Background: Legume antimicrobial peptides (AMPs) mediate Sinorhizobium meliloti bacteroid differentiation.
Results: Cysteine replacements and disulfide bond modifications influence the antimicrobial activity of a legume AMP and its ability to mediate S. meliloti bacteroid differentiation.
Conclusion: Specific changes to legume AMPs influence their activity against S. meliloti.
Significance: Understanding the relationship of AMPs in S. meliloti bacteroid differentiation is fundamental for nitrogen fixation and legume growth.
Keywords: Antimicrobial Peptides; Bacteria; Microbiology; Peptide Conformation; Symbiosis; BacA; Defensins; Nodule-specific, Cysteine-rich Peptides; Rhizobia; Sinorhizobium meliloti
Abstract
The root nodules of certain legumes including Medicago truncatula produce >300 different nodule-specific cysteine-rich (NCR) peptides. Medicago NCR antimicrobial peptides (AMPs) mediate the differentiation of the bacterium, Sinorhizobium meliloti into a nitrogen-fixing bacteroid within the legume root nodules. In vitro, NCR AMPs such as NCR247 induced bacteroid features and exhibited antimicrobial activity against S. meliloti. The bacterial BacA protein is critical to prevent S. meliloti from being hypersensitive toward NCR AMPs. NCR AMPs are cationic and have conserved cysteine residues, which form disulfide (S–S) bridges. However, the natural configuration of NCR AMP S–S bridges and the role of these in the activity of the peptide are unknown. In this study, we found that either cysteine replacements or S–S bond modifications influenced the activity of NCR247 against S. meliloti. Specifically, either substitution of cysteines for serines, changing the S–S bridges from cysteines 1–2, 3–4 to 1–3, 2–4 or oxidation of NCR247 lowered its activity against S. meliloti. We also determined that BacA specifically protected S. meliloti against oxidized NCR247. Due to the large number of different NCRs synthesized by legume root nodules and the importance of bacterial BacA proteins for prolonged host infections, these findings have important implications for analyzing the function of these novel peptides and the protective role of BacA in the bacterial response toward these peptides.
Introduction
Antimicrobial peptides (AMPs)5 are peptides of generally <100 amino acids, produced as part of the host innate immune response (1, 2). The antimicrobial activities of mammalian AMPs against microorganisms have been investigated extensively (1, 3, 4). The root nodules of certain legumes have been found to produce a large family of novel nodule-specific cysteine-rich (NCR) peptides. Transcriptional analysis of the root nodules of the legume Medicago truncatula, which is a model for genomic studies (5), found that they contained >300 different NCR peptides (6–8), and the genome sequence revealed the presence of almost 600 NCR genes (9). These NCR peptides lacked sequence similarity except for the N-terminal signal peptide but were between 50–60 amino acids in length, had 4 or 6 conserved cysteine residues and could be subdivided according to their overall charge into cationic, anionic, and neutral NCRs (6). Cationic M. truncatula NCRs, such as NCR247 (known as NCR AMPs), are similar to mammalian defensins (6, 10) and exhibit antimicrobial activity in vitro against the M. truncatula symbiont, Sinorhizobium meliloti (10, 11).
Within the legume root nodules, NCR AMPs control the differentiation of S. meliloti into persisting, nitrogen-fixing bacteroids (10). S. meliloti enters M. truncatula nodule cells via root hair-derived infection threads, where they are endocytosed into a membrane-bound compartment known as a symbiosome (12). Once inside the symbiosome, S. meliloti are challenged with NCR AMPs, which are targeted to this compartment (10). An M. truncatula dnf1 mutant, which is defective in the transport of NCR peptides to the symbiosome, was unable to mediate S. meliloti bacteroid differentiation and hence was defective in establishing the symbiosis (10, 11, 13). S. meliloti bacteroids are distinctly different from their bacterial form as they are elongated, have extensive genomic DNA amplification, are terminally differentiated (unable to revert back to their bacterial form), and have an increased cytoplasmic membrane permeability relative to their free-living bacterial form (10, 14). The precise in vivo concentration of NCR AMPs is unknown. However, exposure of S. meliloti in vitro toward sublethal levels of a specifically folded NCR AMP, NCR247, with disulfide (S–S) bridges between the 1st and 2nd and the 3rd and 4th cysteine residues (NCR2471–2,3–4), induced a number of features of in planta bacteroids (11). At higher concentrations, NCR2471–2,3–4 exhibited potent antimicrobial activity. Despite the importance of NCR AMPs for the legume symbiosis, no structural requirements for their activity against S. meliloti are known (6, 10, 14). In this paper, we investigated the role of the cysteine residues and S–S bond modifications in NCR AMPs using NCR247 as a model peptide. Because there are nearly 600 different NCR peptides produced within the root nodules of M. truncatula alone (6, 9), the outcome of this study will have important implications for future experiments analyzing other physiological AMPs.
EXPERIMENTAL PROCEDURES
Bacterial Growth
The sequenced S. meliloti Sm1021 strain (parent strain; formerly known as Rm1021) (15) and the SmGF1 BacA-deficient mutant (16) were used in this study. All bacterial strains were grown initially in LB medium (17) supplemented with 2.5 mm MgSO4 and 2.5 mm CaCl2 (LB/MC) followed by subculturing into LB with 500 μg ml−1 streptomycin at 30 °C, 200 rpm. For solid media, 15 g of agar was added per liter of medium.
