Abstract
Background:
Increased Aurora kinase A gene copy number (AURKA-CN) has been reported in metastatic colorectal cancer (mCRC), with unknown relationship to clinical outcome. We correlated increased AURKA-CN in mCRC tumours with KRAS mutation status, overall and progression-free survival (OS, PFS).
Methods:
Sixty-one mCRC tumours were analysed for AURKA-CN using q-PCR, and KRAS mutation status by direct sequencing. Expression of AURKA protein was analysed by immunohistochemistry. Cox-proportional hazard method, Kaplan–Meier curves and log-rank statistics were used to estimate and compare the hazard ratios and median survival between the groups.
Results:
In all, 68% of tumour exhibited high AURKA-CN, and 29% had a KRAS mutation, without correlation between the two. Patients with high AURKA-CN tumours had longer median OS (48.6 vs 18.8 months, P=0.01), with stronger trend among KRAS wild-type tumours (median OS not reached vs 18.8 months, P=0.003). Progression-free survival was longer on first-line or second-line chemotherapy among patients with KRAS wild-type and high vs low AURKA-CN (first: 17.6 vs 5.13 months, P=0.04; second: 10.4 vs 5.1 months, P=0.01). AURKA-CN level did not affect outcomes among patients with KRAS mutant tumours.
Conclusion:
Increased AURKA-CN is common in mCRC tumours and is associated with longer OS and longer PFS during chemotherapy, particularly in KRAS wild-type tumours.
Keywords: colon cancer, aurora kinase A, copy number, KRAS mutation
Aurora kinases (Aurora A, B and C) are important regulatory proteins of the mitotic process (Goepfert and Brinkley, 2000; Sen et al, 2008). Due to their crucial function in cell division, these proteins have been extensively studied in many cancers for their role in carcinogenesis and as potential treatment targets (Zeng et al, 2007; Nadler et al, 2008; Park et al, 2008; Qu et al, 2008; Tanaka et al, 2008; Wan et al, 2008). Aurora kinase A (AURKA) (also known as Aurora-2, BTAK/STK15) regulates mitotic entry, centrosome maturation, bipolar spindle assembly, chromosome alignment, cytokinesis and exit from mitosis (Berdnik and Knoblich, 2002; Hirota et al, 2003; Marumoto et al, 2003; Dutertre et al, 2004). Aberrations in the function of Aurora kinases can result in abnormal cell division and aneuploidy due to losses or gains of whole chromosomes (Marumoto et al, 2003). Amplification of AURKA has been shown to induce the formation of a multipolar mitotic spindle, which results in abnormal chromosome alignment and cell division (Meraldi et al, 2002). Resultant genomic instability, aneuploidy and hyperploidy can promote tumour development.
In colorectal cancer (CRC), AURKA protein overexpression and amplification have been frequently observed. Bischoff et al (1998) were the first to report overexpression of AURKA mRNA in >50% of CRC tumours. Baba et al (2009) later reported overexpression of AURKA protein by immunohistochemistry (IHC) in 19% of CRC samples of various disease stages. In a multivariate analysis, AURKA protein overexpression was associated with chromosomal instability (identified as loss of heterozygosity in 2p, 5q, 17q and 18q) but did not correlate with clinical outcomes. Lam et al (2008) identified AURKA overexpression by IHC in 48.5% of early-stage CRC samples. This finding was associated with well/moderately differentiated tumours (P=0.04), tumours of the distal colon (P=0.01) and non-mucinous histology (P=0.001). However, no association with clinical outcomes was detected.
The group led by Nishida et al (2007) was the first to demonstrate an increase in AURKA gene copy number (AURKA-CN) in 29% of a small sample of colorectal tumours. This study demonstrated a strong correlation between increased AURKA-CN and chromosomal instability, and no association between increased AURKA-CN and KRAS mutation status. Recently, the group led by Zhang et al (2010) reported increased AURKA-CN in 32% of CRC tumour samples and particularly in higher stage tumours, suggesting that AURKA may have a role in tumour progression. To date, the finding of increased AURKA-CN has not been correlated with clinical outcomes of patients with CRC.
In recent years, the KRAS pathway has been proven to have an important predictive role in the treatment of CRC (Cunningham et al, 2004; Van Cutsem et al, 2007; Karapetis et al, 2008; Douillard et al, 2010; Bokemeyer et al, 2011). Anti-EGFR therapy is effective and approved for the management of metastatic CRC (mCRC) without KRAS mutations. Earlier work by our group has demonstrated synergistic lethality achieved by inhibition of both the Aurora and the KRAS pathways on CRC tissue samples (Astsaturov et al, 2010). The goal in this study was to assess the frequency of increased AURKA-CN in archival tumour tissues of patients with metastatic CRC (mCRC) and correlate this finding with progression-free survival (PFS), overall survival (OS), and KRAS mutation status.
Patients and Methods
Patient samples
We analysed 61 consecutive mCRC tumour samples submitted between 2006 and 2009 to the Fox Chase Cancer Center (FCCC) molecular genetic facility for evaluation of KRAS mutation status, with remaining tissue available for AURKA-CN evaluation. Most of the patients had their tumour samples submitted by their treating oncologist for KRAS mutation status evaluation after the test became routinely used in clinical practice (2008). Twenty patients in this cohort were enrolled on an institutional phase II study evaluating the combination of capecitabine, oxaliplatin, cetuximab and bevacizumab in the front line metastatic setting (between 2006 and 2008). These patients submitted tissue samples for future research at time of study enrollment, which was used for this analysis. Overall survival data were available for 59 out of 61 patients. Fifty-three patients had received chemotherapy treatment at FCCC and were evaluable for PFS. Progression-free survival was defined as the time that elapsed from treatment initiation until evidence of progressive disease or death. For the purpose of this analysis, deintensification of therapy for adverse events, or planned treatment breaks were considered as same line of therapy.
