Abstract
Oxysterol binding protein-related proteins, including the yeast proteins encoded by the OSH gene family (OSH1–OSH7), are implicated in the non-vesicular transfer of sterols between intracellular membranes and the plasma membrane. In light of recent studies, we revisited the proposal that Osh proteins are sterol transfer proteins and present new models consistent with known Osh protein functions. These models focus on the role of Osh proteins as sterol-dependent regulators of phosphoinositide and sphingolipid pathways. In contrast to their posited role as non-vesicular sterol transfer proteins, we propose that Osh proteins coordinate lipid signaling and membrane reorganization with the assembly of tethering complexes to promote molecular exchanges at membrane contact sites.
Keywords: Lipid Transport, Membrane Trafficking, Phosphoinositides, Sterol, Yeast, Membrane Contact Sites, Non-vesicular Transport, ORPs, OSH Genes, Oxysterol binding Proteins
Introduction
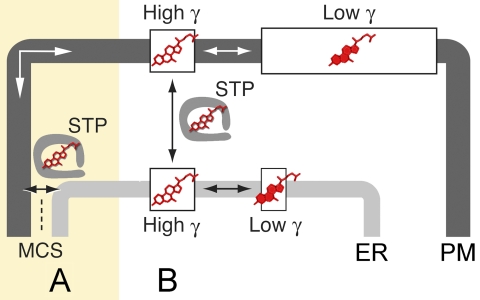
Sterols, including cholesterol in mammalian cells and ergosterol in fungi, constitute ∼30–40% of plasma membrane (PM)5 lipids and play a critical role in the nanoscale organization of the PM bilayer (1). Sterol-enriched membrane domains serve as platforms to cluster and concentrate specific lipid-modified and integral membrane proteins that function in cell signaling, secretory transport, and cytoskeletal organization (1). Sterols are synthesized in the endoplasmic reticulum (ER) and rapidly transported to the PM by a non-vesicular mechanism that is unaffected by drugs or genetic mutations that block vesicle-mediated protein secretion (2, 3). As sterols are largely water-insoluble, their non-vesicular transport between membranes is predicted to require sterol transfer proteins (STPs), which extract a sterol from a donor membrane and, after enclosing the bound sterol in a shielded pocket, transfer the sterol to an acceptor membrane. To exchange sterols between membranes, sterol-loaded STPs might diffuse through the cytoplasm or transfer sterols at membrane contact sites (MCSs), where two membranes are closely apposed (Fig. 1).
FIGURE 1.
Non-vesicular sterol transport. Sterols are largely insoluble in water (<100 nm) (69), and their non-vesicular transport between the ER and PM is proposed to require STPs that either mediate sterol transfer while attached to membranes at MCSs (A) or carry sterols through the cytoplasm (B). Although sterols represent ∼30–40 mol % of all lipids at the PM (70) but only ∼5% of ER lipids (71), the two membranes are at equilibrium with respect to sterol content, i.e. the chemical activity (a) of sterols is similar in both membranes. Chemical activity coefficients (γ) for sterols (where a = γ·c, and c = sterol concentration) in the PM are lower than in the ER due to interactions between sterols and lipids with saturated acyl chains, which are more abundant in the PM (72). The probability that a STP abstracts a sterol from a membrane is proportional to γ. Each membrane is shown as having two pools of sterols characterized by high (open sterols) and low (filled sterols) γ values. The majority of sterol in the PM has low γ. This figure was adapted from Ref. 4. The ergosterol transport rate needed for cell growth is calculated as follows (55). A yeast cell has 108 ergosterol molecules, of which ∼60% are in the PM. A pulse of [3H]ergosterol in the ER equilibrates with the entire PM ergosterol pool with t½ < 4 min (29). This value corresponds to an exchange rate constant (k) > 0.003 s−1 and a rate of ergosterol transport into and out of the PM of >6.5 × 108 molecules/cell/h. There are 40,990 Osh proteins/cell (33), and if all ergosterol transport is due to Osh proteins, then the transport rate is >16,000 ergosterol molecules transferred per Osh protein/h. The ergosterol transport rate needed for one yeast cell doubling is the equivalent of one new PM (6 × 107 ergosterol molecules) per 90 min, which is ∼104 s−1.
One consequence of a rapid non-vesicular transport mechanism is that the membranes involved must be close to equilibrium with respect to sterol levels. To account for the fact that sterols are more concentrated in the PM than elsewhere in the cell, it has been proposed that the lipid environment of the PM sequesters sterols (3–5). In the PM, sphingolipids as well as phospholipids with saturated acyl chains partner with sterols, lowering their chemical activity (or effective concentration) to a level similar to that in the ER (Fig. 1). Thus, even though the anterograde and retrograde flux of sterols between the ER and PM might be equivalent, the PM is enriched in sterols relative to the ER (4, 5).
The identity of yeast STPs is a mystery (6–9). As soluble sterol binding proteins that associate with organelle membranes, oxysterol binding protein (OSBP)-related proteins (ORPs) are potential candidates. Initial reports supported the idea that ORPs are directly involved in sterol transport. However, it is difficult to differentiate between sterol binding proteins that transfer sterols and those that regulate transport without being carriers themselves. Indeed, recent studies focusing on yeast ORPs (see below) suggest that the principal role of ORPs in vivo is to coordinate membrane lipid organization with the assembly of membrane-tethering complexes.
Osh Proteins: Non-vesicular STPs?
OSBP, the canonical mammalian ORP, was originally identified as a cytosolic protein that binds oxysterols (10, 11), which are oxygenated derivatives of cholesterol and are important regulators of cholesterol metabolism (12). OSBP is representative of the larger ORP superfamily that is conserved from yeast to man (13–16), and as discussed below, these proteins bind a variety of sterols. As might be expected of a STP, OSBP shuttles between cellular compartments in response to sterol binding (17–19).
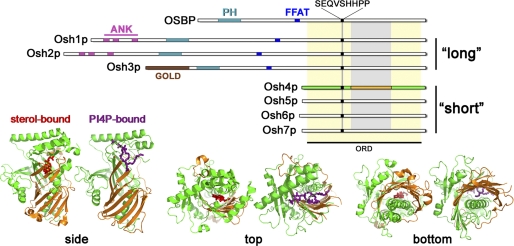
The budding yeast genome encodes seven ORPs (OSH1–OSH7) (Table 1) that can be divided into “short” and “long” classes (Fig. 2). Long ORPs contain N-terminal extensions that can include a phosphoinositide (PIP)-binding pleckstrin homology (PH) domain, a Golgi dynamics domain, ankyrin repeats, and/or FFAT (FF (phenylalanines) in an acidic tract) motifs that bind vesicle-associated membrane protein-associated protein (VAP) homologs (20). All ORPs share homology through a region broadly defined as the ORP-related domain (ORD) motif (21), although the ORD motif is almost the entire length of short ORPs. Yeast Osh proteins can diffuse through the cytoplasm as can most other ORPs (except mammalian ORP5 and ORP8, which contain predicted transmembrane domains) (22), but a variety of different domains (e.g. PH domains in long Osh proteins) confer membrane targeting to all Osh proteins.
