Is Involved in STAT Activation
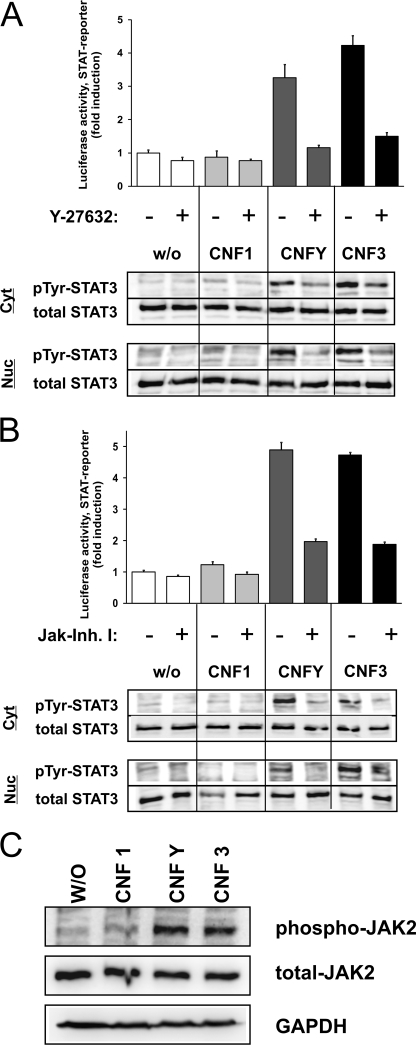
To determine which effector proteins were involved in RhoA-mediated STAT activation, we asked whether Rho kinase (ROCK) is necessary for CNF-induced STAT activation. We treated the cells with the ROCK inhibitor Y-27632, added the different CNFs, and determined STAT activation.
As shown in Fig. 3A, in the presence of the ROCK inhibitor the induction of STAT transcriptional activity (top) as well as phosphorylation of STAT3 (bottom) was nearly completely diminished. These data were verified using a second ROCK inhibitor, H1152 (supplemental Fig. S3).
FIGURE 3.
Rho kinase (ROCK) and Janus kinases (JAKs) are involved. A, ROCK was inhibited with Y-27632. Luciferase assays (top) were performed with lysates from HEK293 cells transfected with pRL-TK and pGRR5-Luc, left untreated or treated overnight with Y-27632 alone (open bars) or together with CNF1 (light gray bars), CNFY (dark gray bars), or CNF3 (black bars), respectively. Relative light unit (RLU) values of firefly luciferase were normalized to those of the internal control pRL-TK and the untreated control was set to one. Results were obtained from one representative experiment performed in triplicates ± S.D. Phosphorylation at Tyr705 (pTyr) of STAT3 (bottom) was determined. Total STAT3 was analyzed as loading control. Images are representatives of at least three independent experiments. B, dependence of Janus kinases. Luciferase assays (top) were performed as described in A with JAK inhibitor I alone (open bars) or together with CNF1 (light gray bars), CNFY (dark gray bars), or CNF3 (black bars), respectively. RLU values of firefly luciferase were normalized to these of the internal control pRL-TK and the untreated control was valued as one. Results were obtained from one representative experiment performed in triplicates ± S.D. Additionally, phosphorylation at Tyr705 of STAT3 (bottom) was determined. STAT3 served as a loading control. C, phosphorylation of JAK2. Cells were untreated or intoxicated overnight with CNF1, CNFY, or CNF3, respectively. Lysates were immunoblotted using a phosphospecific antibody against Tyr1007/Tyr1008 phospho-JAK2. Membranes were stripped and re-probed with a JAK2 antibody or an antibody against GAPDH to show equal protein loading. One representative example out of three is shown.
JAK but Not Src Mediates STAT Activation
Furthermore, we asked whether STAT phosphorylation is mediated through the activity of the Janus kinase family (JAKs) or the family of the Src kinases. Both kinases have been described to phosphorylate STAT proteins (25). Therefore, CNF-intoxicated cells were probed for luciferase activity in the presence and absence of a JAK inhibitor (JAK inhibitor I, Fig. 3B, top). In addition, STAT3 phosphorylation was detected in cytosolic and nuclear fractions (Fig. 3B, bottom). In both assays, a reduction of CNFY- and CNF3-mediated STAT activation by inhibition of Janus kinases was observed by Western blotting with antibodies specific for JAK2 and phosphorylated JAK2. Jak2 phosphorylation per se was not inhibited by JAK inhibitor I. As shown in Fig. 3C, phosphorylation of JAK2 could be detected after CNFY and CNF3 treatment of the cells. In contrast, inhibition of Src and Src-like kinases with PP2 showed no effect (supplemental Fig. S4). These data strongly suggest that JAKs mediate the toxin-induced phosphorylation of STAT3.
