Background: miR-124 is a brain-enriched microRNA that has been shown to be down-regulated in glioma.
Results: miR-124 inhibits glioma cell invasion and tumorigenicity and reduces neurosphere formation, CD133+ cell subpopulation, and stem cell marker expression in part by targeting SNAI2.
Conclusion: Loss of miR-124 enhances the stem-like traits and invasiveness of glioma cells via SNAI2 signaling.
Significance: Therapeutic strategies against glioma can be developed by restoring the level of miR-124.
Keywords: Cancer, Cancer Stem Cells, Head and Neck Cancer, Neural Stem Cell, Stem Cells, SNAI2, Glioma, Invasion, MicroRNA, Self-renewal
Abstract
miR-124 is a brain-enriched microRNA that plays a crucial role in neural development and has been shown to be down-regulated in glioma and medulloblastoma, suggesting its possible involvement in brain tumor progression. Here, we show that miR-124 is down-regulated in a panel of different grades of glioma tissues and in all of the human glioma cell lines we examined. By integrated bioinformatics analysis and experimental confirmation, we identified SNAI2, which is often up-regulated in glioma, as a direct functional target of miR-124. Because SNAI2 has been shown to regulate stem cell functions, we examined the roles of miR-124 and SNAI2 in glioma cell stem-like traits. The results showed that overexpression of miR-124 and knockdown of SNAI2 reduced neurosphere formation, CD133+ cell subpopulation, and stem cell marker (BMI1, Nanog, and Nestin) expression, and these effects could be rescued by re-expression of SNAI2. Furthermore, enhanced miR-124 expression significantly inhibited glioma cell invasion in vitro. Finally, stable overexpression of miR-124 and knockdown of SNAI2 inhibited the tumorigenicity and invasion of glioma cells in vivo. These findings reveal, for the first time, that the tumor suppressor activity of miR-124 could be partly due to its inhibitory effects on glioma stem-like traits and invasiveness through SNAI2.
Introduction
Gliomas are the most lethal primary brain tumors derived from glial tissue. Based on the histopathological and clinical criteria established by the World Health Organization, glioblastoma belongs to grade IV glioma that originates from poorly differentiated astrocytes (1). Glioblastoma is the most common and unfortunately the most aggressive form of brain tumor in adults. The median survival of patients with glioblastoma is <15 months despite the advances in surgical resection, radiotherapy, and chemotherapy. Enormous efforts are being undertaken toward a holistic understanding of the molecular mechanisms responsible for glioblastoma pathogenesis.
Recently, a comprehensive genomic characterization has defined not only the glioblastoma gene signatures (TP53, PTEN, CDKN2A, EGF receptor, IDH1, and IDH2) but also the core signaling pathways (receptor tyrosine kinase/RAS/PI3K, p53, and RB) that are altered in the majority of glioblastoma patients (2). In addition, our group has previously identified cell cycle-related kinase as a novel candidate oncogene in human glioblastoma (3). Although the somatic alterations in protein-coding genes that account for glioblastoma progression have become increasingly clear in recent years, the roles of noncoding genes, particularly microRNAs (miRNAs),5 in glioma pathogenesis are the focus of studies.
Emerging studies have shown that miRNAs are involved in the malignant progression of glioma (4–6). miRNAs, a class of post-transcriptional regulators, are short noncoding RNAs (∼22 nucleotides) that bind to the complementary sequences in the 3′-UTRs of multiple mRNA transcripts, thereby resulting in the silencing of target genes (7). Because miRNA genes are frequently located at the chromosomal fragile sites of cancer genomes (8), miRNAs have been considered as novel classes of oncogenes and tumor suppressors (9). With this in mind, targeting miRNAs, some of which have dysregulated expression in glioblastoma, offers great therapeutic potential for improving the outcome of glioma patients (10, 11).
By miRNA microarray analysis, we previously compared the miRNA expression profiles of normal brain and glioblastoma tissues from Chinese patients. We functionally characterized the roles of two dysregulated miRNAs, miR-15b and miR-146b, in regulating cell cycle progression (12) and inhibiting glioma cell migration and invasion (13), respectively. Our microarray data are concordant with other expression profile studies that reported that miR-124 is significantly down-regulated in glioblastoma samples compared with non-tumor brain tissues (14–16), suggesting that miR-124 may play a critical role in brain tumorigenesis and progression.
miR-124 is a brain-enriched miRNA that has been broadly investigated in physiological neural development (17, 18). However, despites a few studies showing that miR-124 is significantly down-regulated in glioma and medulloblastoma and that miR-124 inhibits the proliferation of glioblastoma cells (16, 19, 20), the roles of miR-124 in central nervous system neoplasia have largely remained unexplored. Here, we investigated the critical role of miR-124 in glioma. We found that miR-124 is frequently down-regulated in both glioma tissues and cell lines. We also demonstrated that miR-124 inhibits glioma stem-like traits and invasiveness and identified SNAI2 as a direct target of miR-124 that mediates the inhibition of these processes.
EXPERIMENTAL PROCEDURES
Cell Lines and Human Tissue Samples
Human glioma cell lines (U87, U118, U138, U373, SW1088, SW1783, and CCF-STTG1) were purchased from American Type Culture Collection (Manassas, VA). U87, U373, U138 and CCF-STTG1 cells were cultured in minimal essential medium, U118 cells in DMEM, and SW1088 and SW1783 cells in Leibovitz's L-15 medium (Invitrogen). All media were supplemented with 10% FBS, 100 units/ml penicillin, and 100 μg/ml streptomycin (all from Invitrogen). Archival frozen human glioma tissue samples and non-glioma patient samples were obtained from the Department of Surgery of The University of Hong Kong. The use of these archival tissues in this study was approved by the Ethics Committee of The University of Hong Kong.
