Background: Neuropilin is an essential cell surface receptor for VEGF-A in angiogenesis.
Results: VEGF-A164 uniquely physically engages neuropilin using two distinct regions.
Conclusion: These data establish the structural basis for selective VEGF-A splice form binding to neuropilin.
Significance: Understanding VEGF receptor binding will advance therapeutic targeting of pathological angiogenesis.
Keywords: Angiogenesis, Neuropilin, Protein Structure, Vascular Endothelial Growth Factor (VEGF), X-ray Crystallography
Abstract
Neuropilin-1 (Nrp1) is an essential receptor for angiogenesis that binds to VEGF-A. Nrp1 binds directly to VEGF-A with high affinity, but the nature of their selective binding has remained unclear. Nrp1 was initially reported to bind to the exon 7-encoded region of VEGF-A and function as an isoform-specific receptor for VEGF-A164/165. Recent data have implicated exon 8-encoded residues, which are found in all proangiogenic VEGF-A isoforms, in Nrp binding. We have determined the crystal structure of the exon 7/8-encoded VEGF-A heparin binding domain in complex with the Nrp1-b1 domain. This structure clearly demonstrates that residues from both exons 7 and 8 physically contribute to Nrp1 binding. Using an in vitro binding assay, we have determined the relative contributions of exon 7- and 8-encoded residues. We demonstrate that the exon 8-encoded C-terminal arginine is essential for the interaction of VEGF-A with Nrp1 and mediates high affinity Nrp binding. Exon 7-encoded electronegative residues make additional interactions with the L1 loop of Nrp1. Although otherwise conserved, the primary sequences of Nrp1 and Nrp2 differ significantly in this region. We further show that VEGF-A164 binds 50-fold more strongly to Nrp1 than Nrp2. Direct repulsion between the electronegative exon 7-encoded residues of the heparin binding domain and the electronegative L1 loop found only in Nrp2 is found to significantly contribute to the observed selectivity. The results reveal the basis for the potent and selective binding of VEGF-A164 to Nrp1.
Introduction
Nrp12 is essential for VEGF-dependent angiogenesis (for review, see Ref. 1). The importance of Nrp1 function in vivo is illustrated by Nrp1-null mice, which show embryonic lethality due to cardiovascular defects (2). In angiogenesis, Nrp1 functions with the vascular endothelial growth factor receptor (VEGFR) family of receptor tyrosine kinases. Nrp1 is necessary for high affinity ligand binding to the cell surface and specifically promotes and stabilizes the active angiogenic signaling complex involving VEGF-A, VEGFR-2, and Nrp1 (for review, see Ref. 3).
The VEGF-A gene is encoded by nine exons. A cystine knot domain, encoded by exons 1–5, is retained in all VEGF-A isoforms. This domain is essential for signaling, mediating homodimerization, and direct interaction with VEGFR (4). Alternative splicing of the remaining introns produces VEGF-A molecules with varying activity, extracellular matrix binding, and diffusibility (5). It has long been recognized that the most potent stimulator of angiogenesis is VEGF-A164/165, named for the total number of amino acid residues in mouse and human proteins, respectively. VEGF-A164 possesses a heparin binding domain (HBD) encoded by exons 7 and 8 (6, 7). It has been demonstrated that Nrp1 binds to the VEGF-A HBD (8) via its b1 coagulation factor domain (9–11). However, the nature and extent of the interaction are not clear and have been the source of considerable study.
It was initially thought that exon 7-encoded residues represented the Nrp binding region of VEGF-A, thus explaining the significant differences in the biological potency of different VEGF-A isoforms (9). In contrast to VEGF-A164, VEGF-A120 differs by exclusion of exon 7. The clear biological role of exon 7-encoded residues in determining the ability of the cytokine to activate endothelial cells has been demonstrated in situ and in vivo (4, 12). However, subsequent studies demonstrated that the conserved exon 8-encoded C terminus of VEGF-A controls Nrp binding. All proangiogenic VEGF-A isoforms retain exon 8 whereas an inhibitory VEGF splice form, VEGF-A165b, replaces exon 8 with exon 9-encoded residues (13). Nrp binding was isolated to the C-terminal portion of the VEGF-A HBD, and a critical role was established for exon 8-encoded residues (14). Further, Tuftsin, an immunostimulatory peptide mimic of VEGF-A exon 8, was found to inhibit VEGF-A binding to Nrp1, although not affecting VEGF-A binding to VEGFR-2 (15). It was further shown that Nrp possesses a specific C-terminal arginine binding pocket located in the Nrp1-b1 domain (16). All known Nrp-binding proteins and peptides have been shown to posses a C-terminal arginine (16–19). Indeed, it was demonstrated convincingly that VEGF-A165 and VEGF-A121 both bind Nrp1, although with different kinetics and affinity (20). This has left the physical role of exon 7 in receptor binding unclear.
There are two Nrp genes in higher vertebrates, Nrp1 and Nrp2, which share 44% identity in their primary sequence and have the same overall domain organization (21). Although Nrp1 and Nrp2 are structurally related, they facilitate activation of functionally distinct pathways utilizing different members of the VEGF family. Nrp1 primarily mediates VEGF-A-dependent angiogenesis (9) whereas Nrp2 primarily mediates VEGF-C-dependent lymphangiogenesis (22). Nrps involvement in multiple physiological processes poses the unique challenge of isolating activation events to prevent inadvertent cross-talk. Differential ligand binding and temporal and tissue-specific expression are important regulatory mechanisms controlling Nrp function (for review, see Ref. 23). Differential ligand binding has been shown to be critical for the specific binding of the VEGF family members to different VEGFRs. For example, VEGF-A binds to VEGFR-1 and VEGFR-2, but with approximately 50-fold higher affinity for VEGFR-1 (24). Although VEGF binding to VEGFR has been characterized, the nature and basis for specific ligand binding to Nrp have not been determined.