Toxin-induced STAT3 Activation Is Dependent on Novel Protein Synthesis and Secretion
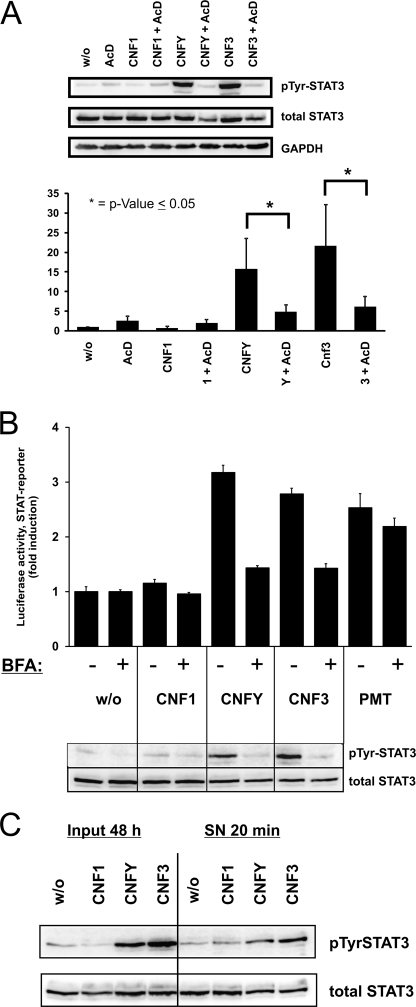
The STAT3 phosphorylation induced by CNFY or CNF3 started after 6–8 h and was maximal after overnight intoxication (Fig. 1B). The delay of about 1 to 2 h between addition of CNF and Rho activation is caused by the receptor-mediated uptake of CNF (26). Considering this delay, activation of STAT3 still occurs after a long time period following activation of RhoA. Therefore, it is likely that protein synthesis is involved in the RhoA-STAT pathway. To test this hypothesis, transcription was inhibited by actinomycin D. In the presence of actinomycin D, STAT3 activation was completely abolished (Fig. 4A). The data were quantified considering the decreased protein amount of STAT3 caused by actinomycin D (Fig. 4A, bottom).
FIGURE 4.
CNF-induced STAT activation is dependent on protein synthesis and secretion. A, inhibition of protein synthesis. HEK293 cells were grown in full medium and treated overnight with actinomycin D (AcD) alone or together with CNF1, CNFY, or CNF3, or left untreated. Phosphorylation at Tyr705 (pTyr) of STAT3 was detected in whole cell lysates by immunoblotting with a phosphospecific antibody and re-probed with total STAT3 antibody. GAPDH was used as loading control. These representative images were repeated at least five times. Quantification (bottom of A) of four immunoblots was performed using MultiGauge software (Fujifilm). The intensity of signals of the phosphorylated protein bands were calculated against the total STAT3 protein bands. Intensities are presented as fold of control (mean ± S.D.). Statistical significance was assessed by using SigmaStat, paired Student's t test with *, p < 0.05. B, inhibition of secretion. Secretion of proteins was blocked with brefeldin A (BFA). Luciferase assays (top) were performed with cell lysates from HEK293 cells transfected with pRL-TK and pGRR5-Luc, then treated overnight with brefeldin A alone or with CNF1, CNFY, CNF3, or left untreated. Relative light unit (RLU) values were normalized to the internal control pRL-TK, signals of brefeldin A-treated samples were calculated against the inhibitor controls set to one. Results were obtained from one representative experiment performed in triplicates ± S.D. Phosphorylation at Tyr705 of STAT3 (bottom) was determined in whole cell extracts. Total STAT3 served as a loading control. Shown is a representative experiment of at least five independent experiments. C, auto/paracrine stimulation. HEK293 cells were intoxicated twice (every 24 h) with CNF1, CNFY, CNF3, or left untreated. After 48 h, supernatants were collected and transferred to fresh cells for 20 min. Lysates of intoxicated cells (48 h input) and stimulated cells (20 min SN) were immunoblotted with a phosphospecific STAT3 antibody (Tyr705). Membranes were stripped and re-probed with a STAT3 antibody. Experiments were repeated independently at least three times.
Next, we analyzed whether secretion is necessary for the CNF-mediated STAT activation. Destruction of the secretory pathway by brefeldin A prevented STAT transcriptional activity as well as phosphorylation of STAT3 induced by CNFY or CNF3 (Fig. 4B). The Pasteurella multocida toxin PMT activates the STAT pathway via direct activation of Gαq (23). We used this toxin as a control. Activation of STAT was only slightly decreased in the presence of brefeldin A.
Our data suggest that RhoA activation by CNFs leads to synthesis and secretion of one or more proteins that activate the STAT pathway in an auto/paracrine manner. To underline this hypothesis, we studied whether STAT phosphorylation is enhanced by adding the conditioned supernatant of toxin-treated cells to unstimulated cells. Therefore, HEK293 cells were stimulated with CNFs for 48 h. Supernatants of the cells were collected and transferred to fresh cells for 20 min. This short time period excludes direct effects of CNF. The subsequent analysis revealed that STAT3 was phosphorylated by adding the supernatant of CNFY or CNF3-treated cells (Fig. 4C).
Transcription Factor AP1 Mediates STAT Activation
The transcription factor AP1 (activator protein 1) composed of a heterodimer of proteins of the c-Jun, c-Fos, ATF, and JDP family is known to be activated by the RhoA-ROCK-JNK signaling axis (27, 28). AP1 induction by CNF-mediated RhoA activation was determined by the luciferase reporter assay with a plasmid containing an AP1 binding site.
