Background: MT1-MMP is a membrane-anchored matrix metalloproteinase involved in pericellular proteolysis and invasion.
Results: MT1-MMP is monoubiquitinated intracellularly at Lys581, modulating intracellular trafficking and cellular invasion.
Conclusion: Regulation of the activity of MT1-MMP by monoubiquitination emphasizes the importance of its intracellular domain during MT1-MMP-driven cellular invasion.
Significance: Given the key role MT1-MMP plays in pathological conditions, a better understanding of the regulation of MT1-MMP is essential for therapeutic intervention.
Keywords: Cell Invasion, Intracellular Trafficking, Matrix Metalloproteinase (MMP), Molecular Cell Biology, Ubiquitin Ligase, Ubiquitination
Abstract
Membrane type 1 matrix metalloproteinase (MT1-MMP/MMP14) is a zinc-dependent type I transmembrane metalloproteinase playing pivotal roles in the regulation of pericellular proteolysis and cellular migration. Elevated expression levels of MT1-MMP have been demonstrated to correlate with a poor prognosis in cancer. MT1-MMP has a short intracellular domain (ICD) that has been shown to play important roles in cellular migration and invasion, although these ICD-mediated mechanisms remain poorly understood. In this study, we report that MT1-MMP is mono-ubiquitinated at its unique lysine residue (Lys581) within the ICD. Our data suggest that this post-translational modification is involved in MT1-MMP trafficking as well as in modulating cellular invasion through type I collagen matrices. By using an MT1-MMP Y573A mutant or the Src family inhibitor PP2, we observed that the previously described Src-dependent MT1-MMP phosphorylation is a prerequisite for ubiquitination. Taken together, these findings show for the first time an additional post-translational modification of MT1-MMP that regulates its trafficking and cellular invasion, which further emphasizes the key role of the MT1-MMP ICD.
Introduction
Matrix metalloproteinases (MMPs)3 are key regulators of normal physiological tissue homeostasis as well as pathologies that include cancer, cardiovascular disease, and arthritis (1–4). Among the MMP family, the membrane-anchored membrane-type 1 MMP (MT1-MMP, MMP14) is of particular interest. The importance of MT1-MMP is demonstrated by the unusually severe phenotype manifest on gene ablation in mice; MT1-MMP−/− mice show defects in vascularization, connective tissue turnover, and bone formation, leading to craniofacial abnormalities, dwarfism, osteopenia, and arthritis (5–7). MT1-MMP has a broad repertoire of substrates, including the extracellular matrix components type I, II, and III collagen, fibronectin, laminins, vitronectin, and aggrecan (8–11) as well as non-extracellular matrix proteins, which include CD44, syndecan-1, intracellular adhesion molecule-1 (ICAM-1), and connective tissue growth factor (CTGF) (12–15). MT1-MMP has been found to be highly expressed in different cancers, and its expression was shown to promote migration, invasion, and metastasis of cancer cells in vitro as well as in vivo (16–22).
MT1-MMP has a 20-amino acid intracellular domain (ICD) that is involved in a plethora of cellular functions that includes the regulation of cell migration, invasion, the induction of intracellular signaling pathways, and the intracellular traffic of the proteinase (23–29). MT1-MMP was shown to be endocytosed by a clathrin-dependent internalization pathway, although additional means of internalization have been described (25, 30, 31). It has been demonstrated that the ICD of MT1-MMP recruits the μ2 subunit of the adaptor protein 2 (AP-2) to its LLY573 motif (26). Interestingly, mutation of the LLY573 motif or deletion of the entire ICD resulted in decreased migration and invasion, suggesting a link between regulated endocytosis and cell locomotion (23, 26, 32). MT1-MMP also was shown to be recycled back to the cell surface in a mechanism dependent on the DKV582 motif (29). Although the mechanism of MT1-MMP recycling remains unclear, DKV582 was also suggested to bind to proteins containing PDZ (postsynaptic density-95/Discs large/zona occludens-1) domains, thus facilitating protein-protein interactions and signal transduction (33). The MT1-MMP ICD further undergoes palmitoylation at Cys574, a lipid post-translational modification that was shown to modulate cell migration and clathrin-mediated internalization (34). Recently, it has been reported that MT1-MMP is phosphorylated within its ICD at Tyr573 in an Src-dependent mechanism (35). Expression of a non-phosphorylable MT1-MMP Tyr573 mutant in fibrosarcoma cells resulted in impaired levels of proliferation within three-dimensional type-I collagen gels (36). Protein kinase C (PKC)-mediated phosphorylation at Thr567 has also been reported as being implicated in the regulation of collagenolytic activity (37).
In this work, we demonstrated that the MT1-MMP ICD is mono-ubiquitinated at Lys581. We were able to show that this modification is dependent on the Src-dependent phosphorylation of the Tyr573, thus changing the intracellular trafficking of the proteinase and increasing the ability of MT1-MMP to invade through collagen I gels.
EXPERIMENTAL PROCEDURES
Cell Culture, Cell Treatments, and Reagents
The human breast cancer cell line MCF-7 was obtained from Cancer Research UK (London, UK) and was routinely cultured in DMEM (Invitrogen) supplemented with 10% (v/v) FCS (Perbio, Northumberland, UK) and 2 mm glutamine (Invitrogen) (complete DMEM). Cells (5 × 105) were seeded and transfected with 1 μg of cDNA of various MT1-MMP cDNAs using FuGENE 6TM (Roche Applied Science, Welwyn, UK) according to the manufacturer's instructions. As indicated, cells were incubated with the phorbol esther phorbol 12-myristate 13-acetate (Sigma-Aldrich) at 100 ng/ml for 30 min. Inhibitors were used as follows: the Src family inhibitor PP2 (Calbiochem, Nottingham, UK) at 20 μm for 18 h and the proteasome inhibitor MG-132 (Calbiochem) at various concentrations for 3 h.
Cloning and Mutagenesis of cDNAs
Full-length MT1-MMP (MT1-WT), a Myc-tagged full-length MT1-MMP (MT1-MYC), MT1-Y573A, and the intracellular domain deleted MT1-MMP (MT1-ΔICD) mutant were described previously (23, 30, 35, 38–40). The MT1-K581R mutant was generated by site-directed mutagenesis as reported previously (39). MT1-MYC-Y573A, MT1-MYC-T567A, MT1-MYC-T567E, MT1-MYCΔICD, and MT1-MYC-K581R were generated by site-directed mutagenesis of MT1-MYC. Rab4-EGFP was a gift from M. Zerial (Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany). Rab6-EGFP was from B. Gould (Institut Pasteur, Paris, France) and Rab7-EGFP was from C. Bucci (Universita del Salento, Lecce, Italy). Rab11-EGFP was from M. Colombo (Universidad National de Cuyo, San Rafael, Argentina). HA-tagged ubiquitin (ubiquitin-HA) was from K. Fujita (University College, London, UK).
Antibodies
The sheep polyclonal antibody (pAb) to MT1-MMP (N175/6) was prepared as described by Remacle et al. (30). The anti-Myc (clone 4A6; immunoblot: 0.5 μg/ml), the anti-early endosome antigen 1 (EEA1; immunofluorescence, 5 μg/ml) and the anti-MT1-MMP (clone LEM-2/15.8; immunoblot, 0.5 μg/ml; FACS, 10 μg/ml; immunofluorescence, 5 μg/ml) monoclonal antibodies (mAbs) were purchased from Millipore (Watford, UK). Anti-HA (immunoblot, 0.2 μg/ml; immunofluorescence, 4 μg/ml), anti-β-actin (immunoblot, 0.2 μg/ml), anti-LAMP-1 (immunofluorescence, 10 μg/ml) and anti-NEDD4-1 (immunoblot, 0.2 μg/ml) pAbs were from Abcam (Cambridge, UK). The anti-Myc pAb (clone 9B11; immunofluorescence, 1 μg/ml) was from Cell Signaling Technology (Hitchin, UK) and the anti-p21/WAF1 pAb (immunoblot, 1 μg/ml) was from Santa Cruz Biotechnology (Heidelberg, Germany). Rabbit and mouse control IgG antibodies were purchased from Dako (Ely, UK). Horseradish peroxidase (HRP) and Cy5-conjugated secondary antibodies were from Jackson ImmunoResearch Laboratories (Newmarket, UK). Species-specific Alexa Fluor® 488 and allophycocyanine-conjugated anti-mouse secondary antibodies were purchased from Invitrogen.
Immunoprecipitation and Protein Immunoblotting
Immunoprecipitation was performed as described previously (41). Briefly, cells were lysed in immunoprecipitation buffer (10 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1% (v/v) Triton X-100, 0.5% (v/v) Nonidet P-40, 1 mm EDTA, 1 mm EGTA, 1 mm sodium vanadate). Antibodies (3 μg) were bound to Dynabeads protein G (Invitrogen) at 4 °C for 16 h in 5 mg/ml BSA in PBS (137 mm NaCl, 4.3 mm Na2HPO4, 2.7 mm KCl, 1.47 mm KH2PO4). Cell lysates were then incubated with antibody-bound Dynabeads for 90 min at 4 °C under constant rotation. Beads were washed 4 × 10 min with immunoprecipitation buffer and resuspended in 2× Laemmli buffer. Denatured proteins were separated on 10% SDS-PAGE and electrotransferred onto nitrocellulose membrane (GE Healthcare). Membranes were blocked in 3% (w/v) lowfat milk in TBS (136.9 mm NaCl, 2.68 mm KCl, 24.76 mm Tris base, pH 8.0) containing 0.1% (v/v) Tween 20 for 1 h at room temperature and probed with the primary antibodies diluted in blocking buffer at 4 °C for 16 h. After washing (3 × 5 min), membranes were incubated for 1 h at room temperature with HRP-conjugated secondary antibodies, and immunoreactive bands were detected by enhanced chemiluminescence (ECL detection kit, GE Healthcare). Band intensities were quantified using NIH ImageJ image processing software.