To elucidate the molecular basis for the potent and specific binding of VEGF-A164 to Nrp, we have determined the structure of the VEGF-A HBD bound to Nrp1. This structure reveals an intermolecular interface with contributions from residues encoded by both exons 7 and 8. We characterize these interactions and show that the exon 8-encoded region determines high affinity interaction with Nrp1. The exon 7-encoded region is found to physically engage Nrp1 uniquely and contribute to binding. Strikingly, the exon 7-mediated interaction is shown to determine the selective binding of VEGF-A164 to Nrp1. These results define the unique physical mechanism underlying VEGF-A binding to Nrp.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Nrp1-b1b2, Nrp2-b1b2, and VEGF-A165 were expressed in Escherichia coli and purified using established methods (16, 19). Nrp point mutants were introduced using the megaprimer method. Nrp point mutants were not structurally deleterious as determined by circular dichroism (data not shown).
For crystallization, a fusion of human Nrp1-b1(274–429) linked to the VEGF-A HBD(115–165) with an intervening Sal1 restriction site and 3X(GS) linker was introduced into the Nde1/EcoR1 sites of pET28b (Novagen). Human and mouse HBD sequences differ by only a single residue in their N terminus, and mouse residue numbering is used throughout for clarity. The fusion protein was produced in Rosetta-gami 2 cells (Novagen). Cells were grown in terrific broth to A600 = 1.5, cold shocked for 15 min in ice, and then grown overnight at 16 °C producing protein that was soluble and correctly folded. Cells were lysed using lysozyme and sonication, and protein was purified by immobilized metal ion chromatography and eluted using buffer supplemented with 300 mm imidazole. The His tag was removed by incubation with 10 units of thrombin/mg of protein for 16 h at 23 °C (ZymoGenetics). Further purification was accomplished by heparin affinity chromatography using a 5-ml HiTrap Heparin HP column (GE Healthcare) with an elution gradient from 200 mm to 1 m NaCl.
Alkaline phosphatase (AP) fusion proteins were produced from pAPtag-5 (GenHunter, Nashville, TN) via large scale transient transfection of Chinese hamster ovary cells according to previously reported methods (19). VEGF-A point mutants expressed near wild-type levels and were secreted as correctly processed disulfide linked dimers as assessed by Western blotting (data not shown). All sequences were verified by DNA sequencing.
Structure Determination
Crystals of the Nrp-VEGF-A HBD fusion protein were produced using hanging-drop vapor-diffusion experiments. Purified protein was concentrated to 5 mg/ml and mixed in a 3:1 ratio of protein to mother liquor containing 1.5 m sodium/potassium phosphate, pH 6.5. Crystals formed in 1–2 weeks at 23 °C with two molecules in the asymmetric unit. Crystals were passed through mother liquor supplemented with 10% glycerol and flash frozen in liquid nitrogen. Diffraction data to 2.65 Å were collected at the SER-CAT ID-22 beamline of the Advanced Proton Source, Argonne National Laboratories (Table 1). Data were processed using HKL2000 (25). Crystal diffraction was somewhat anisotropic, a*, c* = 2.65 Å and b* = 3.1 Å (26).
TABLE 1.
Data collection and refinement statistics
| Data collection | |
| Beamline | APS 22-ID |
| Space group | P43 |
| Wavelength | 1.0000 |
| Unit-cell parameters | 114.97, 114.97, 50.94 |
| Unique reflections | 18,014 |
| Completeness (%) | 91.5 (55.4) |
| Resolution (Å) | 2.65 (2.74–2.65) |
| Rmerge (%) | 12.2 (53.9) |
| Redundancy | 4.3 (1.7) |
| I/σ(I) | 11.4 (2.02) |
| Refinement | |
| Resolution limits (Å) | 20.0–2.65 |
| No. reflections/no. to compute Rfree | 17,074/916 |
| R (Rfree) | 21.2 (26.7) |
| No. protein residues | 432 |
| No. solvent molecules | 19 |
| No. phosphate molecules | 1 |
| Root mean square deviation | |
| Bond, Å | 0.005 |
| Angle, ° | 1.01 |
| Protein geometry | |
| Ramachandran outlier/favored (%) | 0.0/96 |
| Residues with bad bonds/angles (%) | 0.0/0.0 |
| Rotamer outliers (%) | 0.26 |
An initial molecular replacement solution was obtained using PHASER with Nrp1-b1 (Protein Data Bank (PDB) code 2QQI) as the search model (27). Clear electron density for the VEGF-A HBD was observed and manually built. Iterative model building and refinement using COOT (28) and Refmac5 (29) produced a final refined model (Table 1). Protein geometry was analyzed using MolProbity (30), molecular graphics were prepared using PyMOL, and interaction interfaces were analyzed using the PISA interaction server. The atomic coordinates and structure factors have been deposited in the Protein Data Bank (PDB code 4DEQ).