TABLE 1.
Osh proteins and their regulators and effectors
| Protein(s) | Protein/activity | Cellular localization |
|---|---|---|
| Bgl2 | Endo-β-1,3-glucanase | Periplasm/sites of polarized growth, exocytic vesicles |
| Cdc42 | Rho family small GTPase | PM/sites of polarized growth |
| Drs2 | P-type ATPase and aminophospholipid translocase | Golgi |
| Inp51/Sjl1 | PI (4,5)P2 5-phosphatase with Sac1p-like domain; synaptojanin homolog | Cytoplasm, PM/cortical actin |
| Inp52/Sjl2, Inp53/Sjl3 | PIP phosphatases with Sac1p-like domains; synaptojanin homologs | Cytoplasm, PM/cortical actin |
| Osh1/Swh1 | OSBP homolog | ER/NVJ, Golgi |
| Osh2 | OSBP homolog | Cytoplasm, PM/sites of polarized growth, ER |
| Osh3 | OSBP homolog | Cytoplasm, ER |
| Osh4/Kes1 | OSBP homolog | Cytoplasm, Golgi, exocytic vesicles, endosomes |
| Osh5/Hes1 | OSBP homolog | Cytoplasm, ER (?), Golgi (?) |
| Osh6 | OSBP homolog | Cytoplasm, ER, PM |
| Osh7 | OSBP homolog | Cytoplasm, ER |
| Pik1 | PI 4-kinase | Golgi, nucleus |
| Rho1 | Rho family small GTPase | PM/sites of polarized growth |
| Rho3 | Rho family small GTPase | PM/sites of polarized growth |
| Sac1 | PI4P phosphatase | ER, Golgi |
| Scs2 | VAP homolog | ER |
| Sec4 | Rab family small GTPase | PM/sites of polarized growth, Golgi, exocytic vesicles |
| Sec14 | PI/PC transfer protein | Cytoplasm, Golgi |
| Stt4 | PI 4-kinase | PM |
| Ymr1 | PI3P phosphatase | Cytoplasm |
| Ypt31, Ypt32 | Rab family small GTPases | Golgi, vesicles, endosomes |
FIGURE 2.
Yeast Osh protein domains and structure of Osh4p. Upper, domains of the canonical mammalian OSBP compared with all seven yeast Osh proteins. ORPs are defined by the ORD (highlighted in yellow), within which is the SEQVSHHPP signature motif found in all ORPs. The ORP superfamily can be divided into short and long subgroups. The latter contains protein-binding domains including a FFAT motif, ankyrin repeats (ANK), or a Golgi dynamics (GOLD) domain. Long Osh proteins also contain a PH domain that binds PIPs. In Osh4p (green), a conserved region (orange) corresponds to the surface region that associates with anionic lipids, such as PI(4,5)P2. The corresponding sterol-bound (red) and PI4P-bound (purple) structures of Osh4p show the relative positions of these domains on the folded protein (lower).
The structure of the most abundant yeast ORP homolog, Osh4/Kes1p (hereafter referred to as Osh4p), was determined in complexes with oxysterols, cholesterol, and ergosterol, suggesting that all ORPs bind a wide range of sterol ligands (23). Given the modest differences in Osh4p affinities for oxysterols versus non-oxygenated sterols and the relative abundance of ergosterol (cholesterol in mammalian cells) in cellular membranes, ergosterol and cholesterol appear to be the primary sterols bound by ORPs in vivo (23).
The modular architecture of Osh4p is consistent with the presumed structural requisites needed for sterol transfer between membranes (Fig. 2). Osh4p is a β-barrel protein in which the bound sterol is contained head down inside a “beer mug” sealed by a small lid (23). Sterol capture may simply involve placing the Osh4p mug “mouth down” on top of the membrane surface, enabling the sterol to be ensconced in the binding cavity. Within the cavity, the sterol makes van der Waals contacts with Osh4p residues near the mug rim, and the sterol 3-OH headgroup interacts through hydrogen bonds with a number of water molecules inside the Osh4p mug (23). In fact, the Osh4p mug contains 15 water molecules, which provide a surprisingly “watery” environment for containing a hydrophobic lipid. A direct hydrogen bond between Osh4p Gln-96 and the sterol head also contributes to ligand binding (23). The sterol is ultimately enclosed within Osh4p by the flexible N-terminal lid, which might retain the captured sterol (Fig. 2).
The precise function of the lid in sterol binding is unclear. A “lidless” version of Osh4p has essentially the same affinity for cholesterol as wild-type Osh4p (Kd ∼ 0.3 μm) (23), indicating that the lid is not required for sterol retention. However, the lid might have other functions. The Osh4p lid sequence was identified as an ArfGAP1 lipid-packing sensor (ALPS)-like motif, which is an amphipathic helix that preferentially interacts with curved membranes (24). As it is unclear whether other Osh proteins contain bona fide ALPS helices, the broader relevance of the ALPS motif to the Osh protein family is uncertain. Alternatively, Osh4p might use its lid as a “bulldozer” to penetrate the bilayer and capture its sterol ligand. The A10/T4 helix of the phosphatidylinositol (PI)/phosphatidylcholine (PC) transfer protein Sec14p similarly acts like a bulldozer to penetrate the bilayer to the depth of the phospholipid acyl chains embedded in the outer leaflet to help scoop the lipid out of the membrane and into the Sec14p-binding cavity (25). Despite the ambiguities surrounding the role of the Osh4p lid, the structural attributes of Osh4p are generally consistent with the proposed role of ORPs as STPs.
In addition to a sterol, Osh4p can also bind PI4P (26). The crystal structure of Osh4p in a complex with PI4P shows that sterol and PI4P binding is mutually exclusive because the two binding sites overlap (Fig. 2). Indeed, when Osh4p is incubated with vesicles containing both sterols and PI4P, the two lipids compete for extraction by the protein (26). Moreover, the Osh4p-PI4P structure shows that specific but conserved residues bind PI4P, suggesting that all ORPs bind PI4P (26). The binary switch between lipid-bound Osh4p conformers presents new mechanistic possibilities for Osh proteins and ORPs.