Immunocytochemistry
Cells (1 × 105) were seeded on glass coverslips and transfected as described previously. After 18 h, cells were washed in PBS and fixed at room temperature for 20 min with 4% (w/v) PFA in PBS. PFA was quenched using 50 mm glycine (pH 8.0). Cells were subsequently washed in PBS, permeabilized in PBS containing 0.1% (v/v) Triton X-100, and incubated with the primary antibody for 1 h at room temperature. Cells were washed 3 × 5 min with PBS and were incubated with species-specific Alexa Fluor® 488 or Cy5-conjugated secondary antibodies. Slides were mounted using ProLong® Gold antifade reagent containing DAPI (Invitrogen). Series of optical sections were acquired using the Leica Tandem SP5 confocal laser microscope (Leica Microsystems) with a 63× oil immersion objective. Co-localization coefficients were calculated using the Volocity three-dimensional imaging and analysis software (Improvision, PerkinElmer Life Sciences, Coventry, UK).
Flow Cytometry
MCF-7 cells (1.5 × 106) were washed with PBS and detached from the plastic using PBS containing 5 mm EDTA. After a 15-min room temperature fixation in PFA (4% (w/v) in 5 mm EDTA/PBS), cells were incubated with an anti-MT1-MMP mAb (LEM-2/15.8) in PBS for 1 h at room temperature. After three washes (each for 10 min), cells were incubated for 1 h at room temperature with 10 μg/ml allophycocyanine-conjugated anti-mouse secondary antibody. Cells were washed 3 × 10 min with PBS, sieved through a 70-μm filter (BD Biosciences) and analyzed on a FACSCalibur II flow cytometer (BD Biosciences). 104 events were acquired per sample, and the mean fluorescence intensity of four independent experiments was determined using FlowJo flow cytometric analysis software (version 8.8.4, Tree Star, Inc., Olten, Switzerland).
Cell Invasion Assay
Cellular invasion was assessed using a Transwell chamber assay. Millicell hanging tissue culture inserts (Millipore; 8-μm pore size) were assembled in 24-well plates and coated with 100 μl (100 μg/ml) type I collagen (BD Biosciences). The migration of 1 × 105 cells along a serum gradient was assayed. Following an 18-h incubation at 37 °C, the chambers were disassembled, and the side of the filter that was coated with type I collagen was scraped. The migrated cells within the filter were fixed for 20 min with 4% (w/v) PFA in PBS. PFA was quenched using 50 mm glycine (pH 8.0). Cells were washed subsequently in PBS, and filters were mounted using ProLong® Gold antifade reagent containing DAPI (Invitrogen). All cells within five different representative areas of each filter were counted visually. Three biological replicates were used per condition.
Collagen Degradation Assay
The assay was from Wolf et al. (42) and modified as follows. Briefly, MCF-7 cells (1 × 105) were co-polymerized with 50 μl (1.5 mg/ml) non-labeled rat tail type I collagen mixed with 2% FITC-conjugated bovine type I collagen (Exalpha, Biologicals, Inc.) per well of a 96-well plate. 50 μl of medium was added to each well after a 1-h incubation at 37 °C. Following another 40-h incubation at 37 °C, the medium of each well was pelleted by centrifugation at 16,000 × g for 15 min at 4 °C, and the fluorescence of the supernatant was determined at 490/520 nm using a Tecan Infinite M200 plate reader. 100% collagen degradation was determined by incubation with bacterial type IA collagenase from Clostridium histolyticum (0.01 collagen digestion unit/well; Sigma-Aldrich), whereas background values were measured by incubation of cell-free collagen. Five replicates were used per condition.
siRNA Transfection
Sets of four predesigned siRNA constructs directed against NEDD4-1 and Dharmafect 4 transfection reagent were obtained from Dharmacon/Thermo Scientific (Lafayette, CO). MCF-7 cells (5 × 105) were seeded per well of a six-well plate and incubated for 18 h at 37 °C. siRNAs were transfected into MCF-7 cells at a final concentration of 50 nm of each siRNA per well, using Dharmafect 4. Cells were incubated at 37 °C for 54 h, after which the media containing the siRNA mixture was removed and replaced with complete DMEM. The cells were then transfected with MT1-MMP mutant cDNA and ubiquitin-HA as described previously. 18 h after transfection, the cells were lysed and subjected to immunoprecipitation as described previously.
Statistical Analysis
Statistical analysis was performed using the GraphPad Prism® software (version 5, GraphPad Software, Inc., San Diego, CA). Statistical significance was assessed by one-way analysis of variance with a Student's Newman-Keuls post hoc test, unless indicated otherwise. All numerical values shown are the means ± S.E.
RESULTS
MT1-MMP Is Monoubiquitinated
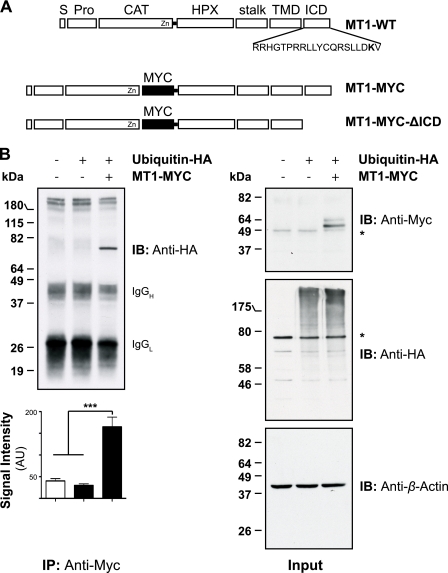
MT1-MMP has a 20-amino acid ICD with a single lysine residue proximal to its C-terminal end (Fig. 1A). Lys581 is part of the previously identified recycling motif DKV582, which is similar to the PDZ domain-binding motifs and is highly conserved among the MT-MMP subfamily (29). However, the precise mechanism underlying the MT1-MMP recycling process remains to be investigated. Because an intracellular lysine is a potential target for ubiquitination (43), we tested whether MT1-MMP undergoes ubiquitination. To assess this, a full-length MT1-MMP construct bearing a Myc tag within its hinge I domain was used (MT1-MYC; Fig. 1A). Initially, MT1-MYC was co-transfected with HA-tagged ubiquitin (ubiquitin-HA) into MCF-7 cells. Cells were lysed and immunoprecipitated with an anti-Myc antibody (clone 4A6, Millipore) and immunoblotted with an anti-HA antibody. As shown in Fig. 1B (top left panel), a confined band corresponding to a monoubiquitinated full-length MT1-MMP (∼72 kDa) was detected. Quantification of four representative immunoprecipitations revealed that significant ubiquitination of MT1-MMP occurred, compared with the controls (Fig. 1B, lower left panel). However, we were unable to detect MT1-MMP polyubiquitination. Input controls demonstrate the expression of MT1-MYC as well as the total increase of ubiquitinated proteins in the cell extracts following exogenous ubiquitin-HA expression (Fig. 1B, right panel).
FIGURE 1.
MT1-MMP is monoubiquitinated at Lys581 in MCF-7 cells. A, schematic representation of the Myc-tagged MT1-MMP cDNAs used. S, signal sequence; Pro, propeptide; CAT, catalytic domain; HPX, hemopexin domain; stalk, stalk region; TMD, transmembrane domain; MYC, N-EQKLISEEDL-C; MT1-WT, wild-type MT1-MMP; MT1-MYC, full-length MT1-MMP with a MYC tag fused into the hinge I region between the catalytic and hemopexin domain; MT1-MYC-ΔICD, MT1-MYC lacking the ICD. The Lys581 residue required for MT1-MMP monoubiquitaination is shown in boldface type. B, MCF-7 cells were transfected with pcDNA3.1, ubiquitin-HA, and MT1-MYC, as indicated, and cell extracts were immunoprecipitated with an anti-Myc antibody (clone 4A6). Immunoprecipitates were immunoblotted with an anti-HA antibody. Input controls were immunoblotted with an anti-Myc, an anti-HA, and an anti-β-actin antibody. The semiquantitative analyses of band intensities of four different immunoblots are shown ± S.E. with p < 0.0001 (***). *, nonspecific band. AU, arbitrary units. C, MCF-7 cells were transfected with pcDNA3.1, ubiquitin-HA, and MT1-MYC as indicated and incubated for 30 min with 10 or 100 μm of hydrogen peroxide. Cell extracts were immunoprecipitated with an anti-Myc antibody and detected with an anti-HA antibody. The input controls were probed with an anti-Myc, anti-HA, and an anti-β-actin antibody. An asterisk indicates detection of a nonspecific band. D, MCF-7 cells were transfected as indicated with either pcDNA3.1, ubiquitin-HA, MT1-MYC-K581R, MT1-MYC, or MT1-MYC-ΔICD. Cell extracts were immunoprecipitated with an anti-Myc antibody. Immunoprecipitates were immunoblotted with an anti-HA antibody. Input controls were immunoblotted with an anti-Myc, an anti-HA, and an anti-β-actin antibody. *, nonspecific band. IB, immunoblot. E, MCF-7 cells were co-transfected with either MT1-WT and ubiquitin-HA or MT1-K581R and ubiquitin-HA. 16 h after transfection, the cells were fixed, permeabilized, and stained with an anti-HA (Alexa Fluor® 488 secondary, green) and an anti-MT1-MMP (N175/6; Cy5 secondary, red) antibody. Inset panels show magnified portions of each merged image as indicated (dashed squares). Arrows indicate regions of MT1-WT and ubiquitin-HA co-localization, whereas open arrowheads point at individual staining of ubiquitin-HA and MT1-K581R. Bars represent a distance of 25 μm. The co-localization between MT1-WT and ubiquitin-HA or MT1-K581R and ubiquitin-HA was determined using the Volocity three-dimensional imaging and analysis software of single confocal optical sections. Data represent the co-localization co-efficients ± S.E. of six optical sections with p = 0.0001 (***).
