Background: Perakine reductase (PR) is an AKR involved in the Rauvolfia alkaloid biosynthetic network.
Results: Three-dimensional structures of PR and the A213W mutant complex with NADPH were solved.
Conclusion: PR folds as an unusual α8/β6 barrel and undergoes unexpected conformational changes upon NADPH binding.
Significance: PR represents the founding member of the new AKR13D subfamily and provides a structural and cofactor binding template for the AKR13 family.
Keywords: Biosynthesis, Enzyme Mechanisms, Enzyme Structure, Plant Biochemistry, Protein Crystallization, Aldo-keto Reductase (AKR), Plant Alkaloids
Abstract
Perakine reductase (PR) catalyzes the NADPH-dependent reduction of the aldehyde perakine to yield the alcohol raucaffrinoline in the biosynthetic pathway of ajmaline in Rauvolfia, a key step in indole alkaloid biosynthesis. Sequence alignment shows that PR is the founder of the new AKR13D subfamily and is designated AKR13D1. The x-ray structure of methylated His6-PR was solved to 2.31 Å. However, the active site of PR was blocked by the connected parts of the neighbor symmetric molecule in the crystal. To break the interactions and obtain the enzyme-ligand complexes, the A213W mutant was generated. The atomic structure of His6-PR-A213W complex with NADPH was determined at 1.77 Å. Overall, PR folds in an unusual α8/β6 barrel that has not been observed in any other AKR protein to date. NADPH binds in an extended pocket, but the nicotinamide riboside moiety is disordered. Upon NADPH binding, dramatic conformational changes and movements were observed: two additional β-strands in the C terminus become ordered to form one α-helix, and a movement of up to 24 Å occurs. This conformational change creates a large space that allows the binding of substrates of variable size for PR and enhances the enzyme activity; as a result cooperative kinetics are observed as NADPH is varied. As the founding member of the new AKR13D subfamily, PR also provides a structural template and model of cofactor binding for the AKR13 family.
Introduction
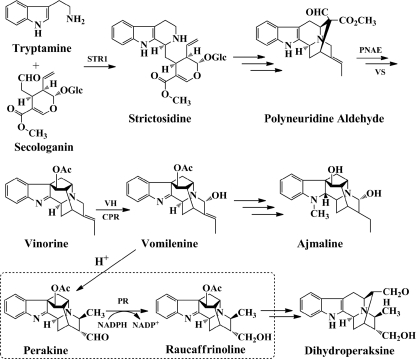
Enzyme-catalyzed biosynthesis and chemo-enzymatic synthesis of pharmacologically active, plant-derived monoterpenoid indole alkaloids have attracted interest in recent years (1–5). A detailed knowledge of the participating enzymes is an indispensable requirement for further progress in this field. One of the largest alkaloid synthetic networks is the Rauvolfia serpentina metabolome. In this network we detected perakine reductase, which is involved in a side route of metabolism that branches from the main ajmaline pathway (Fig. 1) and leads to the biosynthesis of dihydroperaksine (6, 7). As an antiarrhythmic drug, ajmaline is well known to act as an antagonist at sodium channels of the heart muscle (8, 9).
FIGURE 1.
Functional role of PR. PR is located in a side route branching from the ajmaline biosynthetic pathway leading to dihydroperaksine and extends the metabolic network of Rauvolfia monoterpenoid indole alkaloids. (STR1, strictosidine synthase, EC 4.3.3.2; PNAE, polyneuridine aldehyde esterase, EC 3.1.1.78; VS, vinorine synthase, EC 2.3.2.160; VH, vinorine hydroxylase, EC 1.14.13.75; CPR, cytochrome P450 reductase; H+ = transformation under acidic conditions).
Functional overexpression in Escherichia coli together with extended substrate studies recently allowed advanced biochemical characterization of PR (6). The NADPH-dependent PR showed a relaxed substrate preference because three groups of structurally diverse compounds were reduced to their corresponding alcohols: two bulky alkaloids, ten medium-sized cinnamic aldehyde derivatives, and four small (nitro)-benzaldehydes (6). In contrast to PR, other enzymes of Rauvolfia alkaloid biosynthesis exhibit much higher substrate specificity (10). The absence of both a Rossmann fold (11) and the catalytic motif Tyr-Xaa-Xaa-Xaa-Lys (12) excluded PR as a member of the short chain dehydrogenase/reductase superfamily. However, the presence of a functional catalytic tetrad Asp52, Tyr57, Lys84, and His126, as proven by site-directed mutagenesis, and the presence of other residues highly conserved in aldo-keto reductase enzymes (AKRs),2 i.e. Gly20, Gly47, Asp121, Pro133, Gly154, Asn167, Pro192, Gln196, and Ser252, indicate affiliation of PR to the AKR enzyme family (13). Furthermore, sequence alignment and preliminary x-ray analysis of a methylated PR mutant support the view that PR represents a novel member of the AKR superfamily (6, 14). In fact, PR would be the first example of a plant AKR involved in indole alkaloid biosynthesis in contrast to the plant AKR-type codeinone reductase, which is involved in isoquinoline alkaloid metabolism (15).
We now describe x-ray analyses of a methylated N-terminal His6-PR and its A213W mutant complex with the cofactor NADPH. These crystal structures show that PR is an AKR, that it contains an unusual α8/β6 barrel, and that it undergoes a novel conformational change upon binding NADPH not seen previously in AKR members. Extended sequence alignment suggests that PR is the founding member of a new AKR13D subfamily.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
R. serpentina PR cDNA (GenBankTM accession number AY766462) was cloned into pQE-2 vector and expressed as N-terminal His6-tagged fusion protein in E. coli M15 strain (Qiagen). Bacteria were grown at 25 °C for large scale enzyme production, and the conditions for cultivation, induction and purification of recombinant PR were the same as previously reported (6, 14).