Revisiting the Case for Osh Proteins as STPs
In budding yeast, none of the seven OSH genes is required for cell growth, but the deletion of all seven OSH genes results in cell lethality (13). Expressing any single OSH gene alone averts this lethality (13), indicating that each OSH gene is capable of providing the essential function(s) of the entire OSH gene family. When oshΔ osh4-1ts cells (oshΔ refers to the deletion of all OSH genes; osh4-1ts is a temperature-sensitive OSH4 mutation) are grown at elevated temperatures, the last remaining protein encoded by osh4-1ts is inactivated, and free sterols accumulate in internal membranes (27). In addition, the rate of retrograde transfer of exogenously added cholesterol from the PM to the ER and lipid droplets is slowed by ∼3-fold (28, 29). However, it is disconcerting that, in these assays, the elimination of Osh4p alone has no effect on sterol transport, despite the fact that Osh4p is >10-fold more abundant than any of the other Osh proteins (28). Although these results suggest a role for Osh proteins in retrograde sterol transport in vivo, the sterol-trafficking defects in oshΔ osh4-1ts cells might be indirect.
Osh4p was shown to increase the rate of cholesterol exchange between vesicle populations in vitro (28). When cholesterol-containing donor vesicles were prepared with specific PIPs to stimulate transfer, each Osh4p molecule transferred ∼20 cholesterol molecules/h to acceptor vesicles that were present in 10-fold excess (28). PI(4,5)P2 and phosphatidylserine that bind the surface of Osh4p (Fig. 2) were also transferred, albeit at a lower rate (28). Recent in vitro studies with improved temporal resolution showed that Osh4p exchanges sterols from donor vesicles to a 10-fold excess of PI4P-containing acceptor vesicles with an initial transport rate of ∼2000 sterols/Osh4p/h (26). Because PI4P and sterol binding by Osh4p is mutually exclusive, it was proposed that Osh4p acquires PI4P at the PM and exchanges it for a sterol at the ER (26). This vectorial exchange would be driven by PI4P hydrolysis catalyzed by the ER-localized Sac1p PI4P phosphatase (30, 31). However, in vivo, the exchange of sterol between the ER and PM is an equilibrium rather than vectorial (Fig. 1) (3, 4, 29). The fact that an Osh4p mutation that impairs sterol binding does not inactivate the protein, as predicted for a STP, also challenges this model (32).
The in vivo rate of sterol transfer between the ER and PM is >16,000 ergosterol molecules/Osh protein/h (Fig. 1), an order of magnitude greater than the measured in vitro rate. In comparison, in vitro, the mammalian lipid transfer protein STARD4 exchanges dehydroergosterol (DHE) between vesicles at a rate of ∼420 molecules/STARD4/h (9), which is comparable with Osh4p-mediated sterol transport. Because even nonspecific sterol-binding compounds like methyl β-cyclodextrin (MCD) redistribute sterols between liposomes, the relevance of the in vitro liposome assay requires confirmation in vivo. In the case of STARD4, microinjection of MCD into STARD4-silenced cells restores sterol transfer to the ER, indicating that the in vitro assays are relevant to STARD4 activity in vivo (9). Unfortunately for the study of Osh proteins, such experiments are not technically feasible in yeast.
Reservations about rates of Osh4p-mediated sterol transfer in vitro and the weak in vivo dependence of retrograde sterol transport on Osh proteins prompted new studies that revisited the case for Osh proteins as STPs. Using a variety of new assays to monitor retrograde and anterograde transport, sterol transfer between the ER and PM was shown to be essentially unaltered in Osh-deficient cells (29). In a live-cell assay tracking the retrograde transport of DHE, a natural fluorescent sterol, sterol transport was visualized in budding yeast for the first time (29). In hypoxically grown cells, DHE enters the PM, and during an aerobic chase period, it moves to the ER and then to lipid droplets. In Osh-deficient cells, this retrograde transfer is slowed by ∼3-fold. Based on these findings, it is clear that mechanisms other than those involving Osh proteins are required for sterol transport within cells.
Because the bulk of newly synthesized ergosterol is transported from the ER to the PM, another assay tested if Osh proteins affect sterol transfer in the anterograde direction (29). Following de novo synthesis in the ER, pulse-chase-labeled ergosterol was tracked by subcellular fractionation in wild-type and oshΔ osh4-1ts cells. No matter whether Osh proteins were inactivated or not, at the end of the chase, the ergosterol profiles were nearly identical, with most labeled ergosterol concentrated in PM fractions in proportion to the pool of endogenous ergosterol. Other experiments showed that MCD extracts accessible sterols from the outer leaflet of the PM ∼25-fold more efficiently in Osh-deficient cells relative to wild-type cells (29). Instead of a direct role in sterol transfer, these results suggest that Osh proteins affect the organization and sorting of sterols between bilayer leaflets and/or in membrane domains. Consistent with this reasoning, genetic studies indicate a functional interaction between OSH4 and DRS2, which encodes an ATP-dependent phospholipid flippase (34). This interaction might reflect a wider role of Osh proteins as regulators of enzymes and proteins that control the composition and organization of membrane bilayers.
It is a straightforward prediction that a STP would be nonfunctional if stripped of its capacity to bind and thereby transport sterols. As confirmed in vitro, mutations that abolish sterol interactions were designed based on the Osh4p crystal structure (23). Unlike mutations that affect the general association of Osh4p with membranes, the Y97F substitution specifically disrupts water-mediated hydrogen bonding required for Osh4p binding to sterols (23). When tested in vivo, however, elimination of sterol binding by the Osh4p Y97F substitution did not inactivate the protein. Rather, this allele is a gain-of-function mutation that causes lethality when Osh4p Y97F is expressed at levels comparable with wild-type Osh4p (32). A possible molecular explanation for the OSH4Y97F gain-of-function phenotype is provided by the fact that sites for sterol and PI4P binding within Osh4p are mutually exclusive (Fig. 2). If Osh4p function is normally induced by PI4P and repressed by sterols, then preventing sterol binding by Osh4p would end the competition between the lipid ligands for internal binding sites, and PI4P would be predicted to be constitutively bound. Consistent with this model, OSH4 gain-of-function lethality, whether caused by OSH4Y97F or OSH4 overexpression, is suppressed by mutations affecting PI4P metabolism (i.e. sac1Δ) (32). These genetic results suggest that sterols are inhibitory ligands of Osh4p activity, not transported cargo.