To further characterize the ubiquitination of MT1-MMP, cells were transfected with MT1-MYC and ubiquitin-HA as described previously and treated with increasing concentrations of hydrogen peroxide, a known substrate-dependent inducer of mono- and/or polyubiquitination (44–46). Co-transfection of MT1-MYC and ubiquitin-HA led to MT1-MMP monoubiquitination as shown previously (Fig. 1, B and C). Addition of 100 μm H2O2 resulted in a significant induction of total ubiquitination as shown in the anti-HA input control blot as well as in an increase in MT1-MMP monoubiquitination in the immunoprecipitates (Fig. 1C). However, treatment of cells with 10 μm H2O2 led to less MT1-MMP ubiquitination compared with control cells, which may be explained by the reduced amount of total protein used for that immunoprecipitation as shown in the β-actin input control.
MT1-MMP Is Monoubiquitinated at Its Unique ICD Lys581
To further confirm that MT1-MMP is ubiquitinated at its unique lysine residue within the ICD, we generated an MT1-MMP ICD deletion construct (MT1-MYC-ΔICD, Fig. 1A) and an MT1-K581R mutant (MT1-MYC-K581R) within the MT1-MYC backbone. The constructs were co-transfected and co-immunoprecipitated with ubiquitin-HA as described previously (Fig. 1D). MT1-MYC was monoubiquitinated as shown previously (Fig. 1, B–D). However, no ubiquitin-HA was detected following transfection of the ICD deletion mutant or the MT1-MYC-K581R mutant, suggesting that the observed monoubiquitination of MT1-MMP depends on Lys581 (Fig. 1D).
We confirmed this observation by immunofluorescence microscopy. MCF-7 cells were co-transfected with MT1-WT and ubiquitin-HA, fixed, and immunostained using an anti-HA and an anti-MT1-MMP (N175/6) antibody (Fig. 1E). A strong co-localization was observed at or close to the plasma membrane of co-transfected MCF-7 cells (Fig. 1E, arrows within merged inset panel). However, when MCF-7 cells were transfected with ubiquitin-HA and the ubiquitination-deficient MT1-MMP construct (MT1-K581R), a reduced co-localization of both proteins was observed, which was also confirmed by quantification of co-localization (Fig. 1E, arrowheads within merged inset panel and quantification of co-localization). Remaining co-localization was still detected, indicating an overlap in the localization of both proteins although without direct interaction.
MT1-MMP Ubiquitination Is Regulated by Src-mediated Phosphorylation of MT1-MMP
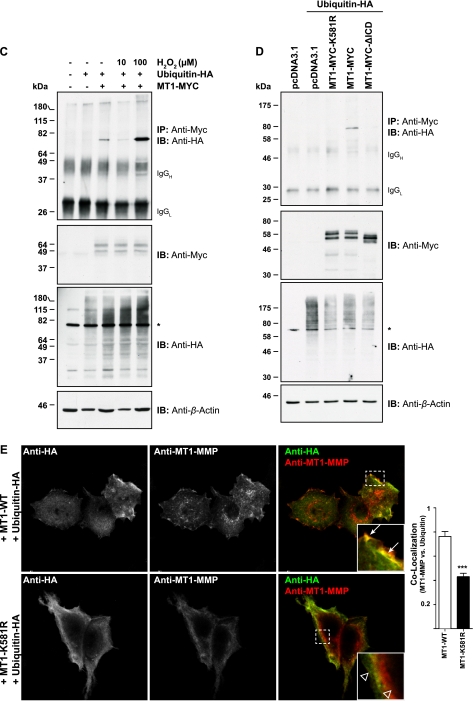
It has been demonstrated previously that MT1-MMP is phosphorylated at Tyr573 and Thr567 in a Src- or PKC-dependent manner (35, 37), which may be a prerequisite for protein ubiquitination (47). To test whether MT1-MMP phosphorylation affects its ubiquitination, MT1-MYC-Y573A and MT1-MYC-T567A mutants were used. We also generated the MT1-MYC-T567E mutant, mimicking constitutive threonine phosphorylation (48). Ubiquitin-HA was co-transfected with the MT1-MYC mutants as indicated, and lysates were co-immunoprecipitated with an anti-Myc antibody. As shown in Fig. 2A, MT1-MYC, MT1-MYC-T567A, and MT1-MYC-T567E were monoubiquitinated (Fig. 2A). However, MT1-MYC-Y573A displayed a significant reduction of ubiquitination compared with the other constructs used, as was also shown by quantification of three representative immunoprecipitations (Fig. 2A).
FIGURE 2.
MT1-MMP ubiquitination is dependent on Src-mediated phosphorylation of Tyr573 within the MT1-MMP ICD. A, MCF-7 cells were transfected as indicated with either pcDNA3.1, ubiquitin-HA, MT1-MYC, MT1-MYC-Y573A, MT1-MYC-T567A, or MT1-MYC-T567E. Cell extracts were immunoprecipitated with an anti-Myc antibody, and immunoprecipitates (IP) were immunoblotted with an anti-HA antibody. Input controls were detected with an anti-Myc, an anti-HA, and an anti-β-actin antibody. The semiquantitative analyses of band intensities of three different immunoblots are shown ± S.E. with p < 0.0001 (***) and p < 0.001 (**). ns, not significant. AU, arbitrary units. B, MCF-7 cells were transfected with ubiquitin-HA and MT1-MYC, and cells were treated for 18 h with 20 μm PP2 or for 30 min with 100 ng/ml phorbol 12-myristate 13-acetate. Cell extracts were immunoprecipitated with an anti-Myc antibody, and immunoprecipitates were immunoblotted with an anti-HA antibody. Input controls were detected with an anti-Myc, an anti-HA, or an anti-β-actin antibody. *, nonspecific band. DMSO, dimethyl sulfoxide; IB, immunoblot.
As mutation of the Tyr573 residue reduced MT1-MMP monoubiquitination, we tested further whether Src-mediated phosphorylation of this residue was required. Cells were co-transfected with ubiquitin-HA and MT1-MYC and treated with 20 μm of the Src family inhibitor PP2. As shown in Fig. 2B, inhibition of Src activity by PP2 significantly reduced the level of MT1-MMP monoubiquitination. In contrast, treatment of transfected cells with 100 ng/ml of the phorbol esther phorbol 12-myristate 13-acetate had little effect on the level of MT1-MMP ubiquitination (Fig. 2B), confirming that activation of PKC and subsequent phosphorylation of Thr567 within the MT1-MMP ICD (Fig. 2A) is not required for its ubiquitination.
NEDD4-1 Is E3 Ubiquitin Ligase Required for MT1-MMP Lys581 Monoubiquitination
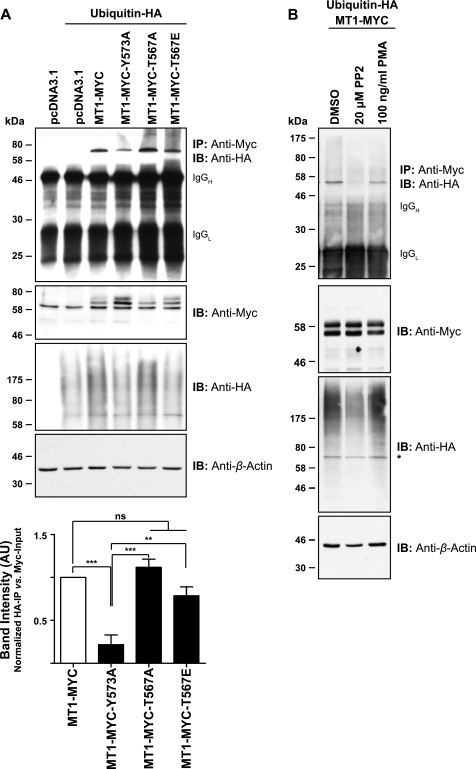
Tomari et al. (49) have recently identified the E3 ubiquitin-protein ligase NEDD4 (NEDD4-1) as an interaction partner of MT1-MMP ICD in a high throughput analysis in A375 cells. To test whether NEDD4-1 is regulating MT1-MMP monoubiquitination, we used siRNA-mediated transcriptional silencing of NEDD4-1. MCF-7 cells were treated with 50 nm siRNA targeting four different regions of the NEDD4-1 mRNA. Cells were transfected subsequently with MT1-MYC and ubiquitin-HA as indicated, and lysates were immunoprecipitated using an anti-Myc antibody. As shown in Fig. 3, NEDD4-1 protein expression could be silenced efficiently with all four siRNAs used. In NEDD4-1 silenced cells, no MT1-MYC monoubiquitination was observed, suggesting that NEDD4-1 ubiquitin E3 ligase is important for MT1-MMP monoubiquitination.