Synthesis of NSR247 and Truncated NCR247 Peptides
The NSR247 and truncated NCR247 peptides were synthesized commercially by GenScript (Piscataway, NJ). Their purity and masses were then determined by RP-HPLC and ESI-MS, respectively. In all cases, the peptides had a purity of >95% (w/v) and ESI-MS confirmed the identity of the peptide.
Synthesis and Folding of NCR247 Peptides
The NCR247 peptide with S–S bridges between the 1st and 2nd and the 3rd and 4th cysteine residues (NCR2471–2,3–4) was assembled and folded exactly as described previously (11). The NCR247 peptide with S–S bridges between the 1st and 3rd and 2nd and 4th cysteine residues (NCR2471–3,2–4) was also assembled as described before (11) except the N-9-fluorenylmethoxycarbonyl amino acid used for the synthesis had the following side chain protecting groups: trityl for cysteines 1 and 3 and acetoamidomethyl for cysteines 2 and 4. The peptides were then purified to homogeneity by semipreparative RP-HPLC, analyzed by ESI-MS, and stored lyophilized until use at 4 °C (11). Prior to use, lyophilized peptides were dissolved in molecular biology grade water at a final concentration of 1 mg ml−1, and aliquots were stored at −20 °C until use.
Reduction of NCR247 Peptide
A 0.3 mg ml−1 solution of NCR2471–2,3–4 peptide in 10 mm sodium phosphate buffer, pH 7.0, was treated with 10 mm dithiothreitol (DTT) and incubated at 37 °C for 5–180 min. At defined time points, 20 μl of the sample was removed and analyzed by RP-HPLC-MS.
RP-HPLC-MS Analysis of Peptides
The NCR247 peptides were analyzed by RP-HPLC-MS using an Agilent 1200 HPLC system equipped with a DAD and an ESI-MS detector. Samples were loaded onto a Phenomenex Jupiter C18, 5-μm, 300-Å (150 × 4.6 mm) HPLC column (Phenomenex) equilibrated with 98% Eluent A (0.18% trifluoroacetic acid (TFA)) and 2% Eluent B (acetonitrile + 0.15% TFA). Peptides were eluted with a linear gradient from 2% to 35% Eluent B in 33 min at 0.8 ml min−1 flow rate and 25 °C. The eluate was monitored at 220 and 280 nm and by positive ion ESI-MS.
Effect of Peptides on Cultured S. meliloti
The effect of stored peptides on either bacterial cell size, and genomic DNA content was determined by flow cytometry exactly as described previously (11). S. meliloti viability after peptide treatment was assessed by determining the colony-forming units/ml as outlined before (11).
RESULTS
Cysteine Residues Are Important for Full Activity of NCR247 against S. meliloti in Vitro
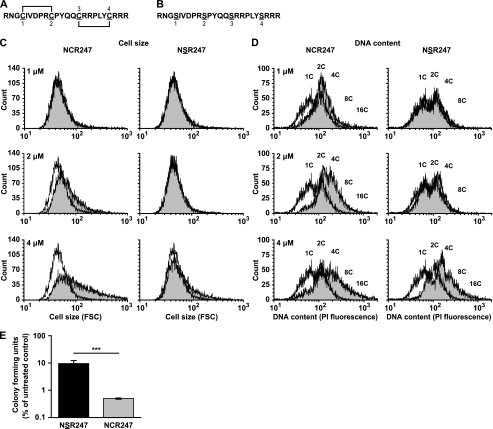
We showed previously that NCR2471–2,3–4 (Fig. 1A), with two defined S–S bridges, induced certain in planta bacteroid features at sublethal concentrations and exhibited antibacterial activity against S. meliloti in vitro at higher concentrations (11). We aimed to investigate whether the cysteine residues of the peptide, and hence potentially the S–S bridges, were important for its full activity (bacteroid-inducing and antimicrobial) against S. meliloti. To do this, a version of the peptide, known as NSR247, was synthesized with greater than 95% (w/v) purity, whereby all 4 cysteine residues were replaced by serines (Fig. 1B). The NSR247 peptide retained the cationic nature of NCR2471–2,3–4 (charge of 5.76 for NSR247 compared with 5.72 for NCR2471–2,3–4 at pH 7.0) but lacked the specific S–S bridges due to the absence of the cysteine residues. The activity of NSR247 against S. meliloti in vitro was then investigated relative to NCR2471–2,3–4. Using flow cytometry to measure bacterial cell size and genomic DNA copy number (10, 11), we compared the ability of different sublethal concentrations of NSR247 and NCR2471–2,3–4 to induce these changes in S. meliloti in vitro (Fig. 1, C and D). We found that, unlike NCR2471–2,3–4, which was capable of increasing S. meliloti cell size at 2 μm, NSR247 only started to increase the cell size of S. meliloti at 4 μm and to a lower extent than NCR2471–2,3–4 (Fig. 1C). Likewise, 1 μm NCR2471–2,3–4 started to induce an increase in the DNA content of S. meliloti, whereas NSR247 only started to affect the DNA content of S. meliloti significantly at 4 μm (Fig. 1D). In addition, we found using higher concentrations of each peptide (10 μm), that the NSR247 peptide had substantially reduced antibacterial activity against S. meliloti in vitro relative to NCR2471–2,3–4 (Fig. 1E). Taken together, these findings showed that the cysteine residues are important for the full activity of NCR247 against S. meliloti.