RNA Extraction, Real-time Quantitative RT-PCR, and Semiquantitative RT-PCR
Total RNA was extracted using TRIzol reagent (Invitrogen). The isolated total RNA was polyadenylated and reverse-transcribed for two-step quantitative RT-PCR (qRT-PCR) using the NCodeTM miRNA first-strand synthesis and qRT-PCR kits (Invitrogen) according to the manufacturer's instructions. The sequence-specific forward primers for mature miR-124 and the U6 internal control were 5′-GCGGTGAATGCCAAAAA-3′ (17 bp, GC = 47.07%, Tm = 54.1 °C) and 5′-CGCAAGGATGACACGCAAATTCGT-3′, respectively. For the analysis of mRNA expression, first-strand cDNA was reverse-transcribed using the SuperScript II reverse transcriptase kit (Invitrogen). The primers used were as follows: SNAI2, 5′-ATTCGGACCCACACATTACC-3′ (forward) and 5′-GGTTTTGGAGCAGTTTTTGC-3′ (reverse) (21); BMI1, 5′-AATCCCCACCTGATGTGTGT-3′ (forward) and 5′-GCTGGTCTCCAGGTAACGAA-3′ (reverse) (22); Nanog, 5′-CTAAGAGGTGGCAGAAAAACA-3′ (forward) and 5′-CTGGTGGTAGGAAGAGTAAAGG-3′ (reverse) (23); and Nestin, 5′-GGCAGCGTTGGAACAGAGGT-3′ (forward) and 5′-CATCTTGAGGTGCGCCAGCT-3′ (reverse) (24). The levels of miR-124 or other mRNAs were detected by qRT-PCR or semiquantitative RT-PCR according to our previously described protocols (13).
Establishment of Glioblastoma Stable Cell Lines
One day before transfection, U87 or U373 cells were seeded onto 6-well plates at ∼60% confluence. Cells were transfected with pLL3.7-miR-124 precursor or pLKO.1-shSNAI2 plasmid (Addgene, Cambridge, MA) or cotransfected with pLL3.7-miR-124 and pBabePuro-SNAI2, which lacks the 3′-UTR of SNAI2 (Addgene) using FuGENE HD transfection reagent (Roche Diagnostics) in the absence of antibiotic. After 48 h, cells were subcultured to 10% confluence in medium containing 1 μg/ml puromycin (Sigma). When all cells in the non-transfected control culture were killed, antibiotic-resistant clones were picked and passaged in medium containing half of the concentration of puromycin as in the first round of selection. The expression of miR-124 and SNAI2 was confirmed by real-time qRT-PCR.
In Vitro Matrigel Invasion Assay
Cell invasiveness was assessed with using BioCoat Matrigel invasion chambers (BD Biosciences). Stable cells (5 × 104) resuspended in 500 μl of serum-free medium were seeded into the rehydrated insert. Medium with 10% FBS was added to the lower chamber as chemoattractant. After 22 h of incubation at 37 °C, non-invading cells on the upper surface of the Matrigel membrane were gently removed with a cotton-tipped swab. After fixing with 100% methanol, staining with 1% toluidine blue (Sigma), and rinsing twice with distilled water, the stained invasive cells on the lower surface of the membrane were photographed under an inverted light microscope (×40 objective) and quantified by manual counting in three randomly selected areas. This experiment was performed in triplicate in three independent experiments.
Western Blotting
Cell or tissue lysates were harvested for immunoblot analysis according to our previously described protocol (12). Goat polyclonal antibodies against human SNAI2 (1:400 dilution; D-19, sc-10437) and GAPDH (1:2500 dilution; FL-335, sc-25778) were bought from Santa Cruz Biotechnology (Santa Cruz, CA).
Dual-Luciferase Reporter Assay
The 3′-UTR sequence of SNAI2 predicted to interact with miR-124 or a mutated sequence with the predicted target sites was synthesized and inserted into the XbaI and FseI sites of the pGL3-Control vector (Promega, Madison, WI). These constructs were named pGL3-SNAI2-wt and pGL3-SNAI2-mut. For reporter assay, U87 cells were plated onto 24-well plates and transfected with 100 ng of pGL3-SNAI2-wt or pGL3-SNAI2-mut and 50 nm pLL3.7-miR-124 or pLL3.7-miR-control vector using FuGENE HD. The Renilla luciferase vector pRL-SV50 (5 ng; Promega) was also cotransfected to normalize the differences in transfection efficiency. After transfection for 48 h, cells were harvested and assayed with the Dual-Luciferase reporter assay system (Promega) according to the manufacturer's instructions. Transfection was repeated three times in triplicate.
Neurosphere Formation Assay
Single cells (1 × 104) were plated onto a 6-well ultra-low attachment plate (Corning, Corning, NY) in serum-free DMEM/F-12 supplemented with 10 ng/ml basic fibroblast growth factor, 20 ng/ml epidermal growth factor, 0.4% bovine serum albumin, and B-27 supplement (1:50 dilution; Invitrogen). After 2–3 weeks of culture, the number of neurospheres (diameter > 40 μm) was manually counted in three randomly selected fields at a magnification of ×40 under an inverted microscope. This assay was performed in triplicate in three independent experiments.
Flow Cytometry
Stable glioma cells or neurosphere cells (1 × 106) were resuspended in 100 μl of staining buffer (eBioscience, San Diego, CA) containing 1% FBS and placed on ice for 20 min to block Fc receptors. After incubation with primary phycoerythrin-conjugated anti-human CD133 antibodies (eBioscience) for another 45 min on ice in the dark, the cells were washed twice with 1 ml of ice-cold staining buffer and centrifuged at 400 × g for 5 min at 4 °C. Cells resuspended in 0.5 ml of 2% formaldehyde fixation buffer were analyzed using a FACSCalibur flow cytometer and CellQuest software (BD Biosciences). All flow cytometry results were obtained from two independent experiments performed in triplicate.