Binding Assays
For binding experiments, conditioned medium of AP-tagged proteins was concentrated and buffer exchanged in binding buffer (20 mm Tris, pH 7.5, 50 mm NaCl). Dilutions were prepared in binding buffer, and protein was incubated in Nrp or VEGF-A affinity plates for 1 h at 25 °C. Wells were washed three times with PBS-T followed by an additional 5-min wash with PBS-T. PBS-T was aspirated, and 100 μl of 1× AP substrate (31) was added. Following evolution of p-nitrophenol, the reaction was quenched with 100 μl of 0.5 n NaOH, and retained AP activity was quantitatively measured at 405 nm using a SpectraMax M5 instrument (Molecular Devices, Sunnyvale, CA). All experiments were performed in triplicate. Results are reported as the mean ± 1 S.D., and statistical significance was calculated using Student's t test.
Nrp affinity plates were produced by adsorption of purified Nrp1-b1b2 or Nrp2-b1b2 proteins to high binding plates (Costar, 9018) with ∼500 ng of bound protein/well (19). For binding experiments, AP-tagged protein concentrations were normalized using AP activity. For quantitative binding experiments, the absolute concentration of AP-VEGF-A164 and AP-VEGF-A120 was determined using the quantitative MMV00 enzyme-linked immunosorbent assay (ELISA) (R&D Systems). Binding of AP-VEGF-A164 and AP-VEGF-A120 to Nrp1 or Nrp2 was measured as a function of retained AP activity. Binding curves were fit using a one-site specific binding mode to determine Kd (GraphPad Prism). Kd error is presented as the 95% confidence interval. VEGF-A affinity plates were produced by adsorption of purified VEGF-A165 protein to high binding plates using the protocol discussed above.
RESULTS
Physical Basis for VEGF-A and Nrp1 Binding
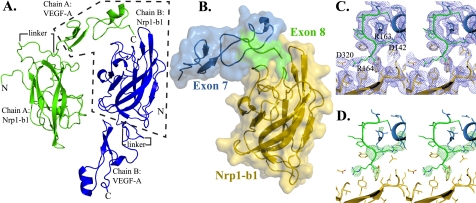
To understand the basis for Nrp binding of VEGF-A, we determined the crystal structure of the VEGF-A HBD in complex with the core ligand binding domain of Nrp1, domain b1. There were two molecules in the asymmetric unit, with intermolecular interactions between the VEGF-A HBD and Nrp-b1 (Fig. 1A). The VEGF-A HBD of chain A fully engages the Nrp1-b1 moiety of chain B and reveals an extended intermolecular interaction interface between VEGF-A and Nrp1 (gold) (Fig. 1B). The intermolecular interface is formed by both exon 7 (blue) and 8 (green)-encoded residues of the VEGF-A HBD (Fig. 1B). Analysis of the HBD–Nrp1-b1 complex reveals that the interface is predominantly hydrophilic in nature and is stabilized by a network of hydrogen bonds and salt bridges. The HBD retains the overall structural architecture determined previously (7), with a root mean square deviation of 2.0 Å (supplemental Fig. 1, overlaying residues 115–164). The orientation of the C-terminal peptide-like exon 8-encoded residues is significantly different from that observed in solution. This unique orientation is due to the presence of an intramolecular salt bridge formed between Asp-142 and Arg-163 as well as its direct association with Nrp1 (Fig. 1, C and D). The Nrp1-b1 domain shows no significant differences from previously determined structures with a root mean square deviation of 0.8 Å (PDB code 2QQI, overlaying residues 274–429).
FIGURE 1.
Crystal structure of VEGF-A HBD in complex with Nrp1. A, chain A (green) and chain B (blue) crystallized in an antiparallel fashion with the chain A VEGF-A HBD fully engaging the Nrp1-b1 domain of chain B, and that of chain B engaged by the symmetry related Nrp1-b1 domain of chain A. B, intermolecular complex enclosed in dashed box. A space-filling model revealing the specific interface with VEGF-A164 exons 7 and 8 (chain A) encoded residues with Nrp1 (chain B). C, stereo view of the 2Fo − Fc electron density map contoured at 1.0 σ of Nrp1 (gold) and exons 7 (blue) and 8 (green) of VEGF-A164. An intramolecular salt bridge between Asp-142 and Arg-163 of VEGF-A164 and an intermolecular salt bridge between Asp-320 of Nrp1 and Arg-164 of VEGF-A164 are observable. D, stereo view of the Fo − Fc omit electron density map for the HBD residues contoured at 3.0 σ.
Isoform-specific Binding of VEGF-A to Nrp1
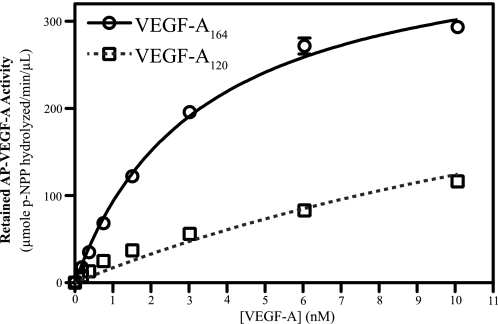
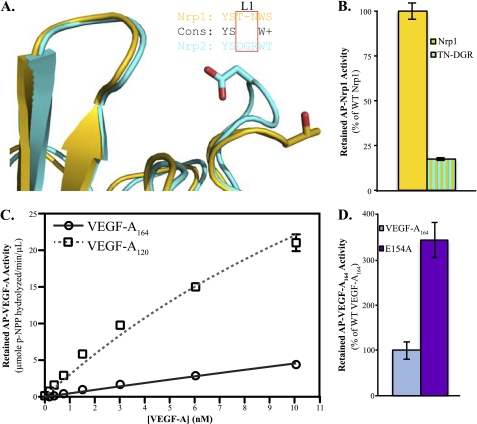
Differences in potency, functionality, and receptor binding have been reported for VEGF-A164 and VEGF-A120, which differ only in the inclusion of exon 7 in VEGF-A164. To delineate the different roles of these exons in Nrp binding, the dose-dependent binding of AP-VEGF-A164 and AP-VEGF-A120 to Nrp1 was analyzed (Fig. 2). VEGF-A164 (black line) bound Nrp1 with high affinity, Kd = 3.0 nm ± 0.2 nm. As expected for high density coupling of Nrp1, the observed binding is consistent with the reported tight cell surface binding (9). VEGF-A120 (gray dashed line) also bound Nrp1, but with lower affinity (Kd = 22 nm ± 1 nm). These data indicate that both exon 8-containing isoforms of VEGF-A are able to bind to Nrp1, but they differ in their affinity.