FIGURE 1.
Cysteine residues are important for NCR2471–2,3–4 activity. A, NCR2471–2,3–4 sequence showing the S–S bridges between cysteines 1 and 2 and cysteines 3 and 4. B, NSR247 peptide sequence. C and D, flow cytometry analysis of S. meliloti Sm1021 cultures treated for 3 h with (shaded histograms) or without (white histograms) 1, 2, and 4 μm NCR2471–2,3–4 or NSR247 peptide, respectively. The forward scatter (FSC) was measured to estimate relative bacterial cell size (C), and the PI fluorescence was measured to estimate the relative bacterial DNA content (D). C indicates number of chromosome copies. Flow cytometry plots are a representative example of at least two independent experiments. E, colony forming ability of S. meliloti Sm1021 assessed after treatment with 10 μm NSR247 (black bar) or NCR2471–2,3–4 (gray bar) for 3 h relative to an untreated control. The dataset shown is representative of trends observed in two independent experiments. Columns represent mean ± S.E. (error bars) (n = 3), and significance was determined using the two-sided unpaired Student's t test on logarithmic data. ***, p ≤ 0.001.
Precise S–S Bridge Configurations Affect the Activity of NCR247
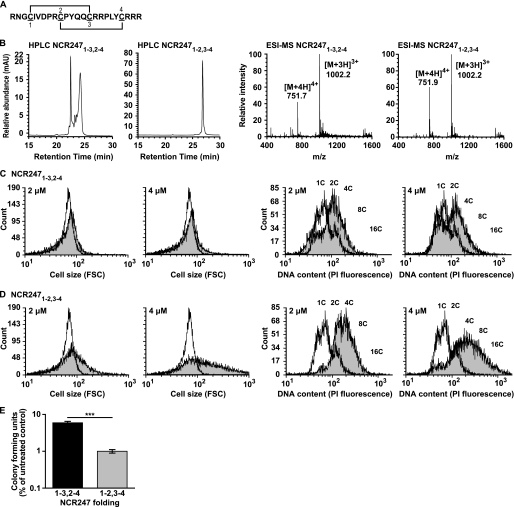
Our finding that the cysteine-containing NCR247 peptide had an enhanced activity relative to the serine-containing NSR247 peptide suggested that the precise S–S bridge configurations were important for the full activity of NCR247 against S. meliloti. The S–S bridge configurations of endogenous NCR peptides are unknown. However, by applying Chou-Fasman rules (18), we previously predicted that the most likely folded configuration of NCR247 was NCR2471–2,3–4 (Fig. 1A) (11). To investigate whether the S–S bridge configuration influenced the stability and biological activity of the peptide, we synthesized an alternatively folded form of NCR247, whereby two S–S bridges were specifically formed between the 1st and 3rd and the 2nd and 4th cysteine residues (NCR2471–3,2–4) (Fig. 2A). This form of the peptide was chosen for synthesis because Chou-Fasman rules predicted that it was the next most likely folded state of NCR247 (data not shown). RP-HPLC of the freshly synthesized and folded NCR2471–3,2–4 peptide showed one major peak, indicating the presence of one pure peptide species (data not shown), as had been observed previously for newly prepared NCR2471–2,3–4 (11). However, upon storage of the lyophilized peptide preparations, RP-HPLC of NCR2471–3,2–4 resulted in multiple peaks whereas one major peak was still observed for NCR2471–2,3–4 (Fig. 2B). ESI-MS analysis of the stored peptides confirmed that they had the correct masses (11), consistent with them being in an oxidized state (Fig. 2B).
FIGURE 2.
S–S bridge configuration is important for NCR247 activity. A, NCR2471–3,2–4 sequence showing the S–S bridges between cysteines 1 and 3 and cysteines 2 and 4. B, RP-HPLC and ESI-MS analysis of NCR247 peptides dissolved in molecular biology grade water at a concentration of 1 mg ml−1. ESI-MS analyses are in agreement with peptides of 3004.5 Da and indicate that the peptides are in the oxidized form. C and D, flow cytometry analysis of S. meliloti Sm1021 cultures treated for 3 h with (shaded histograms) or without (white histograms) 2 and 4 μm NCR2471–3,2–4 (C) or NCR2471–2,3–4 (D) peptide. The forward scatter (FSC) was measured to estimate relative bacterial cell size, and the PI fluorescence was measured to estimate the relative bacterial DNA content. C indicates number of chromosome copies. Flow cytometry plots are a representative example of at least two independent experiments. E, colony-forming ability of S. meliloti Sm1021 assessed after treatment with 10 μm NCR2471–3-2–4 (black) or NCR2471–2,3–4 peptide (gray) for 3 h relative to an untreated control. The dataset shown is representative of trends observed in two independent experiments. Columns represent mean ± S.E. (error bars) (n = 3), and significance was determined using a two-sided unpaired Student's t test on logarithmic data. ***, p ≤ 0.001.