Evaluation of AURKA-CN
We utilised a quantitative genomic PCR method described by Nishida et al (2007) to evaluate the AURKA gene copy number. In all, 20 ng of genomic DNA purified from paraffin-embedded tissue (PET) sections using the QIAamp DNA Mini Kit (QIAGEN, Valencia, CA, USA) was used. A real-time PCR method was used to determine copy number alterations in the AURKA gene at the Fox Chase Genomics and Genetics Facility using an ABI Prism 7900 sequence detection system (Applied Biosystems, Foster City, CA, USA). The primer and probe sequences for genomic real-time PCR for each of the genes were as follows: the Aurora A forward primer: 5′-TCTTTTATAGAAATGTGTGGAAGTTCCT-3′ reverse primer: 5′-CAATAAAAAAGTACAGACGCATAAACCA-3′ probe: 5′-CTGTCCTTAGAAATAACCACTAC-3′. All probes were labelled with FAM as the reporter dye and TAMRA as the quencher. Each PCR amplification reaction was performed in triplicate to ensure accurate results. The RNAse-P used was the endogenous reference gene, for which a set primer probe was purchased from Applied Biosystems. Genomic DNA isolated from DLD1 or HCT116 cell lines served as negative controls for AURKA gene amplification. DNA from the Caco-2 cell line provided a positive control as the AURKA gene locus in this cell line has been reported as four-fold amplified (Nishida et al, 2007).
KRAS mutation status evaluation
Tumour samples were tested for the presence of a KRAS mutation on codons 12 and 13. DNA analyses were performed within the Fox Chase Clinical Molecular Genetics Laboratory. Extraction, isolation and purification of DNA were performed from formalin-fixed PET suitable for molecular analysis using the WaxFree DNA extraction kit (WF-100; TrimGen, Sparks, MD, USA). Ten to fifteen fresh cut unstained slides were used for analysis. DNA (∼10 ng) was amplified by PCR using primer sequences located on either side of the region of the coding exon of interest. PCR products were detected by agarose gel electrophoresis. Mutations were detected by sequencing of the purified PCR amplified product (BigDye Terminator v.1.1 Cycle Sequencing Kit; Applied Biosystems) and evaluated by capillary electrophoresis (ABI 3100; Applied Biosystems).
IHC evaluation of AURKA phosphorylation
Activation of AURKA occurs through autophosphorylation of a threonine-288 residue. We evaluated the hyperactive state of the AURKA by IHC with antibodies specific to the autophosphorylated threonine-288 residue (Aurora A phospho-T288, Abcam (Cambridge, MA, USA), rabbit primer, Cat. AB58494) (Walter et al, 2000). Immunohistochemical staining was performed on 5 μm sections. After deparaffinisation and rehydration, sections were subjected to heat-induced epitope retrieval by immersion in a 0.01 M citrate buffer (pH 6.0). Endogenous peroxidase activity was blocked for 15 min in 3% hydrogen peroxide in methanol. Non-specific binding was blocked by treatment with a blocking reagent (Protein Block Serum-Free; Dako, Carpinteria, CA, USA) for 30 min at room temperature. Immunodetection was performed with the Dako Envision+ system. The antigen–antibody immunoreaction was visualised using 3-3′-diaminobenzidine as the chromogen. The slides were washed, counterstained with haematoxylin, dehydrated with alcohol, cleared in xylene and mounted. Patient samples that were shown previously to express high levels of phospho-T288 Aurora A (pAurora A) were used as positive controls. The negative control was performed by replacing the primary antibody with negative control rabbit IgG.
Evaluation of the staining intensity and positive cell numbers of pAurora A was performed by a pathologist (KC). Tumours with an intermediate or strong nuclear pAurora A immunoreaction in >10% of the tumour cells were scored as pAurora positive. Lack of staining or staining <10% of tumour cells was scored as negative (Lam et al, 2008).
Clinical data collection
A clinicopathological database was created and included demographic information, stage at presentation, treatment regimens, response to therapies and survival. Clinical data were obtained through medical record review using the electronic medical record at FCCC. All patient data were coded and all identifiers were removed before analysis. The study was approved by FCCC's Institutional Review Board.
Statistical analysis
Sixty-one tissue samples of mCRC patients at FCCC were included in this analysis. The distribution of AURKA-CN was examined. Full survival information was available for 59 patients. The distribution of AURKA-CN was examined. There are outliers with very high values. To avoid the large influence from the outliers while studying the association between AURKA-CN and survival, we delineated the values by quartile. Kaplan–Meier curves were constructed and log-rank test analysis showed a significant association (results not shown). After confirming this association, we conducted an analysis to search for the most sensitive cutoff for the definition of ‘high’ AURKA-CN based on our samples. From the Kaplan–Meier curves stratified by quartiles, we found that only the group with AURKA-CN in the first quartile showed a large separation from the remainder of the cohort. The distribution of AURKA-CN was then summarised based on the percentile of the values and used to create data sets with various cutoff points. We examined the data sets with cutoffs from the 25th percentile to 50th percentile. For each data set, Kaplan–Meier curves were created and compared using log-rank statistics and Cox-proportional hazard model. Considering our outcome was longitudinal and traditional area under the ROC curve analyses do not taken into account the varied follow-up time, we utilised the C index (Harrell et al, 1982, 1996) which is a member of the ‘Kendall family’ of rank parameters and is constantly used to estimate the concordance probability with censored data. Like AUC, a value of 0.5 implies complete discordance. Higher values suggest higher concordance between the data and the predicted values from the model. This statistical analysis enabled us to determine the best cutoff points by comprehensively considering hazard ratios (HRs), P-values, separation between Kaplan-Meier curves and C index. After we determined the best cutoff points, we compared the baseline demographics and treatment using a t-test or χ2-test depending on whether the variable under consideration was continuous or categorical. As the groups were well balanced, no further adjustment for confounding factors was performed.