Osh Proteins at Organelle MCSs
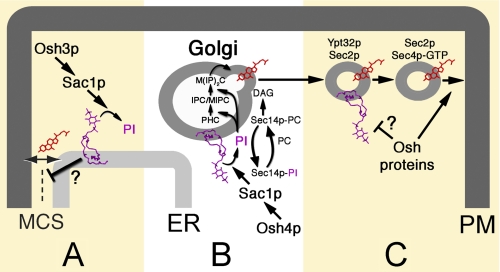
The simple model that Osh proteins are STPs does not appear to be consistent with all the in vivo evidence. However, these proteins might play an indirect role in the intermembrane exchange of sterols and other lipids by regulating the assembly of contact sites between organelle membranes where sterol transfer might occur (Fig. 1). The best example of such a site is the nucleus-vacuole junction (NVJ), which is where Osh1p is localized (35, 36). The NVJ is a MCS involved in piecemeal microautophagy of the nucleus, in which portions of the yeast nucleus are directly transferred into the vacuolar lumen for degradation (37). Inactivation of all OSH genes inhibits piecemeal microautophagy of the nucleus (36), suggesting that NVJ assembly is a shared function among all Osh proteins. Of relevance to sterol transfer is the possibility that PM/ER MCSs might mediate molecular exchanges. Osh2p and Osh3p each contain a FFAT motif and a PIP-binding PH domain (Fig. 2). The FFAT motif is recognized by the ER membrane protein Scs2p (38). Binding to both Scs2p and PIPs might enable Osh2p and Osh3p to promote and/or stabilize contacts between the ER and PM. Consistent with this idea, the deletion of SCS2 disrupts the association of the cortical ER with the PM (39). Although Osh2p and Osh3p are found in the cytoplasm, a significant fraction of Osh3p can be detected on the cortical ER closest to the PM, and SCS2 overexpression drives even more of these proteins to the cortical ER (38–42). Osh6p and Osh7p are also implicated in PM/ER MCS formation (40, 41), but neither protein contains a FFAT motif required for Scs2p-mediated recruitment to the cortical ER, and other results suggest that these short Osh proteins are associated with endosomal membranes (43). Despite the localization of certain Osh proteins to MCSs, there is no direct evidence to support the idea that they promote lipid exchange at those sites.
Osh Proteins Promote Vesicle/PM Tethering
Osh proteins share an overlapping role in promoting membrane contact between exocytic vesicles and the PM (Fig. 3). During polarized exocytosis in yeast, vesicles are targeted to the PM, where localized membrane expansion occurs to support formation of the bud. At the PM, the docking of exocytic vesicles involves the association of vesicle- and PM-localized subunits of the exocyst complex (44), which effectively form a MCS. The assembly of the exocyst complex is regulated in part by the Rab GTPase Sec4p and the Rho family GTPases Cdc42p, Rho1p, and Rho3p (45). After the exocyst complex attaches a vesicle to the PM, membrane fusion ensues. In the absence of all OSH gene function, undocked vesicles accumulate within buds, as observed either by electron microscopy (27) or when vesicles marked with GFP-Sec4p are tracked in vivo (32, 46). An in vivo assay of polarized exocytosis, using β-1,3-glucanase (Bgl2p) as a marker of vesicular transport (47), affirmed that major defects in polarized exocytosis result after Osh protein inactivation (46). More specifically, genetic and physical interactions directly implicate Osh proteins in the regulation of exocyst complex assembly (32, 46). Many OSH genes are allele-specific suppressors of cdc42 polarized cell growth defects, and OSH genes interact with other genes encoding exocyst complex subunits. In addition, Osh4p co-precipitates with the assembled exocyst complex isolated from yeast cell extracts, and the polarized localization of Cdc42p and Rho1p is lost as a result of Osh protein inactivation (32, 46). Of the eight exocyst complex subunits and the many ancillary regulatory proteins, it has yet to be determined which component makes specific contact with Osh4p, although SEC6 mutations inhibit Osh4p binding to Rho1p and block Osh4p association with vesicles (32). It is not known if vesicle tethering to the PM represents a unique Osh protein activity or is indicative of a wider involvement of ORPs in MCS formation.
FIGURE 3.
Roles of Osh proteins in membrane trafficking. A, at PM/ER MCSs, Osh3p is proposed to activate Sac1p-dependent dephosphorylation of PI4P (purple) to coordinate PIP signaling in both membranes (41). B, in the Golgi, Sec14p-PC and Sec14p-PI act on separate pathways to increase diacylglycerol (DAG) levels for secretory vesicle biogenesis (54). Opposing this positive regulatory control of vesicle biogenesis, Osh4p stimulates PI4P dephosphorylation by Sac1p, and the resulting PI is consumed in the production of complex sphingolipids (inositol phosphoceramide/mannosylinositol phosphoceramide (IPC/MIPC) and mannosyldiinositol phosphoceramide (M(IP)2C)) at the expense of diacylglycerol synthesis (see Footnote 6). Sterols (red) are enriched with complex sphingolipids in membrane domains that are sorted into nascent vesicles (56). The binding and inactivation of Osh4p by sterols enriched in sterol/sphingolipid membrane domains might lead to Sac1p feedback inhibition. C, as newly formed exocytic vesicles move to the PM, PI4P levels in their membranes decrease, triggering an exchange of Rab GTPases that bind Sec2p wherein Ypt32p is swapped for Sec4p (58). As the GEF for Sec4p, Sec2p activates Sec4p, and vesicle-bound exocyst complex subunits are recruited for later docking with their counterparts on the PM. To reduce PI4P levels on vesicles, Osh proteins might sequester PI4P or activate a PI4P phosphatase for PI4P turnover and Ypt32p/Sec4p exchange.
Osh Proteins as Regulators of PIP and Sphingolipid Metabolism
Osh1p, Osh2p, and Osh3p contain bona fide PIP-binding PH domains contained within their extra N-terminal extensions, which are not found in the other short Osh proteins (Fig. 1) (13, 48, 49). Despite the absence of a canonical PH domain, Osh4p interacts with a variety of PIPs (50–52) and specifically binds PI4P as discussed above (26). In addition, a PIP-binding domain was mapped to a surface region originally described as an atypical PH domain (50), but from the Osh4p structure, it corresponds to a surface region that binds anionic lipids (Fig. 2). Point mutations in this PIP-binding region inhibit the association of Osh4p with membranes in vitro and in vivo (50, 52). Because this region is conserved, it suggests that all ORPs, long or short, have a general capacity to associate with PIPs.
Although Osh4p is the focus of most studies analyzing ORP regulation of PIPs, Osh4p has a unique role in PIP metabolism distinct from that of other Osh proteins. Osh4p is observed in the cytoplasm, on endosomes, in post-Golgi vesicles, and on the Golgi, where it is involved in regulating PI4P metabolism (32, 41, 50, 52, 53). Inactivation of the Golgi-specific PI 4-kinase Pik1p blocks PI4P synthesis and decreases Osh4p association with the Golgi, suggesting that PI4P is required in vivo for Osh4p membrane association (50). The deletion of OSH4 also suppresses mutations in PI4P kinase PIK1 and SEC14 (which encodes a PI/PC transfer protein), both of which affect PI4P synthesis and vesicle biogenesis in the Golgi (50, 53). Like the elimination of Sac1p (the ER/Golgi-localized PI4P phosphatase), the deletion of OSH4 restores PI4P levels in cells with conditional PIK1 or SEC14 mutations (50, 53). Indeed all OSH genes share an overlapping function in activating Sac1p because, in Osh-deficient cells, there is an ∼20-fold increase in PI4P levels, well beyond that observed when just OSH4 is deleted (41).