Tumor Xenografts in Nude Mice
Following our previous protocol (3), glioma tumor xenografts were established in female BALB/c athymic mice by subcutaneous injection of stable U87-124, U87-shSNAI2, or U87-124-SNAI2 cells or their respective control cells (1.5 × 106) into the back flanks of the mice (n = six mice per group). Tumor size was measured weekly for 6 weeks. Paraffin sections of tumors were subjected to standard H&E staining. This animal study was approved by the Department of Health of the Government of the Hong Kong Special Administrative Region and by the Committee on the Use of Live Animals in Teaching and Research of The University of Hong Kong.
Statistical Analysis
Experimental data are presented as the mean ± S.D. All statistical analyses were performed using Student's two-tailed t test (SPSS 12.0, SPSS Inc., Chicago, IL). Differences were considered to be statistically significant at p < 0.05.
RESULTS
miR-124 Is Down-regulated in Human Glioma Tissues and Cell Lines
To explore the functional role of miR-124 in glioma carcinogenesis, we first analyzed the expression of miR-124 in 27 cases of glioma patient samples and 20 cases of non-glioma patient samples, which included two non-tumor brain tissues, by real-time qRT-PCR. Compared with non-glioma brain tissues, the expression of miR-124 was significantly lower by ∼2-fold in all glioma samples examined (Fig. 1A). Concurrent with this finding, remarkable down-regulation of miR-124 could also be observed in various glioma cell lines (Fig. 1B). Of note, according to a previous study that characterized the invasive property of these cell lines (25), miR-124 expression was lower in highly invasive glioma cells (U87, U118, U138, and U373) than in glioma cells with low invasiveness (SW1088, SW1783, and CCF-STTG1) (Fig. 1B). These results suggest that the decrease in miR-124 expression may account for glioma carcinogenesis and its invasive propensity.
FIGURE 1.
Expression of miR-124 in human glioma tissue samples and cell lines. miR-124 expression was measured by real-time qRT-PCR. A, relative expression level of miR-124 in glioma samples and non-tumor brain tissues. B, miR-124 expression in seven glioma cell lines. The invasiveness of these cell lines was shown according to the data of de Ridder et al. (25). The expression of miR-124 in glioma tissues and cell lines was significantly lower than that in non-tumor tissues (p < 0.05).
SNAI2 Is a Direct Target of miR-124
To elucidate the molecular mechanisms by which miR-124 inhibits glioblastoma cell invasion, we predicted its downstream targets using five prediction algorithms: TargetScan, miRDB, DIANA-microT, miRNAMap, and miRNAviewer. Among the candidate target genes that were commonly predicted by all five algorithms, eight genes had been shown to be down-regulated by miR-124 in microarray analysis (Table 1) (26). LAMC1 and PTBP1 have been reported as targets of miR-124 in neural development (27, 28).
TABLE 1.
Predicted targets of miR-124
| Gene symbol | Gene name | Ref. |
|---|---|---|
| Predicted targets of miR-124a | ||
| CHSY1 | Chondroitin sulfate synthase 1 | |
| EYA4 | Eyes absent homolog 4 | |
| LAMC1 | Laminin, gamma 1 (formerly LAMB2) | 27 |
| LRRC1 | Leucine-rich repeat-containing 1 | |
| PTBP1 | Polypyrimidine tract-binding protein 1 | 28 |
| PTPN12 | Protein-tyrosine phosphatase, non-receptor type 12 | 47 |
| SNAI2 | Snail homolog 2 | 39 |
| TARBP1 | TAR (HIV-1) RNA-binding protein 1 | |
| Other reported targets of miR-124b | ||
| ACTL6A | Actin-like 6A (also known as BRG1-associated factor, BAF53A) | 48 |
| CDK6 | Cyclin-dependent kinase 6 | 16 |
| ITGB1 | Integrin, beta 1 | 27 |
| SCP1 | Small C-terminal domain phosphatase 1 | 49 |
| SLC16A1 | Solute carrier family 16, member 1 | 19 |
| SOX9 | SRY (sex-determining region Y)-box 9 | 17 |
a Eight genes were commonly predicted by TargetScan, miRDB, DIANA-microT, miRNAMap, and miRNAviewer and were down-regulated by miR-124 as detected by previous microarray profiling (26).
b Another six experimentally validated targets of miR-124 are shown for reference.
In this study, we specifically focused on SNAI2, a member of the Snail family of zinc finger transcription factors, because it has been implicated in epithelial-mesenchymal transition and tumor metastasis (29–31). To determine whether SNAI2 is a downstream target of miR-124, we detected the expression of SNAI2 in our established glioblastoma cell lines stably overexpressing miR-124. qRT-PCR analysis showed that the SNAI2 mRNA level was significantly reduced in U87-124 and U373-124 cells (Fig. 2A). By transient transfection, overexpression of miR-124 in U87 cells also down-regulated the endogenous mRNA and protein expression of SNAI2 (Fig. 2B).
FIGURE 2.
SNAI2 is a direct downstream target of miR-124. A, the level of SNAI2 expression in stable U87 and U373 cells with miR-124 overexpression was analyzed by real-time qRT-PCR. NC refers to the non-treated cells. B, RT-PCR and Western blot (WB) analyses of SNAI2 expression in pLL3.7-miR-control- and pLL3.7-miR-124-transfected U87 cells at 48 h post-transfection. GAPDH was used as an internal control. C, three predicted target sequences of miR-124 within the 3′-UTR of SNAI2 mRNA. Several nucleotides within the seed region were mutated in the 3′-UTR of SNAI2. hsa refers to Homo sapiens. D, effect of miR-124 on SNAI2 expression by luciferase reporter assay. The data were normalized to the ratio of firefly and Renilla luciferase activities measured at 48 h post-transfection. Values represent the mean ± S.D. from three independent transfection experiments. *, p < 0.05 compared with the pLL3.7-miR-control group.