Results
Patient and treatment characteristics
Table 1 summarises the characteristics and treatment of patients whose tumours were included in this cohort. The majority of patients in this analysis had initially presented with metastatic disease and were treated with FOLFOX (infusional and bolus 5-fluoruracil, leucovorin and oxaliplatin) plus bevacizumab. All characteristics were well balanced between high and low AURKA-CN cohorts (Table 1). The use of various chemotherapeutic agents was also well balanced between the groups with the exception of bevacizumab, which was used more commonly in the low AURKA-CN group in the second-line setting.
Table 1. Patient and treatment characteristics.
| Full cohort (N=59) | Low Aurora CN ⩽2.6 (N=18) | High Aurora CN >2.6 (N=41) | P-value | |
|---|---|---|---|---|
| Age, mean (s.d.) | 58.6 (1.44) | 58.6 (3.2) | 58.7 (1.58) | 0.97 |
| Gender, N (%) | 0.68 | |||
| Male | 37 (62.7) | 12 (66.7) | 25 (61.0) | |
| Female | 22 (37.3) | 6 (33.3) | 16 (39.0) | |
| Race, N (%) | ||||
| Caucasian | 51 (87.9) | 14 (82.3) | 37 (90.2) | 0.41 |
| AA | 7 (12.1) | 3 (17.7) | 4 (9.8) | |
| Stage at diagnosis, N (%) | 0.80 | |||
| 2 | 1 (1.9) | 0 (0) | 1 (2.7) | |
| 3 | 10 (8.9) | 3 (18.8) | 7 (18.9) | |
| 4 | 42 (79.3) | 13 (81.2) | 29 (78.4) | |
| Clinical and treatment data for 53 patients n(%) | ||||
| (N=53) | (N=16) | (N=37) | ||
| No. of metastatic sites | ||||
| 1 | 30 (56.7) | 11 (51.4) | 19 (68.8) | 0.28 |
| 2 | 20 (37.7) | 16 (43.25) | 4 (25.0) | |
| 3 | 2 (3.8) | 1 (2.7) | 1 (6.25) | |
| 4 | 1 (1.8) | 1 (2.7) | 0 (0.0) | |
| First-line therapy (all patients received 5-FU or capecitabine with initial therapy) | ||||
| Oxaliplatin, N (%) | 45 (84.9) | 11 (81.3) | 34 (86.5) | 0.63 |
| Irinotecan, N (%) | 5 (9.4) | 2 (12.5) | 3 (8.1) | 0.63 |
| Cetuximab, N (%) | 19 (35.9) | 7 (43.8) | 12 (32.4) | 0.54 |
| Bevacizumab, N (%) | 35 (66.0) | 10 (62.5) | 25 (67.6) | 0.76 |
| Bevacizumb+Cetuximab, N (%) | 8 (15.0) | 4 (25) | 4 (10.8) | 0.19 |
| Second-line therapy | ||||
| (N=33) | (N=11) | (N=22) | ||
| 5-FU or Capecitabine, N (%) | 14 (42.4) | 7 (63.6) | 7 (31.8) | 0.14 |
| Irinotecan, N (%) | 26 (78.7) | 9 (81.8) | 17 (77.3) | 0.57 |
| Oxaliplatin, N (%) | 2 (6) | 1 (9) | 1 (4.5) | 0.56 |
| Bevacizumab, N (%) | 11 (33.3) | 8 (72.7) | 3 (13.6) | 0.001 |
| Cetuximab/Panitumumab, N (%) | 17 (51.5) | 6 (54.5) | 11 (50) | 0.55 |
| Third-line therapy | 9 (27.2) | 2 (18.2) | 7 (31.8) | 0.35 |
Abbreviations: AA=African American; CN=copy number.
Determination and frequency of high AURKA-CN and KRAS
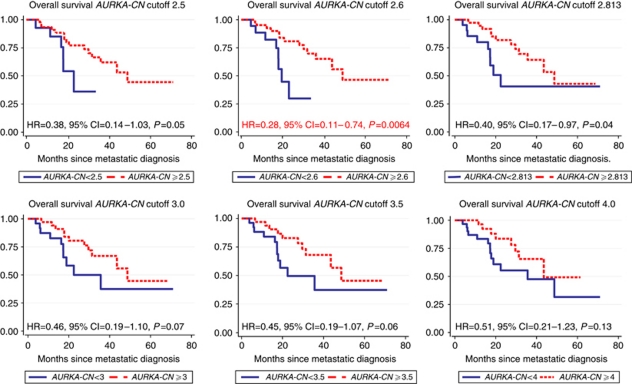
Of the 61 tissue samples obtained for AURKA-CN analysis, 62% originated from primary tumours, while 38% originated from metastatic sites. This distribution was similar between the high AURKA-CN group and the low AURKA-CN group (P=0.925). In the analysis of AURKA-CN, we viewed any value >2 as representing abnormal expression of the AURKA gene. We thus created multiple data sets using values selected from percentile tables. These data sets were each analysed using various cutoff points to determine the one most predictive of clinical outcomes. An OS analysis demonstrated statistically significant differences in OS using multiple cutoff values <3.0 (Table 2; Figure 1). The percentile analysis demonstrated a cutoff of 2.6 (defining approximately the 30th percentile; Table 2) as the most sensitive and this was selected as the cutoff value used to define high vs low AURKA-CN for further analyses.