As reported for the activation of STAT3, a similar activation pattern of AP1-dependent transcription was found. In contrast to CNFY and CNF3, CNF1 did not induce AP1 activation. As expected, also the catalytically inactive mutants of CNF1 (CNF1-CS), CNFY (CNFY-CS), and CNF3 (CNF3-CS) had no effect (Fig. 5A). It has been shown that ATF2 is phosphorylated after transfection of constitutive active RhoA in NIH3T3 fibroblasts (28). Therefore, we analyzed ATF2 phosphorylation caused by toxin-induced RhoA activation in a time course. As shown in Fig. 5B, CNF1 induces a transient phosphorylation of ATF2. In contrast, CNFY and CNF3 cause a long-lasting ATF2 phosphorylation in HEK293 cells. This is comparable with the effects on RhoA activation by these toxins (compare Fig. 1C). These data support the hypothesis that transient activation of RhoA by CNF1 is the cause for the missing STAT activation.
FIGURE 5.
AP1 proteins are involved. A, CNF intoxication acts on AP1 activation similar to STAT activation. AP1 and STAT transcriptional activities were quantified by luciferase assays. HEK293 cells were transfected with pRL-TK, pGRR5-Luc (black bars), or pAP1-Luc (gray bars), respectively. Cells were left untreated or treated overnight with CNF1, CNFY, CNF3, or the biologically inactive mutants CNF1-C866S, CNFY-C866S, or CNF3-C865S, as indicated. Relative light unit values of firefly luciferase were normalized to those of Renilla luciferase. Results are shown as fold-induction obtained from one representative experiment out of at least three performed in triplicates ± S.D. B, time course phosphorylation of the AP1 protein ATF2. HEK293 cells were starved in serum-free medium, and treated with the toxins as indicated. Lysates were immunoblotted with a phosphospecific ATF2 antibody (Thr71) and re-probed with a GAPDH antibody. In addition, cell lysates were immunoblotted with an ATF2 antibody. Shown is a representative experiment of at least three independent experiments. C, AP1 activity induced by expression of RhoA. HEK293 cells were cotransfected with pRL-TK and pAP1-Luc with an expression plasmid for RhoA-wild type (RhoA-wt), the constitutive active mutant RhoA-Q63L, RhoA-dCAAX lacking the isoprenylation motif, or empty vector. 24 h after transfection, cells were lysed and luciferase activity was determined. Values for mock transfected cells were set to one. Levels of protein expression were analyzed by immunoblotting with a RhoA-specific antibody. The shown experiment was repeated independently three times. D, AP1 transcriptional activity and phosphorylation of ATF2 is prohibited by the ROCK inhibitor Y-27632. Luciferase activity (top) was measured in cell lysates from HEK293 cells transfected with pRL-TK and either with pGRR5-Luc (black bars) or pAP1-Luc (gray bars), treated overnight with Y-27632 alone, or together with CNF1, CNFY, CNF3, or left untreated. After normalization to the internal control pRL-TK, signals of inhibitor-treated samples were calculated against the inhibitor controls and shown as fold-induction. Results were obtained from one representative experiment performed in triplicates ± S.D. For detection of ATF-2 phosphorylation, cells were serum-starved, pretreated with 20 μm Y-27632 for 30 min before intoxication either with CNF1 for 2 h or CNFY or CNF3 for 4 h. Cell extracts were immunoblotted with a Thr71 phosphospecific ATF2 antibody. GAPDH served as loading control. In addition, cell lysates were immunoblotted with an ATF2 antibody. E, involvement of Jun kinase (JNK) luciferase assays (top) were performed with cell lysates from HEK293 cells transfected with pRL-TK and either with pGRR5-Luc (black bars) or pAP1-Luc (gray bars), and then treated overnight with SP600125 alone or together with the toxins as indicated. Relative light unit values of lysates were normalized to those of the internal control pRL-TK and given as fold-induction to untreated controls. Results were obtained from one representative experiment out of three performed in triplicates ± S.D. Phosphorylation at Tyr705 of STAT3 (bottom) was determined in whole cell extracts. Blots were re-probed for total STAT3.
AP1 Activation Is Downstream of RhoA and ROCK and Upstream of STAT Activation
To show that the RhoA-ROCK pathway is necessary for AP1 induction, we performed the same studies as described above for STAT activation.
Expression of wild-type or active RhoA stimulated AP1 transcriptional activity (Fig. 5C), whereas inhibition of the Rho kinase by Y-27632 (Fig. 5D, top) blocked CNF-induced AP1- and STAT-dependent transcription. In addition, the CNF-induced phosphorylation of ATF2 was prevented by inhibition of ROCK (Fig. 5D, bottom), suggesting ROCK activation upstream of ATF2.
However, in contrast to STAT activation, inhibition of the secretory pathway could not block toxin-induced AP1 transcriptional activity (supplemental Fig. S5). Moreover, expression of a constitutive active form of STAT3 is not able to induce the transcriptional activity measured by the AP1 reporter plasmid (supplemental Fig. S6).
To test whether AP1 activation is necessary for CNF-induced STAT activation, we inhibited the AP1 kinase JNK with SP600125. In the presence of the inhibitor, AP1 activation as well as STAT activation by CNFY or CNF3 was reduced (Fig. 5E). Moreover, AP1 activation occurs much earlier as compared with STAT activation. All these data suggest AP1 activation upstream of auto/paracrine STAT activation. However, the question of a direct connection of AP1 and CCL1 production (see below) remains to be analyzed. By blocking protein synthesis or secretion, we show that STAT stimulation is indirect and requires the (probably AP1-triggered) synthesis of one or more proteins, which are secreted and lead to phosphorylation and activation of STAT3 and STAT5 in an auto/paracrine manner.