FIGURE 3.
Silencing of the ubiquitin E3 ligase NEDD4-1 reduces levels of MT1-MMP monoubiquitination. MCF-7 cells were transfected as indicated with siRNAs targeting four different regions of the NEDD4-1 coding region (siRNA 1–4) or a non-targeting siRNA (NT siRNA). Following 48 h, siRNA-transfected cells were co-transfected with ubiquitin-HA and MT1-MYC. Furthermore, MCF-7 cells were transfected with pcDNA3.1, ubiquitin-HA, and MT1-MYC, without prior siRNA treatment, and cultured for 24 h. Cell extracts were immunoprecipitated with an anti-Myc antibody, and immunoprecipitates were immunoblotted with an anti-HA antibody. Input controls were detected with an anti-Myc, an anti-HA, an anti-NEDD4-1, or an anti-β-actin antibody. *, nonspecific band; IB, immunoblot.
Ablation of MT1-MMP Ubiquitination Decreased Expression of Proteinase at Cell Surface
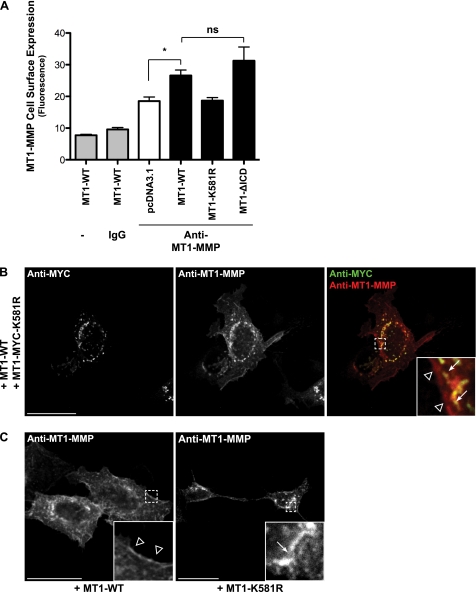
MT1-MMP cell surface expression has been demonstrated to be highly regulated, at least partly by controlled protein internalization (30, 31). Given the key role that monoubiquitination plays in protein endocytosis (50), we assessed the MT1-MMP cell surface levels of MT1-WT, MT1-K581R, and MT1-ΔICD by flow cytometry. Cells were transfected with the different MT1-MMP constructs, fixed, and stained with an anti-MT1-MMP mAb (LEM-2/15.8) directed against the catalytic domain. As controls, MT1-MMP-transfected cells either remained unstained or cells were stained with an anti-IgG control antibody (Fig. 4A). As expected, cells expressing MT1-WT showed a significantly enhanced level of MT1-MMP cell surface staining compared with the cells transfected with the vector control (pcDNA3.1; Fig. 4A). The MT1-MMP cell surface expression of MT1-ΔICD expressing cells was further increased, although not significantly, compared with MT1-WT transfectants. This confirmed previous observations (26). However, no difference in MT1-MMP cell surface staining was observed between the vector control and cells expressing MT1-K581R (Fig. 4A).
FIGURE 4.
Mutation of the MT1-MMP ubiquitination site at Lys581 decreases MT1-MMP cell surface expression. A, MCF-7 cells expressing pcDNA3.1, MT1-WT, MT1-K581R, or MT1-ΔICD were fixed, and MT1-MMP cell surface expression was detected by flow cytometry with an antibody against the MT1-MMP catalytic domain (LEM-2/15.8). Furthermore, MT1-WT expressing cells remained unstained as control. Data represent the mean fluorescence (n = 3 ± S.E.) with p < 0.05 (*). ns, not significant. B and C, MCF-7 cells were either co-transfected with MT1-WT and MT1-MYC-K581R (B) or MT1-WT and MT1-K581R were transfected individually (C). Cells were subsequently fixed, permeabilized, and immunostained with an anti-Myc (Alexa Fluor® 488 secondary, green) and an anti-MT1-MMP antibody (N175/6; Cy5 secondary, red) (B) or with an anti-MT1-MMP antibody (N175/6; Alexa Fluor 488® secondary, green) (C). Arrows point at intracellular staining of the MT1-K581R mutants, whereas arrowheads indicate staining at or close to the plasma membrane of MT1-WT. Bars represent a distance of 25 μm.
The lack of MT1-MMP-K581R expression at the cell surface was also demonstrated by immunofluorescence microscopy. Wild-type MT1-MMP and the ubiquitination-deficient Myc-tagged MT1-Lys581 mutant were co-transfected into MCF-7 cells, permeabilized, and subsequently co-stained with the anti-Myc and anti-MT1-MMP antibodies. Both constructs were detected with the anti-MT1-MMP antibody (N175/6, directed against the extracellular domain), whereas the anti-Myc antibody only detected the MT1-MYC-K581R mutant. As shown in Fig. 4B, the staining of both antibodies overlapped in the intracellular compartment (arrows), whereas only MT1-WT was detected at or close to the cell membrane (arrowheads), confirming the decreased cell surface localization of the MT1-K581R mutant as observed by flow cytometry. The same staining pattern was observed when cells were transfected either with MT1-WT or with MT1-K581R and stained with the anti-MT1-MMP antibody (N175/6), indicating that the different staining pattern shown in Fig. 4B was not an artifact of cDNA co-transfection (Fig. 4C).
Although proteasomal degradation of ubiquitinated proteins is linked mainly to Lys48 polyubiquitination (51), we tested whether inhibition of the proteasome affects MT1-MMP protein levels present in the cell. Therefore, MCF-7 cells were transfected with ubiquitin-HA and MT1-MYC, as indicated, and the cells were treated with increasing concentrations of MG-132 for 4 h. As shown in supplemental Fig. 1A, proteasome inhibition resulted in an increase in the amount of total ubiquitinated proteins, as well as in an enrichment of cyclin-dependent kinase inhibitor 1 (CDKN1A/CIP1/p21/WAF-1), an established marker of protein degradation by the proteasome (52). However, we observed a decrease of ubiquitinated MT1-MMP and no induction of MT1-MMP degradation upon inhibition of proteasome function, suggesting that MT1-MMP ubiquitination is not required for proteasomal degradation. Furthermore, no increase in cellular MT1-MMP levels was observed in MT1-K581R expressing cells compared with MT1-WT transfectants, indicating that MT1-MMP ubiquitination is not followed by lysosomal degradation (supplemental Fig. 1B).
MT1-MMP Monoubiquitination Modulates Trafficking and Cell Surface Recycling of Protease
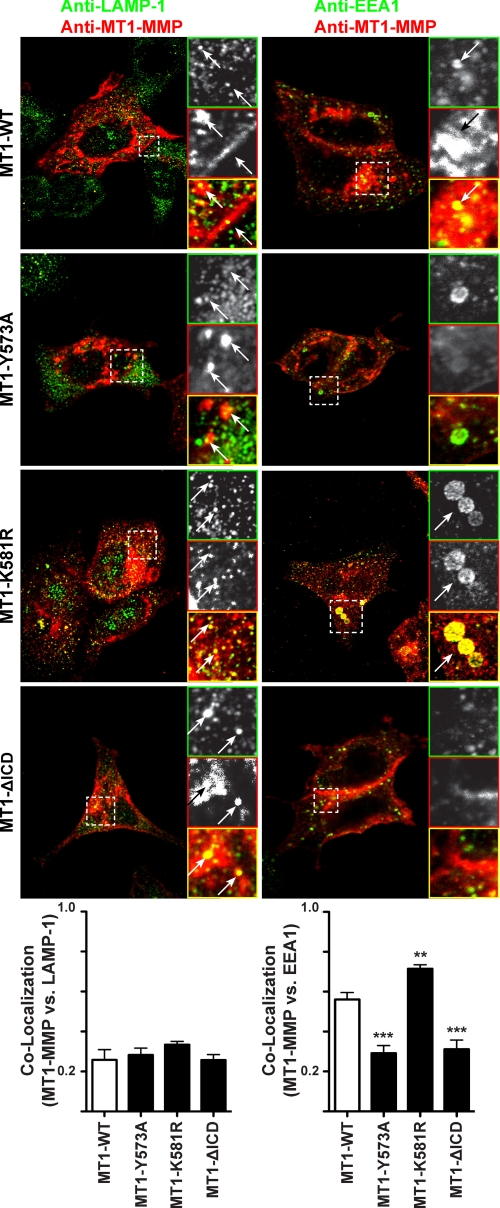
Given that MT1-MMP monoubiquitination was not found to enhance MT1-MMP cell surface localization or modulate protein degradation, we next tested how the K581R mutation affected trafficking of the proteinase. Therefore, MT1-WT, MT1-Y573A, MT1-K581R, or MT1-ΔICD were transfected into MCF-7 cells. The expression of MT1-MMP constructs and various markers of the endosomal sorting machinery were visualized by immunofluorescence. As shown in Fig. 5, MT1-WT and all MT1-MMP mutants partly co-localized with the endogenous lysosomal marker LAMP-1. Quantification of the co-localization coefficients revealed no significant differences in co-localization between MT1-MMP constructs and LAMP-1 (Fig. 5, lower left panel), confirming that MT1-MMP monoubiquitination is not followed by lysosomal degradation as shown in supplemental Fig. 1B. In contrast, MT1-WT clearly co-localized with the EEA1 as described previously (25), which was reduced upon mutation of the Tyr573 residue and deletion of the ICD (Fig. 5). These observations were expected because the Y573A mutation has been reported to inhibit endocytosis of MT1-MMP by 40% and thus is less present in early endosomes (26). Uekita et al. (26) also reported a decrease in MT1-MMP internalization when the ICD was deleted. In contrast, the MT1-K581R mutant exhibited a significantly stronger co-localization with EEA1 than MT1-WT. Rab4 (Ras-related protein 4)-EGFP and Rab11-EGFP have been co-transfected with the MT1-MMP mutants to assess the localization of MT1-MMP with respect to recycling endosomes. However, no difference in staining patterns could be observed (supplemental Fig. 2). The same results were obtained upon co-transfection with the Golgi marker Rab6-EGFP or the marker of the late endosomes Rab7-EGFP (supplemental Fig. 2). Taken together, these data demonstrate that the mutation of Lys581 within the MT1-MMP ICD disrupts the trafficking of the protease and leads to an accumulation of the proteinase in EEA1-positive early endosomes.