Table 2. Survival analysis utilising various AURKA-CN cutoff values for the entire cohort.
| Cutoff | Percentage of low AURKA-CN | HR | s.e. | Z | P>∣z∣ | 95% Confidence interval | Harrell's C index | Log-rank P-value |
|---|---|---|---|---|---|---|---|---|
| 2.5 | 24 | 0.377 | 0.193 | −1.9 | 0.057 | 0.138–1.030 | 0.59 | 0.05 |
| 2.6 | 31 | 0.280 | 0.138 | −2.57 | 0.01 | 0.106–0.738 | 0.63 | 0.0064 |
| 2.813 | 36 | 0.401 | 0.180 | −2.03 | 0.042 | 0.166–0.969 | 0.64 | 0.04 |
| 3 | 41 | 0.461 | 0.205 | −1.74 | 0.081 | 0.193–1.101 | 0.62 | 0.07 |
| 3.5 | 44 | 0.447 | 0.199 | −1.8 | 0.071 | 0.187–1.072 | 0.62 | 0.06 |
| 4 | 52 | 0.507 | 0.229 | −1.5 | 0.133 | 0.210–1.228 | 0.61 | 0.13 |
| 5 | 64 | 0.633 | 0.307 | −0.94 | 0.347 | 0.245–1.640 | 0.58 | 0.34 |
Abbreviation: AURKA-CN=Aurora kinase A gene copy number.
Figure 1.
Kaplan–Meier survival estimates by AURKA-CN and various cutoff values.
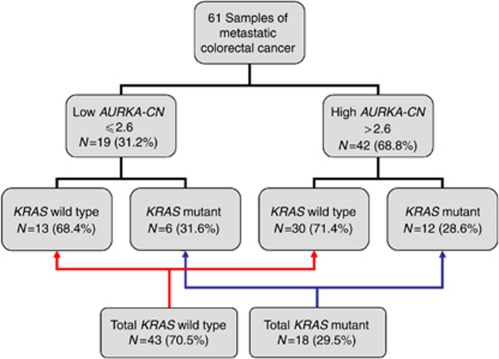
By these criteria, 42 of 61 (68.85%) samples expressed increased AURKA-CN. AURKA-CN values ranged from 0.122 to 23.73 (mean 4.875, s.d. 3.96). A KRAS mutation was identified in 18 of 61 samples (29.5%). The most common mutation noted was in codon 12 (16 samples, 89%) with two patient tumours having a mutation in codon 13. No correlation was noted between KRAS mutation status and the presence of high or low AURKA-CN (P=0.81; Figure 2).
Figure 2.
Frequency of KRAS mutations by AURKA-CN.
AURKA-CN, KRAS and PFS
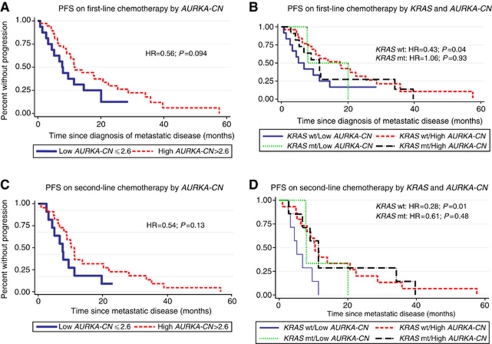
Complete treatment information was available for 53 out of 61 patients, while eight patients were lost to follow-up or elected to receive treatment at an outside institution. The median PFS on first-line chemotherapy was 11.4 months for the full cohort. For patients with high AURKA-CN tumours, the median PFS on first-line chemotherapy was 11.5 months vs 7.7 months for patients with low AURKA-CN tumours (HR=0.56, 95% CI: 0.28–1.1, P=0.094; Figure 3A). Among patients with KRAS wild-type tumours, those with high AURKA-CN tumours had prolonged PFS compared with patients with low AURKA-CN tumours (HR=0.43, 95% CI: 0.19–0.94, P=0.04; Figure 3B). Conversely, PFS did not differ by AURKA-CN expression among patients with KRAS mutant tumours (HR=1.06, 95% CI: 0.28–3.93, P=0.93), although the small sample size limits definitive conclusions regarding this population.
Figure 3.
Progression-free survival (PFS) of metastatic colorectal cancer patients receiving first-line and second-line chemotherapy by AURKA-CN and KRAS mutation status. (A) PFS on first-line chemotherapy by AURKA-CN; (B) PFS on first-line chemotherapy by AURKA-CN and KRAS mutation status; (C) PFS on second-line chemotherapy by AURKA-CN; and (D) PFS on second-line chemotherapy by AURKA-CN and KRAS mutation status.
Thirty-three patients in our cohort who received second-line chemotherapy were evaluable for PFS. The median PFS on second-line chemotherapy was 9.1 months for the full cohort. No statistically significant difference in PFS was noted among patients with high vs low AURKA-CN tumours in the group overall (10.4 months vs 7.7 months; HR=0.54, 95% CI: 0.24–1.19, P=0.13; Figure 3C). However, as with response to first-line treatment, patients with high AURKA-CN and KRAS wild-type status had an improved PFS compared with patients with low AURKA-CN and KRAS wild-type status (10.4 months vs 5.1 months; HR=0.28, 95% CI: 0.11–0.74, P=0.01; Figure 3D).
We next performed an exploratory analysis to determine if the relationship of AURKA-CN to PFS was more pronounced in patients receiving the anti-EGFR antibody cetuximab. Table 3 outlines median PFS and HRs for high and low AURKA-CN tumours by KRAS mutational status and the use of cetuximab. Interestingly, among patients with KRAS wild-type tumours, high AURKA-CN appeared to have the largest association with outcome among patients who did not receive cetuximab. However, small numbers preclude any definitive conclusion.