Identification of CCL1 As a Relevant Factor
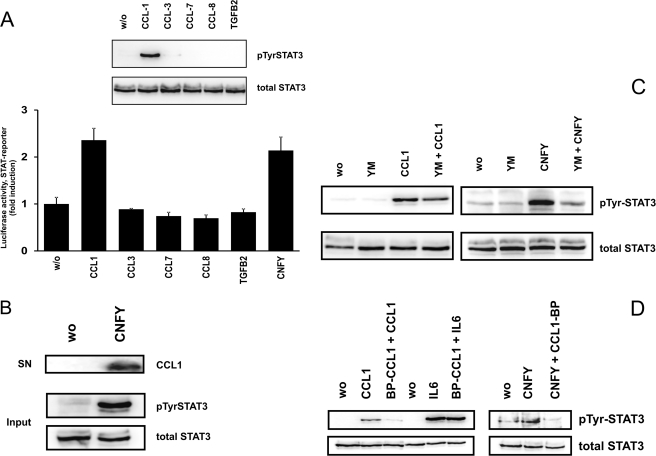
To screen for possible cytokines and chemokines, which are up-regulated due to RhoA activation, we performed two different commercial PCR arrays (“Common Cytokines” and “Chemokines and Receptors,” from Qiagen) with cDNA from CNFY-treated and control cells. In these screens the mRNAs of 14 of 160 different genes were found to be enhanced in CNFY-treated cells as compared with controls (supplemental Table S1). Up-regulation of several of these mRNAs could be subsequently verified by real time PCR. IL-8, IL-24, CCL1, CCL-3, CCL-7, CCL-8, and TGFβ2 were selected for further evaluation of their role for RhoA-induced STAT signaling. As shown in Fig. 6, stimulation of HEK293 cells with the recombinant cyto/chemokines (100 ng/ml of CCL or 10 ng/ml of TGFβ2) for 15 min, led to the identification of only one chemokine (CCL1), which stimulates STAT phosphorylation (Fig. 6A, top). In line with that, recombinant CCL1 as well as CNFY stimulated the production of luciferase under control of a STAT3 responsive promotor in HEK293 cells (treatment overnight, Fig. 6A, bottom). Testing different concentrations of recombinant CCL1 revealed that 20 ng/ml are required and sufficient for detectable STAT3 phosphorylation in HEK cells (supplemental Fig. S7). We tested whether we could detect CCL1 in the precipitated supernatant of toxin-treated cells by Western blotting. The TCA-precipitated proteins from 5 ml of culture medium were probed for the presence of CCL1. Only in the supernatant of CNFY-treated cells CCL1 was detectable (Fig. 6B). CCL1 signals through its receptor CCR8, a G-protein-coupled receptor. Blocking Gαq with the specific inhibitor YM-254890 decreased CCL1-induced STAT3 phosphorylation, indicating that CCR8 signaling is mediated by Gαq (Fig. 6C, left) (29, 30). In line with CCL1 as one of the crucial STAT3 activators of CNFY-treated cells, YM-254890 also decreased STAT3 phosphorylation in toxin-treated cells to nearly background levels (Fig. 6C, right). To further support the role of CCL1 for RhoA-mediated STAT activation, we used a C-terminal peptide of CCL1 (blocking peptide CCL1 (BP-CCL1)). As shown in Fig. 6D, preincubation with BP-CCL1 considerably reduced CCL1- and CNFY-mediated phosphorylation of STAT3. As control we screened for IL6-induced STAT activation, which was not influenced by the peptide. This indicates that toxin-induced STAT activation requires CCR8 and that CCL1 plays an important role in this pathway.
FIGURE 6.
CCL1 mediates RhoA-dependent STAT activation. A, CCL-1-induced phosphorylation and activation of STAT3. HEK293 cells were treated with cytokines or chemokines (100 ng/ml of CCLs and 10 ng/ml of TGFβ2, respectively) as indicated for 15 min. Lysates were immunoblotted using a specific antibody against Tyr705 phospho-STAT3 (pTyr). Membranes were stripped and re-probed with a STAT3 antibody to show an equal amount of protein. One representative example out of three is shown (top). Luciferase activity was measured in cell lysates from cyto/chemokine-treated HEK293 cells previously transfected with pRL-TK and pGRR5-Luc. Relative light unit values of firefly luciferase were normalized to those of the internal control (pRL-TK). The luciferase level of the untreated control was set to one. Results were obtained from several representative experiments performed in triplicates ± S.D. (bottom). B, CCL1 is detectable in the supernatant of CNFY-treated cells. HEK293 cells growing in serum-free medium were treated with or without CNFY (200 ng/ml every day for 2 days) and the supernatant of the cells (SN) was collected. Proteins were precipitated from 5 ml of culture medium with TCA (trichloroacetic acid). Western blotting was performed using a specific antibody against CCL1 (top). Lysates of the respective cells (Input) were blotted and analyzed for total and phosphorylated STAT3 (bottom). Shown is a typical result of two independent experiments. C, an inhibitor of Gαq (YM-2548890) blocks CCL1- and CNFY-induced STAT phosphorylation. HEK293 cells were pretreated with YM-2548890 (1 μm) for 1 h as indicated and then treated with CCL1 (40 ng/ml) for 15 min (left) or incubated with YM-2548890 (1 μm) and CNFY overnight as indicated in the figure (right). Phosphorylation at Tyr705 of STAT3 was determined in whole cell extracts. Blots were re-probed for total STAT3. D, blocking peptide BP-CCL1 inhibits CCL1- and CNFY-induced STAT phosphorylation. HEK293 cells were starved overnight in serum-free medium, pre-treated with BP-CCL1 (200 ng/μl) for 1.5 h, and then treated with CCL1 (40 ng/ml) or IL6 (20 ng/ml) for 10 min, respectively (left). CNFY (200 ng/ml) was incubated for 3 h before addition of the blocking peptide and further incubation for 8 h (right). Phosphorylation at Tyr705 of STAT3 was determined in whole cell extracts. Blots were re-probed for total STAT3. Shown is a typical result of three independent experiments.