To validate that SNAI2 is indeed directly targeted by miR-124, we investigated whether miR-124 recognizes the 3′-UTR of SNAI2 mRNA by Dual-Luciferase reporter assay. According to the predicted target sites from TargetScan (Fig. 2C), we cloned the wild-type 3′-UTR fragment containing these predicted sites into the pGL3 luciferase reporter vector (pGL3-SNAI2-wt). Another 3′-UTR fragment with four nucleotides mutated within each seed region was also cloned as a control (pGL3-SNAI2-mut). We found in U87 cells that transfection of pLL3.7-miR-124 significantly suppressed the luciferase activity of the pGL3-SNAI2-wt vector by 30% (Fig. 2D). By contrast, the repressive effect of pLL3.7-miR-124 on luciferase activity was abrogated by mutations in the pGL3-SNAI2-mut vector, confirming SNAI2 as a direct downstream target of miR-124.
Neurospheres Have Down-regulated miR-124 Levels, Enriched CD133+ Subpopulation, and Up-regulated Stem Cell Marker Expression
SNAI2 has been shown to control key aspects of stem cell function in human and mouse (30); therefore, we speculated that miR-124, as a negative regulator of SNAI2, may control the stem-like traits of the glioma cell subpopulation. We assessed the self-renewal ability of glioblastoma cells by means of neurosphere formation, which is considered a hallmark of cancer stem-like cells. When culturing in the presence of suitable factors, U87 cells formed typical neurospheres (U87-spheres) after 2–3 weeks (Fig. 3A). Flow cytometry analysis showed that the neurospheres had an enriched subpopulation of CD133+ cells compared with parental cells (Fig. 3B), indicative of the induction of stem-like cell traits. In addition, qRT-PCR revealed that miR-124 was down-regulated in U87-spheres, with its downstream target SNAI2 up-regulated (Fig. 3C). Importantly, several stem cell markers, BMI1 (22), Nanog (32), and Nestin (33), were all up-regulated in U87-spheres. These results suggest that miR-124 may play an important role in regulating the stem-like traits of glioma cells.
FIGURE 3.
Expression of miR-124 and stem cell markers in neurospheres. A, image of a typical neurosphere (U87-spheres) derived from its parental U87 cells (magnification ×40). B, flow cytometry analysis of the CD133+ cell population in parental U87 cells and U87-spheres. PE, phycoerythrin. C, analysis of expression of miR-124, SNAI2, and stem cell markers (BMI1, Nanog, and Nestin) in U87-spheres by real-time qRT-PCR. mRNA levels in U87 cells were normalized to 1. *, p < 0.05; **, p < 0.01 compared with U87 cells.
miR-124 Restricts Stem-like Properties of Glioma Cells via SNAI2
To better understand the potential role of miR-124 and SNAI2 in controlling the stemness of glioma cells, we analyzed the neurosphere-forming ability, CD133+ cell population, and stem cell marker expression in our established stable cells with miR-124 overexpression, SNAI2 suppression, or SNAI2 re-expression. In the sphere formation assay, overexpression of miR-124 and knockdown of SNAI2 significantly reduced the number of neurospheres (Fig. 4A). By contrast, constitutive expression of SNAI2 mRNA, which is resistant to miR-124 inhibition, partly restored the neurosphere-forming ability of miR-124-overexpressing cells. This was confirmed by the results obtained from flow cytometry analysis of CD133+ cells, in which overexpression of miR-124 and knockdown of SNAI2 resulted in loss of the CD133+ cell subpopulation, and this loss could be compensated by SNAI2 re-expression (Fig. 4B). In addition, the expression of BMI1, Nanog, and Nestin stem cell markers were down-regulated by miR-124 overexpression and SNAI2 knockdown and rescued by SNAI1 restoration (Fig. 4C). These results collectively indicate that miR-124 limits the in vitro stem-like characteristics of glioma cells by targeting SNAI2.
FIGURE 4.
Effects of miR-124 and SNAI2 on glioma cell stem-like traits in vitro. A, neurosphere formation assay. The bar graph indicates the number of neurospheres (mean ± S.D.) generated after 2–3 weeks of single-cell culture. B, flow cytometry analysis of CD133+ cell distribution in established stable cells. PE, phycoerythrin. C, qRT-PCR analysis of BMI1, Nanog, and Nestin stem cell markers. The expression in U87-control cells was arbitrarily set to 1.
miR-124 Inhibits Glioblastoma Cell Invasion in Vitro
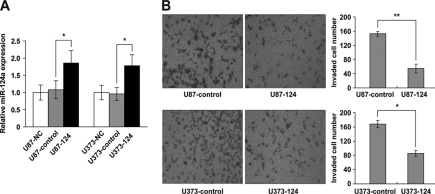
Emerging evidence has suggested that cancer stem-like cells may possess higher invasive activity compared with mesenchymal and differentiated cancer cells. In light of the above observation that miR-124 expression is inversely associated with the invasiveness of glioma cells, we hypothesized that miR-124 inhibits glioma cell invasion. To test this hypothesis, we established stable U87 and U373 glioblastoma cell lines that overexpress miR-124. The overexpression of miR-124 in U87-124 and U373-124 cells was first confirmed by qRT-PCR (Fig. 5A). Matrigel invasion assay revealed that miR-124 overexpression significantly restrained U87-124 and U373-124 cells from invading through the Matrigel membrane (Fig. 5B), suggesting that miR-124 inhibits glioblastoma cell invasion in vitro.
FIGURE 5.
Effect of miR-124 overexpression on glioma cell invasion. A, qRT-PCR analysis of miR-124 levels in U87 and U373 cells stably overexpressing miR-124 or plasmid controls. NC refers to the non-treated cells. B, Matrigel invasion assay. The bar graphs indicate the mean number of invaded cells ± S.D. counted in three randomly selected fields under a microscope (magnification, ×40). *, p < 0.05; **, p < 0.01 compared with control cells.