FIGURE 2.
VEGF-A164 binds to Nrp1 with high affinity. VEGF-A164 (black line) binds Nrp1 with a Kd = 3.0 nm ± 0.2 nm. VEGF-A120 (gray dashed line) binds Nrp1 with a Kd = 22 nm ± 1 nm.
Exon 8-encoded Residues Are Essential for High Affinity VEGF-A Binding to Nrp1
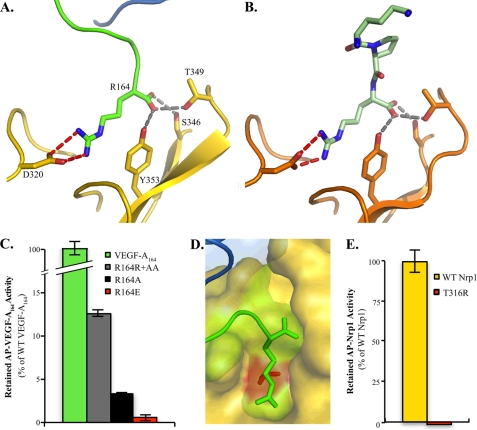
Close examination of the residues of exon 8 that interact with Nrp1 reveals that VEGF-A164 utilizes a C-terminal arginine (Arg-164) to bind Nrp1 (Fig. 3A). Analysis of the intermolecular interface in this region indicates that Arg-164 contributes a majority of the interaction with Nrp1. Although the electron density associated with the side chain indicates some disorder (Fig. 1, C and D), the binding mode is seen in both molecules in the asymmetric unit (supplemental Fig. 2) and is very similar to the previously determined structure with the inhibitory peptide Tuftsin (TKPR), a peptide mimic of VEGF-A exon 8 (Fig. 3B). Arg-164 is fully engaged by b1, burying 247 Å2 in the binding pocket. A salt bridge is formed with Asp-320, and the free C terminus forms hydrogen bonds with Ser-346, Thr-349, and Tyr-353 of the b1 domain (Fig. 3A).
FIGURE 3.
Exon 8-encoded C-terminal arginine of VEGF-A mediates high affinity Nrp1 binding. A, Arg-164 forms specific contacts with the b1 binding pocket of Nrp1. The guanidinium moiety forms a salt bridge with Asp-320 carboxylate oxygens (dashed red lines, 3.08 Å and 3.32 Å). The C terminus forms hydrogen bonds with three Nrp1-b1 residues (dashed gray lines, 3.08 Å, 2.95 Å, and 3.13 Å to Ser-346, Thr-349, and Tyr-353, respectively). B, Tuftsin binds to the Nrp1-b1 domain (PDB code 2ORZ) utilizing the same C-terminal arginine binding mode. C, mutagenesis demonstrates a specific role for the side chain (R164A, black bar) and C terminus (R164R + AA, gray bar) in Nrp1 binding. Charge reversal (R164E, red bar) completely abolishes binding to Nrp1. Statistical comparison of mean wild-type and mutant binding demonstrates significant differences, p < 0.0002, between all constructs. AP-tagged wild-type and mutant VEGF were used at an activity of 25 μmol of pNPP hydrolyzed/min/μl. D, surface representation of VEGF-A bound to Nrp1 reveals the critical location of Thr-316 (red) and illustrates the mechanism by which mutation to arginine would occlude binding. E, occlusion of the Nrp1 binding pocket in the Thr-Arg Nrp1 mutant (T316R, red bar) completely abolishes binding to VEGF-A164. AP-tagged wild-type and mutant Nrp1 were used at an activity of 1 μmol of pNPP hydrolyzed/min per μl. Error bars, S.D.
To characterize the contribution of the exon 8-encoded C-terminal arginine to binding, we analyzed site-directed mutants of both VEGF-A and Nrp1. To determine the role of the salt bridge, the C-terminal arginine was first mutated to alanine (Fig. 3C). Retention of R164A by Nrp1 (black bar) was reduced by 97% relative to WT VEGF-A164. To determine the role of the C-terminal hydrogen bond network, a VEGF-A164 construct with a C-terminal dialanine addition was generated. Retention of R164R + AA by Nrp1 (gray bar) was reduced by 87% relative to WT VEGF-A164. The observed contributions from both the side chain and C terminus emphasize the unique requirement for a C-terminal arginine in high affinity Nrp ligands. Lastly, a charge reversal was produced. R164E (red bar) showed no significant binding to Nrp1. These data demonstrate the essential role of the C-terminal arginine of VEGF.
To complement these results we examined the amount of VEGF-A164 binding retained when the Nrp1 binding pocket was occluded. Thr-316, which sits at the base of the Nrp1 binding pocket adjacent to Asp-320 (Fig. 3D), was mutated to arginine to generate a binding-deficient Nrp1 mutant. The binding of AP-Nrp1 or AP-T316R to VEGF-A164 was determined (Fig. 3E). Strikingly, occlusion of the Nrp1-b1 binding pocket in AP-T316R completely abolished binding with VEGF-A164. These data demonstrate the essential role of the Nrp1-b1 C-terminal arginine binding pocket in mediating high affinity VEGF-A binding.