Site-directed Mutagenesis
Site-directed mutagenesis was performed to generate the PR-A213W point mutant by using the QuikChangeTM site-directed mutagenesis kit (Stratagene, La Jolla, CA). The wild type PR was cloned into pQE-2 as a template, and the following oligonucleotides were used as primers to introduce the mutation: A213W forward primer, 5′-d-GGT CTT TTT TGG GGG AAG GCC-3′, and A213W reverse primer, 5′-d-GGC CTT CCC CCA AAA AAG ACC-3′.
Protein Methylation
Because of unsuccessful crystallization of native PR, reductive methylation was chosen to modify the protein surface. The purified enzyme was dialyzed against potassium phosphate (KPi) buffer (50 mm, pH 7.0) and concentrated to 10 mg/ml for the subsequent methylation. Methylation was performed by dimethylamine-borane and formaldehyde as previously reported (14, 16).
NADPH Kinetic Behavior
The kinetic properties of NADPH were determined by monitoring product formation by HPLC with seven different NADPH concentrations (2–80 μm) in the presence of substrate (4-nitrobenzaldehyde) (0.2 mm) under standard conditions with 5 μg of purified His6-PR in KPi buffer (50 mm, pH 7.0) in a total volume of 200 μl. The enzymatic reaction was initiated by adding NADPH. After 15 min of incubation at 30 °C, the reaction was terminated by adding 200 μl of methanol, and the sample was subjected to HPLC to monitor product formation at 254 nm (Waters 2487 detector) with Lichrospher 60 RP-select B column (250 × 4 mm). Acetonitrile/water (1:4, pH 2.3) were used as mobile phase at a flow rate of 1 ml/min.
Size Exclusion Chromatography
Determination of relative molecular weight and oligomerization state of recombinant methylated and unmethylated His6-PR (6 mg/ml applied) was performed on HiPrep Sephacryl S-200 column using KPi buffer (50 mm, pH 7.0) at a flow rate of 0.5 ml/min. For calibration, the following proteins were used: glucose oxidase (Aspergillus niger) (150 kDa), BSA (67 kDa), ovalbumin (45 kDa), and α-chymotrypsin (25 kDa).
Crystallization
Crystallization of methylated His6-PR was accomplished by the hanging drop vapor diffusion method. The drops were set up by mixing 2 μl of methylated enzyme samples (5.5 mg/ml, 10 mm Tris-HCl buffer, pH 7.0, 1 mm DTT, 10 mm EDTA) with 2 μl of reservoir solution (25% v/v PEG 4000, 0.1 mm sodium citrate, pH 5.6) and equilibrated against 1 ml of reservoir solution at 20 °C for 7 days. Methylated His6-PR-A213W was also crystallized under the same conditions as methylated His6-PR.
The crystals of methylated His6-PR-A213W complex with NADPH were obtained by cocrystallization. Prior to crystallization, the enzyme solutions were incubated with a 3-fold molar excess of NADPH for 2 h at 4 °C to form the complex. The crystallization of the methylated His6-PR-A213W and NADPH complex was carried out under the same conditions as methylated His6-PR. The same crystallization conditions were used for the unmethylated His6-PR-A213W and NADPH complex, with the exception that the enzyme samples were in KPi buffer (50 mm, pH 7.0).
X-ray Data Collection and Processing
All the PR crystals were cryoprotected with 20% glycerol added to the reservoir solution before flash cooling in a stream of nitrogen at 100 K.
Diffraction data of methylated His6-PR crystals were collected at BW7B beamline at the European Molecular Biology Laboratory Outstation (Hamburg, Germany). The data were integrated, scaled, and merged with the HKL2000 software package (17). A platinum derivative was derived after the incubation of the methylated His6-PR crystals with 21% PEG 4000, 0.1 m sodium citrate, pH 5.6, and 10 mm K2PtCl4 for 20 min. The multiwavelength anomalous diffraction data collection around the Pt-LIII edge was performed at the MX-beamline BL14–2 of Helmholtz Zentrum Berlin at BESSY II using a SX165 CCD detector (Rayonix, Evanston, IL) and DTB goniostat (Marresearch, Norderstedt, Germany) at 100 K (supplemental Table S1A). The data processing was performed by XDS (18).
The crystals of the methylated His6-PR-A213W, its complex with NADPH, and the unmethylated His6-PR-A213W complex with NADPH were all measured at PX III beamline at Swiss Light Source (Villigen, Switzerland). Program XDS (18) was applied for data processing.
Structure Solution and Refinement of the Methylated His6-PR
The crystal structure of the methylated His6-PR was solved with multiwavelength anomalous diffraction method using the program SHELXC/D/E (19). The initial model (60% of the full structure) was built using RESOLVE (20). This model was used as a starting model in the MRSAD protocol of Auto-Rickshaw (21) against the peak data set for the phase improvement and for the model completion. 85% of the full structure was built automatically. First refinement was performed using REFMAC (22). The model was further improved in several rounds of refinement using automated, restrained refinement with the program PHENIX (23) and by interactive modeling with Coot (24).
Structure Determination of the Methylated/Unmethylated His6-PR-A213W Complex with NADPH and apo His6-PR-A213W
The internal processing script at SLS was utilized for initial data processing that uses XDS (18), Labelit (25), MOSFLM (26), and phenix.xtriage (23). Molecular replacement was performed with MOLREP within the CCP4 suite (27) using the refined methylated His6-PR structure as a search model. The refinement was performed as described above. NADPH was identified in the first electron density map based on the phase of the molecular replacement solution and was included in the model of the methylated and unmethylated His6-PR-A213W. The refinement statistics are listed in Table 1 and supplemental Table S1B. The final model was analyzed with the program MolProbity (28). All of the figures were prepared using PyMOL (29).
TABLE 1.