Table 3. Progression-free survival on first-line chemotherapy by KRAS mutation status and receipt of cetuximab.
| N | Low AURKA-CN median PFS (months) | High AURKA-CN median PFS (months) | HR | P-value | |
|---|---|---|---|---|---|
| First-line chemotherapy | |||||
| All patients | 53 | 7.7 (95% CI: 3.3–14.5) | 11.5 (95% CI: 8.9–20.6) | 0.57 (95% CI: 0.29–1.1) | 0.10 |
| No cetuximab exposure | 34 | 5.1 (95% CI: 1.4–14.5) | 11.5 (95% CI: 8.9–20.7) | 0.47 (95% CI: 0.20–1.11) | 0.08 |
| With cetuximab exposure | 19 | 7.9 (95% CI: 0.7–XXa) | 10.2 (95% CI: 4.5–XXa) | 0.68 (95% CI: 0.21–2.17) | 0.51 |
| Odds ratio for cetuximab | 0.70 (0.23–2.18) P=0.54 | 0.90 (0.39–2.06) P=0.80 | |||
| First-line chemotherapy – KRAS wild type | |||||
| All patients | 38 | 5.13 (95% CI: 1.4–14.5) | 17.6 (95% CI: 9.1–25.3) | 0.43 (95% CI: 0.19–0.94) | 0.04 |
| No cetuximab exposure | 24 | 4.4 (95% CI: 1.4–11.4) | 14.0 (95% CI: 6.2–22.4) | 0.24 (95% CI: 0.08–0.67) | 0.01 |
| With cetuximab exposure | 14 | 9.6 (95% CI: 0.7–XXa) | 17.6 (95% CI: 2.5–XXa) | 0.71 (95% CI: 0.17–3.04) | 0.65 |
| Odds ratio for cetuximab | 0.39 (0.10–1.55) P=0.18 | 0.78 (0.29–2.09) P=0.62 | |||
| First-line chemotherapy – KRAS mutant | |||||
| All patients | 15 | 7.9 (95% CI: 7.7–XXa) | 11.4 (95% CI: 4.2–34.2) | 1.06 (95% CI: 0.29–3.93) | 0.93 |
| No cetuximab exposure | 10 | 20.1 (95% CI: XXa–XXa) | 11.4 (95% CI: 4.2–34.1) | 2.5 (95% CI: 0.29–20.9) | 0.46 |
| With cetuximab exposure | 5 | 7.7 (95% CI: 7.7–XXa) | 5.3 (95% CI: 2.8–XXa) | 1.1 (95% CI: 0.15–7.9) | 0.95 |
| Odds ratio for cetuximab | Number too small to estimate | 1.36 (0.27–6.84) P=0.71 | |||
Abbreviations: AURKA-CN=Aurora kinase A gene copy number; CI=confidence interval; HR=hazard ratio; PFS=progression-free survival.
Not enough data to estimate the complete 95% CI.
AURKA-CN, KRAS and OS
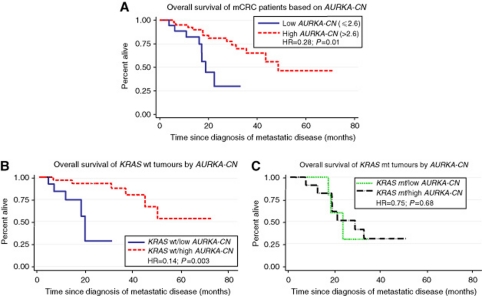
The median follow-up of patients in this cohort was 742 days (range 173–3229 days). When using quartiles, one quartile increase of AURKA-CN reduced the hazard by 0.63 (95% CI: 0.40–0.97, P=0.04). The C index is the highest at a cutoff of 2.813. However, the P-values for Cox model and log-rank tests were barely significant. The C index for cutoff at 2.6 is very similar while the HR is much lower. Thus, we decided to use a cutoff of 2.6 for the rest of the analysis. A significantly longer OS was noted among patients with high vs low AURKA-CN (median OS 48.6 months for patients with high AURKA-CN tumours compared with 18.8 months for patients with low AURKA-CN tumours, HR=0.28, 95% CI: 0.10–0.73, P=0.01; Figure 4A). In all, 1-year and 2-year survival were also longer among patients with high AURKA-CN tumours compared with those with low AURKA-CN tumours (1 year: 92.5% vs 82.2% P<0.001; 2 year: 80.9% vs 29.9%, P<0.001).
Figure 4.
Overall survival of metastatic colorectal cancer patients by AURKA-CN for entire cohort (A), the KRAS wild-type population (B) and the KRAS mutant population (C).
Similarly to PFS, the longer OS for patients with high AURKA-CN tumours was particularly pronounced in the KRAS wild-type patient population (Figure 4B). Among patients with KRAS wild-type tumours, the median OS was not reached for high AURKA-CN compared with 18.8 months for the group with low AURKA-CN tumours (HR=0.14, 95% CI: 0.038–0.514, P=0.003). Similarly, increased 1-year and 2-year survival was seen among those patients with KRAS wild-type tumours and high AURKA-CN compared with patients with KRAS wild-type tumours and low AURKA-CN (1 year: 96% vs 75% P<0.0001; 2 year: 93% vs 28% P<0.0001). No difference in survival was noted by AURKA-CN status in the KRAS mutated population (Figure 4C, HR=0.75, 95% CI: 0.18–2.98, P=0.68).
IHC evaluation of AURKA hyperactivity
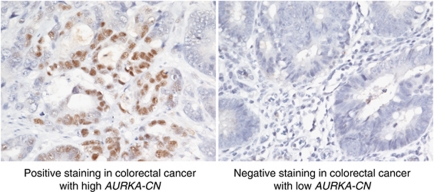
Immunohistochemical evaluation of phosphorylated AURKA protein was performed on 27 available tumour samples (20 with high AURKA-CN and 7 with low AURKA-CN). Among the seven tumours with low AURKA-CN, none were positive for phosphorylated T-288. However, among the 20 tumours with high AURKA-CN, 9 were found to have positive staining (P=0.06 for difference between high and low AURKA-CN tumours). A representative image of positive and negative phosphorylated AURKA staining is provided in Figure 5.
Figure 5.
Immunohistochemistry staining for phosphorylated AURKA in metastatic colorectal cancer samples. Activated state of the Aurora A kinase was evaluated using specific antibodies to the phosphorylated threonine-288 residue (Aurora A phospho-T288). Positive staining (>10% nuclear staining) is displayed on the left and negative staining is displayed on the right.