FIGURE 5.
Ablation of MT1-MMP ubiquitination increases the localization of MT1-MMP in EEA1-positive early endosomes. MCF-7 cells were transfected with MT1-WT, MT1-Y573A, MT1-K581R, or MT1-ΔICD. 16 h after transfection, the cells were fixed, permeabilized, and stained with an anti-LAMP1 or anti-EEA1 (Alexa Fluor® 488 secondary, green) and an anti-MT1-MMP (N175/6; Cy5 secondary, red) antibody. Inset panels show magnified portions of each merged image as indicated (dashed squares). Arrows indicate regions of MT1-MMP and LAMP-1 or MT1-MMP and EEA1 co-localization. Bars represent a distance of 10 μm. The co-localization of the different MT1-MMP constructs with LAMP-1 or EEA1 was determined using the Volocity three-dimensional imaging and analysis software of single confocal optical sections. Data represent the co-localization coefficients ± S.E. of six optical sections with p < 0.0001 (***) and p < 0.001 (**).
Mutation of MT1-MMP Ubiquitination Site Decreases Degradation of and Invasion through Type I Collagen Gels
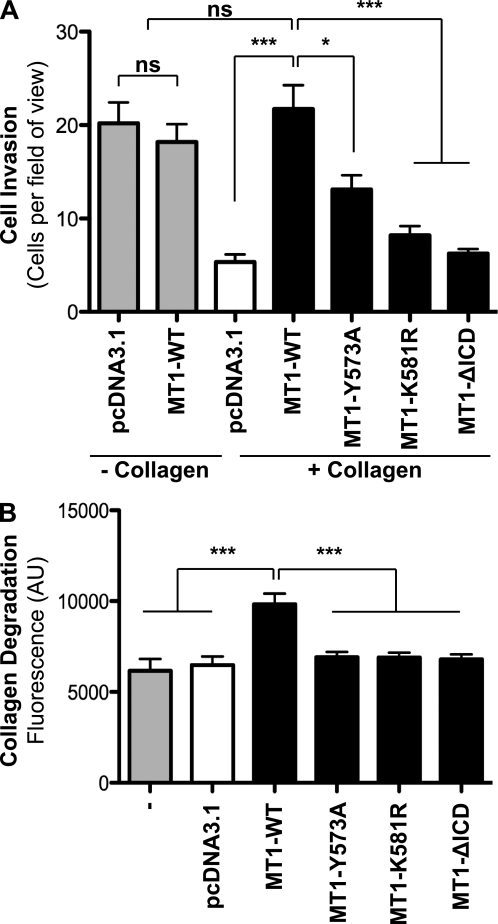
To test whether mutation of the MT1-MMP Lys581 residue affects the ability of MT1-MMP to invade through type I collagen, we used a modified Boyden chamber assay. Untransfected cells or cells expressing various MT1-MMP constructs were tested for their ability to migrate through uncoated filters (chemotaxis) or through type I collagen-coated filters (100 μl of 100 μg/ml) (chemoinvasion). MCF-7 cells expressing either pcDNA3.1, MT1-WT, MT1-Y573A, MT1-K581R, or MT1-ΔICD were seeded in serum-free medium in the top chamber. A serum gradient was applied by filling the lower chamber with medium containing 10% serum. The cellular invasiveness of MCF-7 cells expressing pcDNA3.1 vector control was reduced when transwells were coated with type I collagen (Fig. 6A). However, this effect was rescued upon expression of MT1-WT, demonstrating that the activity of MT1-MMP is sufficient for collagen degradation, as demonstrated previously (53, 54). Reduced invasion was observed for cells expressing MT1-Y573A, as described for invasion through fibronectin in HT1080 cells (35). Expression of MT1-ΔICD in MCF-7 cells resulted in a slight increase, although not significant, of invasion through collagen compared with vector control (Fig. 6A). Similarly, Uekita et al. reported a significant decrease in cellular invasion through Matrigel in MT1-ΔICD transfectants compared with MT1-WT expressing CHO-K1 cells (26). Expression of MT1-ΔICD has been shown to result in an increased cell surface expression level due to reduced protein internalization (Fig. 4A), and it has been proposed that a defect in MT1-MMP internalization perturbs the ability of the protein to induce cellular invasion (26). Expression of MT1-K581R in MCF-7 cells also led to a significant decrease of cell invasion through collagen compared with MT1-WT expressing cells (Fig. 6A), which may be explained by the decreased cell surface expression of this mutant (Fig. 4A). Accordingly, the ability of MT1-Y573A, MT1-K581R, and MT1-ΔICD to degrade FITC-labeled type I collagen was decreased compared with MT1-WT-expressing cells (Fig. 6B). These results are in agreement with the decreased expression of MT1-K581R at the cell surface of MCF-7 cells as detected by flow cytometry and immunofluorescence (Fig. 4, A–C).
FIGURE 6.
Ablation of MT1-MMP ubiquitination reduces cell invasion through type I collagen. A, MCF-7 cells were transfected with either pcDNA3.1, MT1-WT, MT1-Y573A, MT1-K581R, or MT1-ΔICD, and cell migration was quantified using a modified Boyden chamber assay. Data represent mean number of cells per field of view of three independent biological replicates with p < 0.0001 (***) and p < 0.01 (*). ns, not significant. B, wild-type MCF-7 cells or MCF-7 cells expressing pcDNA3.1, MT1-WT, MT1-Y573A, MT1-K581R, or MT1-ΔICD were co-polymerized with 1.5 mg/ml non-labeled rat tail type I collagen mixed with 2% FITC-conjugated bovine type I collagen. Following a 40-h incubation, the fluorescence of the supernatant was determined. Data represent mean fluorescence of five independent biological replicates with p < 0.0001 (***). AU, arbitrary units.
Given the variance between cellular invasion and degradation results often obtained in a two-dimensional versus a three-dimensional setting, we employed a three-dimensional spheroid assay to assess the role of the MT1-MMP Lys581 residue in type I collagen degradation and invasion in three dimensions. Hereby, untransfected MCF-7 cells or cells transfected with pcDNA3.1, MT1-WT, MT1-Y573A, MT1-K581R, or MT1-ΔICD were cultured to form spheroids as described in the supplemental “Experimental Procedures.” Following a 40-h incubation, the spheroids were fixed, and the area of invasion was measured. As expected, the area of the spheroids, representing the ability of the cells to degrade and invade through type I collagen, was enhanced significantly in MT1-WT expressing cells compared with the vector control or the MCF-7 wild-type cells (supplemental Fig. 3). The area of spheroid growth was decreased in MT1-Y573A expressing cells, similar to the results observed in the two-dimensional invasion assay (Fig. 4A). Interestingly, we detected that spheroids containing MT1-K581R- or MT1-ΔICD-expressing cells showed the same area of invasion as spheroids with MT1-WT-expressing cells (supplemental Fig. 3).
DISCUSSION
Recent evidence showed that the MT1-MMP ICD is pivotal for the MT1-MMP-driven cellular invasion in a number of tumor cell lines (24, 26, 55, 56) as well as in primary endothelial cells (57). ICD deletion mutants exhibit no effect on MT1-MMP cell surface proteolytic activity but reduce MT1-MMP-driven cell migration, thus establishing a link between the ICD and the proteolytic activity of the enzyme, its localization, as well as efficient cellular migration (24, 26, 56, 57). Given the numerous roles of the ICD in biological functions induced by MT1-MMP, the reduction in cell migration upon expression of an ICD deletion mutant may be related to perturbed endocytosis, reduced recycling or an disruption of the extracellular signal-regulated protein kinase (ERK) signaling cascade (56).
In this work, we demonstrate for the first time the ubiquitination of MT1-MMP at Lys581. In MT1-MYC tranfected MCF-7 cells, we detected a well defined signal corresponding to monoubiquitinated full-length MT1-MMP. By employing either an ICD deletion or a K581R mutant, we were able to confine the site of MT1-MMP ubiquitination to the Lys581 residue. An immunocytochemical approach supported these observations as a strong co-localization between wild-type MT1-MMP and ubiquitin-HA was observed, whereas this co-localization was reduced significantly upon expression of the MT1-K581R mutant.
Nyalendo and co-workers (35) have reported recently the phosphorylation of MT1-MMP by Src at the intracellular Tyr573 residue. Given the extensive cross-talk between post-translational modifications, especially the link between phosphorylation and subsequent ubiquitination (47), we found a consistent requirement for the Tyr573 residue as well as Src activity for MT1-MMP monoubiquitination. This observation has a profound impact, as we have shown that the presence of MT1-MMP in MCF-7 cells per se induces the activity of Src (41), thus inducing a positive feedback loop resulting in increased MT1-MMP phosphorylation, ubiquitination, and, hence, increased cell migration and invasion.