Osh4p and Sac1p play dual roles during vesicle biogenesis at the Golgi by decreasing PI4P but also by increasing PI levels (Fig. 3) (50, 53). Lower PI4P levels inhibit Sec14p-dependent production of diacylglycerol, which is required for vesicle budding from the Golgi (54). In addition, Osh4p and Sac1p promote PI4P dephosphorylation, and the resulting pool of PI is used for complex sphingolipid synthesis in the Golgi. Consistent with the notion that Osh4p activates Sac1p to produce this PI pool, complex sphingolipid levels are decreased in osh4Δ cells.6 By antagonizing Sec14p, Osh4p might delay vesicle formation long enough for PI produced from Sac1p PI4P dephosphorylation to be incorporated into complex sphingolipids. Because sterols and sphingolipids are enriched in polarized exocytic vesicles (56), this mechanism might coordinate lipid biosynthesis with the sorting of sterol/sphingolipid membrane domains into nascent vesicles. Indeed, in osh4Δ cells, some proteins that partition into sterol/sphingolipid domains are missorted and not properly delivered from the Golgi to the PM (57).6 If sterol binding causes Osh4p inhibition, which is alleviated by the activated Y97F mutation that abolishes Osh4p sterol binding, then sterol-enriched membrane domains might favor Osh4p inactivation. In a negative feedback model, the generation of sterol/sphingolipid domains induces Osh4p exchange of PI4P for a sterol, and sterol-bound Osh4p cannot stimulate Sac1p-dependent PI4P turnover. As a result, PI4P levels increase, and Sec14p-dependent vesicle budding proceeds.
Sac1p and PI4P Are Effectors of Osh Proteins for Membrane Tethering
New functional assays and genetic analyses also support a model for Osh proteins as a novel class of sterol-dependent regulators of PIP4 signaling and metabolism. In this model, the Sac1p PI4P phosphatase appears to be particularly important as a presumptive effector for several Osh proteins. For example, the deletion of SAC1 suppresses both the lethality of the Osh4p Y97F gain-of-function mutation and most growth defects of cells in which Osh4p is overexpressed (32). This result suggests that Osh4p is an upstream regulator of Sac1p signaling. Indeed, much of the functional diversity of Osh proteins can be explained if their primary activity is to sequester and present PI4P to cognate signaling proteins and enzymes, such as Sac1p.
The overlapping shared role of all Osh proteins in exocyst complex-mediated vesicle docking is likely to involve, in part, PI4P regulation (Fig. 3). En route to the PM, PI4P levels decrease in exocytic vesicles, thereby triggering the release of the Ypt32p GTPase from Sec2p. Once released, Sec2p acts as a guanine nucleotide exchange factor (GEF), directly activating Sec4p to initiate the first steps in exocyst complex formation (58). By promoting Sac1p-dependent PI4P hydrolysis or by directly sequestering PI4P themselves, Osh proteins might promote this Ypt32p/Sec4p GTPase swap. Indeed, OSH4 (KES1) was originally identified as a suppressor of kre11-1, a mutation affecting the Kre11/Trs65p subunit of the TRAPP (transport protein particle) II complex, which acts as a GEF for Ypt32p activation (59, 60). Thus, Osh4p and potentially other Osh proteins are functionally linked to the TRAPP II complex and Ypt32p.
The role of Osh proteins at PM/ER MCSs is also proposed to involve Sac1p regulation (Fig. 3). Localization of Osh3p to PM/ER MCSs depends on PI4P levels, and many Osh proteins, including Osh3p, induce Sac1p PI4P phosphatase activity (41). Osh3p and Osh7p physically interact with Sac1p, albeit weakly, as complexes are detectable only after incubating isolated membrane fractions with chemical cross-linkers and by fusing a lengthy 13-Myc epitope onto Sac1p to boost antibody binding (41). However, in vivo, the interaction might involve a larger complex that includes Scs2p, a scaffolding protein that is required for full Sac1p phosphatase activity (41). In Drosophila melanogaster, Sac1 interacts with VAP-33-1 (a fly member of the VAP/Scs2 protein family), which in turn interacts with an ORP homolog (61). However, the inferred role for Sac1p in PM/ER MCS formation has not been established. For that matter, it is still to be determined whether Osh3p, with or without other Osh proteins, is required for PM/ER MCS formation.
Although important, Sac1p cannot be the only effector of Osh protein regulation because the cellular effects of SAC1 and OSH mutations are not the same (32). These findings indicate that other downstream effectors of Osh proteins must exist. The physical interaction of Osh4p with the exocyst complex suggests that one of these effectors might be an exocyst complex subunit (32). Other potential Osh protein effectors include other Sac1-domain phosphatases, such as Inp51/Sjl1p, Inp52/Sjl2p, and Inp53/Sjl3p. Consistent with this idea, OSH7 overexpression suppresses the growth defects of a ymr1ts inp52/sjl2Δ inp53/sjl3Δ triple phosphatase mutant (62).
Do Osh Proteins Provide a Valid Model for ORP Functions in Other Organisms?
Osh4p is the best understood yeast Osh protein and arguably the best studied ORP in general. However, as it actually represents a fungus-specific clade of the ORP superfamily (20), it is valid to question whether yeast Osh proteins (Osh4p in particular) truly represent the activities of ORPs in other organisms. Not only do the 12 human ORP genes share sequence homology with yeast OSH genes, but several can functionally replace them. When expressed in yeast, human ORP1S rescues the inviability of Osh-deficient cells (63). In addition, expression of human ORP1S or ORP9S in yeast complements osh4Δ phenotypes with respect to the suppression of SEC14 mutations (51, 63, 64). These results suggest that ORPs share common functions even between species. Apart from any role as STPs, certainly yeast and metazoan ORPs have been implicated in many different signaling pathways, as reviewed previously (63, 65).
It remains to be seen if the proposed role of Osh4p and other Osh proteins in coordinating lipid signaling and biosynthesis with secretory trafficking applies to mammalian ORPs. Like the long Osh proteins, many ORPs interact with PIPs through PH domains. Also like Osh4p, the canonical mammalian OSBP coordinates sterol and sphingolipid synthesis between compartments in a PI4P-dependent manner (66, 67). The cellular compartmentalization of lipid biosynthesis is sometimes different in yeast compared with mammalian cells, which is the case for complex sphingolipids. Nevertheless, OSBP couples sterol binding with the regulation of PI 4-kinase IIa and the PI4P-dependent recruitment of CERT (ceramide transport protein) to the Golgi (67). Unlike yeast cells that lack CERT, CERT-mediated transfer of ceramide to the Golgi is critical for the metabolism of complex sphingolipids in mammalian cells. Despite the differences between yeast and mammalian cells, the general paradigm of ORPs as sterol- and PI4P-dependent regulators of lipid metabolism and signaling appears to be applicable.