PR X-ray data collection and refinement statistics
| Structure | mPRa | mPR-A213W + NADPHb |
|---|---|---|
| Data collection | ||
| Wavelength (Å) | 0.8148 | 1.0 |
| Total reflections | 213,159 | 337,656 |
| Unique reflections | 16,975 | 37,054 |
| Mosaicity | 0.908 | 0.183 |
| Resolution (Å) | 20-2.31 (2.43-2.31) | 48-1.77 (1.82-1.77) |
| Completeness (%) | 96.8 (93.0) | 97.6 (80.0) |
| I/σ(I) | 33.5 (5.4) | 49.4 (3.38) |
| Rmerge (%)c | 7.9 (34.9) | 4.6 (37.4) |
| Space group | C2221 | P3221 |
| Unit cell (Å) | a = 58.8, b = 93.0, c = 143.0 | a = b = 55.1, c = 209.8 |
| Refinement statistics | ||
| Rworkd/Rfree (%)e | 18.9/23.7 | 19.1/21.1 |
| No. of protein atoms | 2480 | 2227 |
| No. of water molecules | 54 | 358 |
| No. of NADP+ atoms | 31 | |
| RMSD bond length (Å) | 0.015 | 0.012 |
| RMSD bond angle (°) | 1.513 | 1.494 |
| Average B-factors (Å2) | ||
| Protein | 55.3 | 23.7 |
| Water | 50.4 | 36.6 |
| NADP+ | 26.8 | |
| Ramachandran analysis (%) | ||
| Most favored region | 96.7 | 96.8 |
| Allowed | 3.3 | 3.2 |
| Disallowed | 0.0 | 0.0 |
| Protein Data Bank code | 3UYI | 3V0S |
a mPR, methylated His6-PR wild type.
b mPR A213W + NADPH, methylated His6-PR-A213W complexed with NADPH.
c Rmerge = |Ii−<Ii>|Ii, where Ii is the average intensity value of the equivalent reflections.
d Rwork = Σ(|Fo − Fc|)/Σ|Fo|, where Fo and Fc are the observed and calculated structure factors, respectively.
e Rfree was calculated using 5% randomly excluded data from refinement.
Supplemental Data
The following materials are available in the online version of this article: supplemental Table S1A showing multiwavelength anomalous diffraction data collection and phasing statistics of methylated His6-PR, supplemental Table S1B showing x-ray data collection and refinement statistics of methylated apo His6-PR-A213W and unmethylated His6-PR-A213W complex with NADPH, supplemental Fig. S1 showing residue Ala213 of methylated His6-PR and its nearby region, supplemental Fig. S2 showing size exclusion chromatogram of methylated and unmethylated His6-PR, supplemental Fig. S3 showing NADPH binding site, supplemental Fig. S4 showing two symmetric holo His6-PR-A213W molecules in the crystal packing, and supplemental Fig. S5 showing structural comparison of PR and a “generic” AKR to display the diversity in the C terminus.
RESULTS AND DISCUSSION
Overall Structure of PR
The final three-dimensional structure model of apo methylated PR was derived following refinement of the 2.31 Å data set with 18.9% Rwork and 23.7% Rfree (Table 1) and contained 312 amino acid residues and 54 water molecules. There was no defined electron density for two loops, which are residues 26–30 and 221–239.
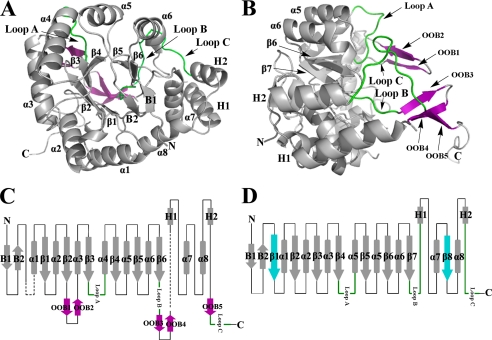
Methylated apo PR folds into an unusual α/β-barrel consisting unexpectedly of only six β-strands (β1–β6) with eight α-helices that pack along the outside of the β-strands (Fig. 2, A and B). Because of the two missing β-strands, the last two α-helices, α7 and α8, are directly connected by a loop (Fig. 2C). Up to now, all other three-dimensional structures identified for AKR members fold as (α/β)8-barrels with eight parallel β-strands and eight α-helices, also known as a triose phosphate isomerase barrel (13, 30). Some typical AKR structural features, such as the β-hairpins (B1 and B2), which cover the top of the barrel at the N terminus, the two helices (H1 and H2), which pack together along the outer barrel, and three large loops (Loops A, B, and C) at the C terminus, are conserved in PR; however, there are also some striking differences when PR is compared with other AKR members (Fig. 2, C and D). Different from other AKR members, the barrel structure of PR starts with an α-helix instead of a β-strand. In addition, the decoration of the structure with five β-strands, named OOB1–5 (out of barrel) (Fig. 2B), on the C-terminal side is a second notable difference. OOB1–2 are localized between β2 and α3, OOB3–4 are found between Loop B and H1, and OOB5 is directly upstream of Loop C (Fig. 2C). Two pairs of these β-strands, OOB1–2 and OOB3–4, form a clamp that covers the C-terminal base of the barrel, whereas the two connecting loops serve as the closing lids. The unique topology of the PR overall architecture is shown in Fig. 2C, and the topology of a representative AKR is shown in Fig. 2D.
FIGURE 2.