SNAI2 Is Up-regulated in Glioma and Recues miR-124-inhibited Glioblastoma Cell Invasion
SNAI2 is barely detectable in adult human brain tissues. Previous gene expression profile analysis showed that the SNAI2 expression level is significantly higher in primary glioblastoma than in the white matter of non-neoplastic brain tissue (30, 34). However, the pathological significance of SNAI2 in glioma is still unclear. To explore the potential role of SNAI2 in glioma, we detected the expression of SNAI2 in a panel of glioma tissue and non-glioma patient samples and seven glioma cell lines. In line with the abovementioned profile analysis, semiquantitative RT-PCR and Western blot analyses revealed that both the mRNA and protein levels of SNAI2 were significantly up-regulated in glioma cell lines compared with noncancerous brain tissues (Fig. 6A). Concurrent with this finding, compared with non-glioma brain tissues, the expression of SNAI2 was significantly higher by ∼2-fold in all glioma samples examined (Fig. 1A).
FIGURE 6.
Expression of SNAI2 and its role in glioma cell invasion. A, RT-PCR and Western blot (WB) analyses of SNAI2 expression in seven glioma cell lines and two non-tumor brain tissues (N1 and N2). B, relative expression level of SNAI2 in glioma samples and non-tumor brain tissues. SNAI2 expression was measured by qRT-PCR. C, qRT-PCR analysis of miR-124 and SNAI2 levels in stable U87-124 cells re-expressing SNAI2 that lacks the 3′-UTR (left panel) and in U87 cells stably suppressing endogenous SNAI2 expression by SNAI2 shRNA (right panel). NC refers to the non-treated cells. D, in vitro Matrigel invasion assay in U87-124 cells with reintroduced SNAI2 and in U87 cells with SNAI2 knockdown. Quantification of invaded cells (mean ± S.D.) in three randomly selected fields (magnification, ×40) is shown. *, p < 0.05; **, p < 0.01 compared with control cells.
Because SNAI2 is a direct target of miR-124, we hypothesized that miR-124 inhibits glioma cell invasion via SNAI2. To this end, we examined the role of SNAI2 in glioma cell invasion by loss-of-function and gain-of-function studies. We used short hairpin RNA targeting SNAI2 mRNA to specifically suppress the expression of SNAI2 in U87 cells (Fig. 6B). Stable knockdown of endogenous SNAI2 expression resulted in dramatic inhibition of glioblastoma cell invasion (Fig. 6C), consistent with the effect of miR-124 overexpression (Fig. 5B). To test whether miR-124 acts through SNAI2 to inhibit glioblastoma cell invasion, we reintroduced SNAI2 lacking the 3′-UTR into U87-124 cells such that the ectopic SNAI2 mRNA was refractory to miR-124. Restoration of the SNAI2 level in this stable cell line (U87-124-SNAI2) was confirmed by qRT-PCR analysis (Fig. 6B). We demonstrated by Matrigel invasion assay that restoration of SNAI2 significantly abrogated the inhibition of glioblastoma cell invasion by miR-124 (Fig. 6C). These results suggest that miR-124 inhibits glioblastoma cell invasion by targeting SNAI2.
Stable Overexpression of miR-124 and Knockdown of SNAI2 Suppress Tumorigenicity and Invasiveness of Glioblastoma Cells in Vivo
To substantiate the roles of miR-124 and SNAI2 in regulating glioma cell stem-like traits, we assessed the effects of miR-124 overexpression and SNAI2 knockdown on tumor initiation and growth in vivo. U87-124, U87-shSNAI2, and U87-124-SNAI2 cells and their respective control cells were inoculated into the back flanks of nude mice by subcutaneous injection. At 42 days post-injection, the mean volumes of tumors generated from U87-124 and U87-shSNAI2 cells were significantly smaller than those originating from the U87-miR-control and U87-shControl cells, respectively (Fig. 7, A and B). U87-124-SNAI2 cells with SNAI2 re-expression produced tumors of significantly larger sizes compared with U87-124-control cells. Further H&E staining of glioma xenografts indicated that U87-124 cells with miR-124 overexpression or U87-shSNAI2 cells with SNAI2 knockdown were less invasive than their control counterparts (Fig. 7C). By contrast, U87-124-SNAI2 cells re-expressing SNAI2 could invade non-tumor tissues. These data on in vivo tumorigenicity and invasiveness support the effects of miR-124 in suppressing the stem-like traits and invasion of glioma cells via SNAI2.
FIGURE 7.
Effects of miR-124 and SNAI2 on tumorigenicity and glioma cell invasion in vivo. A, representative images of nude mice (n = 6) subcutaneously injected with stable glioma cells overexpressing miR-124 (upper panel), suppressing SNAI2 (middle panel), and re-expressing SNAI2 (lower panel). B, tumor volumes. *, p < 0.05. C, H&E staining of tumor tissues. The arrows indicate the invasion of glioma cells into non-tumor tissues (magnification, ×40).
DISCUSSION
Cancer stem cells have recently been identified in human glioma (35, 36). These cells are self-renewable and highly tumorigenic and can be enriched by sorting for the expression of the surface marker CD133. Several signaling pathways important for glioma stem cell self-renewal have also been delineated. These include the bone morphogenetic protein, Hedgehog (Hh), Notch, and Wnt pathways (37). Recently, miR-128 has been shown to target the BMI1 oncogene/stem cell renewal factor and inhibit glioma proliferation and self-renewal (38), highlighting the roles of miRNAs in regulating the cancer stem cell subpopulation and thereby the stemness of glioma cells.
On the basis of our previous miRNA microarray analysis, which revealed a significant down-regulation in glioblastoma patient samples, we chose miR-124 for detailed investigation in this study. We observed that miR-124 is commonly down-regulated in glioma tissues and cell lines compared with non-neoplastic brain tissues. This expression pattern is consistent with previous reports showing a decreased expression level of miR-124 in anaplastic astrocytoma, glioblastoma (16), and medulloblastoma (19). We further demonstrated that miR-124 inhibits glioblastoma cell stemness and invasion both in vitro and in vivo. These results suggest that the down-regulation of miR-124 in glioblastoma may impair its inhibitory effect on glioma stem-like traits and cell invasion, thereby leading to the uprising of invasive growth.