Exon 7 Residues Directly Physically Engage the L1 Loop of Nrp1
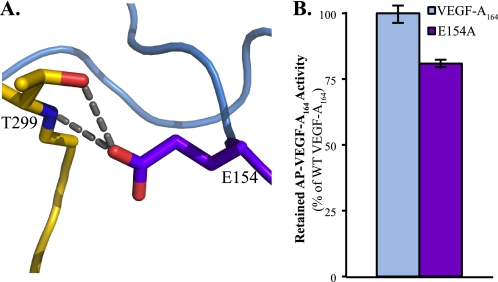
The enhanced affinity of VEGF-A164 for Nrp1 versus that of VEGF-A120 suggests a role for the exon 7-encoded residues in the interaction with Nrp1. The reported structure reveals that specific exon 7-encoded residues also engage Nrp1 directly. The interface with exon 7-encoded residues is more extended and involves Lys-146, Glu-151, and Glu-154. The residue with the largest interface contribution is Glu-154, with 67 Å2 buried surface area at the interface. The side chain of Glu-154 forms a hydrogen bond with both the backbone amide and side chain hydroxyl of Thr-299 in the Nrp1 L1 loop (Fig. 4A). The role of Glu-154 in Nrp binding was examined (Fig. 4B). An E154A (purple bar) mutant showed reduced Nrp1 binding, but the reduction was not as pronounced as that observed for Arg-164 mutants.
FIGURE 4.
Glu-154 of VEGF-A164 exon 7 contributes to Nrp1 binding. A, Glu-154 interacts with the side chain hydroxyl (bond distance = 2.73 Å) and backbone amide (bond distance = 3.16 Å) of Thr-299 of the Nrp1 L1 loop. B, mutation of Glu-154 to alanine (E154A, purple bar) reduces binding to Nrp1 (p < 0.0004). AP-tagged wild-type and mutant VEGF were used at an activity of 25 μmol of pNPP hydrolyzed/min per μl. Error bars, S.D.
Selective VEGF-A Binding to Nrp1
Nrp1 and Nrp2 b1 domains are structurally highly homologous (Fig. 5A). However, there are regions that are divergent. The L1 loops of Nrp1 and Nrp2 have distinct amino acid composition: the Nrp1 L1 loop Thr-299 is replaced by Asp-301 in the Nrp2 L1 loop. This replacement would be expected to result in electrostatic repulsion of Glu-154 of VEGF-A164. We generated a Nrp1 L1 loop chimeric mutant, replacing the L1 loop of Nrp1 (299TN300) with the Nrp2 loop (301DGR303) and assessed its ability to bind VEGF-A164 (Fig. 5B). This mutant shows a 75% reduction in its ability to bind VEGF-A164 (gold/cyan bar) relative to WT Nrp1 (gold bar). These data directly demonstrate that the VEGF-A164 Glu-154 interaction with the L1 loop of Nrp1 contributes to binding.
FIGURE 5.
Exon 7-encoded residues of VEGF-A164 are responsible for Nrp1 binding selectivity. A, superimposition of Nrp1 (PDB code 1KEX) and Nrp2 b1 domains (PDB code 2QQJ, residues 276–427) reveals a similar overall architecture, root mean square deviation 0.7Å, but unique amino acid composition of the L1 loop, with Nrp1-T299 and Nrp2-D301 highlighted in gold and cyan, respectively. B, chimeric Nrp1, containing the L1 loop of Nrp2 (gold/cyan), loses >75% binding to VEGF-A164 relative to WT Nrp1 (gold) (p < 0.000004). AP-tagged wild-type and mutant Nrp1 were used at an activity of 1 μmol of pNPP hydrolyzed/min per μl. C, VEGF-A120, which lacks exon 7, has essentially the same affinity for Nrp2 (gray dashed line, Kd = 23 nm ± 1 nm) as it does for Nrp1. VEGF-A164 retains exon 7 and has dramatically reduced affinity for Nrp2 (black line, Kd = 150 nm ± 4 nm) compared with Nrp1. D, Nrp2 shows 3-fold higher retention of E154A (purple bar) relative to WT VEGF-A164 (blue bar) (p < 0.0003). AP-tagged wild-type and mutant VEGF were used at an activity of 25 μmol of pNPP hydrolyzed/min per μl. Error bars, S.D.
As observed, substitution of the L1 loop of Nrp2 into Nrp1 dramatically reduces Nrp1 binding to VEGF-A164. This led us to consider whether the physical interaction we describe may be directly reflected in decreased affinity of VEGF-A164 for Nrp2. To test this, we assayed the binding of VEGF-A164 to Nrp2. In fact, VEGF-A164 shows dramatically weaker binding to Nrp2, with approximately 50-fold lower affinity (Fig. 5C, black line, Kd = 150 nm ± 4 nm) relative to Nrp1 (Fig. 2, black line, Kd = 3 nm). Our data suggest that the observed binding selectivity may be due to the exon 7-encoded residues. To test this, we compared the binding of VEGF-A120 to the two Nrp receptors. Indeed, binding of VEGF-A120 to Nrp2 is unchanged (Fig. 5C, gray dashed line, Kd = 23 nm ± 1 nm) from that of Nrp1 (Fig. 2, gray dashed line, Kd = 22 nm).