Overall structure of methylated His6-PR. A, the structure of methylated His6-PR displays an unusual α8/β6 barrel fold for an AKR enzyme. B, to show the conformation of OOB1-OOB5 (purple) and Loop A–C, Fig. 2A is rotated by 90°. The long C-terminal tail is pointing away from the molecule. C, topology of the methylated His6-PR structure. The disordered residues are in black dashed lines. D, topology of a representative AKR (3α-hydroxysteroid dehydrogenase, AKR1C9). α1–α8, the α-helices; β1–β6, the β-strands in PR; β1–β8, the β-strands in a representative AKR (β1 and β8 (blue) are missing in PR); B1 and B2, the two N-terminal β-strands of the hairpin; H1 and H2, the two additional α-helices. The three typical loops (Loops A, B, and C) for AKR enzymes are in green. All the other loops are shown as black lines. The five additional out of barrel β-strands OOB1–5 at the C terminus are in purple. The N and C termini are marked with N and C.
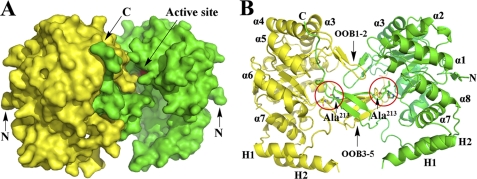
In the asymmetric unit cell there is only one PR molecule, but the two symmetric molecules are strongly connected in the crystal packing (Fig. 3A). The interactions between the two molecules occur at their C-terminal regions. The additional five β-strands (OOB1–5), Loop C, and the C-terminal tail appear to be tightly interwoven with a symmetry molecule (Fig. 3B) and block the active site (Fig. 3A). To break the interactions between the two molecules, the small residue Ala213 located in the interface region between the two symmetric molecules was mutated (supplemental Fig. S1 and Fig. 3B). A bulky Trp was selected, and the PR-A213W mutant, which is still functional, was generated to prepare PR complexes with cofactor and/or substrate bound.
FIGURE 3.
Two symmetric methylated His6-PR molecules in crystal. A, the two symmetric molecules from the crystal packing are shown in yellow and green surface presentation. The catalytic tetrad (Asp52, Tyr57, Lys84, and His126) of the molecule is in red (arrow). B, cartoon presentation of A. The additional five out of barrel β-strands are grouped together as OOB1–2 and OOB3–5. The N and C termini are marked with N and C.
Two symmetric methylated PR molecules are packed together tightly in the crystal form, but size exclusion studies indicated that methylated PR occurs exclusively as a monomer in solution (supplemental Fig. S2). Moreover, 80% of native (unmethylated) wild type PR seems to exist as a monomer in solution, whereas only 20% are in the dimeric state (supplemental Fig. S2).
Description of PR-Cofactor Complex
The final structure of methylated His6-PR-A213W complex with NADPH was refined to 1.77 Å resolution to an Rwork and Rfree of 19.1% and 21.1%, respectively (Table 1). The model is comprised of 289 amino acids (residues 1–24, 32–219, 243–310, and 328–336), NADPH, and 358 water molecules. Compared with the apo form of PR, there are more disordered residues in the NADPH complex model, especially in the C-terminal region. The strong interactions between the C-terminal residues of the two symmetric molecules found in the crystal packing of the apo form explain the good electron density in this form. Mutation of Ala213 to Trp disrupted these interactions and made the cofactor-binding site available for ligand binding. However, in the absence of these intermolecular interactions, some of the C-terminal residues (residues 311–327), which folded as Loop C and OOB5 in the apo form, are now more flexible in the new complex, and as such there is insufficient electron density to permit complete structural determination in this region.
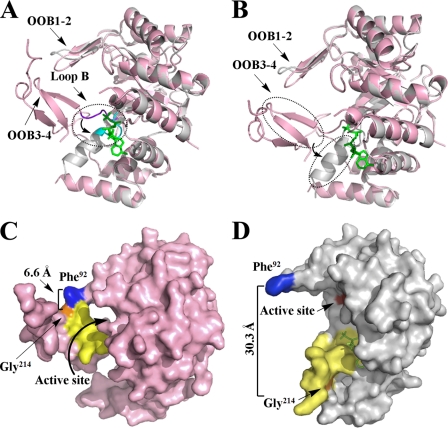
For the portion of the PR complex with NADPH that provides a structure, there is similarity in the protein fold with the PR apo form. For example the α8/β6 barrel of the two PR forms can be superimposed with RMSD < 0.3 Å, but there are striking differences when the C-terminal regions of both PR forms are compared (Fig. 4). The most significant conformation change occurs from residues 205 to 219. Residues 205–208 reside in Loop B, which is an important part of the cofactor-binding site. Loop B bends inward to the center of the barrel when NADPH is not bound (Fig. 4A). However, it points outward and away from the center of the barrel when NADPH is bound. When the two structures are superimposed in this region, an RMSD of the main chains of 2.02 Å with a maximum deviation of 3.94 Å is observed. The similar movement of Loop B is the most frequently observed conformational change in AKRs upon cofactor binding (13, 31, 32).
FIGURE 4.
Structure comparison of apo (pink) and holo (gray) forms of PR. A, superimposed apo and holo forms of PR to show the movement of Loop B upon NADPH binding. Loop B is shown in purple in the PR apo form and in cyan in the holo form. B, superimposed apo and holo forms of PR to show the conformational change and movement of two β-strands (OOB3–4), which switched to one α-helix upon NADPH binding. C, surface presentation of the apo form of methylated PR displays the “closed” conformation. D, surface presentation of the holo form of methylated PR illustrates the “open” conformation upon NADPH (green) binding. The residues that mark protein movement Phe92 and Gly214, are in blue and orange. The residues that change position and conformation (residues 205–219) are in yellow. The active site (Asp52, Tyr57, Lys84, and His126) is in red.
More severe conformational changes and accompanying movements were observed in the immediate region downstream of Loop B, residues 209–219. The secondary structure in this region contains two β-strands (OOB3–4) with a connecting loop in the apo form, but this changed surprisingly to one α-helix in the NADPH bound (holo) form (Fig. 4B). The RMSD of the main chains of this part of the structure between the apo and holo form is 5.73 Å, and the maximum deviation is 12.5 Å. A second AKR enzyme with significant conformational changes induced by NADPH binding is 2,5-diketo-d-gluconic acid reductase A, which shows remarkable structural rearrangements (up to 8 Å) in the catalytic pocket upon cofactor binding accompanied by a switch of an α-helix to an extended β-strand (33). However, the conformational changes discussed herein for PR are significantly more pronounced than those in 2,5-diketo-d-gluconic acid reductase A.