To determine whether the S–S bridge configurations of NCR247 affected its activity, we compared the activity of NCR2471–3,2–4 with NCR2471–2,3–4 in inducing bacteroid changes in S. meliloti in vitro (11) (Fig. 2, C and D, respectively). We found that, although 2 and 4 μm NCR2471–3,2–4 increased the cell size and DNA content of S. meliloti relative to the untreated control, it was significantly less active than NCR2471–2,3–4 at the same concentrations. In addition, we determined that stored NCR2471–3,2–4 was less effective than NCR2471–2,3–4 at reducing the colony forming ability of S. meliloti in vitro (Fig. 2E). Combined, these findings showed that NCR2471–2,3–4 was more active against S. meliloti in vitro than NCR2471–3,2–4 in terms of inducing bacteroid-like features and exhibiting antimicrobial activity. Therefore, this demonstrated that the precise S–S bridge configurations of NCR247 are important for the stability and full activity of this peptide against S. meliloti.
Full-length Peptide Is Important for Full Activity of NCR247 against S. meliloti
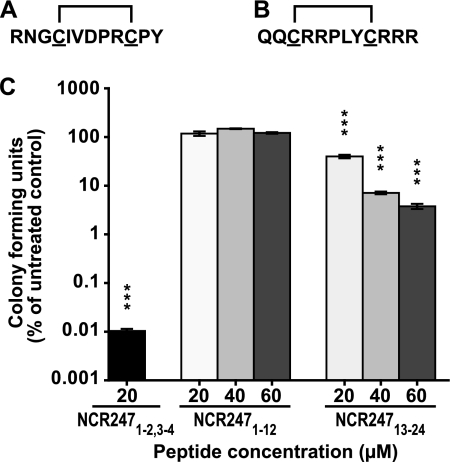
To gain insights into the importance of cationic residues for the activity of NCR247 against S. meliloti, we commercially synthesized two truncated versions of this peptide, NCR2471–12 (lacking the last 12 amino acids from the C-terminal end) and NCR24713–24 (lacking the first 12 amino acids from the N terminus of the peptide) (Fig. 3, A and B, respectively). These truncated peptides were synthesized with >95% (w/v) purity (data not shown), and each of them contained one S–S bridge (Fig. 3, A and B). NCR2471–12 and NCR24713–24 had charges at pH 7.0 of 0.74 and 4.74, respectively, thereby enabling us to understand how the cationic nature of the peptide affected its activity. We found no antimicrobial activity of the neutral NCR2471–12 peptide against S. meliloti, even when the concentration was increased to 60 μm (Fig. 3C). In contrast, although the cationic NCR24713–24 peptide was substantially reduced in its activity relative to the full-length NCR247 peptide, it retained some antibacterial activity against S. meliloti (Fig. 3C). No significant changes in cell size and DNA content were observed in flow cytometry experiments of S. meliloti treated with 1–4 μm truncated peptides (data not shown). Therefore, these data showed that the full-length peptide was important for the full activity of NCR247 against S. meliloti. However, because the cationic, truncated peptide was still capable of killing S. meliloti, this provided evidence for the importance of the cationic residues for the antimicrobial activity of the peptide.
FIGURE 3.
Truncated NCR247 peptides show reduced antimicrobial activity against S. meliloti. A and B, NCR2471–12 (A) and NCR24713–24 (B) peptide sequences showing disulfide bridges. C, colony forming ability of S. meliloti Sm1021 assessed after treatment with the respective NCR peptide for 3 h relative to an untreated control. Columns represent mean ± S.E. (error bars) (n = 3). The dataset shown is representative of trends observed in two independent experiments. Significant differences compared with the untreated control were determined with one-way ANOVA and Bonferroni's post test on logarithmic data. ***, p ≤ 0.001.
Reduction of NCR247 S–S Bridges Enhances Its Antibacterial Activity against S. meliloti
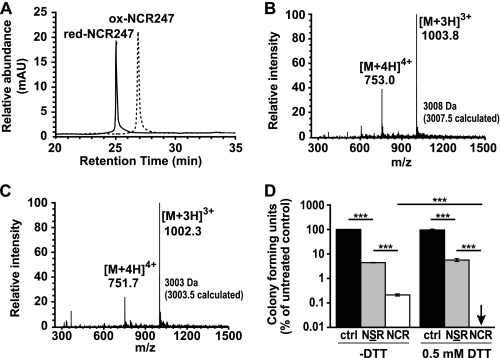
The experiments conducted thus far with the NCR247 peptides used oxidized forms of the peptide with defined S–S bridges. However, it was shown recently that the antimicrobial activity of a cysteine-rich β-defensin was dramatically enhanced by reduction of this peptide (19). To investigate whether the antimicrobial activity of NCR247 was affected by reduction, the oxidized NCR2471–2,3–4 peptide was treated with 10 mm reducing agent, dithiothreitol (DTT), and incubated from 5 to 180 min. The oxidized and reduced forms of the peptides were then separated by RP-HPLC and their peptide masses determined by ESI-MS. We found using RP-HPLC that the NCR2471–2,3–4 peptide was all reduced by DTT within 5 min of treatment and that the reduced peptide had a lower retention time than the oxidized peptide (Fig. 4A), indicating that it was more hydrophilic. This finding is in line with the presence of SH groups in the reduced peptide, which are more polar than the S–S bridges in the oxidized peptide. ESI-MS analysis of the peptides confirmed that the reduced peptide had an increased mass equivalent to four protons relative to the oxidized peptide (compare Fig. 4, B and C, respectively).
FIGURE 4.