Discussion
Although five classes of systemic agents are available for the treatment of advanced CRC, the majority of patients are not cured and median survival has plateaued near 2 years (Saltz et al, 2008; Hecht et al, 2009; Tol et al, 2009; Jemal et al, 2010; American Cancer Society, 2010). Thus, the search for novel targets such as AURKA is of critical importance. Better understanding of the role of the Aurora kinases and their activity in CRC would support the development of new treatment modalities targeting this pathway. Previous groups have studied the frequency of increased AURKA-CN in CRC samples using various cutoff values for defining high AURKA-CN. We performed a novel statistical analysis incorporating the HRs, P-values and Kaplan–Meier survival curves to determine the most sensitive cutoff value. Our analysis demonstrated the most sensitive cutoff value to be 2.6, which resulted in identification of 68% of mCRC samples as having high AURKA-CN. The groups led by Nishida et al (2007) and Zhang et al (2010) reported increased AURKA-CN in ∼30% of CRC tumours. The differences in frequency may simply reflect alternate methodology and cutoffs. The two aforementioned studies were conducted in an Asian patient population with all stages of disease. Differences in patient population and stage of disease studied may thus also account for differences in AURKA-CN frequency.
The role of Aurora kinase in tumourigenesis is well documented, although its utility as a prognostic marker is still under investigation in many cancers. Nadler et al (2008) reported a high expression of Aurora A by AQUA to be a poor prognostic marker in patients with breast cancer. Similarly overexpression of Aurora kinase B was found to be a poor prognostic marker in non-small cell lung cancer (Smith et al, 2005). In contrast, our report in mCRC is the first demonstrating an association between high AURKA-CN and longer OS. A few groups have previously been unable to demonstrate a correlation between AURKA protein expression by IHC and clinical outcome. Similarly to our findings, Lam and colleagues have demonstrated higher detection of the AURKA protein by IHC in low-grade (well or moderately differentiated) CRC samples compared with high-grade (poorly differentiated) tumours. Therefore, high AURKA-CN may be a marker of a less aggressive biology with improved clinical outcome. Alternatively, increased AURKA-CN may predict for improved benefit from chemotherapy by rendering the cells more susceptible, due to abnormal cell divisions. Our data demonstrating increased PFS for patients with high AURKA-CN tumours support this hypothesis. However, as our sample population did not include untreated patients, this hypothesis requires further evaluation.
The improved OS for patients with high AURKA-CN was most pronounced in our study among patients with KRAS wild-type tumours. Furthermore, the improvement in PFS on first-line and second-line chemotherapy was most pronounced in this specific patient population. The small sample size of this subset analysis, while demonstrating statistical significance, limits definitive conclusion and requires further validation in an independent data set. We did not observe any correlation between the presence of KRAS mutations and AURKA-CN, suggesting these may represent two independent pathways in the biology of CRC. By combining these two biomarkers, we may be able to identify a subgroup of patients with mCRC who exhibit increased response to therapy and superior outcomes. The improved response of patients with KRAS wild-type tumours treated with anti-EGFR therapy has been well documented in the literature (Cunningham et al, 2004; Karapetis et al, 2008; Van Cutsem et al, 2009; Douillard et al, 2010; Peeters et al, 2010; Bokemeyer et al, 2011). However in our study, among patients with KRAS wild-type tumours, the impact of high AURKA-CN level was most pronounced in patients who did not receive cetuximab. One unifying hypothesis may be that Aurora A overexpression confers sensitivity to chemotherapy and not to anti-EGFR therapy. Alternatively, the relatively small number of patients available for this subgroup analysis may preclude a meaningful assessment of this association. In-vitro work has demonstrated strong synergy between anti-EGFR drugs and novel agents targeting the Aurora pathway, which may potentially become a therapeutic approach (Astsaturov et al, 2010). Further research to evaluate the response to EGFR-targeted therapy in patients with high and low AURKA-CN may further define the interaction between these two pathways.
Many groups have studied AURKA protein expression by IHC in CRC and other cancers. In CRC, increased expression of AURKA protein by IHC has been noted in 19–30% of patients (Bischoff et al, 1998; Lam et al, 2008; Baba et al, 2009). In our study, IHC analysis was performed on approximately one third of the available samples, of which a third displayed positive staining (n=9; 33%). Our results demonstrated lack of staining of samples with low AURKA-CN, and positive staining in less than half of the samples with high AURKA-CN. Increased gene copy number is expected to result in increased activity of the product protein. Thus, the fact that most of the samples with high AURKA-CN were not found to have a positive stain suggests the possibility that despite high copy number AURKA may in fact be inactive in these cells due to post-transcriptional and post-translational regulation. Alternatively, this observation may raise questions regarding the sensitivity of IHC staining. IHC carries many challenges, most notably a variable pattern of staining and the presence of artifacts related to the fixation process. Differences in the preservation process between the samples may also result in various rates of protein degradation, which may affect the IHC protein expression. Furthermore, this assay is user dependent and may suffer from significant inter-reader variability. A PCR-based method is likely to be more accurate and sensitive in detecting increased activity of a pathway at the gene level.
Our study does have several limitations. As a retrospective review, we were limited by specimen availability, which may have introduced unexpected bias into the results. However, by using all specimens available during a specific timeframe, we attempted to minimise this impact. The small sample size limits our ability to draw clear conclusions regarding some of the analyses conducted in the study, such as the interaction between AURKA-CN level and the use of cetuximab. We also did not evaluate a validation cohort to support our chosen cutoff for the analysis of AURKA-CN level. However, all cutoffs selected demonstrated improved clinical outcome for patients with high compared with low AURKA-CN.
In summary, we demonstrated a high frequency of increased AURKA-CN in mCRC samples using a novel statistical methodology to evaluate the most appropriate cutoff for analysis. Moreover, our study is the first to demonstrate an association between high AURKA-CN and improved clinical outcome among patients with mCRC receiving chemotherapy, with a more pronounced association noted among patients with KRAS wild-type tumours. Additional study utilising tissue from larger randomised studies to distinguish the prognostic and predictive impact of AURKA-CN is warranted. Implications of these findings for future development of agents targeting Aurora kinase should be considered.