To better illustrate the space produced by this movement, residue Phe92 in the connecting loop between OOB1 and OOB2, which is located in the middle of the relatively immobile upper lid of the “clamp” (made of OOB1–2 and OOB3–4), was selected as a marker of a stable position. Residue Gly214, which directly faces Phe92, was selected as a marker of a movable position. The distance between the two Cα atoms of Phe92 and Gly214 was 6.6 Å in the apo form (Fig. 4C), but upon NADPH binding, the distance dramatically increased by ∼5-fold to 30.3 Å (Fig. 4D). Consequently the “clamp” was completely open. This conformation change creates enough space around the catalytic site for substrate binding, e.g. the bulky alkaloids can be accommodated.
To exclude the possibility that mutation Ala213 to Trp induces the conformational changes and movements of residues 205–219, the crystallization of apo methylated His6-PR-A213W was carried out. The x-ray structure of apo methylated His6-PR-A213W was solved with 2.20 Å resolution (supplemental Table S1B). In the absence of the intermolecular interactions that were disrupted by the mutation of Ala213 to Trp, the C-terminal segments (residues 205–219 and 311–337) of the apo A213W mutant were much flexible in the crystals, and there is insufficient density for the structure elucidation of above mentioned residues. When NADPH was bound to the A213W mutant, there was clear density corresponding for residues 205–219. The structural difference between apo and holo forms of the mutant indicates that residues 205–219 are involved in cofactor binding. Upon NADPH binding, this part became ordered to form as an α-helix and was fixed to keep the open form and provide enough space for the catalytic reaction to proceed. This observation also confirms that NADPH binding is responsible for the positional and conformational changes.
Crystallography was also performed on the unmethylated His6-PR-A213W complex with NADPH (supplemental Table S1B). The structure obtained at 2.33 Å had a RMSD of 0.7 Å when the methylated and unmethylated PR forms were compared and demonstrates that methylation had no significant influence on the three-dimensional structures.
Binding Mode of NADPH
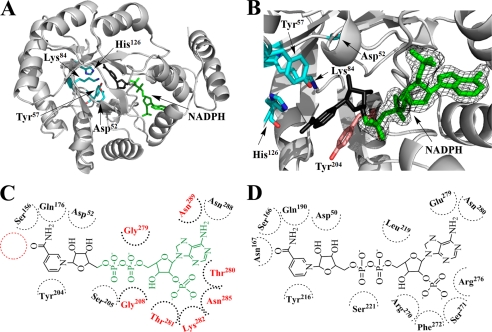
The cofactor NADPH was bound in the C-terminal core of the barrel of PR and points toward the active site, which is formed by the catalytic tetrad Asp52, Tyr57, Lys84, and His126 at the center of the barrel, as in most AKR enzymes (34, 35) (Fig. 5A). Because NADPH is easily oxidized to NADP+, the crystal structures of PR complexes likely contain NADP+. However, there was no clear density for the nicotinamide riboside moiety. The final structural model derived from the 1.77 Å diffraction data set clearly defined the binding pattern of this moiety of NADPH in PR (Fig. 5B and supplemental Fig. S3). The density (corresponding to the adenine ring, the ribose ring, the adenosine phosphate, and pyrophosphate) of NADPH lies in an extended groove that stretches from the center to the edge of the barrel and is formed by the surface of residues 205–208 from Loop B, residues 277–282 from the loop linking α7 and α8, and residues 285, 288, and 289 in α8. Many AKR enzymes share a similar mode of cofactor binding across the superfamily (13, 36, 37). However, except for the conservation of Ser205 and Asn289 in PR, all of the other residues directly hydrogen-bonded with the cofactor are “nonclassical” when compared with other AKR members (Fig. 5, C and D) (13, 38). As the first example of a structure from the AKR13 family, the PR structural model may provide valuable information on understanding the role of cofactor binding residues across this family.
FIGURE 5.
Cofactor binding mode of PR. A, overall structure of methylated His6-PR-A213W complexed with NADPH (green) is shown in a similar orientation as in Fig. 2A. The nicotinamide riboside moiety of NADPH (black) is disordered in the crystal. The catalytic tetrad (Asp52, Tyr57, Lys84, and His126) is in stick presentation (cyan). B, a close-up of the catalytic site of methylated His6-PR-A213W with bound NADPH (green). The observed electron density (final 2Fo − Fc) for NADPH was contoured at 1 σ. Tyr204 (pink) is the conserved planar aromatic amino acid that in other AKRs stacks with the nicotinamide ring (compared with A, rotated by 180°). C, NADPH binding in PR. The modeled nicotinamide riboside moiety of NADPH is in black. The position of Asn, which is strictly conserved in other AKRs but absent in PR, is shown with a red circle. The amino acids which are involved in cofactor binding of PR but differ from the highly conserved amino acids found in other AKRs, are in red. D, NADPH binding in a typical AKR (3α-hydroxysteroid dehydrogenase) (38).
The disordered nicotinamide riboside portion was therefore modeled in its usual anti-conformation (Fig. 5, A and B) as seen in other AKR members (36, 39). Sequence alignment shows that several typical AKR cofactor binding residues, e.g. Asp52, Ser156, Gln176, and Tyr204, were conserved in PR (Fig. 5, C and D). These amino acids reside in the space surrounding the active site, and the modeled nicotinamide ring supports their important role in binding the cofactor. The asparagine next to Ser156 is strictly conserved in the AKR superfamily and is an important residue in cofactor binding. The hydrogen in δ-N of this asparagine forms a hydrogen bond with the oxygen atom of the carboxamide group in the nicotinamide ring, thereby stabilizing the whole ring. However, in PR this asparagine is replaced by a glutamic acid, which could account for the disorder observed in the binding of the nicotinamide ring.