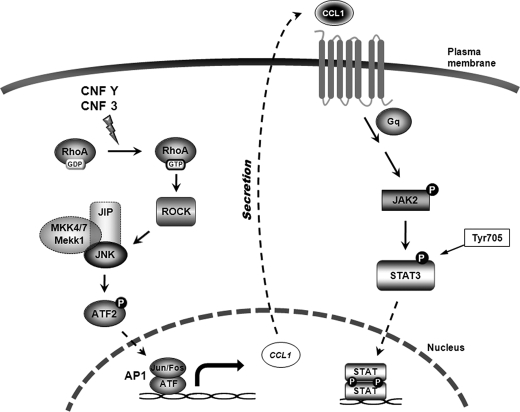
However, it remains to be analyzed whether exclusively CCL1 or several factors are responsible for RhoA-induced auto/paracrine STAT stimulation. Our data are summarized in the model of RhoA-induced STAT stimulation in Fig. 7.
FIGURE 7.
Model of the RhoA signaling cascade leading to STAT activation induced by CNFY and CNF3 intoxication. CNFY and CNF3 intoxication of mammalian host cells lead to constitutive activation of RhoA by deamidation of Gln63. RhoA activates the direct effector ROCK, known to bind to the MKK1/4/7-JIP-JNK complex to activate JNK. JNK activation leads to phosphorylation of ATF2, a member of the AP1 family, which then triggers AP1-dependent transcription. After synthesis and secretion of proteins, CCL1 (besides others) stimulates receptor-associated JAK activation. JAK proteins phosphorylate STAT3 at Tyr705. Once they are activated, STAT3 dimers are imported into the nucleus.
DISCUSSION
STAT proteins are known to mediate cellular responses elicted by cytokines (31). The discovery that STATs are also activated via growth factor receptors and nonreceptor tyrosine kinases like Src and Abl put transcription factors in the light of cancer progression (5, 25, 32). In several cancer cells STAT3 and STAT5 are found activated, and especially STAT3 has been shown to regulate key processes of cancer progression like angiogenesis and immune suppression (5). Therefore, the development of STAT inhibitors is one promising aspect of cancer therapy (25, 32). In this view, it is of interest to analyze the signaling pathways, which lead to STAT activation. Rho GTPases are found activated in several tumors, which is mainly based on overexpression of the GTPases or their activators, the guanine nucleotide exchange factors (33). Several reports show that Rho GTPases mediate STAT activation or inhibition (3, 4, 7, 34, 35). These studies show that Rho proteins are involved in tumor progression due to stimulation of STAT-dependent transcription. Activation of STAT due to RhoA activation and CCL1 production could represent a link between these tumorigenic pathways within cancer cells or between tumor and surrounding cells. Because the microenvironment of tumor cells consists of several cells of different origin, including immune cells, this connection is not necessarily combined within the tumor cell but may also represent intercellular communication between immune cells and cancer cells. Therefore, the pathways that link diverse Rho GTPases with STATs have to be analyzed. Data from a number of laboratories show that activated Rac or Cdc42 leads to NFκB-dependent STAT3 activation (for review, see Ref. 33).
Here, we show that direct activation of RhoA by bacterial toxins CNFY or CNF3 or expression of RhoA is sufficient to induce STAT3 phosphorylation and activation. STAT5 is also activated. However, we could not detect phosphorylation of STAT5 using different antibodies. STAT1 and STAT2 are not activated by RhoA. RhoA stimulates the direct effector ROCK, which we suggest is involved in STAT phosphorylation, because ROCK inhibitors prevent STAT activation. ROCK is known to bind to a protein complex including MKK1/4/7, the Jun interacting protein (JIP) and the Jun kinase to activate JNK (27). JNK is also an accepted effector of Rac and Cdc42. These GTPases are strongly activated by CNF1 but not by CNFY and hardly by CNF3 (14). Therefore, activation of Rac or Cdc42 is unlikely to be involved in JNK-dependent STAT activation in HEK293 cells. In line with this, overexpression of wild-type Rac or dominant-active Rac did not induce STAT-dependent transcription, although activated Rac leads to JNK activation (36).