To elucidate the molecular mechanism by which miR-124 inhibits glioma stem-like traits and cell invasion, we identified SNAI2 as a direct and functional target of miR-124. While we were preparing this manuscript, SNAI2 was shown to be a specific target of miR-124 during cell migration for embryoid body formation (39). SNAI2 is a neurogenic transcription factor belonging to the Snail family of zinc finger transcription factors. Several studies have indicated the critical role of SNAI2 in epithelial-mesenchymal transition and cell survival. SNAI2 down-regulation by RNA interference can facilitate apoptosis and inhibit invasive growth in neuroblastoma preclinical models (40). In agreement with the report by Scrideli et al. (34), we found that, in contrast to down-regulation of miR-124, SNAI2 is frequently up-regulated in glioma tissues and cell lines. Moreover, knockdown of SNAI2 inhibits glioma cell invasion, whereas restoration of SNAI2 significantly rescues miR-124-inhibited glioma cell invasion. These findings indicate that miR-124 inhibits glioma cell invasion by repressing SNAI2 post-transcriptionally.
Given that SNAI2 controls several key aspects of stem cell function, such as cancer stem cell properties in human and mouse (29–31), we provided evidence that miR-124 inhibits glioma cell stem-like traits by targeting SNAI2. This conclusion was based on both in vitro and in vivo assays, in which miR-124 overexpression and SNAI2 knockdown reduce neurosphere formation, CD133+ cell population, stem cell marker expression, and tumorigenicity of glioma cells in a xenograft mouse model. In addition, re-expression of SNAI2 abolishes this inhibition, strongly suggesting that miR-124 inhibits glioma cell stem-like traits by targeting SNAI2.
Invasive growth is a defining hallmark of high-grade gliomas that greatly affects the survival of patients (41). Increasing studies have shed light on the mechanisms by which glioma cells infiltrate normal brain. Glioma invasion is a multistep process that involves detachment from the tumor mass, remodeling of the extracellular matrix, and migration of glioma cells (42, 43). Several factors, including CD44, neural cell adhesion molecule, cadherins, integrins, matrix metalloproteinases, and the EGF receptor, have been shown to mediate these processes (42, 44). In addition, noncoding miRNAs have recently been reported to play a fundamental role in glioma invasion. For example, we (13) and others (45) have shown that miR-146b inhibits, whereas miR-21 promotes, glioma cell invasion, respectively. More recently, our collaborators also demonstrated an important role of miR-124 in the regulation of invasion and metastasis of hepatocellular carcinoma cells aggressiveness by repressing ROCK2 and EZH2 (46). Identifying the factors and mechanisms associated with glioma invasion provides a better understanding of the molecular pathways that lead to the invasive progression of glioma.
This work demonstrates the functions of miR-124 in inhibiting the stem-like traits and invasive propensity of glioma cells. miR-124 controls these phenotypes via one of its downstream targets, SNAI2. In view of its frequent down-regulation in glioblastoma, it is likely that brain-enriched miR-124 may not only be important for neural development but may also serve as a putative tumor suppressor miRNA that prevents glioma carcinogenesis by inhibiting the stem-like traits and invasiveness. Our results open up a new avenue for elucidating the molecular mechanism of glioma cell invasion and developing therapeutic strategies against glioma by restoring the level of miR-124 and suppressing SNAI2.
This work was supported by Grants 467109 and 470911 from the Research Grants Council of the Hong Kong Special Administrative Region, China, and by Grants 2010CB529400 and 2010CB912800 from the National Basic Research Program of China (973 Program).
- miRNA
- microRNA
- qRT-PCR
- quantitative RT-PCR.
REFERENCES
- 1. Louis D. N., Ohgaki H., Wiestler O. D., Cavenee W. K., Burger P. C., Jouvet A., Scheithauer B. W., Kleihues P. (2007) The 2007 WHO classification of tumors of the central nervous system. Acta Neuropathol. 114, 97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cancer Genome Atlas Research Network (2008) Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ng S. S., Cheung Y. T., An X. M., Chen Y. C., Li M., Li G. H., Cheung W., Sze J., Lai L., Peng Y., Xia H. H., Wong B. C., Leung S. Y., Xie D., He M. L., Kung H. F., Lin M. C. (2007) Cell cycle-related kinase: a novel candidate oncogene in human glioblastoma. J. Natl. Cancer Inst. 99, 936–948 [DOI] [PubMed] [Google Scholar]
- 4. Wong J. W. (2010) MicroRNA-induced silencing of glioma progression. J. Neurosci. 30, 3868–3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lawler S., Chiocca E. A. (2009) Emerging functions of microRNAs in glioblastoma. J. Neurooncol. 92, 297–306 [DOI] [PubMed] [Google Scholar]
- 6. Novakova J., Slaby O., Vyzula R., Michalek J. (2009) MicroRNA involvement in glioblastoma pathogenesis. Biochem. Biophys. Res. Commun. 386, 1–5 [DOI] [PubMed] [Google Scholar]