Reduced NCR247 shows increased antimicrobial activity against S. meliloti. A, RP-HPLC analysis of NCR2471–2,3–4 treated with (solid line) and without (dashed line) 10 mm DTT for 5 min in sodium phosphate buffer. B, ESI-MS analysis of the peak eluted between 24.7 and 25.5 min, consistent with a fully reduced NCR247 peptide. C, ESI-MS analysis of the peak eluted between 26.7 and 27.2 min, consistent with the folded, oxidized form of NCR247. D, colony forming ability of S. meliloti Sm1021 assessed after treatment with 10 μm NCR2471–2,3–4 and NSR247 peptide for 3 h in the presence and absence of DTT. The dataset shown is representative of trends observed in two independent experiments. Columns represent mean ± S.E. (error bars) (n = 3), and significance was determined with one-way ANOVA and Bonferroni's post test on logarithmic data. ***, p ≤, 0.001.
Because we found that the NCR2471–2,3–4 peptide was completely reduced after a 5-min incubation with DTT, we analyzed the effect of this reduced peptide relative to the oxidized form on the viability of S. meliloti. We determined that reduction of NCR247 significantly enhanced its antibacterial activity against S. meliloti (Fig. 4D). In contrast, DTT addition itself had no effect on the viability of S. meliloti or on the antibacterial activity of the NSR247 peptide against S. meliloti (Fig. 4D). Together, these findings showed that reduction of the cysteine residues in NCR247 enhanced its antimicrobial activity against S. meliloti.
The BacA Protein Specifically Protects S. meliloti against Oxidized NCR247 Peptides
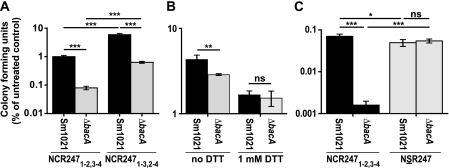
To date, the BacA protein is the only bacterial factor known to be critical for protection of S. meliloti against the antibacterial activity of NCR2471–2,3–4 (11). Hence, to gain further insights into the mechanism of BacA-mediated protection in S. meliloti and the structural requirements for NCR peptides in this process, we investigated the effect of different versions/states of the NCR247 peptide on the viability of an S. meliloti BacA-deficient mutant relative to the parent strain. We found, consistent with our previous observations using oxidized NCR2471–2,3–4 (11), that the S. meliloti BacA-deficient mutant was hypersensitive toward both oxidized and specifically folded forms of NCR247 (Fig. 5A). In contrast, the BacA-deficient mutant had the same viability as the parent strain toward either the reduced form of NCR247 (Fig. 5B) or the NSR247 peptide (Fig. 5C). Consequently, these data showed that the S. meliloti BacA-deficient mutant was only hypersensitive toward the oxidized NCR247 peptides but not reduced NCR247 or the NSR247 peptide.
FIGURE 5.
NCR247 folding is important for BacA-mediated protection. A–C, colony forming ability of S. meliloti Sm1021 parent and the BacA-deficient mutant assessed after peptide treatment. A, 10 μm NCR2471–2,3–4 and NCR2471–3,2–4. B, 5 μm NCR2471–2,3–4 without (oxidized) and with (reduced) 1 mm DTT. C, 20 μm NCR2471–2,3–4 and NSR247. The datasets shown are representative of trends observed in two independent experiments each. Columns represent mean ± S.E. (error bars) (n = 3). Significance was determined with one-way ANOVA and Bonferroni's post test; ns, not significant. *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001.
DISCUSSION
Why Is the NCR2471–3,2–4 Peptide Unstable?
Our analysis showed that the NCR2471–2,3–4 peptide remained stable after storage at +4 °C in the lyophilized form and −20 °C when dissolved in water, providing further evidence that this is the naturally folded, stable state of the peptide. In contrast, although a single pure RP-HPLC peak was found for freshly synthesized NCR2471–3,2–4 peptide, after storage, we observed multiple RP-HPLC peaks indicative of an unstable peptide. Because these experiments were performed under oxidizing conditions, the additional peaks are not due to reduction of the S–S bonds. This was also confirmed by ESI-MS analysis, which showed that the stored peptides were in their oxidized states. Instead, they are likely due to a complex mixture of peptide polymers, which form by the strained S–S bridges in NCR2471–3,2–4 opening up to form radicals (R-S•) and/or thiolate groups (R-S−) via photochemical processes or by the action of nucleophilic or basic trace impurities, respectively. These reactive intermediates then trigger inter- and intramolecular disulfide isomerizations forming new less constrained intermolecular S–S bridges (20). Hence, these findings demonstrate that the precise S–S bridge configurations of NCR247 are important for peptide stability upon storage. Because there are nearly 600 different NCR peptides predicted within the root nodules of M. truncatula alone (6–9), our data suggest that Chou-Fasman rules will be very useful in predicting the most likely stably folded state of these peptides to investigate their functions in vitro.
Why Do Differently Folded NCR247 Peptides Exhibit Different Levels of Activities against S. meliloti?
In this study, we showed that the cysteine residues are important for the full activity of NCR247 against S. meliloti in vitro. We found that the NCR2471–2,3–4 peptide was significantly more active than NCR2471–3,2–4, against S. meliloti in vitro in all of the assays performed. Because we proposed that the stored NCR2471–3,2–4 peptide consists of a complex mixture of polymers, these might not be as biologically active against S. meliloti as the smaller, stably folded version of the peptide or may mask activity of an active fraction in the preparation. In addition, the specifically folded NCR2471–2,3–4 peptide could be important for interactions with specific targets in S. meliloti. It is known from previous studies (10, 11) and further demonstrated in this work that NCR247 disrupts S. meliloti membrane integrity. However, further genetic and biochemical studies will be necessary to identify potential S. meliloti targets of NCR AMPs to understand how changes in peptide folding could affect these interactions.