It is obvious that the cofactor and substrate bind in different regions of the enzyme and converge at the active site. Thereby, the substrate of PR probably occupies the cleft made up of OOB1–2 and the α-helix (switched from OOB3–4). Mutation of A213W completely breaks up the interactions between the OOB3–5, Loop C, and the C-terminal tail and successfully opens the cofactor-binding site, but at the same time, interactions from OOB1–2 still exist. The crystallization pattern of PR complex with NADPH shows that the loop bridging OOB1 and OOB2 from the neighboring molecule partly occupies the active site and the substrate-binding site (supplemental Fig. S4). Our observations may explain why our trials to obtain the ternary complexes of PR, cofactor, and substrates failed.
Kinetics of PR and Structural Changes
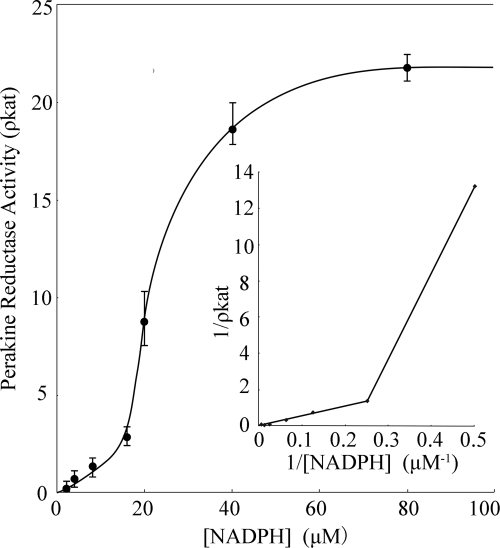
PR does not display classical Michaelis-Menten kinetics. When the activity of wild type PR was measured at constant substrate (4-nitrobenzaldehyde) concentration and NADPH concentrations were varied, a sigmoidal velocity (v) versus [substrate] plot was obtained (Fig. 6). This result points toward cooperative binding of NADPH (Hill coefficient > 1), which may lead to the enhanced reductase activity of the enzyme. The increase in v may be accounted for the significant conformational change of PR, which we observed for the NADPH-bound structure (Fig. 4). The cofactor likely opens the entry to the active center of PR, which rapidly and immediately triggers the reduction of the substrate. The NADPH-induced conformational changes are consistent with an ordered bi-bi reaction mechanism, (in which cofactor binds first and leaves last), which is observed in other AKRs (13, 30). Once the second substrate binds, reduction likely utilizes the conserved catalytic tetrad of Tyr, Lys, His, and Asp that lies at the base of the substrate-binding site (40).
FIGURE 6.
Plot of initial velocity (v) versus NADPH concentration observed for the reduction of 4-nitrobenzaldehyde catalyzed by PR.
Classification and Aspects of Evolution
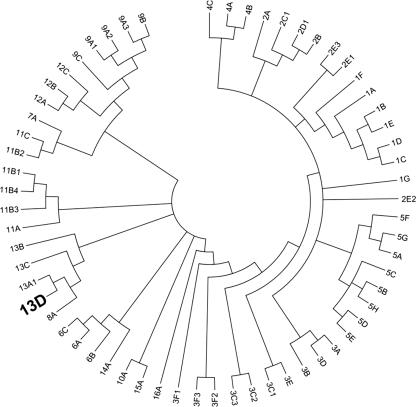
Multiple sequence alignment and phylogenetic analyses established PR as an AKR13 family member, founding the new subgroup of AKR13D (Fig. 7). PR is also the first plant enzyme in this family that up until now only contained proteins of bacterial and fungal origin. Blast analyses indicate that PR type proteins are absent from the animal kingdom. Furthermore, screening of the NCBI database identified putative PR homologs in a diversity of higher plant species, e.g. Medicago truncatula, Arabidopsis thaliana, Oryza sativa, Zea mays, and Picea sitchensis. Alignment of R. serpentina PR and these putative homologs showed high sequence conservation with ∼70% of the residues being identical. In comparison, strictosidine synthase (STR1), the first enzyme in the biosynthetic pathway to monoterpenoid indole alkaloids (Fig. 1) (41), appears to be far less conserved. Sequence identity between well characterized Rauvolfia STR1 and its putative homologs in the species listed above was only ∼40%. However, because of the high sequence diversity of proteins such as STR1 that feature a six-bladed β-propeller structure and their lack of functional characterization, these putative homologous proteins identified in silico may not be functional STR1 orthologs (42). Consequently, STR1 and the strictosidine pathway would be absent in those species that otherwise seem to harbor a well conserved PR. These observations and the wide spectrum of PR substrates (6) suggest that although PR is involved in the secondary metabolism of strictosidine in Rauvolfia, it may play additional roles in other well conserved biosynthesis pathways in higher plants.
FIGURE 7.
Evolutionary relationship of new AKR13D from R. serpentina to annotated AKR families and subfamilies. The dendrogram shows the topological relationships between AKR subfamilies. The new AKR13D from R. serpentina is most closely related to other members of the AKR13 family and AKR8. The evolutionary history of all annotated AKRs including the new AKR from R. serpentina was inferred using the neighbor joining method (43). A bootstrap consensus tree was inferred from 1000 replicates (44), with subfamily branches collapsed where possible. Branches corresponding to partitions reproduced in less than 30% bootstrap replicates were also collapsed. The evolutionary distances were computed using the Poisson correction method (45) and are in the units of the number of amino acid substitutions per site. All of the positions containing alignment gaps and missing data were eliminated only in pairwise sequence comparisons (pairwise deletion option). There were a total of 607 positions in the final data set. Phylogenetic analyses were conducted in MEGA4 (46).