- 7. Bartel D. P. (2009) MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Calin G. A., Sevignani C., Dumitru C. D., Hyslop T., Noch E., Yendamuri S., Shimizu M., Rattan S., Bullrich F., Negrini M., Croce C. M. (2004) Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc. Natl. Acad. Sci. U.S.A. 101, 2999–3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen C. Z. (2005) MicroRNAs as oncogenes and tumor suppressors. N. Engl. J. Med. 353, 1768–1771 [DOI] [PubMed] [Google Scholar]
- 10. Corsten M. F., Miranda R., Kasmieh R., Krichevsky A. M., Weissleder R., Shah K. (2007) MicroRNA-21 knockdown disrupts glioma growth in vivo and displays synergistic cytotoxicity with neural precursor cell-delivered S-TRAIL in human gliomas. Cancer Res. 67, 8994–9000 [DOI] [PubMed] [Google Scholar]
- 11. Bastian M., Marietta W., Franziska L., Michael G., Kai S., Helmut E. M., Guido R. (2009) Identification and functional characterization of microRNAs involved in the malignant progression of gliomas. Brain Pathol. 20, 539–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xia H., Qi Y., Ng S. S., Chen X., Chen S., Fang M., Li D., Zhao Y., Ge R., Li G., Chen Y., He M. L., Kung H. F., Lai L., Lin M. C. (2009) MicroRNA-15b regulates cell cycle progression by targeting cyclins in glioma cells. Biochem. Biophys. Res. Commun. 380, 205–210 [DOI] [PubMed] [Google Scholar]
- 13. Xia H., Qi Y., Ng S. S., Chen X., Li D., Chen S., Ge R., Jiang S., Li G., Chen Y., He M. L., Kung H. F., Lai L., Lin M. C. (2009) microRNA-146b inhibits glioma cell migration and invasion by targeting MMPs. Brain Res. 1269, 158–165 [DOI] [PubMed] [Google Scholar]
- 14. Godlewski J., Nowicki M. O., Bronisz A., Nuovo G., Palatini J., De Lay M., Van Brocklyn J., Ostrowski M. C., Chiocca E. A., Lawler S. E. (2010) MicroRNA-451 regulates LKB1/AMPK signaling and allows adaptation to metabolic stress in glioma cells. Mol. Cell 37, 620–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Silber J., James C. D., Hodgson J. G. (2009) MicroRNAs in gliomas: small regulators of a big problem. Neuromolecular Med. 11, 208–222 [DOI] [PubMed] [Google Scholar]
- 16. Silber J., Lim D. A., Petritsch C., Persson A. I., Maunakea A. K., Yu M., Vandenberg S. R., Ginzinger D. G., James C. D., Costello J. F., Bergers G., Weiss W. A., Alvarez-Buylla A., Hodgson J. G. (2008) miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 6, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cheng L. C., Pastrana E., Tavazoie M., Doetsch F. (2009) miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat. Neurosci. 12, 399–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yoo A. S., Sun A. X., Li L., Shcheglovitov A., Portmann T., Li Y., Lee-Messer C., Dolmetsch R. E., Tsien R. W., Crabtree G. R. (2011) MicroRNA-mediated conversion of human fibroblasts to neurons. Nature 476, 228–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li K. K., Pang J. C., Ching A. K., Wong C. K., Kong X., Wang Y., Zhou L., Chen Z., Ng H. K. (2009) miR-124 is frequently down-regulated in medulloblastoma and is a negative regulator of SLC16A1. Hum. Pathol. 40, 1234–1243 [DOI] [PubMed] [Google Scholar]
- 20. Pierson J., Hostager B., Fan R., Vibhakar R. (2008) Regulation of cyclin-dependent kinase 6 by microRNA-124 in medulloblastoma. J. Neurooncol. 90, 1–7 [DOI] [PubMed] [Google Scholar]
- 21. Gupta P. B., Kuperwasser C., Brunet J. P., Ramaswamy S., Kuo W. L., Gray J. W., Naber S. P., Weinberg R. A. (2005) The melanocyte differentiation program predisposes to metastasis after neoplastic transformation. Nat. Genet. 37, 1047–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Abdouh M., Facchino S., Chatoo W., Balasingam V., Ferreira J., Bernier G. (2009) BMI1 sustains human glioblastoma multiforme stem cell renewal. J. Neurosci. 29, 8884–8896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hart A. H., Hartley L., Parker K., Ibrahim M., Looijenga L. H., Pauchnik M., Chow C. W., Robb L. (2005) The pluripotency homeobox gene NANOG is expressed in human germ cell tumors. Cancer 104, 2092–2098 [DOI] [PubMed] [Google Scholar]
- 24. Wagner N., Wagner K. D., Scholz H., Kirschner K. M., Schedl A. (2006) Intermediate filament protein Nestin is expressed in developing kidney and heart and might be regulated by the Wilms tumor suppressor Wt1. Am. J. Physiol. Regul. Integr. Comp. Physiol. 291, R779–R787 [DOI] [PubMed] [Google Scholar]
- 25. de Ridder L. I., Laerum O. D., Mørk S. J., Bigner D. D. (1987) Invasiveness of human glioma cell lines in vitro: relation to tumorigenicity in athymic mice. Acta Neuropathol. 72, 207–213 [DOI] [PubMed] [Google Scholar]
- 26. Lim L. P., Lau N. C., Garrett-Engele P., Grimson A., Schelter J. M., Castle J., Bartel D. P., Linsley P. S., Johnson J. M. (2005) Microarray analysis shows that some microRNAs down-regulate large numbers of target mRNAs. Nature 433, 769–773 [DOI] [PubMed] [Google Scholar]
- 27. Cao X., Pfaff S. L., Gage F. H. (2007) A functional study of miR-124 in the developing neural tube. Genes Dev. 21, 531–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Makeyev E. V., Zhang J., Carrasco M. A., Maniatis T. (2007) The microRNA miR-124 promotes neuronal differentiation by triggering brain-specific alternative pre-mRNA splicing. Mol. Cell 27, 435–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Alves C. C., Carneiro F., Hoefler H., Becker K. F. (2009) Role of the epithelial-mesenchymal transition regulator Slug in primary human cancers. Front. Biosci. 14, 3035–3050 [DOI] [PubMed] [Google Scholar]