Why Is Positive Charge Important for the Activity of NCR247 against S. meliloti?
Although the cysteineless, positively charged NSR247 peptide had a reduced activity relative to NCR247, we found that it was still capable of inducing bacteroid features and reducing the viability of S. meliloti in vitro, demonstrating that the positive charge is also important for the activity of this AMP. It is well established that other cationic AMPs act via interacting with negatively charged bacterial lipopolysaccharide leading to their insertion into the outer membrane, which eventually leads to disruption of the outer membrane, followed by inner membrane disruption (21). Hence, NCR AMPs are likely to function in a similar manner. Consistent with this, we found that the cationic truncated NCR24713–24 peptide, which lacked the first 12 amino acids from the N terminus, retained some antimicrobial activity against S. meliloti. In contrast, a neutral NCR247 peptide, which lacked the last 12 amino acids, was inactive against S. meliloti. Therefore, because the truncated cationic version of the peptide possessed some activity against S. meliloti, this provided further evidence for the importance of the positive charge for activity.
What Are in Vivo Consequences of Reduced NCR247 Peptide Exhibiting Enhanced Antibacterial Activity against S. meliloti?
We found that the reduced NCR247 peptide was more potent as an antimicrobial agent against S. meliloti in vitro relative to its oxidized form. Because no difference was observed in the activity of NSR247 by DTT treatment, these findings showed that the reduced cysteine residues contributed to the antibacterial action of the peptide. A similar increase in antimicrobial activity was also observed recently when a mammalian cysteine-rich β-defensin from the colon was reduced (19). Within the colon, it was suggested that the β-defensin is stored in its inactive oxidized form and then activated through reduction by host thioredoxin proteins to release its potent antimicrobial activity (19). Specific thioredoxins are also produced within the root nodules of M. truncatula (22). These thioredoxins possess signal peptides (22) and are thus potentially targeted to the symbiosomes, similar to the NCR AMPs. Thus, these nodule-specific thioredoxins may serve to reduce endogenous NCR AMPs, which could then dramatically increase their antimicrobial activity. Because the S–S bonds of bacterial proteins are also oxidized in the periplasm (23), exposure of NCR AMPs to this cellular compartment might also influence their antimicrobial activity against S. meliloti. However, because we found that the reduced form of NCR247 was more potent than the oxidized form of this peptide against S. meliloti, this suggests that it is exerting its toxic effect before entering the periplasm and potentially being reoxidized. Conversely, NCRs are processed and transported toward the symbiosomes in the endoplasmic reticulum (10), and the signal peptides of the thioredoxins strongly suggest that these proteins also pass through the endoplasmic reticulum. Because the endoplasmic reticulum is an oxidizing environment (24) it could be envisaged that the thioredoxins mediate oxidation rather than reduction of the NCR peptides. In this scenario, the role of the thioredoxins could thus be to prevent NCR reduction and the formation of potentially harmful reduced peptides. The thioredoxins could also promote the recycling of misfolded NCR peptides with nonnative disulfide bridges. This situation is also in agreement with the observation that the BacA protein only provides protection against the oxidized peptides (see below). Because BacA is required in vivo for protecting S. meliloti against the antimicrobial activity of NCR peptides (11), this is a strong indication that in nodules the peptides are in the oxidized state. In any case, further studies will be necessary to determine whether NCRs co-localize with thioredoxins and are thioredoxin substrates.
What Is the Role of BacA in Protection of S. meliloti against NCR247?
We showed that the BacA protein specifically protected S. meliloti in vitro against the oxidized NCR247 peptide but not the reduced form of this peptide or the NSR247 peptide. Although the precise mechanism by which BacA protects S. meliloti against NCR247 remains to be discovered, it is interesting to speculate that BacA may influence the oxidation state of the NCR247 peptide leading to protection. Because the NCR247 peptide is more active in its reduced state, BacA may either directly or indirectly influence the balance between oxidized and reduced peptide, favoring the oxidized state and thereby limiting the amount of NCR247-induced inner membrane damage. Therefore, in a S. meliloti BacA-deficient mutant, more peptide may become reduced within the periplasm leading to a higher degree of cytoplasmic membrane damage. Because BacA is known to mediate peptide transport in S. meliloti (25, 26), this transport activity may indirectly lead to a change in the oxidation state of the periplasm. Alternatively, because BacA only conferred protection against the specifically folded forms of the peptides there may be a direct interaction between BacA and the specifically folded states of NCR247.
In conclusion, we have shown the importance of the cysteine residues and S–S bond configuration on the activity of NCR247 against S. meliloti. Because there are almost 600 different NCR peptides predicted within M. truncatula root nodules alone (9), these studies will provide a platform for the future analysis of these and other legume nodule NCR peptides. BacA proteins are also found in diverse bacterial symbionts and pathogens (11, 27–29). Therefore, the results of this study also have important implications for the roles of these proteins in the bacterial response toward host cysteine-rich AMPs.