Supplementary Material
Acknowledgments
We thank Dr. Yi Jin (Research Assistant Professor at the Perleman School of Medicine) for a careful read of the manuscript. We thank the staff members of Shanghai Synchrotron Radiation Facility.
This work was supported by funds from Fonds der Chemischen Industrie (Frankfurt/Main, Germany), Deutsche Forschungsgemeinschaft (Bad Godesberg, Germany), Zhejiang University K.P. Chao's Hi-Tech Foundation (Hangzhou, China; to J. S.), and China Postdoctoral Science Foundation Grant 20100471745 (to L. S.).

This article contains supplemental Table S1 and Figs. S1–S5.
The atomic coordinates and structure factors (codes 3UYI and 3V0S) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- AKR
- aldo-keto reductase
- PR
- perakine reductase
- KPi
- potassium phosphate
- STR1
- strictosidine synthase
- RMSD
- root mean square deviation.
REFERENCES
- 1. Ruppert M., Ma X., Stöckigt J. (2005) Alkaloid biosynthesis in Rauvolfia-cDNA cloning of major enzymes of the ajmaline pathway. Curr. Org. Chem. 9, 1431–1444 [Google Scholar]
- 2. Lee H. Y., Yerkes N., O'Connor S. E. (2009) Aza-tryptamine substrates in monoterpene indole alkaloid biosynthesis. Chem. Biol. 16, 1225–1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Runguphan W., Qu X., O'Connor S. E. (2010) Integrating carbon-halogen bond formation into medicinal plant metabolism. Nature 468, 461–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Loris E. A., Panjikar S., Ruppert M., Barleben L., Unger M., Schübel H., Stöckigt J. (2007) Structure-based engineering of strictosidine synthase: auxiliary for alkaloid libraries. Chem. Biol. 14, 979–985 [DOI] [PubMed] [Google Scholar]
- 5. Zou H. B., Zhu H. J., Zhang L., Yang L. Q., Yu Y. P., Stöckigt J. (2010) A facile chemoenzymatic approach: one-step syntheses of monoterpenoid indole alkaloids. Chem. Asian J. 5, 2400–2404 [DOI] [PubMed] [Google Scholar]
- 6. Sun L., Ruppert M., Sheludko Y., Warzecha H., Zhao Y., Stöckigt J. (2008) Purification, cloning, functional expression and characterization of perakine reductase: the first example from the AKR enzyme family, extending the alkaloidal network of the plant Rauvolfia. Plant Mol. Biol. 67, 455–467 [DOI] [PubMed] [Google Scholar]
- 7. Edwankar R. V., Edwankar C. R., Deschamps J., Cook J. M. (2011) Regiospecific, enantiospecific total synthesis of C-19 methyl substituted sarpagine alkaloids dihydroperaksine-17-al and dihydroperaksine. Org. Lett. 13, 5216–5219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cordell G. A. (1981) Introduction to Alkaloids: A Biogenetic Approach, John Wiley and Sons, New York [Google Scholar]
- 9. Kiesecker C., Zitron E., Lück S., Bloehs R., Scholz E. P., Kathöfer S., Thomas D., Kreye V. A. W., Katus H. A., Schoels W., Karle C. A., Kiehn J. (2004) Class la anti-arrhythmic drug ajmaline blocks HERG potassium channels: model of action. Naunyn-Schmiedebergs Arch. Pharmacol. 370, 423–435 [DOI] [PubMed] [Google Scholar]
- 10. Yang L., Hill M., Wang M., Panjikar S., Stöckigt J. (2009) Structural basis and enzymatic mechanism of the biosynthesis of C9 from C10 monoterpenoid indole alkaloids. Angew. Chem. Int. Ed. Engl. 48, 5211–5213 [DOI] [PubMed] [Google Scholar]
- 11. Rao S. T., Rossmann M. G. (1973) Comparison of super-secondary structures in proteins. J. Mol. Biol. 76, 241–256 [DOI] [PubMed] [Google Scholar]
- 12. Persson B., Krook M., Jörnvall H. (1991) Characteristics of short-chain alcohol dehydrogenases and related enzymes. Eur. J. Biochem. 200, 537–543 [DOI] [PubMed] [Google Scholar]
- 13. Jez J. M., Bennett M. J., Schlegel B. P., Lewis M., Penning T. M. (1997) Comparative anatomy of the aldo-keto reductase superfamily. Biochem. J. 326, 625–636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rosenthal C., Mueller U., Panjikar S., Sun L., Ruppert M., Zhao Y., Stöckigt J. (2006) Expression, purification, crystallization and preliminary x-ray analysis of perakine reductase, a new member of the aldo-keto reductase enzyme superfamily from higher plants. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 62, 1286–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Unterlinner B., Lenz R., Kutchan T. M. (1999) Molecular cloning and functional expression of codeinone reductase: the penultimate enzyme in morphine biosynthesis in the opium poppy Papaver somniferum. Plant J. 18, 465–475 [DOI] [PubMed] [Google Scholar]
- 16. Rypniewski W. R., Holden H. M., Rayment I. (1993) Structural consequences of reductive methylation of lysine residues in hen egg white lysozyme. An X-ray analysis at 1.8-. ANG. resolution. Biochemistry 32, 9851–9858 [DOI] [PubMed] [Google Scholar]
- 17. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 18. Kabsch W. (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr. 26, 795–800 [Google Scholar]
- 19. Sheldrick G. M. (2002) Macromolecular phasing with SHELXE. Z. Kristallogr 217, 644–650 [Google Scholar]
- 20. Terwilliger T. C. (2000) Maximum-likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 56, 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Panjikar S., Parthasarathy V., Lamzin V. S., Weiss M. S., Tucker P. A. (2005) Auto-rickshaw: an automated crystal structure determination platform as an efficient tool for the validation of an X-ray diffraction experiment. Acta Crystallogr. D Biol. Crystallogr. 61, 449–457 [DOI] [PubMed] [Google Scholar]