An interesting question is the difference between Rac-mediated and RhoA-dependent JNK activation. The answer may lie in the formation of different signaling units with only JIP recruiting ROCK, JNK, and ATF2. JNK activation in this signaling unit may lead to phosphorylation of ATF2, a member of the AP1 family, which then triggers AP1-dependent transcription. Rac activation may lead to the formation of other signaling complexes. Therefore, scaffold proteins could be important mediators of cell or cell state-dependent decisions of signal transduction. Our studies suggest AP1-dependent activation of CCL1 transcription. In line with this, we identified 9 binding sites for AP1 transcription factors in the human sequence of the ccl1 gene (chromosome 17, 32.695.252–32.682399) using bioinformatics analysis (Genomatix MatInspector, based on weight matrix library 8.4, 2011). However, further studies are needed to verify a direct connection between AP1 and CCL1 synthesis. Despite this, we identified chemokine CCL1 to be expressed and secreted following RhoA activation and that this chemokine in turn is sufficient to activate STAT3-dependent transcription. To our knowledge, this is the first report showing that CCL1 leads to STAT3 activation.
CCL1 has originally been identified as a chemokine expressed in activated T cells. It binds to CCR8 on type 2 T helper (Th2) cells and macrophages (37). CCR8 is also expressed in smooth muscle cells and causes migration and secretion of metalloproteases (38). In line with STAT3 activation, it has been shown that CCL1 is a potent pro-survival factor for cells, for example, for thymocytes (39). The blocking peptide BP-CCL1 considerably inhibited toxin-induced STAT phosphorylation, showing that CCR8 is required for this effect. However, we cannot exclude that other factors are also required for sufficient STAT3 activation.
Detection of these factors could be difficult because they may be expressed in low amounts. We need a long stimulation of the cells (48 h) to allow phosphorylation of STAT3 in fresh cells with the conditioned medium of toxin-treated cells. Phosphorylation of STAT in fresh cells was already detected after 10 to 20 min, indicating that it is the direct action of the secreted factors that induce STAT activation. Moreover, the main portion of the factors produced may directly bind to receptors on the same cell (autocrine stimulation) and only a few molecules may be released into the culture medium. Most interestingly, CCL1 is encoded by an open reading frame of the human herpesvirus 8 (HHV-8), which is associated with the endothelial tumor Karposi sarcoma. It has been reported that v-CCL1 enhances endothelial survival (40). One may speculate that the virus-produced CCL1 could play a role in promoting tumor formation by activating migration of tumor cells and by inducing STAT signaling. Besides CCL1, IL-24 was up-regulated upon RhoA activation in HEK293 cells. However, addition of recombinant IL-24 did not induce STAT3 phosphorylation. Interestingly, a mainly autocrine action has been shown for IL-24 (belongs to IL-10 family of cytokines), which kills melanoma cells following delivery by viral transduction, whereas recombinant IL-24 had no effect (41). IL-10 cytokines are also produced by epithelial cells and are important for the integrity of tissue epithelial layers (for review, see Ref. 42). Thus the production of cyto/chemokines could be a defense mechanism of epithelial cells against pathogens by enhancing the barrier function of the tissue. However, the proteins, which are produced upon RhoA activation and stimulate STAT phosphorylation are not necessarily cytokines, because growth factors also can stimulate STAT phosphorylation and activation (25).
Altogether, our study connects two protein families, which are known to be activated in several tumors: Rho GTPases and STATs. It will be interesting to analyze whether the activation of only one of the protein families is necessary and sufficient for efficient tumor progression and if CCL1 plays a role for the migration and/or metastasis of certain tumor cells.
Supplementary Material
Acknowledgments
We thank Jürgen Dumbach for excellent technical assistance and Dr. Masatoshi Taniguchi from Astellas (Japan) for YM-254890.
This work was supported by Deutsche Forschungsgemeinschaft Grants Schm 1385/5-1 and SFB850.

This article contains supplemental Figs. S1–S7 and Table S1.