This work was supported by Medical Research Council New Investigator Grant G0501107 (to G. P. F.).
- AMP
- antimicrobial peptide
- ESI
- electrospray ionization
- NCR
- nodule-specific cysteine-rich
- RP
- reverse phase
- PI
- propidium iodide.
REFERENCES
- 1. Maróti G., Kereszt A., Kondorosi E., Mergaert P. (2011) Natural roles of antimicrobial peptides in microbes, plants and animals. Res. Microbiol. 162, 363–374 [DOI] [PubMed] [Google Scholar]
- 2. Ganz T. (2003) Defensins: antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 3, 710–720 [DOI] [PubMed] [Google Scholar]
- 3. Lehrer R. I., Ganz T. (1999) Antimicrobial peptides in mammalian and insect host defense. Curr. Opin. Immunol. 11, 23–27 [DOI] [PubMed] [Google Scholar]
- 4. Wiesner J., Vilcinskas A. (2010) Antimicrobial peptides: the ancient arm of the human immune system. Virulence 1, 440–464 [DOI] [PubMed] [Google Scholar]
- 5. Barker D., Bianchi S., Blondon F., Dattée Y., Duc G., Essad S., Flament P., Gallusci P., Génier G., Guy P., Muel X., Tourneur J., Dénarié J., Huguet T. (1990) Medicago truncatula, a model plant for studying the molecular genetics of the Rhizobium-legume symbiosis. Plant Mol. Biol. Rep. 8, 40–49 [Google Scholar]
- 6. Mergaert P., Nikovics K., Kelemen Z., Maunoury N., Vaubert D., Kondorosi A., Kondorosi E. (2003) A novel family in Medicago truncatula consisting of more than 300 nodule-specific genes coding for small, secreted polypeptides with conserved cysteine motifs. Plant Physiol. 132, 161–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Graham M. A., Silverstein K. A., Cannon S. B., VandenBosch K. A. (2004) Computational identification and characterization of novel genes from legumes. Plant Physiol. 135, 1179–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Alunni B., Kevei Z., Redondo-Nieto M., Kondorosi A., Mergaert P., Kondorosi E. (2007) Genomic organization and evolutionary insights on GRP and NCR genes, two large nodule-specific gene families in Medicago truncatula. Mol. Plant Microbe Interact. 20, 1138–1148 [DOI] [PubMed] [Google Scholar]
- 9. Young N. D., Debellé F., Oldroyd G. E., Geurts R., Cannon S. B., Udvardi M. K., Benedito V. A., Mayer K. F., Gouzy J., Schoof H., Van de Peer Y., Proost S., Cook D. R., Meyers B. C., Spannagl M., Cheung F., De Mita S., Krishnakumar V., Gundlach H., Zhou S., Mudge J., Bharti A. K., Murray J. D., Naoumkina M. A., Rosen B., Silverstein K. A., Tang H., Rombauts S., Zhao P. X., Zhou P., Barbe V., Bardou P., Bechner M., Bellec A., Berger A., Bergès H., Bidwell S., Bisseling T., Choisne N., Couloux A., Denny R., Deshpande S., Dai X., Doyle J. J., Dudez A. M., Farmer A. D., Fouteau S., Franken C., Gibelin C., Gish J., Goldstein S., Gonzalez A. J., Green P. J., Hallab A., Hartog M., Hua A., Humphray S. J., Jeong D. H., Jing Y., Jocker A., Kenton S. M., Kim D. J., Klee K., Lai H., Lang C., Lin S., Macmil S. L., Magdelenat G., Matthews L., McCorrison J., Monaghan E. L., Mun J. H., Najar F. Z., Nicholson C., Noirot C., O'Bleness M., Paule C. R., Poulain J., Prion F., Qin B., Qu C., Retzel E. F., Riddle C., Sallet E., Samain S., Samson N., Sanders I., Saurat O., Scarpelli C., Schiex T., Segurens B., Severin A. J., Sherrier D. J., Shi R., Sims S., Singer S. R., Sinharoy S., Sterck L., Viollet A., Wang B. B., Wang K., Wang M., Wang X., Warfsmann J., Weissenbach J., White D. D., White J. D., Wiley G. B., Wincker P., Xing Y., Yang L., Yao Z., Ying F., Zhai J., Zhou L., Zuber A., Denarie J., Dixon R. A., May G. D., Schwartz D. C., Rogers J., Quetier F., Town C. D., Roe B. A. (2011) The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480, 520–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Van de Velde W., Zehirov G., Szatmari A., Debreczeny M., Ishihara H., Kevei Z., Farkas A., Mikulass K., Nagy A., Tiricz H., Satiat-Jeunemaître B., Alunni B., Bourge M., Kucho K., Abe M., Kereszt A., Maroti G., Uchiumi T., Kondorosi E., Mergaert P. (2010) Plant peptides govern terminal differentiation of bacteria in symbiosis. Science 327, 1122–1126 [DOI] [PubMed] [Google Scholar]