Acknowledgments
This study was supported by AMGEN Medical Oncology Fellowship Award (Grant number: 3840610) and by the core grant of the Comprehensive Cancer Center Program at Fox Chase – 3P30CA006927-47S4. Dr Golemis is supported by NIH funding (Grant number: R01CA63366).
Footnotes
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
References
- American Cancer Society (2010) Cancer Facts & Figures 2010. American Cancer Society [Google Scholar]
- Astsaturov I, Ratushny V, Sukhanova A, Einarson MB, Bagnyukova T, Zhou Y, Devarajan K, Silverman JS, Tikhmyanova N, Skobeleva N, Pecherskaya A, Nasto RE, Sharma C, Jablonski SA, Serebriiskii IG, Weiner LM, Golemis EA (2010) Synthetic lethal screen of an EGFR-centered network to improve targeted therapies. Sci Signal 3: ra67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba Y, Nosho K, Shima K, Irahara N, Kure S, Toyoda S, Kirkner GJ, Goel A, Fuchs CS, Ogino S (2009) Aurora-A expression is independently associated with chromosomal instability in colorectal cancer. Neoplasia 11: 418–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdnik D, Knoblich JA (2002) Drosophila Aurora-A is required for centrosome maturation and actin-dependent asymmetric protein localization during mitosis. Curr Biol 12: 640–647 [DOI] [PubMed] [Google Scholar]
- Bischoff JR, Anderson L, Zhu Y, Mossie K, Ng L, Souza B, Schryver B, Flanagan P, Clairvoyant F, Ginther C, Chan CS, Novotny M, Slamon DJ, Plowman GD (1998) A homologue of Drosophila aurora kinase is oncogenic and amplified in human colorectal cancers. EMBO J 17: 3052–3065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokemeyer C, Bondarenko I, Hartmann JT, de Braud F, Schuch G, Zubel A, Celik I, Schlichting M, Koralewski P (2011) Efficacy according to biomarker status of cetuximab plus FOLFOX-4 as first-line treatment for metastatic colorectal cancer: the OPUS study. Ann Oncol 22: 1535–1546 [DOI] [PubMed] [Google Scholar]
- Cunningham D, Humblet Y, Siena S, Khayat D, Bleiberg H, Santoro A, Bets D, Mueser M, Harstrick A, Verslype C, Chau I, Van Cutsem E (2004) Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med 351: 337–345 [DOI] [PubMed] [Google Scholar]
- Douillard JY, Siena S, Cassidy J, Tabernero J, Burkes R, Barugel M, Humblet Y, Bodoky G, Cunningham D, Jassem J, Rivera F, Kocakova I, Ruff P, Blasinska-Morawiec M, Smakal M, Canon JL, Rother M, Oliner KS, Wolf M, Gansert J (2010) Randomized, phase III trial of panitumumab with infusional fluorouracil, leucovorin, and oxaliplatin (FOLFOX4) vs FOLFOX4 alone as first-line treatment in patients with previously untreated metastatic colorectal cancer: the PRIME study. J Clin Oncol 28: 4697–4705 [DOI] [PubMed] [Google Scholar]
- Dutertre S, Cazales M, Quaranta M, Froment C, Trabut V, Dozier C, Mirey G, Bouche JP, Theis-Febvre N, Schmitt E, Monsarrat B, Prigent C, Ducommun B (2004) Phosphorylation of CDC25B by Aurora-A at the centrosome contributes to the G2-M transition. J Cell Sci 117: 2523–2531 [DOI] [PubMed] [Google Scholar]
- Goepfert TM, Brinkley BR (2000) The centrosome-associated Aurora/Ipl-like kinase family. Curr Top Dev Biol 49: 331–342 [DOI] [PubMed] [Google Scholar]
- Harrell Jr FE, Califf RM, Pryor DB, Lee KL, Rosati RA (1982) Evaluating the yield of medical tests. JAMA 247: 2543–2546 [PubMed] [Google Scholar]
- Harrell Jr FE, Lee KL, Mark DB (1996) Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med 15: 361–387 [DOI] [PubMed] [Google Scholar]
- Hecht JR, Mitchell E, Chidiac T, Scroggin C, Hagenstad C, Spigel D, Marshall J, Cohn A, Mccollum D, Stella P, Deeter R, Shahin S, Amado RG (2009) A randomized phase IIIB trial of chemotherapy, bevacizumab, and panitumumab compared with chemotherapy and bevacizumab alone for metastatic colorectal cancer. J Clin Oncol 27: 672–680 [DOI] [PubMed] [Google Scholar]
- Hirota T, Kunitoku N, Sasayama T, Marumoto T, Zhang D, Nitta M, Hatakeyama K, Saya H (2003) Aurora-A and an interacting activator, the LIM protein Ajuba, are required for mitotic commitment in human cells. Cell 114: 585–598 [DOI] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Xu J, Ward E (2010) Cancer statistics, 2010. CA Cancer J Clin 60: 277–300 [DOI] [PubMed] [Google Scholar]
- Karapetis CS, Khambata-Ford S, Jonker DJ, O’callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD, Robitaille S, Price TJ, Shepherd L, Au HJ, Langer C, Moore MJ, Zalcberg JR (2008) K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med 359: 1757–1765 [DOI] [PubMed] [Google Scholar]
- Lam AK, Ong K, Ho YH (2008) Aurora kinase expression in colorectal adenocarcinoma: correlations with clinicopathological features, p16 expression, and telomerase activity. Hum Pathol 39: 599–604 [DOI] [PubMed] [Google Scholar]
- Marumoto T, Honda S, Hara T, Nitta M, Hirota T, Kohmura E, Saya H (2003) Aurora-A kinase maintains the fidelity of early and late mitotic events in HeLa cells. J Biol Chem 278: 51786–51795 [DOI] [PubMed] [Google Scholar]