Understanding the functional roles of ORPs has taken on a new imperative with the recent discovery that human ORPs are specific targets of ORPphilins, a diverse group of nanomolar inhibitors that prevent cancer cell growth (68). In some cases, these compounds vaguely resemble a sterol fused to a PIP, implying that they confer inhibition by simultaneously occupying both sterol- and PI4P-binding sites. How these drugs affect ORP activities is still unknown, but studies of yeast Osh proteins suggest that Sac1p and PI4P regulation might be the ultimate targets.
This work was supported in part by grants from the Canadian Cancer Society Research Institute and the Natural Sciences and Engineering Research Council of Canada (to C. T. B.).
M. A. LeBlanc, G. D. Fairn, S. E. Brice, L. A. Cowart, and C. R. McMaster, submitted for publication.
- PM
- plasma membrane
- ER
- endoplasmic reticulum
- STP
- sterol transfer protein
- MCS
- membrane contact site
- OSBP
- oxysterol binding protein
- ORP
- OSBP-related protein
- PIP
- phosphoinositide
- PH
- pleckstrin homology
- VAP
- vesicle-associated membrane protein-associated protein
- ORD
- ORP-related domain
- ALPS
- ArfGAP1 lipid-packing sensor
- PI
- phosphatidylinositol
- PC
- phosphatidylcholine
- DHE
- dehydroergosterol
- MCD
- methyl β-cyclodextrin
- NVJ
- nucleus-vacuole junction
- GEF
- guanine nucleotide exchange factor.
REFERENCES
- 1. Lingwood D., Simons K. (2010) Lipid rafts as a membrane-organizing principle. Science 327, 46–50 [DOI] [PubMed] [Google Scholar]
- 2. Urbani L., Simoni R. D. (1990) Cholesterol and vesicular stomatitis virus G protein take separate routes from the endoplasmic reticulum to the plasma membrane. J. Biol. Chem. 265, 1919–1923 [PubMed] [Google Scholar]
- 3. Baumann N. A., Sullivan D. P., Ohvo-Rekilä H., Simonot C., Pottekat A., Klaassen Z., Beh C. T., Menon A. K. (2005) Transport of newly synthesized sterol to the sterol-enriched plasma membrane occurs via non-vesicular equilibration. Biochemistry 44, 5816–5826 [DOI] [PubMed] [Google Scholar]
- 4. Maxfield F. R., Menon A. K. (2006) Intracellular sterol transport and distribution. Curr. Opin. Cell Biol. 18, 379–385 [DOI] [PubMed] [Google Scholar]
- 5. Mesmin B., Maxfield F. R. (2009) Intracellular sterol dynamics. Biochim. Biophys. Acta 1791, 636–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Reiner S., Micolod D., Zellnig G., Schneiter R. (2006) A genome-wide screen reveals a role of mitochondria in anaerobic uptake of sterols in yeast. Mol. Biol. Cell 17, 90–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fei W., Alfaro G., Muthusamy B. P., Klaassen Z., Graham T. R., Yang H., Beh C. T. (2008) Genome-wide analysis of sterol-lipid storage and trafficking in Saccharomyces cerevisiae. Eukaryot. Cell 7, 401–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sullivan D. P., Georgiev A., Menon A. K. (2009) Tritium suicide selection identifies proteins involved in the uptake and intracellular transport of sterols in Saccharomyces cerevisiae. Eukaryot. Cell 8, 161–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mesmin B., Pipalia N. H., Lund F. W., Ramlall T. F., Sokolov A., Eliezer D., Maxfield F. R. (2011) STARD4 abundance regulates sterol transport and sensing. Mol. Biol. Cell 22, 4004–4015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kandutsch A. A., Chen H. W., Shown E. P. (1977) Binding of 25-hydroxycholesterol and cholesterol to different cytoplasmic proteins. Proc. Natl. Acad. Sci. U.S.A. 74, 2500–2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dawson P. A., Van der Westhuyzen D. R., Goldstein J. L., Brown M. S. (1989) Purification of oxysterol binding protein from hamster liver cytosol. J. Biol. Chem. 264, 9046–9052 [PubMed] [Google Scholar]
- 12. Schroepfer G. J., Jr. (2000) Oxysterols: modulators of cholesterol metabolism and other processes. Physiol. Rev. 80, 361–554 [DOI] [PubMed] [Google Scholar]
- 13. Beh C. T., Cool L., Phillips J., Rine J. (2001) Overlapping functions of the yeast oxysterol binding protein homologs. Genetics 157, 1117–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jaworski C. J., Moreira E., Li A., Lee R., Rodriguez I. R. (2001) A family of 12 human genes containing oxysterol-binding domains. Genomics 78, 185–196 [DOI] [PubMed] [Google Scholar]
- 15. Lehto M., Laitinen S., Chinetti G., Johansson M., Ehnholm C., Staels B., Ikonen E., Olkkonen V. M. (2001) The OSBP-related protein family in humans. J. Lipid Res. 42, 1203–1213 [PubMed] [Google Scholar]
- 16. Anniss A. M., Apostolopoulos J., Dworkin S., Purton L. E., Sparrow R. L. (2002) An oxysterol binding protein family identified in the mouse. DNA Cell Biol. 21, 571–580 [DOI] [PubMed] [Google Scholar]
- 17. Ridgway N. D., Dawson P. A., Ho Y. K., Brown M. S., Goldstein J. L. (1992) Translocation of oxysterol binding protein to Golgi apparatus triggered by ligand binding. J. Cell Biol. 116, 307–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ridgway N. D., Lagace T. A., Cook H. W., Byers D. M. (1998) Differential effects of sphingomyelin hydrolysis and cholesterol transport on oxysterol binding protein phosphorylation and Golgi localization. J. Biol. Chem. 273, 31621–31628 [DOI] [PubMed] [Google Scholar]
- 19. Wyles J. P., McMaster C. R., Ridgway N. D. (2002) Vesicle-associated membrane protein-associated protein-A (VAP-A) interacts with the oxysterol binding protein to modify export from the endoplasmic reticulum. J. Biol. Chem. 277, 29908–29918 [DOI] [PubMed] [Google Scholar]
- 20. Raychaudhuri S., Prinz W. A. (2010) The diverse functions of oxysterol binding proteins. Annu. Rev. Cell Dev. Biol. 26, 157–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lehto M., Olkkonen V. M. (2003) The OSBP-related proteins: a novel protein family involved in vesicle transport, cellular lipid metabolism, and cell signaling. Biochim. Biophys. Acta 1631, 1–11 [DOI] [PubMed] [Google Scholar]
- 22. Olkkonen V. M., Levine T. P. (2004) Oxysterol-binding proteins: in more than one place at one time? Biochem. Cell Biol. 82, 87–98 [DOI] [PubMed] [Google Scholar]