- 11. Haag A. F., Baloban M., Sani M., Kerscher B., Pierre O., Farkas A., Longhi R., Boncompagni E., Hérouart D., Dall'angelo S., Kondorosi E., Zanda M., Mergaert P., Ferguson G. P. (2011) Protection of Sinorhizobium against host cysteine-rich antimicrobial peptides is critical for symbiosis. PLoS Biol. 9, e1001169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jones K. M., Kobayashi H., Davies B. W., Taga M. E., Walker G. C. (2007) How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat. Rev. Microbiol. 5, 619–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang D., Griffitts J., Starker C., Fedorova E., Limpens E., Ivanov S., Bisseling T., Long S. (2010) A nodule-specific protein secretory pathway required for nitrogen-fixing symbiosis. Science 327, 1126–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mergaert P., Uchiumi T., Alunni B., Evanno G., Cheron A., Catrice O., Mausset A. E., Barloy-Hubler F., Galibert F., Kondorosi A., Kondorosi E. (2006) Eukaryotic control on bacterial cell cycle and differentiation in the Rhizobium-legume symbiosis. Proc. Natl. Acad. Sci. U.S.A. 103, 5230–5235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Galibert F., Finan T. M., Long S. R., Puhler A., Abola P., Ampe F., Barloy-Hubler F., Barnett M. J., Becker A., Boistard P., Bothe G., Boutry M., Bowser L., Buhrmester J., Cadieu E., Capela D., Chain P., Cowie A., Davis R. W., Dreano S., Federspiel N. A., Fisher R. F., Gloux S., Godrie T., Goffeau A., Golding B., Gouzy J., Gurjal M., Hernandez-Lucas I., Hong A., Huizar L., Hyman R. W., Jones T., Kahn D., Kahn M. L., Kalman S., Keating D. H., Kiss E., Komp C., Lelaure V., Masuy D., Palm C., Peck M. C., Pohl T. M., Portetelle D., Purnelle B., Ramsperger U., Surzycki R., Thebault P., Vandenbol M., Vorholter F. J., Weidner S., Wells D. H., Wong K., Yeh K. C., Batut J. (2001) The composite genome of the legume symbiont Sinorhizobium meliloti. Science 293, 668–672 [DOI] [PubMed] [Google Scholar]
- 16. Ferguson G. P., Roop R. M., 2nd, Walker G. C. (2002) Deficiency of a Sinorhizobium meliloti BacA mutant in alfalfa symbiosis correlates with alteration of the cell envelope. J. Bacteriol. 184, 5625–5632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., p. A.1, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 18. Chou P. Y., Fasman G. D. (1978) Empirical predictions of protein conformation. Annu. Rev. Biochem. 47, 251–276 [DOI] [PubMed] [Google Scholar]
- 19. Schroeder B. O., Wu Z., Nuding S., Groscurth S., Marcinowski M., Beisner J., Buchner J., Schaller M., Stange E. F., Wehkamp J. (2011) Reduction of disulphide bonds unmasks potent antimicrobial activity of human β-defensin 1. Nature 469, 419–423 [DOI] [PubMed] [Google Scholar]
- 20. Alegre-Cebollada J., Kosuri P., Rivas-Pardo J. A., Fernández J. M. (2011) Direct observation of disulfide isomerization in a single protein. Nat. Chem. 3, 882–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sochacki K. A., Barns K. J., Bucki R., Weisshaar J. C. (2011) Real-time attack on single Escherichia coli cells by the human antimicrobial peptide LL-37. Proc. Natl. Acad. Sci. U.S.A. 108, E77–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Alkhalfioui F., Renard M., Frendo P., Keichinger C., Meyer Y., Gelhaye E., Hirasawa M., Knaff D. B., Ritzenthaler C., Montrichard F. (2008) A novel type of thioredoxin dedicated to symbiosis in legumes. Plant Physiol. 148, 424–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Missiakas D., Raina S. (1997) Protein folding in the bacterial periplasm. J. Bacteriol. 179, 2465–2471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Riemer J., Bulleid N., Herrmann J. M. (2009) Disulfide formation in the ER and mitochondria: two solutions to a common process. Science 324, 1284–1287 [DOI] [PubMed] [Google Scholar]
- 25. Marlow V. L., Haag A. F., Kobayashi H., Fletcher V., Scocchi M., Walker G. C., Ferguson G. P. (2009) Essential role for the BacA protein in the uptake of a truncated eukaryotic peptide in Sinorhizobium meliloti. J. Bacteriol. 191, 1519–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wehmeier S., Arnold M. F., Marlow V. L., Aouida M., Myka K. K., Fletcher V., Benincasa M., Scocchi M., Ramotar D., Ferguson G. P. (2010) Internalization of a thiazole-modified peptide in Sinorhizobium meliloti occurs by BacA-dependent and -independent mechanisms. Microbiology 156, 2702–2713 [DOI] [PubMed] [Google Scholar]
- 27. Saeki K. (2011) Rhizobial measures to evade host defense strategies and endogenous threats to persistent symbiotic nitrogen fixation: a focus on two legume-rhizobium model systems. Cell Mol. Life Sci. 68, 1327–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. LeVier K., Phillips R. W., Grippe V. K., Roop R. M., 2nd, Walker G. C. (2000) Similar requirements of a plant symbiont and a mammalian pathogen for prolonged intracellular survival. Science 287, 2492–2493 [DOI] [PubMed] [Google Scholar]
- 29. Domenech P., Kobayashi H., LeVier K., Walker G. C., Barry C. E., 3rd (2009) BacA, an ABC transporter involved in maintenance of chronic murine infections with Mycobacterium tuberculosis. J. Bacteriol. 191, 477–485 [DOI] [PMC free article] [PubMed] [Google Scholar]