- Meraldi P, Honda R, Nigg EA (2002) Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53−/− cells. EMBO J 21: 483–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadler Y, Camp RL, Schwartz C, Rimm DL, Kluger HM, Kluger Y (2008) Expression of Aurora A (but not Aurora B) is predictive of survival in breast cancer. Clin Cancer Res 14: 4455–4462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida N, Nagasaka T, Kashiwagi K, Boland CR, Goel A (2007) High copy amplification of the Aurora-A gene is associated with chromosomal instability phenotype in human colorectal cancers. Cancer Biol Ther 6: 525–533 [DOI] [PubMed] [Google Scholar]
- Park HS, Park WS, Bondaruk J, Tanaka N, Katayama H, Lee S, Spiess PE, Steinberg JR, Wang Z, Katz RL, Dinney C, Elias KJ, Lotan Y, Naeem RC, Baggerly K, Sen S, Grossman HB, Czerniak B (2008) Quantitation of Aurora kinase A gene copy number in urine sediments and bladder cancer detection. J Natl Cancer Inst 100: 1401–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peeters M, Price TJ, Cervantes A, Sobrero AF, Ducreux M, Hotko Y, Andre T, Chan E, Lordick F, Punt CJ, Strickland AH, Wilson G, Ciuleanu TE, Roman L, Van Cutsem E, Tzekova V, Collins S, Oliner KS, Rong A, Gansert J (2010) Randomized phase III study of panitumumab with fluorouracil, leucovorin, and irinotecan (FOLFIRI) compared with FOLFIRI alone as second-line treatment in patients with metastatic colorectal cancer. J Clin Oncol 28: 4706–4713 [DOI] [PubMed] [Google Scholar]
- Qu Y, Zhang L, Mao M, Zhao F, Huang X, Yang C, Xiong Y, Mu D (2008) Effects of DNAzymes targeting Aurora kinase A on the growth of human prostate cancer. Cancer Gene Ther 15: 517–525 [DOI] [PubMed] [Google Scholar]
- Saltz LB, Clarke S, Diaz-Rubio E, Scheithauer W, Figer A, Wong R, Koski S, Lichinitser M, Yang TS, Rivera F, Couture F, Sirzen F, Cassidy J (2008) Bevacizumab in combination with oxaliplatin-based chemotherapy as first-line therapy in metastatic colorectal cancer: a randomized phase III study. J Clin Oncol 26: 2013–2019 [DOI] [PubMed] [Google Scholar]
- Sen S, Katayama H, Sasai K (2008) Functional significance of Aurora kinase A in centrosome amplification and genomic instability. Adv Exp Med Biol 617: 99–108 [DOI] [PubMed] [Google Scholar]
- Smith SL, Bowers NL, Betticher DC, Gautschi O, Ratschiller D, Hoban PR, Booton R, Santibanez-Koref MF, Heighway J (2005) Overexpression of aurora B kinase (AURKB) in primary non-small cell lung carcinoma is frequent, generally driven from one allele, and correlates with the level of genetic instability. Br J Cancer 93: 719–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S, Arii S, Yasen M, Mogushi K, Su NT, Zhao C, Imoto I, Eishi Y, Inazawa J, Miki Y, Tanaka H (2008) Aurora kinase B is a predictive factor for the aggressive recurrence of hepatocellular carcinoma after curative hepatectomy. Br J Surg 95: 611–619 [DOI] [PubMed] [Google Scholar]
- Tol J, Koopman M, Cats A, Rodenburg CJ, Creemers GJ, Schrama JG, Erdkamp FL, Vos AH, Van Groeningen CJ, Sinnige HA, Richel DJ, Voest EE, Dijkstra JR, Vink-Borger ME, Antonini NF, Mol L, Van Krieken JH, Dalesio O, Punt CJ (2009) Chemotherapy, bevacizumab, and cetuximab in metastatic colorectal cancer. N Engl J Med 360: 563–572 [DOI] [PubMed] [Google Scholar]
- Van Cutsem E, Kohne CH, Hitre E, Zaluski J, Chang Chien CR, Makhson A, D’haens G, Pinter T, Lim R, Bodoky G, Roh JK, Folprecht G, Ruff P, Stroh C, Tejpar S, Schlichting M, Nippgen J, Rougier P (2009) Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med 360: 1408–1417 [DOI] [PubMed] [Google Scholar]
- Van Cutsem E, Peeters M, Siena S, Humblet Y, Hendlisz A, Neyns B, Canon JL, Van Laethem JL, Maurel J, Richardson G, Wolf M, Amado RG (2007) Open-label phase III trial of panitumumab plus best supportive care compared with best supportive care alone in patients with chemotherapy-refractory metastatic colorectal cancer. J Clin Oncol 25: 1658–1664 [DOI] [PubMed] [Google Scholar]
- Walter AO, Seghezzi W, Korver W, Sheung J, Lees E (2000) The mitotic serine/threonine kinase Aurora2/AIK is regulated by phosphorylation and degradation. Oncogene 19: 4906–4916 [DOI] [PubMed] [Google Scholar]
- Wan XB, Long ZJ, Yan M, Xu J, Xia LP, Liu L, Zhao Y, Huang XF, Wang XR, Zhu XF, Hong MH, Liu Q (2008) Inhibition of Aurora-A suppresses epithelial-mesenchymal transition and invasion by downregulating MAPK in nasopharyngeal carcinoma cells. Carcinogenesis 29: 1930–1937 [DOI] [PubMed] [Google Scholar]
- Zeng WF, Navaratne K, Prayson RA, Weil RJ (2007) Aurora B expression correlates with aggressive behaviour in glioblastoma multiforme. J Clin Pathol 60: 218–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Fang Z, Xiong Y, Li J, Liu L, Li M, Zhang W, Wan J (2010) Copy number increase of aurora kinase A in colorectal cancers: a correlation with tumor progression. Acta Biochim Biophys Sin (Shanghai) 42: 834–838 [DOI] [PubMed] [Google Scholar]