Ubiquitination is traditionally associated with targeting cargo proteins for (proteasomal) degradation (51); however, it has become increasingly apparent that ubiquitination has further non-proteolytic functions, including the regulation of trafficking and localization, thus modulating protein activity (58–62). MT1-MMP is known to relocalize to local invasive structures, including lammellipodia and podosomes, and it has been shown that this constant redistribution and adaptation during cellular invasion is dependent on a balanced interplay between protein endocytosis and recycling. MT1-MMP protein turnover depends on its ICD; the LLY573 motif has been shown to recruit the μ2 subunit of AP-2, thus facilitating clathrin-dependent endocytosis (23, 26). The recycling of MT1-MMP back to the cell surface is a process that is dependent on the C-terminal DKV582 motif (29). Here, we have shown that mutation of the Lys581 residue, and therefore the ablation of MT1-MMP monoubiquitination, led to decreased levels of the enzyme at the cell surface. An analysis of the localization of MT1-MMP and the ubiquitin-deficient mutant with respect to various markers of the trafficking and recycling machinery showed a significant increase in co-localization between the MT1-K581R mutant and EEA1. The observed reduction in cell surface levels and the increase in localization of MT1-MMP in early endosomes in cells expressing the ubiquitin-deficient mutant may be explained by a modulation of either protein recycling or exocytosis.
Although we have established the ubiquitination of MT1-MMP Lys581, it is essential to bear in mind that the Lys581 residue is part of the previously reported motif required for protein recycling (29). Wang et al. (29) have unequivocally shown that deletion of the entire ICD or truncation of the ICD at Leu579 (thus deleting the DKV582 motif) reduced MT1-MMP recycling to the cell surface, albeit showing a normal internalization process. MT1-MMP recycling was hypothesized to require the interaction of DKV582 with adaptor proteins containing PDZ motifs, as has been shown for the recycling of MT5-MMP depending on the adaptor protein Mint-3 (63). However, no mechanism regulating MT1-MMP recycling has been identified to date. The most pressing issue that needs to be addressed as a result of the present study is to test for the effect single mutations of the encompassing residues (Asp580 and Val582) induce on intracellular MT1-MMP localization.
The trafficking and intracellular compartmentalisation of MT1-MMP depends on the extracellular matrix surrounding the cell. It has been shown previously that MT1-MMP is recycled back to the cell surface in Rab4-positive recycling endosomes in HT1080 cells, in a setting where MT1-MMP is not involved in extracellular matrix degradation (30). However, when MDA-MB-231 cells were cultured in a three-dimensional type I collagen matrix, MT1-MMP was recruited from an intracellular storage compartment, different from the recycling endosomes, in Rab8-positive exocytic vesicles (64). Accordingly, we observed a difference in cellular invasion and migration in cells cultured in two dimensions versus three dimensions, with a reduction of cell invasion in the MT1-K581R expressing cells in two dimensions and no apparent change in spheroid outgrowth compared with MT1-WT expressing cells (supplemental Fig. 3). These differences observed were expected as the migratory capability of cells not only depends on the right localization of MT1-MMP at the cell surface but also on the substrate surrounding the cell. Furthermore, it has been demonstrated that only the migratory capabilities of the outer cellular layer of multicellular spheroids are measured in three-dimensional migration assays, introducing a bias in the quantification (65). Given that cells expressing MT1-K581R or MT1-WT exhibited similar migratory capabilities, we believe that MT1-MMP monoubiquitination might be required for protein recycling as being the most probable explanation. Indeed, how monoubiquitination is regulating the recycling of MT1-MMP back to the cell surface is a question we intend to address in future studies. A possible mechanism that induces recycling of ubiquitinated cargo is employed by the well characterized mechanism of epidermal growth factor receptor (EGFR) trafficking. EGFR is targeted for lysosomal degradation by ubiquitin-mediated interactions with the ESCRTs (endosomal-sorting complexes required for transport) machinery. However, Aoh et al. (66) have demonstrated previously that the tetraspanin integral membrane-protein SCAMP3 (secretory carrier membrane protein 3) is competing with members of the ESCRT family for binding of ubiquitinated cargo, thus increasing EGFR recycling to the cell surface. SCAMP proteins are present in the cell surface recycling system including the membranes of early, late, and recycling endosomes, in the trans-Golgi network and the plasma membrane (67), thus introducing an interesting mechanism that may explain MT1-MMP recycling depending on ubiquitination. Interestingly, in this study, we identified NEDD4-1 (neural precursor cell expressed developmentally down-regulated protein 4) as the E3 ubiquitin ligase that is required for MT1-MMP monoubiquitination. NEDD4-1 is a member of the WW domain containing HECT (homologous to E6-AP carboxyl terminus) E3 ubiquitin ligases (68) that has been shown to ubiquitinate various target genes, including EGFR (66, 69), vascular endothelial growth factor receptor-2 (VEGFR-2) (70) as well as components of the ESCRT system (71). Helliwell et al. (72) recently have reported that the trafficking of general amino acid permease GAP1 is regulated by NEDD4-1-induced ubiquitination. Therefore, monoubiquitination induces the trafficking of the protein to the cell surface. However, the exact mechanism of NEDD4-1 induced MT1-MMP monoubiquitination in the intracellular trafficking of the proteinase remains to be investigated.
In conclusion, we have shown that MT1-MMP is monoubiquitinated at its unique lysine residue within its ICD, thus modulating protein recycling and subsequently cellular invasion and migration. The work presented here also offers a potential explanation for the question of how MT1-MMP recycling is regulated and what the role of the DKV582 motif is during that process.
Supplementary Material
Acknowledgments
We thank the Cancer Research UK Cambridge Research Institute core facilities for advice and assistance. We are also grateful to Julia Freede and Helen Gillingham (University of Cambridge) for technical support and to K. Fujita (University College, London, UK) for providing reagents.
This work was supported by Cancer Research UK and Hutchison Whampoa Ltd. (to P. A. E., C. R., and G. M.), European Union Framework Programme 6, LSHC-CT-2003-503297 (to G. M. and P. A. E.), the German National Academic Foundation (Studienstiftung des DeutschenVolkes) (to P. A. E.), and the Portuguese Foundation for Science and Technology (Fundaçãopara a Ciência e Tecnologia) (to P. C. d. S.).

This article contains supplemental “Experimental Procedures” and Figs. 1–3.
- MMP
- matrix metalloproteinase
- EEA1
- early endosome antigen 1
- LAMP1
- lysosome-associated membraneglycoprotein 1
- MT1-MMP
- membrane type-1 MMP
- ICD
- intracellular domain
- EGFP
- enhanced GFP
- EGFR
- epidermal growth factor receptor
- PFA
- paraformaldehyde.