- 30. Cobaleda C., Pérez-Caro M., Vicente-Dueñas C., Sánchez-García I. (2007) Function of the zinc-finger transcription factor SNAI2 in cancer and development. Annu. Rev. Genet. 41, 41–61 [DOI] [PubMed] [Google Scholar]
- 31. Pérez-Mancera P. A., González-Herrero I., Pérez-Caro M., Gutiérrez-Cianca N., Flores T., Gutiérrez-Adán A., Pintado B., Sánchez-Martín M., Sánchez-García I. (2005) SLUG in cancer development. Oncogene 24, 3073–3082 [DOI] [PubMed] [Google Scholar]
- 32. Pan G., Thomson J. A. (2007) Nanog and transcriptional networks in embryonic stem cell pluripotency. Cell Res. 17, 42–49 [DOI] [PubMed] [Google Scholar]
- 33. Mangiola A., Lama G., Giannitelli C., De Bonis P., Anile C., Lauriola L., La Torre G., Sabatino G., Maira G., Jhanwar-Uniyal M., Sica G. (2007) Stem cell marker Nestin and c-Jun NH2-terminal kinases in tumor and peritumor areas of glioblastoma multiforme: possible prognostic implications. Clin. Cancer Res. 13, 6970–6977 [DOI] [PubMed] [Google Scholar]
- 34. Scrideli C. A., Carlotti C. G., Jr., Okamoto O. K., Andrade V. S., Cortez M. A., Motta F. J., Lucio-Eterovic A. K., Neder L., Rosemberg S., Oba-Shinjo S. M., Marie S. K., Tone L. G. (2008) Gene expression profile analysis of primary glioblastomas and non-neoplastic brain tissue: identification of potential target genes by oligonucleotide microarray and real-time quantitative PCR. J. Neurooncol. 88, 281–291 [DOI] [PubMed] [Google Scholar]
- 35. Singh S. K., Hawkins C., Clarke I. D., Squire J. A., Bayani J., Hide T., Henkelman R. M., Cusimano M. D., Dirks P. B. (2004) Identification of human brain tumor-initiating cells. Nature 432, 396–401 [DOI] [PubMed] [Google Scholar]
- 36. Singh S. K., Clarke I. D., Terasaki M., Bonn V. E., Hawkins C., Squire J., Dirks P. B. (2003) Identification of a cancer stem cell in human brain tumors. Cancer Res. 63, 5821–5828 [PubMed] [Google Scholar]
- 37. Xie Z. (2009) Brain tumor stem cells. Neurochem. Res. 34, 2055–2066 [DOI] [PubMed] [Google Scholar]
- 38. Godlewski J., Nowicki M. O., Bronisz A., Williams S., Otsuki A., Nuovo G., Raychaudhury A., Newton H. B., Chiocca E. A., Lawler S. (2008) Targeting of the Bmi1 oncogene/stem cell renewal factor by microRNA-128 inhibits glioma proliferation and self-renewal. Cancer Res. 68, 9125–9130 [DOI] [PubMed] [Google Scholar]
- 39. Lee M. R., Kim J. S, Kim K. S. (2010) miR-124a is important for migratory cell fate transition during gastrulation of human embryonic stem cells. Stem Cells 28, 1550–1559 [DOI] [PubMed] [Google Scholar]
- 40. Vitali R., Mancini C., Cesi V., Tanno B., Mancuso M., Bossi G., Zhang Y., Martinez R. V., Calabretta B., Dominici C., Raschellà G. (2008) Slug (SNAI2) down-regulation by RNA interference facilitates apoptosis and inhibits invasive growth in neuroblastoma preclinical models. Clin. Cancer Res. 14, 4622–4630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hoelzinger D. B., Demuth T., Berens M. E. (2007) Autocrine factors that sustain glioma invasion and paracrine biology in the brain microenvironment. J. Natl. Cancer Inst. 99, 1583–1593 [DOI] [PubMed] [Google Scholar]
- 42. Demuth T., Berens M. E. (2004) Molecular mechanisms of glioma cell migration and invasion. J. Neurooncol. 70, 217–228 [DOI] [PubMed] [Google Scholar]
- 43. Mareel M., Leroy A. (2003) Clinical, cellular, and molecular aspects of cancer invasion. Physiol. Rev. 83, 337–376 [DOI] [PubMed] [Google Scholar]
- 44. Fillmore H. L., VanMeter T. E., Broaddus W. C. (2001) Membrane-type matrix metalloproteinases (MT-MMPs): expression and function during glioma invasion. J. Neurooncol. 53, 187–202 [DOI] [PubMed] [Google Scholar]
- 45. Gabriely G., Wurdinger T., Kesari S., Esau C. C., Burchard J., Linsley P. S., Krichevsky A. M. (2008) MicroRNA-21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol. Cell. Biol. 28, 5369–5380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zheng F., Liao Y. J., Cai M. Y., Liu Y. H., Liu T. H., Chen S. P., Bian X. W., Guan X. Y., Lin M. C., Zeng Y. X. (2012) The putative tumor suppressor microRNA-124 modulates hepatocellular carcinoma cell aggressiveness by repressing ROCK2 and EZH2. Gut 61, 278–289 [DOI] [PubMed] [Google Scholar]
- 47. Sun T., Aceto N., Meerbrey K. L., Kessler J. D., Zhou C., Migliaccio I., Nguyen D. X., Pavlova N. N., Botero M., Huang J., Bernardi R. J., Schmitt E., Hu G., Li M. Z., Dephoure N., Gygi S. P., Rao M., Creighton C. J., Hilsenbeck S. G., Shaw C. A., Muzny D., Gibbs R. A., Wheeler D. A., Osborne C. K., Schiff R., Bentires-Alj M., Elledge S. J., Westbrook T. F. (2011) Activation of multiple proto-oncogenic tyrosine kinases in breast cancer via loss of the PTPN12 phosphatase. Cell 144, 703–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yoo A. S., Staahl B. T., Chen L., Crabtree G. R. (2009) MicroRNA-mediated switching of chromatin-remodeling complexes in neural development. Nature 460, 642–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Visvanathan J., Lee S., Lee B., Lee J. W., Lee S. K. (2007) The microRNA miR-124 antagonizes the anti-neural REST/SCP1 pathway during embryonic CNS development. Genes Dev. 21, 744–749 [DOI] [PMC free article] [PubMed] [Google Scholar]