- 22. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 23. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 24. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 25. Sauter N. K., Poon B. K. (2010) Autoindexing with outlier rejection and identification of superimposed lattices. J. Appl. Crystallogr. 43, 611–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Leslie A. G. W. (2006) The integration of macromolecular diffraction data. Acta Crystallogr. D Biol. Crystallogr. 62, 48–57 [DOI] [PubMed] [Google Scholar]
- 27. Potterton L., McNicholas S., Krissinel E., Gruber J., Cowtan K., Emsley P., Murshudov G. N., Cohen S., Perrakis A., Noble M. (2004) Developments in the CCP4 molecular-graphics project. Acta Crystallogr. D Biol. Crystallogr. 60, 2288–2294 [DOI] [PubMed] [Google Scholar]
- 28. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C., (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA, U.S.A [Google Scholar]
- 30. Jin Y., Penning T. M. (2007) Aldo-keto reductases and bioactivation/detoxication. Annu. Rev. Pharmacol. Toxicol. 47, 263–292 [DOI] [PubMed] [Google Scholar]
- 31. Richter N., Breicha K., Hummel W., Niefind K. (2010) The three-dimensional structure of AKR11B4, a glycerol dehydrogenase from Gluconobacter oxydans, reveals a tryptophan residue as an accelerator of reaction turnover. J. Mol. Biol. 404, 353–362 [DOI] [PubMed] [Google Scholar]
- 32. Olsen J. G., Pedersen L., Christensen C. L., Olsen O., Henriksen A. (2008) Barley aldose reductase: structure, cofactor binding, and substrate recognition in the aldo/keto reductase 4C family. Proteins 71, 1572–1581 [DOI] [PubMed] [Google Scholar]
- 33. Sanli G., Blaber M. (2001) Structural assembly of the active site in an aldo-keto reductase by NADPH cofactor. J. Mol. Biol. 309, 1209–1218 [DOI] [PubMed] [Google Scholar]
- 34. Simpson P. J., Tantitadapitak C., Reed A. M., Mather O. C., Bunce C. M., White S. A., Ride J. P. (2009) Characterization of two novel aldo-keto reductases from Arabidopsis: expression patterns, broad substrate specificity, and an open active-site structure suggest a role in toxicant metabolism following stress. J. Mol. Biol. 392, 465–480 [DOI] [PubMed] [Google Scholar]
- 35. Scoble J., McAlister A. D., Fulton Z., Troy S., Byres E., Vivian J. P., Brammananth R., Wilce M. C. J., Le Nours J., Zaker-Tabrizi L., Coppel R. L., Crellin P. K., Rossjohn J., Beddoe T. (2010) Crystal structure and comparative functional analyses of a Mycobacterium aldo-keto reductase. J. Mol. Biol. 398, 26–39 [DOI] [PubMed] [Google Scholar]
- 36. Sundaram K., Dhagat U., Endo S., Chung R., Matsunaga T., Hara A., El-Kabbani O. (2011) Structure of rat aldose reductase-like protein AKR1B14 holoenzyme: probing the role of His269 in coenzyme binding by site-directed mutagenesis. Bioorg. Med. Chem. Lett. 21, 801–804 [DOI] [PubMed] [Google Scholar]
- 37. Di Costanzo L., Drury J. E., Penning T. M., Christianson D. W. (2008) Crystal structure of human liver Δ4–3-ketosteroid 5β-reductase (AKR1D1) and implications for substrate binding and catalysis. J. Biol. Chem. 283, 16830–16839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bennett M. J., Schlegel B. P., Jez J. M., Penning T. M., Lewis M. (1996) Structure of 3α-hydroxysteroid/dihydrodiol dehydrogenase complexed with NADP+. Biochemistry 35, 10702–10711 [DOI] [PubMed] [Google Scholar]
- 39. Faucher F., Pereira de Jésus-Tran K., Cantin L., Luu-The V., Labrie F., Breton R. (2006) Crystal structures of mouse 17α-hydroxysteroid dehydrogenase (apoenzyme and enzyme-NADP(H) binary complex): identification of molecular determinants responsible for the unique 17α-reductive activity of this enzyme. J. Mol. Biol. 364, 747–763 [DOI] [PubMed] [Google Scholar]
- 40. Schlegel B. P., Jez J. M., Penning T. M. (1998) Mutagenesis of 3α-hydroxysteroid dehydrogenase reveals a “push-pull” mechanism for proton transfer in aldo-keto reductases. Biochemistry 37, 3538–3548 [DOI] [PubMed] [Google Scholar]
- 41. Stöckigt J., Barleben L., Panjikar S., Loris E. A. (2008) 3D-Structure and function of strictosidine synthase: the key enzyme of monoterpenoid indole alkaloid biosynthesis. Plant Physiol. Biochem. 46, 340–355 [DOI] [PubMed] [Google Scholar]
- 42. Hicks M. A., Barber A. E., 2nd, Giddings L. A., Caldwell J., O'Connor S. E., Babbitt P. C. (2011) The evolution of function in strictosidine synthase-like proteins. Proteins 79, 3082–3098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Saitou N., Nei M. (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 [DOI] [PubMed] [Google Scholar]
- 44. Felsenstein J. (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39, 783–791 [DOI] [PubMed] [Google Scholar]
- 45. Zuckerkandl E., Pauling L. (1965) “Evolutionary divergence and convergence in proteins,” in Evolving Genes and Proteins, pp. 97–166, Academic Press, New York [Google Scholar]
- 46. Tamura K., Dudley J., Nei M., Kumar S. (2007) MEGA4: molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.