REFERENCES
- 1. Egeblad M., Werb Z. (2002) New functions for the matrix metalloproteinases in cancer progression. Nat. Rev. Cancer 2, 161–174 [DOI] [PubMed] [Google Scholar]
- 2. Janssens S., Lijnen H. R. (2006) What has been learned about the cardiovascular effects of matrix metalloproteinases from mouse models? Cardiovasc. Res. 69, 585–594 [DOI] [PubMed] [Google Scholar]
- 3. Overall C. M., Kleifeld O. (2006) Tumor microenvironment–opinion: Validating matrix metalloproteinases as drug targets and anti-targets for cancer therapy. Nat. Rev. Cancer 6, 227–239 [DOI] [PubMed] [Google Scholar]
- 4. Burrage P. S., Mix K. S., Brinckerhoff C. E. (2006) Matrix metalloproteinases: Role in arthritis. Front. Biosci. 11, 529–543 [DOI] [PubMed] [Google Scholar]
- 5. Holmbeck K., Bianco P., Caterina J., Yamada S., Kromer M., Kuznetsov S. A., Mankani M., Robey P. G., Poole A. R., Pidoux I., Ward J. M., Birkedal-Hansen H. (1999) MT1-MMP-deficient mice develop dwarfism, osteopenia, arthritis, and connective tissue disease due to inadequate collagen turnover. Cell 99, 81–92 [DOI] [PubMed] [Google Scholar]
- 6. Zhou Z., Apte S. S., Soininen R., Cao R., Baaklini G. Y., Rauser R. W., Wang J., Cao Y., Tryggvason K. (2000) Impaired endochondral ossification and angiogenesis in mice deficient in membrane-type matrix metalloproteinase I. Proc. Natl. Acad. Sci. U.S.A. 97, 4052–4057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lehti K., Allen E., Birkedal-Hansen H., Holmbeck K., Miyake Y., Chun T. H., Weiss S. J. (2005) An MT1-MMP-PDGF receptor-β axis regulates mural cell investment of the microvasculature. Genes Dev. 19, 979–991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. d'Ortho M. P., Will H., Atkinson S., Butler G., Messent A., Gavrilovic J., Smith B., Timpl R., Zardi L., Murphy G. (1997) Membrane-type matrix metalloproteinases 1 and 2 exhibit broad-spectrum proteolytic capacities comparable to many matrix metalloproteinases. Eur. J. Biochem. 250, 751–757 [DOI] [PubMed] [Google Scholar]
- 9. Ohuchi E., Imai K., Fujii Y., Sato H., Seiki M., Okada Y. (1997) Membrane type 1 matrix metalloproteinase digests interstitial collagens and other extracellular matrix macromolecules. J. Biol. Chem. 272, 2446–2451 [DOI] [PubMed] [Google Scholar]
- 10. Fosang A. J., Last K., Fujii Y., Seiki M., Okada Y. (1998) Membrane-type 1 MMP (MMP-14) cleaves at three sites in the aggrecan interglobular domain. FEBS Lett. 430, 186–190 [DOI] [PubMed] [Google Scholar]
- 11. Koshikawa N., Giannelli G., Cirulli V., Miyazaki K., Quaranta V. (2000) Role of cell surface metalloprotease MT1-MMP in epithelial cell migration over laminin-5. J. Cell Biol. 148, 615–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sithu S. D., English W. R., Olson P., Krubasik D., Baker A. H., Murphy G., D'Souza S. E. (2007) Membrane-type 1-matrix metalloproteinase regulates intracellular adhesion molecule-1 (ICAM-1)-mediated monocyte transmigration. J. Biol. Chem. 282, 25010–25019 [DOI] [PubMed] [Google Scholar]
- 13. Kajita M., Itoh Y., Chiba T., Mori H., Okada A., Kinoh H., Seiki M. (2001) Membrane-type 1 matrix metalloproteinase cleaves CD44 and promotes cell migration. J. Cell Biol. 153, 893–904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Endo K., Takino T., Miyamori H., Kinsen H., Yoshizaki T., Furukawa M., Sato H. (2003) Cleavage of syndecan-1 by membrane type matrix metalloproteinase-1 stimulates cell migration. J. Biol. Chem. 278, 40764–40770 [DOI] [PubMed] [Google Scholar]
- 15. Tam E. M., Morrison C. J., Wu Y. I., Stack M. S., Overall C. M. (2004) Membrane protease proteomics: Isotope-coded affinity tag MS identification of undescribed MT1-matrix metalloproteinase substrates. Proc. Natl. Acad. Sci. U.S.A. 101, 6917–6922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang W., Matrisian L. M., Holmbeck K., Vick C. C., Rosenthal E. L. (2006) Fibroblast-derived MT1-MMP promotes tumor progression in vitro and in vivo. BMC Cancer 6, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nomura H., Sato H., Seiki M., Mai M., Okada Y. (1995) Expression of membrane-type matrix metalloproteinase in human gastric carcinomas. Cancer Res. 55, 3263–3266 [PubMed] [Google Scholar]
- 18. Ueno H., Nakamura H., Inoue M., Imai K., Noguchi M., Sato H., Seiki M., Okada Y. (1997) Expression and tissue localization of membrane types 1, 2, and 3 matrix metalloproteinases in human invasive breast carcinomas. Cancer Res. 57, 2055–2060 [PubMed] [Google Scholar]
- 19. Tokuraku M., Sato H., Murakami S., Okada Y., Watanabe Y., Seiki M. (1995) Activation of the precursor of gelatinase A/72-kDa type IV collagenase/MMP-2 in lung carcinomas correlates with the expression of membrane-type matrix metalloproteinase (MT-MMP) and with lymph node metastasis. Int. J. Cancer 64, 355–359 [DOI] [PubMed] [Google Scholar]
- 20. Yoshizaki T., Sato H., Maruyama Y., Murono S., Furukawa M., Park C. S., Seiki M. (1997) Increased expression of membrane type 1-matrix metalloproteinase in head and neck carcinoma. Cancer 79, 139–144 [DOI] [PubMed] [Google Scholar]
- 21. Soulié P., Carrozzino F., Pepper M. S., Strongin A. Y., Poupon M. F., Montesano R. (2005) Membrane-type-1 matrix metalloproteinase confers tumorigenicity on nonmalignant epithelial cells. Oncogene 24, 1689–1697 [DOI] [PubMed] [Google Scholar]
- 22. Sounni N. E., Devy L., Hajitou A., Frankenne F., Munaut C., Gilles C., Deroanne C., Thompson E. W., Foidart J. M., Noel A. (2002) MT1-MMP expression promotes tumor growth and angiogenesis through an up-regulation of vascular endothelial growth factor expression. FASEB J. 16, 555–564 [DOI] [PubMed] [Google Scholar]
- 23. Roghi C., Jones L., Gratian M., English W. R., Murphy G. (2010) Golgi reassembly stacking protein 55 interacts with membrane-type (MT) 1-matrix metalloprotease (MMP) and furin and plays a role in the activation of the MT1-MMP zymogen. FEBS J. 277, 3158–3175 [DOI] [PubMed] [Google Scholar]
- 24. Lehti K., Valtanen H., Wickström S. A., Lohi J., Keski-Oja J. (2000) Regulation of membrane-type-1 matrix metalloproteinase activity by its cytoplasmic domain. J. Biol. Chem. 275, 15006–15013 [DOI] [PubMed] [Google Scholar]
- 25. Jiang A., Lehti K., Wang X., Weiss S. J., Keski-Oja J., Pei D. (2001) Regulation of membrane-type matrix metalloproteinase 1 activity by dynamin-mediated endocytosis. Proc. Natl. Acad. Sci. U.S.A. 98, 13693–13698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Uekita T., Itoh Y., Yana I., Ohno H., Seiki M. (2001) Cytoplasmic tail-dependent internalization of membrane-type 1 matrix metalloproteinase is important for its invasion-promoting activity. J. Cell Biol. 155, 1345–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nakahara H., Howard L., Thompson E. W., Sato H., Seiki M., Yeh Y., Chen W. T. (1997) Transmembrane/cytoplasmic domain-mediated membrane type 1-matrix metalloprotease docking to invadopodia is required for cell invasion. Proc. Natl. Acad. Sci. U.S.A. 94, 7959–7964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rozanov D. V., Ghebrehiwet B., Ratnikov B., Monosov E. Z., Deryugina E. I., Strongin A. Y. (2002) The cytoplasmic tail peptide sequence of membrane type-1 matrix metalloproteinase (MT1-MMP) directly binds to gC1qR, a compartment-specific chaperone-like regulatory protein. FEBS Lett. 527, 51–57 [DOI] [PubMed] [Google Scholar]
- 29. Wang X., Ma D., Keski-Oja J., Pei D. (2004) Co-recycling of MT1-MMP and MT3-MMP through the trans-Golgi network. Identification of DKV582 as a recycling signal. J. Biol. Chem. 279, 9331–9336 [DOI] [PubMed] [Google Scholar]
- 30. Remacle A., Murphy G., Roghi C. (2003) Membrane type I-matrix metalloproteinase (MT1-MMP) is internalized by two different pathways and is recycled to the cell surface. J. Cell Sci. 116, 3905–3916 [DOI] [PubMed] [Google Scholar]
- 31. Gálvez B. G., Matías-Román S., Yáñez-Mó M., Vicente-Manzanares M., Sánchez-Madrid F., Arroyo A. G. (2004) Caveolae are a novel pathway for membrane-type 1 matrix metalloproteinase traffic in human endothelial cells. Mol. Biol. Cell 15, 678–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wickramasinghe R. D., Ko Ferrigno P., Roghi C. (2010) Peptide aptamers as new tools to modulate clathrin-mediated internalization–inhibition of MT1-MMP internalization. BMC Cell Biol. 11, 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stricker N. L., Christopherson K. S., Yi B. A., Schatz P. J., Raab R. W., Dawes G., Bassett D. E., Jr., Bredt D. S., Li M. (1997) PDZ domain of neuronal nitric oxide synthase recognizes novel C-terminal peptide sequences. Nat. Biotechnol. 15, 336–342 [DOI] [PubMed] [Google Scholar]
- 34. Anilkumar N., Uekita T., Couchman J. R., Nagase H., Seiki M., Itoh Y. (2005) Palmitoylation at Cys574 is essential for MT1-MMP to promote cell migration. FASEB J. 19, 1326–1328 [DOI] [PubMed] [Google Scholar]
- 35. Nyalendo C., Michaud M., Beaulieu E., Roghi C., Murphy G., Gingras D., Béliveau R. (2007) Src-dependent phosphorylation of membrane type I matrix metalloproteinase on cytoplasmic tyrosine 573: Role in endothelial and tumor cell migration. J. Biol. Chem. 282, 15690–15699 [DOI] [PubMed] [Google Scholar]