To confirm that the observed binding selectivity involves electrostatic repulsion between Glu-154 and Nrp2, we tested the binding of E154A to Nrp2 (Fig. 5D). Indeed, E154A (purple bar) shows 3-fold higher retention by Nrp2 relative to WT VEGF-A164 (blue).
DISCUSSION
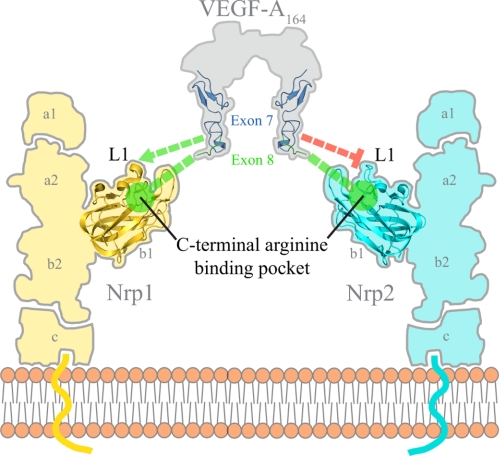
Together, these data demonstrate a mechanism for selective VEGF binding to Nrp. We report the first detailed picture of the structural basis for the binding of Nrp1 and VEGF-A. The interface is shown to involve regions encoded by both exons 7 and 8 and is found to determine splice form-specific receptor binding and selectivity (Fig. 6). Together with mutagenesis of key interfacial residues, our data define the specific contribution of different regions of VEGF-A. The exon 8-encoded C terminus of VEGF-A is confirmed to be necessary for high affinity Nrp binding. The C-terminal arginine of VEGF-A is shown to engage the Nrp1-b1 domain binding pocket utilized by all known ligands (16–19). A number of mutations to residues in the C-terminal arginine binding pocket have been reported (32). These mutations modulate binding to Nrp1 and show different signaling properties. We report here that T316R represents a true binding-deficient Nrp1 mutant that will be useful for future studies.
FIGURE 6.
HBD of VEGF-A is responsible for selective binding to the Nrp1 b1 domain. Exon 8-encoded residues mediate high affinity binding whereas exon 7-encoded residues primarily govern selectivity.
Exon 7-encoded residues are also found to interact physically with Nrp1 directly. In particular, we show that Glu-154 physically engages Thr-299 directly in the L1 loop of Nrp1, resulting in enhanced and selective Nrp1 binding. The importance of this interaction interface is further underlined by the recently reported VEGF blocking antibody, anti-Nrp1B (33). Surprisingly, this antibody was shown not to block the expected C-terminal arginine binding site of the b1 domain. Instead, it binds to residues in the nearby loops including Thr-299 of the Nrp1 L1 loop (34). These data reveal that anti-Nrp1B engages a binding site on Nrp1 that is shared with VEGF-A164, thus explaining its ability to potently inhibit VEGF-A binding to Nrp1.
It is interesting to note that the other VEGF family members that signal via Nrp1 also possess conserved electronegative residues at positions analogous to Glu-154: Asp-158 in VEGF-B167 and Glu-194 in placental growth factor 3 isoform-3. This suggests that electrostatic repulsion between VEGF family members and Nrp2 may be a general mechanism governing ligand binding selectivity.
Although the Nrp2 L1 loop chimera resulted in increased VEGF-A164 binding (data not shown), the observed binding is still significantly lower than that observed for wild-type Nrp1. Consistent with this observation, mutating Arg-287 and Asn-290 of Nrp2, which are immediately N-terminal to the L1 loop, has been reported to enhance the binding of VEGF-A to Nrp2 (35). Because the L1 loop of Nrp2 is highly conserved, it will also be interesting to explore whether there are distinct mechanisms that promote selective Nrp2 binding by its in vivo ligands VEGF-C and VEGF-D. Indeed, there may be Nrp-dependent physical mechanisms differentiating Nrp1-dependent angiogenesis and Nrp2-dependent lymphangiogenesis. This also suggests that design of specific Nrp inhibitors that exploit the difference in the L1 loop may be attainable.
Our data also suggest a physical basis for the observed functional differences of VEGF-A isoforms. VEGF-A164 is well documented as the most potent VEGF-A isoform in stimulating angiogenesis. It is well recognized that VEGFR dimerization, although necessary, is insufficient for activation. Indeed, a specific dimeric organization of the juxtamembrane domain of VEGFR-2 has been shown to be critical to couple ligand binding to intracellular receptor activation (36, 37). It is possible that the observed binding imposes specific steric constraints, allowing a stable organization of the heterohexameric VEGF-A–Nrp1–VEGFR-2 signaling complex. Indeed, the heparin-binding residues of the HBD (38) and Nrp1-b1 domain (16) are positioned spatially close together in the complex. This spatial orientation would allow binding of a single glucosaminoglycan/heparin chain by the protein complex and further reinforce the specific orientation of the signaling complex. Our data establish the unique physical engagement of VEGF-A164 by Nrp1 and open up new avenues to explore the specific physical mechanism of Nrp in angiogenesis.
Supplementary Material
Acknowledgments
We thank Drs. Chris Rife, Louis Hersh, David Rodgers, Matthew Gentry, and Hou-Fu Guo for valuable help and discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant R01GM094155 and Training Grant T32HL072743 (to M. W. P.). This work was also supported by the Kentucky Lung Cancer Research Program.

This article contains supplemental Figs. 1 and 2.
The atomic coordinates and structure factors (code 4DEQ) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- Nrp
- neuropilin
- AP
- alkaline phosphatase
- HBD
- heparin binding domain
- PDB
- Protein Data Bank
- pNPP
- p-nitrophenyl phosphate
- VEGFR
- vascular endothelial growth factor receptor.