- STAT
- signal transducer and activator of transcription
- CNF
- cytotoxic necrotizing factor
- AP1
- activator protein 1
- JIP
- Jun interacting protein
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1. Etienne-Manneville S., Hall A. (2002) Rho GTPases in cell biology. Nature 420, 629–635 [DOI] [PubMed] [Google Scholar]
- 2. Mackay D. J., Hall A. (1998) Rho GTPases. J. Biol. Chem. 273, 20685–20688 [DOI] [PubMed] [Google Scholar]
- 3. Kreiselmeier N. E., Kraynack N. C., Corey D. A., Kelley T. J. (2003) Statin-mediated correction of STAT1 signaling and inducible nitric-oxide synthase expression in cystic fibrosis epithelial cells. Am. J. Physiol. Lung Cell Mol. Physiol 285, L1286–L1295 [DOI] [PubMed] [Google Scholar]
- 4. Loucks F. A., Le S. S., Zimmermann A. K., Ryan K. R., Barth H., Aktories K., Linseman D. A. (2006) Rho family GTPase inhibition reveals opposing effects of mitogen-activated protein kinase kinase/extracellular signal-regulated kinase and Janus kinase/signal transducer and activator of transcription signaling cascades on neuronal survival. J. Neurochem. 97, 957–967 [DOI] [PubMed] [Google Scholar]
- 5. Bowman T., Garcia R., Turkson J., Jove R. (2000) STATs in oncogenesis. Oncogene 19, 2474–2488 [DOI] [PubMed] [Google Scholar]
- 6. Pfeiffer M., Hartmann T. N., Leick M., Catusse J., Schmitt-Graeff A., Burger M. (2009) Alternative implication of CXCR4 in JAK2/STAT3 activation in small cell lung cancer. Br. J. Cancer 100, 1949–1956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fujimoto Y., Ochi H., Maekawa T., Abe H., Hayes C. N., Kumada H., Nakamura Y., Chayama K. (2011) A single nucleotide polymorphism in activated Cdc42 associated tyrosine kinase 1 influences the interferon therapy in hepatitis C patients. J. Hepatol. 54, 629–639 [DOI] [PubMed] [Google Scholar]
- 8. Turkson J., Bowman T., Adnane J., Zhang Y., Djeu J. Y., Sekharam M., Frank D. A., Holzman L. B., Wu J., Sebti S., Jove R. (1999) Requirement for Ras/Rac1-mediated p38 and c-Jun N-terminal kinase signaling in STAT3 transcriptional activity induced by the Src oncoprotein. Mol. Cell. Biol. 19, 7519–7528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gündogdu M. S., Liu H., Metzdorf D., Hildebrand D., Aigner M., Aktories K., Heeg K., Kubatzky K. F. (2010) The haematopoietic GTPase RhoH modulates IL3 signaling through regulation of STAT activity and IL3 receptor expression. Mol. Cancer 9, 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schmidt G., Sehr P., Wilm M., Selzer J., Mann M., Aktories K. (1997) Gln-63 of Rho is deamidated by Escherichia coli cytotoxic necrotizing factor-1. Nature 387, 725–729 [DOI] [PubMed] [Google Scholar]
- 11. Flatau G., Lemichez E., Gauthier M., Chardin P., Paris S., Fiorentini C., Boquet P. (1997) Toxin-induced activation of the G protein p21 Rho by deamidation of glutamine. Nature 387, 729–733 [DOI] [PubMed] [Google Scholar]
- 12. Hoffmann C., Pop M., Leemhuis J., Schirmer J., Aktories K., Schmidt G. (2004) The Yersinia pseudotuberculosis cytotoxic necrotizing factor (CNFY) selectively activates RhoA. J. Biol. Chem. 279, 16026–16032 [DOI] [PubMed] [Google Scholar]
- 13. De Rycke J., Milon A., Oswald E. (1999) Necrotoxic Escherichia coli (NTEC). Two emerging categories of human and animal pathogens. Vet. Res. 30, 221–233 [PubMed] [Google Scholar]
- 14. Stoll T., Markwirth G., Reipschläger S., Schmidt G. (2009) A new member of a growing toxin family–Escherichia coli cytotoxic necrotizing factor 3 (CNF3). Toxicon 54, 745–753 [DOI] [PubMed] [Google Scholar]
- 15. De Rycke J., González E. A., Blanco J., Oswald E., Blanco M., Boivin R. (1990) Evidence for two types of cytotoxic necrotizing factor in human and animal clinical isolates of Escherichia coli. J. Clin. Microbiol. 28, 694–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deleted in proof
- 17. Sotiropoulos A., Gineitis D., Copeland J., Treisman R. (1999) Signal-regulated activation of serum response factor is mediated by changes in actin dynamics. Cell 98, 159–169 [DOI] [PubMed] [Google Scholar]
- 18. Dumoutier L., Tounsi A., Michiels T., Sommereyns C., Kotenko S. V., Renauld J. C. (2004) Role of the interleukin (IL)-28 receptor tyrosine residues for antiviral and antiproliferative activity of IL-29/interferon-λ 1. Similarities with type I interferon signaling. J. Biol. Chem. 279, 32269–32274 [DOI] [PubMed] [Google Scholar]
- 19. Dalpke A. H., Eckerle S., Frey M., Heeg K. (2003) Triggering of Toll-like receptors modulates IFN-γ signaling. Involvement of serine 727 STAT1 phosphorylation and suppressors of cytokine signaling. Eur. J. Immunol. 33, 1776–1787 [DOI] [PubMed] [Google Scholar]
- 20. Herrmann A., Vogt M., Mönnigmann M., Clahsen T., Sommer U., Haan S., Poli V., Heinrich P. C., Müller-Newen G. (2007) Nucleocytoplasmic shuttling of persistently activated STAT3. J. Cell Sci. 120, 3249–3261 [DOI] [PubMed] [Google Scholar]
- 21. Sotiropoulos A., Moutoussamy S., Renaudie F., Clauss M., Kayser C., Gouilleux F., Kelly P. A., Finidori J. (1996) Differential activation of Stat3 and Stat5 by distinct regions of the growth hormone receptor. Mol. Endocrinol. 10, 998–1009 [DOI] [PubMed] [Google Scholar]