- 36. Nyalendo C., Beaulieu E., Sartelet H., Michaud M., Fontaine N., Gingras D., Béliveau R. (2008) Impaired tyrosine phosphorylation of membrane type 1-matrix metalloproteinase reduces tumor cell proliferation in three-dimensional matrices and abrogates tumor growth in mice. Carcinogenesis 29, 1655–1664 [DOI] [PubMed] [Google Scholar]
- 37. Moss N. M., Wu Y. I., Liu Y., Munshi H. G., Stack M. S. (2009) Modulation of the membrane type 1 matrix metalloproteinase cytoplasmic tail enhances tumor cell invasion and proliferation in three-dimensional collagen matrices. J. Biol. Chem. 284, 19791–19799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Atkinson S. J., Roghi C., Murphy G. (2006) MT1-MMP hemopexin domain exchange with MT4-MMP blocks enzyme maturation and trafficking to the plasma membrane in MCF7 cells. Biochem. J. 398, 15–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Labrecque L., Nyalendo C., Langlois S., Durocher Y., Roghi C., Murphy G., Gingras D., Béliveau R. (2004) Src-mediated tyrosine phosphorylation of caveolin-1 induces its association with membrane type 1 matrix metalloproteinase. J. Biol. Chem. 279, 52132–52140 [DOI] [PubMed] [Google Scholar]
- 40. Sounni N. E., Roghi C., Chabottaux V., Janssen M., Munaut C., Maquoi E., Galvez B. G., Gilles C., Frankenne F., Murphy G., Foidart J. M., Noel A. (2004) Up-regulation of vascular endothelial growth factor-A by active membrane-type 1 matrix metalloproteinase through activation of Src-tyrosine kinases. J. Biol. Chem. 279, 13564–13574 [DOI] [PubMed] [Google Scholar]
- 41. Eisenach P. A., Roghi C., Fogarasi M., Murphy G., English W. R. (2010) MT1-MMP regulates VEGF-A expression through a complex with VEGFR-2 and Src. J. Cell Sci. 123, 4182–4193 [DOI] [PubMed] [Google Scholar]
- 42. Wolf K., Müller R., Borgmann S., Bröcker E. B., Friedl P. (2003) Amoeboid shape change and contact guidance: T-lymphocyte crawling through fibrillar collagen is independent of matrix remodeling by MMPs and other proteases. Blood 102, 3262–3269 [DOI] [PubMed] [Google Scholar]
- 43. Li W., Ye Y. (2008) Polyubiquitin chains: Functions, structures, and mechanisms. Cell. Mol. Life Sci. 65, 2397–2406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Onishi Y., Hirasaka K., Ishihara I., Oarada M., Goto J., Ogawa T., Suzue N., Nakano S., Furochi H., Ishidoh K., Kishi K., Nikawa T. (2005) Identification of mono-ubiquitinated LDH-A in skeletal muscle cells exposed to oxidative stress. Biochem. Biophys. Res. Commun. 336, 799–806 [DOI] [PubMed] [Google Scholar]
- 45. Cao C., Li Y., Leng Y., Li P., Ma Q., Kufe D. (2005) Ubiquitination and degradation of the Arg tyrosine kinase is regulated by oxidative stress. Oncogene 24, 2433–2440 [DOI] [PubMed] [Google Scholar]
- 46. van der Horst A., de Vries-Smits A. M., Brenkman A. B., van Triest M. H., van den Broek N., Colland F., Maurice M. M., Burgering B. M. (2006) FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat. Cell Biol. 8, 1064–1073 [DOI] [PubMed] [Google Scholar]
- 47. Hunter T. (2007) The age of cross-talk: Phosphorylation, ubiquitination, and beyond. Mol. Cell 28, 730–738 [DOI] [PubMed] [Google Scholar]
- 48. Moss N. M., Barbolina M. V., Liu Y., Sun L., Munshi H. G., Stack M. S. (2009) Ovarian cancer cell detachment and multicellular aggregate formation are regulated by membrane type 1 matrix metalloproteinase: A potential role in I.p. metastatic dissemination. Cancer Res. 69, 7121–7129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tomari T., Koshikawa N., Uematsu T., Shinkawa T., Hoshino D., Egawa N., Isobe T., Seiki M. (2009) High throughput analysis of proteins associating with a proinvasive MT1-MMP in human malignant melanoma A375 cells. Cancer Sci. 100, 1284–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Sigismund S., Polo S., Di Fiore P. P. (2004) Signaling through monoubiquitination. Curr. Top Microbiol. Immunol. 286, 149–185 [DOI] [PubMed] [Google Scholar]
- 51. Pickart C. M. (2001) Mechanisms underlying ubiquitination. Ann. Rev. Biochem. 70, 503–533 [DOI] [PubMed] [Google Scholar]
- 52. An W. G., Hwang S. G., Trepel J. B., Blagosklonny M. V. (2000) Protease inhibitor-induced apoptosis: Accumulation of wt p53, p21WAF1/CIP1, and induction of apoptosis are independent markers of proteasome inhibition. Leukemia 14, 1276–1283 [DOI] [PubMed] [Google Scholar]
- 53. Hotary K., Allen E., Punturieri A., Yana I., Weiss S. J. (2000) Regulation of cell invasion and morphogenesis in a three-dimensional type I collagen matrix by membrane-type matrix metalloproteinases 1, 2, and 3. J. Cell Biol. 149, 1309–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sabeh F., Ota I., Holmbeck K., Birkedal-Hansen H., Soloway P., Balbin M., Lopez-Otin C., Shapiro S., Inada M., Krane S., Allen E., Chung D., Weiss S. J. (2004) Tumor cell traffic through the extracellular matrix is controlled by the membrane-anchored collagenase MT1-MMP. J. Cell Biol. 167, 769–781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Rozanov D. V., Deryugina E. I., Ratnikov B. I., Monosov E. Z., Marchenko G. N., Quigley J. P., Strongin A. Y. (2001) Mutation analysis of membrane type-1 matrix metalloproteinase (MT1-MMP). The role of the cytoplasmic tail Cys574, the active site Glu240, and furin cleavage motifs in oligomerization, processing, and self-proteolysis of MT1-MMP expressed in breast carcinoma cells. J. Biol. Chem. 276, 25705–25714 [DOI] [PubMed] [Google Scholar]
- 56. Gingras D., Bousquet-Gagnon N., Langlois S., Lachambre M. P., Annabi B., Béliveau R. (2001) Activation of the extracellular signal-regulated protein kinase (ERK) cascade by membrane-type-1 matrix metalloproteinase (MT1-MMP). FEBS Lett. 507, 231–236 [DOI] [PubMed] [Google Scholar]
- 57. Langlois S., Gingras D., Béliveau R. (2004) Membrane type 1-matrix metalloproteinase (MT1-MMP) cooperates with sphingosine 1-phosphate to induce endothelial cell migration and morphogenic differentiation. Blood 103, 3020–3028 [DOI] [PubMed] [Google Scholar]
- 58. Giasson B. I., Lee V. M. (2003) Are ubiquitination pathways central to Parkinson disease? Cell 114, 1–8 [DOI] [PubMed] [Google Scholar]
- 59. Bhoj V. G., Chen Z. J. (2009) Ubiquitylation in innate and adaptive immunity. Nature 458, 430–437 [DOI] [PubMed] [Google Scholar]
- 60. Chen Z. J., Sun L. J. (2009) Nonproteolytic functions of ubiquitin in cell signaling. Mol. Cell 33, 275–286 [DOI] [PubMed] [Google Scholar]
- 61. Hoeller D., Dikic I. (2009) Targeting the ubiquitin system in cancer therapy. Nature 458, 438–444 [DOI] [PubMed] [Google Scholar]
- 62. Raiborg C., Stenmark H. (2009) The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 458, 445–452 [DOI] [PubMed] [Google Scholar]
- 63. Wang P., Wang X., Pei D. (2004) Mint-3 regulates the retrieval of the internalized membrane-type matrix metalloproteinase, MT5-MMP, to the plasma membrane by binding to its carboxyl end motif EWV. J. Biol. Chem. 279, 20461–20470 [DOI] [PubMed] [Google Scholar]
- 64. Bravo-Cordero J. J., Marrero-Diaz R., Megias D., Genis L., Garcia-Grande A., Garcia M. A., Arroyo A. G., Montoya M. C. (2007) EMBO J. 26, 1499–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Pettet G. J., Please C. P., Tindall M. J., McElwain D. L. (2001) The migration of cells in multicell tumor spheroids. Bull Math Biol. 63, 231–257 [DOI] [PubMed] [Google Scholar]
- 66. Aoh Q. L., Castle A. M., Hubbard C. H., Katsumata O., Castle J. D. (2009) SCAMP3 negatively regulates epidermal growth factor receptor degradation and promotes receptor recycling. Mol. Biol. Cell 20, 1816–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Castle A., Castle D. (2005) Ubiquitously expressed secretory carrier membrane proteins (SCAMPs) 1–4 mark different pathways and exhibit limited constitutive trafficking to and from the cell surface. J. Cell Sci. 118, 3769–3780 [DOI] [PubMed] [Google Scholar]
- 68. Shearwin-Whyatt L., Dalton H. E., Foot N., Kumar S. (2006) Regulation of functional diversity within the Nedd4 family by accessory and adaptor proteins. Bioessays 28, 617–628 [DOI] [PubMed] [Google Scholar]
- 69. Katz M., Shtiegman K., Tal-Or P., Yakir L., Mosesson Y., Harari D., Machluf Y., Asao H., Jovin T., Sugamura K., Yarden Y. (2002) Ligand-independent degradation of epidermal growth factor receptor involves receptor ubiquitylation and Hgs, an adaptor whose ubiquitin-interacting motif targets ubiquitylation by Nedd4. Traffic 3, 740–751 [DOI] [PubMed] [Google Scholar]
- 70. Murdaca J., Treins C., Monthouël-Kartmann M. N., Pontier-Bres R., Kumar S., Van Obberghen E., Giorgetti-Peraldi S. (2004) Grb10 prevents Nedd4-mediated vascular endothelial growth factor receptor-2 degradation. J. Biol. Chem. 279, 26754–26761 [DOI] [PubMed] [Google Scholar]
- 71. Usami Y., Popov S., Popova E., Inoue M., Weissenhorn W., G Göttlinger H. (2009) The ESCRT pathway and HIV-1 budding. Biochem. Soc. Trans. 37, 181–184 [DOI] [PubMed] [Google Scholar]
- 72. Helliwell S. B., Losko S., Kaiser C. A. (2001) Components of a ubiquitin ligase complex specify polyubiquitination and intracellular trafficking of the general amino acid permease. J. Cell Biol. 153, 649–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.