REFERENCES
- 1. Pellet-Many C., Frankel P., Jia H., Zachary I. (2008) Neuropilins: structure, function and role in disease. Biochem. J. 411, 211–226 [DOI] [PubMed] [Google Scholar]
- 2. Kitsukawa T., Shimizu M., Sanbo M., Hirata T., Taniguchi M., Bekku Y., Yagi T., Fujisawa H. (1997) Neuropilin-semaphorin III/D-mediated chemorepulsive signals play a crucial role in peripheral nerve projection in mice. Neuron 19, 995–1005 [DOI] [PubMed] [Google Scholar]
- 3. Grünewald F. S., Prota A. E., Giese A., Ballmer-Hofer K. (2010) Structure-function analysis of VEGF receptor activation and the role of coreceptors in angiogenic signaling. Biochim. Biophys. Acta 1804, 567–580 [DOI] [PubMed] [Google Scholar]
- 4. Keyt B. A., Berleau L. T., Nguyen H. V., Chen H., Heinsohn H., Vandlen R., Ferrara N. (1996) The carboxyl-terminal domain(111–165) of vascular endothelial growth factor is critical for its mitogenic potency. J. Biol. Chem. 271, 7788–7795 [DOI] [PubMed] [Google Scholar]
- 5. Houck K. A., Leung D. W., Rowland A. M., Winer J., Ferrara N. (1992) Dual regulation of vascular endothelial growth factor bioavailability by genetic and proteolytic mechanisms. J. Biol. Chem. 267, 26031–26037 [PubMed] [Google Scholar]
- 6. Leung D. W., Cachianes G., Kuang W. J., Goeddel D. V., Ferrara N. (1989) Vascular endothelial growth factor is a secreted angiogenic mitogen. Science 246, 1306–1309 [DOI] [PubMed] [Google Scholar]
- 7. Fairbrother W. J., Champe M. A., Christinger H. W., Keyt B. A., Starovasnik M. A. (1998) Solution structure of the heparin-binding domain of vascular endothelial growth factor. Structure 6, 637–648 [DOI] [PubMed] [Google Scholar]
- 8. Soker S., Fidder H., Neufeld G., Klagsbrun M. (1996) Characterization of novel vascular endothelial growth factor (VEGF) receptors on tumor cells that bind VEGF165 via its exon 7-encoded domain. J. Biol. Chem. 271, 5761–5767 [DOI] [PubMed] [Google Scholar]
- 9. Soker S., Takashima S., Miao H. Q., Neufeld G., Klagsbrun M. (1998) Neuropilin-1 is expressed by endothelial and tumor cells as an isoform-specific receptor for vascular endothelial growth factor. Cell 92, 735–745 [DOI] [PubMed] [Google Scholar]
- 10. Gu C., Limberg B. J., Whitaker G. B., Perman B., Leahy D. J., Rosenbaum J. S., Ginty D. D., Kolodkin A. L. (2002) Characterization of neuropilin-1 structural features that confer binding to semaphorin 3A and vascular endothelial growth factor 165. J. Biol. Chem. 277, 18069–18076 [DOI] [PubMed] [Google Scholar]
- 11. Mamluk R., Gechtman Z., Kutcher M. E., Gasiunas N., Gallagher J., Klagsbrun M. (2002) Neuropilin-1 binds vascular endothelial growth factor 165, placenta growth factor-2, and heparin via its b1b2 domain. J. Biol. Chem. 277, 24818–24825 [DOI] [PubMed] [Google Scholar]
- 12. Soker S., Gollamudi-Payne S., Fidder H., Charmahelli H., Klagsbrun M. (1997) Inhibition of vascular endothelial growth factor (VEGF)-induced endothelial cell proliferation by a peptide corresponding to the exon 7-encoded domain of VEGF165. J. Biol. Chem. 272, 31582–31588 [DOI] [PubMed] [Google Scholar]
- 13. Bates D. O., Cui T. G., Doughty J. M., Winkler M., Sugiono M., Shields J. D., Peat D., Gillatt D., Harper S. J. (2002) VEGF165b, an inhibitory splice variant of vascular endothelial growth factor, is down-regulated in renal cell carcinoma. Cancer Res. 62, 4123–4131 [PubMed] [Google Scholar]
- 14. Jia H., Bagherzadeh A., Hartzoulakis B., Jarvis A., Löhr M., Shaikh S., Aqil R., Cheng L., Tickner M., Esposito D., Harris R., Driscoll P. C., Selwood D. L., Zachary I. C. (2006) Characterization of a bicyclic peptide neuropilin-1 (NP-1) antagonist (EG3287) reveals importance of vascular endothelial growth factor exon 8 for NP-1 binding and role of NP-1 in KDR signaling. J. Biol. Chem. 281, 13493–13502 [DOI] [PubMed] [Google Scholar]
- 15. von Wronski M. A., Raju N., Pillai R., Bogdan N. J., Marinelli E. R., Nanjappan P., Ramalingam K., Arunachalam T., Eaton S., Linder K. E., Yan F., Pochon S., Tweedle M. F., Nunn A. D. (2006) Tuftsin binds neuropilin-1 through a sequence similar to that encoded by exon 8 of vascular endothelial growth factor. J. Biol. Chem. 281, 5702–5710 [DOI] [PubMed] [Google Scholar]
- 16. Vander Kooi C. W., Jusino M. A., Perman B., Neau D. B., Bellamy H. D., Leahy D. J. (2007) Structural basis for ligand and heparin binding to neuropilin B domains. Proc. Natl. Acad. Sci. U.S.A. 104, 6152–6157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Starzec A., Ladam P., Vassy R., Badache S., Bouchemal N., Navaza A., du Penhoat C. H., Perret G. Y. (2007) Structure-function analysis of the antiangiogenic ATWLPPR peptide inhibiting VEGF(165) binding to neuropilin-1 and molecular dynamics simulations of the ATWLPPR/neuropilin-1 complex. Peptides 28, 2397–2402 [DOI] [PubMed] [Google Scholar]