- 22. Bromberg J. F., Horvath C. M., Besser D., Lathem W. W., Darnell J. E., Jr. (1998) Stat3 activation is required for cellular transformation by v-src. Mol. Cell. Biol. 18, 2553–2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Orth J. H., Aktories K., Kubatzky K. F. (2007) Modulation of host cell gene expression through activation of STAT transcription factors by Pasteurella multocida toxin. J. Biol. Chem. 282, 3050–3057 [DOI] [PubMed] [Google Scholar]
- 24. Koch G., Benz C., Schmidt G., Olenik C., Aktories K. (1997) Role of Rho protein in lovastatin-induced breakdown of actin cytoskeleton. J. Pharmacol. Exp. Ther. 283, 901–909 [PubMed] [Google Scholar]
- 25. Kortylewski M., Jove R., Yu H. (2005) Targeting STAT3 affects melanoma on multiple fronts. Cancer Metastasis Rev. 24, 315–327 [DOI] [PubMed] [Google Scholar]
- 26. Blumenthal B., Hoffmann C., Aktories K., Backert S., Schmidt G. (2007) The cytotoxic necrotizing factors from Yersinia pseudotuberculosis and from Escherichia coli bind to different cellular receptors but take the same route to the cytosol. Infect. Immun. 75, 3344–3353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ongusaha P. P., Qi H. H., Raj L., Kim Y. B., Aaronson S. A., Davis R. J., Shi Y., Liao J. K., Lee S. W. (2008) Identification of ROCK1 as an upstream activator of the JIP-3 to JNK signaling axis in response to UVB damage. Sci. Signal 1, ra14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Marinissen M. J., Chiariello M., Tanos T., Bernard O., Narumiya S., Gutkind J. S. (2004) The small GTP-binding protein RhoA regulates c-jun by a ROCK-JNK signaling axis. Mol. Cell 14, 29–41 [DOI] [PubMed] [Google Scholar]
- 29. Takasaki J., Saito T., Taniguchi M., Kawasaki T., Moritani Y., Hayashi K., Kobori M. (2004) A novel Gαq/11-selective inhibitor. J. Biol. Chem. 279, 47438–47445 [DOI] [PubMed] [Google Scholar]
- 30. Orth J. H., Lang S., Taniguchi M., Aktories K. (2005) Pasteurella multocida toxin-induced activation of RhoA is mediated via two families of Gα proteins, Gαq and Gα12/13. J. Biol. Chem. 280, 36701–36707 [DOI] [PubMed] [Google Scholar]
- 31. Darnell J. E., Jr. (1997) STATs and gene regulation. Science 277, 1630–1635 [DOI] [PubMed] [Google Scholar]
- 32. Yue P., Turkson J. (2009) Targeting STAT3 in cancer. How successful are we? Expert. Opin. Investig. Drugs 18, 45–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Raptis L., Arulanandam R., Geletu M., Turkson J. (2011) The R(h)oads to Stat3. Stat3 activation by the Rho GTPases. Exp. Cell Res. 317, 1787–1795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lee S. J., Qin H., Benveniste E. N. (2007) Simvastatin inhibits IFN-γ-induced CD40 gene expression by suppressing STAT-1α. J. Leukoc. Biol. 82, 436–447 [DOI] [PubMed] [Google Scholar]
- 35. Banes-Berceli A. K., Shaw S., Ma G., Brands M., Eaton D. C., Stern D. M., Fulton D., Caldwell R. W., Marrero M. B. (2006) Effect of simvastatin on high glucose- and angiotensin II-induced activation of the JAK/STAT pathway in mesangial cells. Am. J. Physiol. Renal Physiol. 291, F116–F121 [DOI] [PubMed] [Google Scholar]
- 36. Lerm M., Selzer J., Hoffmeyer A., Rapp U. R., Aktories K., Schmidt G. (1999) Deamidation of Cdc42 and Rac by Escherichia coli cytotoxic necrotizing factor 1. Activation of c-Jun N-terminal kinase in HeLa cells. Infect. Immun. 67, 496–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tiffany H. L., Lautens L. L., Gao J. L., Pease J., Locati M., Combadiere C., Modi W., Bonner T. I., Murphy P. M. (1997) Identification of CCR8. A human monocyte and thymus receptor for the CC chemokine I-309. J. Exp. Med. 186, 165–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Haque N. S., Fallon J. T., Pan J. J., Taubman M. B., Harpel P. C. (2004) Chemokine receptor-8 (CCR8) mediates human vascular smooth muscle cell chemotaxis and metalloproteinase-2 secretion. Blood 103, 1296–1304 [DOI] [PubMed] [Google Scholar]
- 39. Homey B., Meller S., Savinko T., Alenius H., Lauerma A. (2007) Modulation of chemokines by staphylococcal superantigen in atopic dermatitis. Chem. Immunol. Allergy 93, 181–194 [DOI] [PubMed] [Google Scholar]
- 40. Choi Y. B., Nicholas J. (2008) Autocrine and paracrine promotion of cell survival and virus replication by human herpesvirus 8 chemokines. J. Virol. 82, 6501–6513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Emdad L., Lebedeva I. V., Su Z. Z., Gupta P., Sauane M., Dash R., Grant S., Dent P., Curiel D. T., Sarkar D., Fisher P. B. (2009) Historical perspective and recent insights into our understanding of the molecular and biochemical basis of the antitumor properties of mda-7/IL-24. Cancer Biol. Ther. 8, 391–400 [DOI] [PubMed] [Google Scholar]
- 42. Ouyang W., Rutz S., Crellin N. K., Valdez P. A., Hymowitz S. G. (2011) Regulation and functions of the IL-10 family of cytokines in inflammation and disease. Annu. Rev. Immunol. 29, 71–109 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.