- 23. Im Y. J., Raychaudhuri S., Prinz W. A., Hurley J. H. (2005) Structural mechanism for sterol sensing and transport by OSBP-related proteins. Nature 437, 154–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Drin G., Casella J. F., Gautier R., Boehmer T., Schwartz T. U., Antonny B. (2007) A general amphipathic α-helical motif for sensing membrane curvature. Nat. Struct. Mol. Biol. 14, 138–146 [DOI] [PubMed] [Google Scholar]
- 25. Sha B., Phillips S. E., Bankaitis V. A., Luo M. (1998) Crystal structure of the Saccharomyces cerevisiae phosphatidylinositol transfer protein. Nature 391, 506–510 [DOI] [PubMed] [Google Scholar]
- 26. de Saint-Jean M., Delfosse V., Douguet D., Chicanne G., Payrastre B., Bourguet W., Antonny B., Drin G. (2011) Osh4p exchanges sterols for phosphatidylinositol 4-phosphate between lipid bilayers. J. Cell Biol. 195, 965–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Beh C. T., Rine J. (2004) A role for yeast oxysterol binding protein homologs in endocytosis and in the maintenance of intracellular sterol-lipid distribution. J. Cell Sci. 117, 2983–2996 [DOI] [PubMed] [Google Scholar]
- 28. Raychaudhuri S., Im Y. J., Hurley J. H., Prinz W. A. (2006) Non-vesicular sterol movement from plasma membrane to ER requires oxysterol binding protein-related proteins and phosphoinositides. J. Cell Biol. 173, 107–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Georgiev A. G., Sullivan D. P., Kersting M. C., Dittman J. S., Beh C. T., Menon A. K. (2011) Osh proteins regulate membrane sterol organization but are not required for sterol movement between the ER and PM. Traffic 12, 1341–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Whitters E. A., Cleves A. E., McGee T. P., Skinner H. B., Bankaitis V. A. (1993) Sac1p is an integral membrane protein that influences the cellular requirement for phospholipid transfer protein function and inositol in yeast. J. Cell Biol. 122, 79–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tahirovic S., Schorr M., Mayinger P. (2005) Regulation of intracellular phosphatidylinositol 4-phosphate by the Sac1 lipid phosphatase. Traffic 6, 116–130 [DOI] [PubMed] [Google Scholar]
- 32. Alfaro G., Johansen J., Dighe S. A., Duamel G., Kozminski K. G., Beh C. T. (2011) The sterol binding protein Kes1/Osh4p is a regulator of polarized exocytosis. Traffic 12, 1521–1536 [DOI] [PubMed] [Google Scholar]
- 33. Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., Weissman J. S. (2003) Global analysis of protein expression in yeast. Nature 425, 737–741 [DOI] [PubMed] [Google Scholar]
- 34. Muthusamy B. P., Raychaudhuri S., Natarajan P., Abe F., Liu K., Prinz W. A., Graham T. R. (2009) Control of protein and sterol trafficking by antagonistic activities of a type IV P-type ATPase and oxysterol binding protein homolog. Mol. Biol. Cell 20, 2920–2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Levine T. P., Munro S. (2001) Dual targeting of Osh1p, a yeast homolog of oxysterol binding protein, to both the Golgi and the nucleus-vacuole junction. Mol. Biol. Cell 12, 1633–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kvam E., Goldfarb D. S. (2004) Nvj1p is the outer nuclear membrane receptor for oxysterol binding protein homolog Osh1p in Saccharomyces cerevisiae. J. Cell Sci. 117, 4959–4968 [DOI] [PubMed] [Google Scholar]
- 37. Roberts P., Moshitch-Moshkovitz S., Kvam E., O'Toole E., Winey M., Goldfarb D. S. (2003) Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol. Biol. Cell 14, 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Loewen C. J., Roy A., Levine T. P. (2003) A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO J. 22, 2025–2035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Loewen C. J., Young B. P., Tavassoli S., Levine T. P. (2007) Inheritance of cortical ER in yeast is required for normal septin organization. J. Cell Biol. 179, 467–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schulz T. A., Choi M. G., Raychaudhuri S., Mears J. A., Ghirlando R., Hinshaw J. E, Prinz W. A. (2009) Lipid-regulated sterol transfer between closely apposed membranes by oxysterol binding protein homologs. J. Cell Biol. 187, 889–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stefan C. J., Manford A. G., Baird D., Yamada-Hanff J., Mao Y., Emr S. D. (2011) Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell 144, 389–401 [DOI] [PubMed] [Google Scholar]
- 42. Toulmay A., Prinz W. A. (2011) Lipid transfer and signaling at organelle contact sites: the tip of the iceberg. Curr. Opin. Cell Biol. 23, 458–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang P., Zhang Y., Li H., Chieu H. K., Munn A. L., Yang H. (2005) AAA ATPases regulate membrane association of yeast oxysterol binding proteins and sterol metabolism. EMBO J. 24, 2989–2999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Boyd C., Hughes T., Pypaert M., Novick P. (2004) Vesicles carry most exocyst subunits to exocytic sites marked by the remaining two subunits, Sec3p and Exo70p. J. Cell Biol. 167, 889–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Park H. O., Bi E. (2007) Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol. Mol. Biol. Rev. 71, 48–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kozminski K. G., Alfaro G., Dighe S., Beh C. T. (2006) Homologs of oxysterol binding proteins affect Cdc42p- and Rho1p-mediated cell polarization in Saccharomyces cerevisiae. Traffic 7, 1224–1242 [DOI] [PubMed] [Google Scholar]
- 47. Harsay E., Bretscher A. (1995) Parallel secretory pathways to the cell surface in yeast. J. Cell Biol. 131, 297–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Levine T. P., Munro S. (1998) The pleckstrin homology domain of oxysterol binding protein recognizes a determinant specific to Golgi membranes. Curr. Biol. 8, 729–739 [DOI] [PubMed] [Google Scholar]
- 49. Roy A., Levine T. P. (2004) Multiple pools of phosphatidylinositol 4-phosphate detected using the pleckstrin homology domain of Osh2p. J. Biol. Chem. 279, 44683–44689 [DOI] [PubMed] [Google Scholar]