- 18. Teesalu T., Sugahara K. N., Kotamraju V. R., Ruoslahti E. (2009) C-end rule peptides mediate neuropilin-1-dependent cell, vascular, and tissue penetration. Proc. Natl. Acad. Sci. U.S.A. 106, 16157–16162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Parker M. W., Hellman L. M., Xu P., Fried M. G., Vander Kooi C. W. (2010) Furin processing of semaphorin 3F determines its anti-angiogenic activity by regulating direct binding and competition for neuropilin. Biochemistry 49, 4068–4075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pan Q., Chathery Y., Wu Y., Rathore N., Tong R. K., Peale F., Bagri A., Tessier-Lavigne M., Koch A. W., Watts R. J. (2007) Neuropilin-1 binds to VEGF121 and regulates endothelial cell migration and sprouting. J. Biol. Chem. 282, 24049–24056 [DOI] [PubMed] [Google Scholar]
- 21. Kolodkin A. L., Levengood D. V., Rowe E. G., Tai Y. T., Giger R. J., Ginty D. D. (1997) Neuropilin is a semaphorin III receptor. Cell 90, 753–762 [DOI] [PubMed] [Google Scholar]
- 22. Karpanen T., Egeblad M., Karkkainen M. J., Kubo H., Ylä-Herttuala S., Jäättelä M., Alitalo K. (2001) Vascular endothelial growth factor C promotes tumor lymphangiogenesis and intralymphatic tumor growth. Cancer Res. 61, 1786–1790 [PubMed] [Google Scholar]
- 23. Staton C. A., Kumar I., Reed M. W., Brown N. J. (2007) Neuropilins in physiological and pathological angiogenesis. J. Pathol. 212, 237–248 [DOI] [PubMed] [Google Scholar]
- 24. Waltenberger J., Claesson-Welsh L., Siegbahn A., Shibuya M., Heldin C. H. (1994) Different signal transduction properties of KDR and Flt1, two receptors for vascular endothelial growth factor. J. Biol. Chem. 269, 26988–26995 [PubMed] [Google Scholar]
- 25. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 26. Strong M., Sawaya M. R., Wang S., Phillips M., Cascio D., Eisenberg D. (2006) Toward the structural genomics of complexes: crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 103, 8060–8065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of COOT. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Murshudov G. N. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 30. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Jardin B. A., Zhao Y., Selvaraj M., Montes J., Tran R., Prakash S., Elias C. B. (2008) Expression of SEAP (secreted alkaline phosphatase) by baculovirus-mediated transduction of HEK 293 cells in a hollow fiber bioreactor system. J. Biotechnol. 135, 272–280 [DOI] [PubMed] [Google Scholar]
- 32. Herzog B., Pellet-Many C., Britton G., Hartzoulakis B., Zachary I. C. (2011) VEGF binding to NRP1 is essential for VEGF stimulation of endothelial cell migration, complex formation between NRP1 and VEGFR2, and signaling via FAK Tyr-407 phosphorylation. Mol. Biol. Cell 22, 2766–2776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liang W. C., Dennis M. S., Stawicki S., Chanthery Y., Pan Q., Chen Y., Eigenbrot C., Yin J., Koch A. W., Wu X., Ferrara N., Bagri A., Tessier-Lavigne M., Watts R. J., Wu Y. (2007) Function blocking antibodies to neuropilin-1 generated from a designed human synthetic antibody phage library. J. Mol. Biol. 366, 815–829 [DOI] [PubMed] [Google Scholar]
- 34. Appleton B. A., Wu P., Maloney J., Yin J., Liang W. C., Stawicki S., Mortara K., Bowman K. K., Elliott J. M., Desmarais W., Bazan J. F., Bagri A., Tessier-Lavigne M., Koch A. W., Wu Y., Watts R. J., Wiesmann C. (2007) Structural studies of neuropilin/antibody complexes provide insights into semaphorin and VEGF binding. EMBO J. 26, 4902–4912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Geretti E., Shimizu A., Kurschat P., Klagsbrun M. (2007) Site-directed mutagenesis in the B-neuropilin-2 domain selectively enhances its affinity to VEGF165, but not to semaphorin 3F. J. Biol. Chem. 282, 25698–25707 [DOI] [PubMed] [Google Scholar]
- 36. Yang Y., Xie P., Opatowsky Y., Schlessinger J. (2010) Direct contacts between extracellular membrane-proximal domains are required for VEGF receptor activation and cell signaling. Proc. Natl. Acad. Sci. U.S.A. 107, 1906–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dosch D. D., Ballmer-Hofer K. (2010) Transmembrane domain-mediated orientation of receptor monomers in active VEGFR-2 dimers. FASEB J. 24, 32–38 [DOI] [PubMed] [Google Scholar]
- 38. Krilleke D., DeErkenez A., Schubert W., Giri I., Robinson G. S., Ng Y. S., Shima D. T. (2007) Molecular mapping and functional characterization of the VEGF164 heparin-binding domain. J. Biol. Chem. 282, 28045–28056 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.