- 50. Li X., Rivas M. P., Fang M., Marchena J., Mehrotra B., Chaudhary A., Feng L., Prestwich G. D., Bankaitis V. A. (2002) Analysis of oxysterol binding protein homolog Kes1p function in regulation of Sec14p-dependent protein transport from the yeast Golgi complex. J. Cell Biol. 157, 63–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Fairn G. D., McMaster C. R. (2005) Identification and assessment of the role of a nominal phospholipid-binding region of ORP1S (oxysterol binding protein-related protein 1 short) in the regulation of vesicular transport. Biochem. J. 387, 889–896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. LeBlanc M. A., McMaster C. R. (2010) Lipid binding requirements for oxysterol binding protein Kes1 inhibition of autophagy and endosome-trans-Golgi trafficking pathways. J. Biol. Chem. 285, 33875–33884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Fairn G. D., Curwin A. J., Stefan C. J., McMaster C. R. (2007) The oxysterol binding protein Kes1p regulates Golgi apparatus phosphatidylinositol 4-phosphate function. Proc. Natl. Acad. Sci. U.S.A. 104, 15352–15357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kearns B. G., McGee T. P., Mayinger P., Gedvilaite A., Phillips S. E., Kagiwada S., Bankaitis V. A. (1997) Essential role for diacylglycerol in protein transport from the yeast Golgi complex. Nature 387, 101–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Sullivan D. P., Ohvo-Rekilä H., Baumann N. A., Beh C. T., Menon A. K. (2006) Sterol trafficking between the endoplasmic reticulum and plasma membrane in yeast. Biochem. Soc. Trans. 34, 356–358 [DOI] [PubMed] [Google Scholar]
- 56. Klemm R. W., Ejsing C. S., Surma M. A., Kaiser H. J., Gerl M. J., Sampaio J. L., de Robillard Q., Ferguson C., Proszynski T. J., Shevchenko A., Simons K. (2009) Segregation of sphingolipids and sterols during formation of secretory vesicles at the trans-Golgi network. J. Cell Biol. 185, 601–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Proszynski T. J., Klemm R. W., Gravert M., Hsu P. P., Gloor Y., Wagner J., Kozak K., Grabner H., Walzer K., Bagnat M., Simons K., Walch-Solimena C. (2005) A genome-wide visual screen reveals a role for sphingolipids and ergosterol in cell surface delivery in yeast. Proc. Natl. Acad. Sci. U.S.A. 102, 17981–17986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mizuno-Yamasaki E., Medkova M., Coleman J., Novick P. (2010) Phosphatidylinositol 4-phosphate controls both membrane recruitment and a regulatory switch of the Rab GEF Sec2p. Dev. Cell 18, 828–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Jiang B., Brown J. L., Sheraton J., Fortin N., Bussey H. (1994) A new family of yeast genes implicated in ergosterol synthesis is related to the human oxysterol binding protein. Yeast 10, 341–353 [DOI] [PubMed] [Google Scholar]
- 60. Jones S., Newman C., Liu F., Segev N. (2000) The TRAPP complex is a nucleotide exchanger for Ypt1 and Ypt31/32. Mol. Biol. Cell 11, 4403–4411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Giot L., Bader J. S., Brouwer C., Chaudhuri A., Kuang B., Li Y., Hao Y. L., Ooi C. E., Godwin B., Vitols E., Vijayadamodar G., Pochart P., Machineni H., Welsh M., Kong Y., Zerhusen B., Malcolm R., Varrone Z., Collis A., Minto M., Burgess S., McDaniel L., Stimpson E., Spriggs F., Williams J., Neurath K., Ioime N., Agee M., Voss E., Furtak K., Renzulli R., Aanensen N., Carrolla S., Bickelhaupt E., Lazovatsky Y., DaSilva A., Zhong J., Stanyon C. A., Finley R. L., Jr., White K. P., Braverman M., Jarvie T., Gold S., Leach M., Knight J., Shimkets R. A., McKenna M. P., Chant J., Rothberg J. M. (2003) A protein interaction map of Drosophila melanogaster. Science 302, 1727–1736 [DOI] [PubMed] [Google Scholar]
- 62. Parrish W. R., Stefan C. J., Emr S. D. (2005) PtdIns(3)P accumulation in triple lipid phosphatase deletion mutants triggers lethal hyperactivation of the Rho1p/Pkc1p cell integrity MAP kinase pathway. J. Cell Sci. 118, 5589–5601 [DOI] [PubMed] [Google Scholar]
- 63. Fairn G. D., McMaster C. R. (2008) Emerging roles of the oxysterol binding protein family in metabolism, transport, and signaling. Cell. Mol. Life Sci. 65, 228–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Fairn G. D., McMaster C. R. (2005) The roles of the human lipid binding proteins ORP9S and ORP10S in vesicular transport. Biochem. Cell Biol. 83, 631–636 [DOI] [PubMed] [Google Scholar]
- 65. Yan D., Olkkonen V.M. (2008) Characteristics of oxysterol binding proteins. Int. Rev. Cytol. 265, 253–285 [DOI] [PubMed] [Google Scholar]
- 66. Perry R. J., Ridgway N. D. (2006) Oxysterol binding protein and vesicle-associated membrane protein-associated protein are required for sterol-dependent activation of the ceramide transport protein. Mol. Biol. Cell 17, 2604–2616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Banerji S., Ngo M., Lane C. F., Robinson C. A., Minogue S., Ridgway N. D. (2010) Oxysterol binding protein-dependent activation of sphingomyelin synthesis in the Golgi apparatus requires phosphatidylinositol 4-kinase IIα. Mol. Biol. Cell 21, 4141–4150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Burgett A. W., Poulsen T. B., Wangkanont K., Anderson D. R., Kikuchi C., Shimada K., Okubo S., Fortner K. C., Mimaki Y., Kuroda M., Murphy J. P., Schwalb D. J., Petrella E. C., Cornella-Taracido I., Schirle M., Tallarico J. A., Shair M. D. (2011) Natural products reveal cancer cell dependence on oxysterol binding proteins. Nat. Chem. Biol. 7, 639–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Saad H. Y., Higuchi W. I. (1965) Water solubility of cholesterol. J. Pharm. Sci. 54, 1205–1206 [DOI] [PubMed] [Google Scholar]
- 70. Yeagle P. L. (1985) Lanosterol and cholesterol have different effects on phospholipid acyl chain ordering. Biochim. Biophys. Acta 815, 33–36 [DOI] [PubMed] [Google Scholar]
- 71. Radhakrishnan A., Goldstein J. L., McDonald J. G., Brown M. S. (2008) Switch-like control of SREBP-2 transport triggered by small changes in ER cholesterol: a delicate balance. Cell Metab. 8, 512–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Maxfield F. R., van Meer G. (2010) Cholesterol, the central lipid of mammalian cells. Curr. Opin. Cell Biol. 22, 422–429 [DOI] [PMC free article] [PubMed] [Google Scholar]