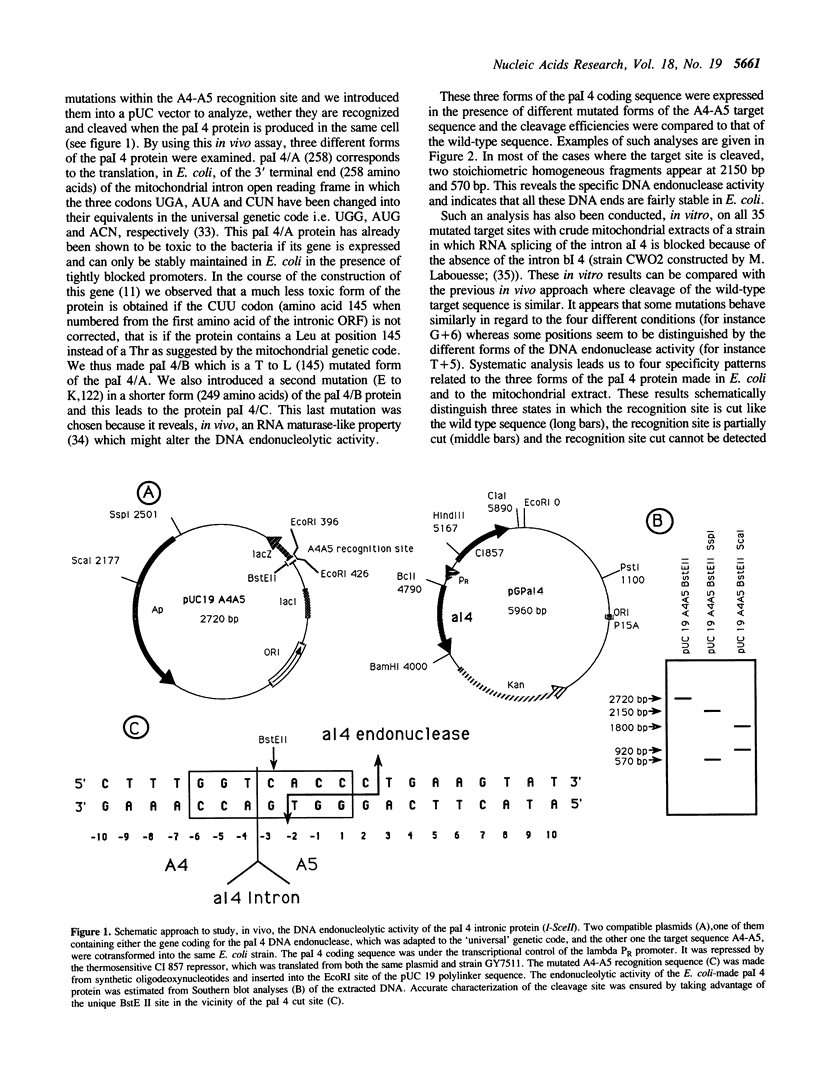
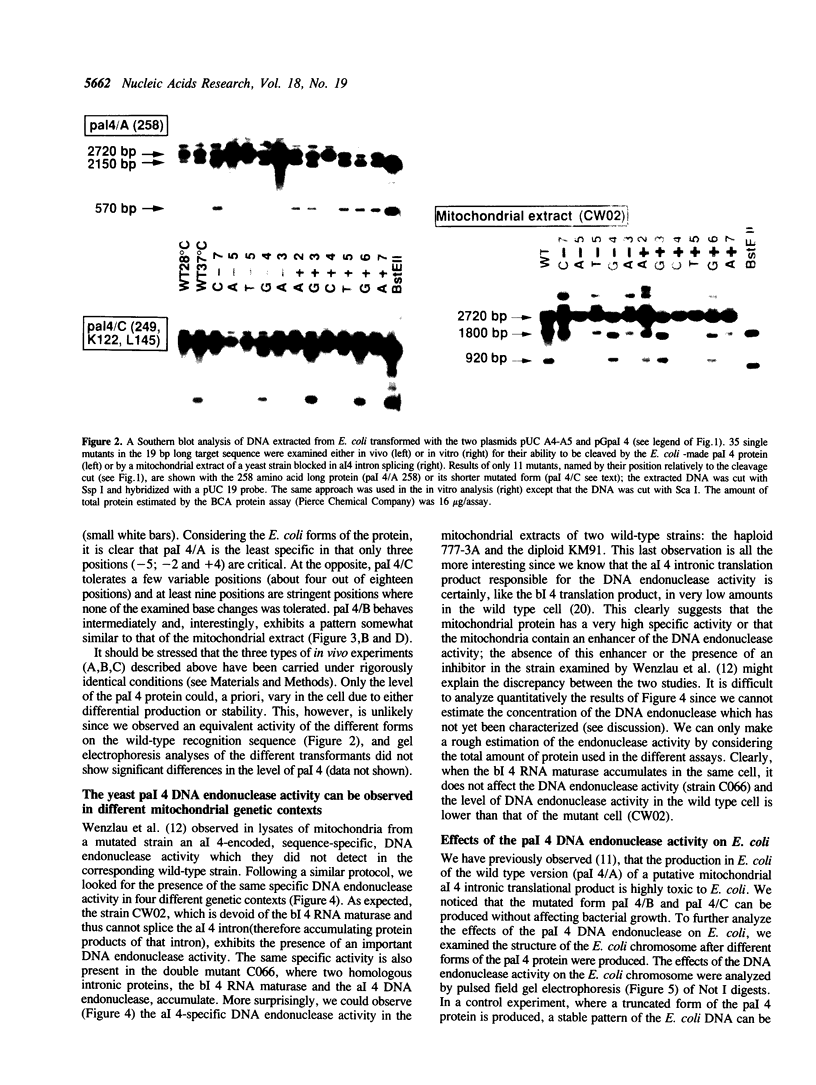
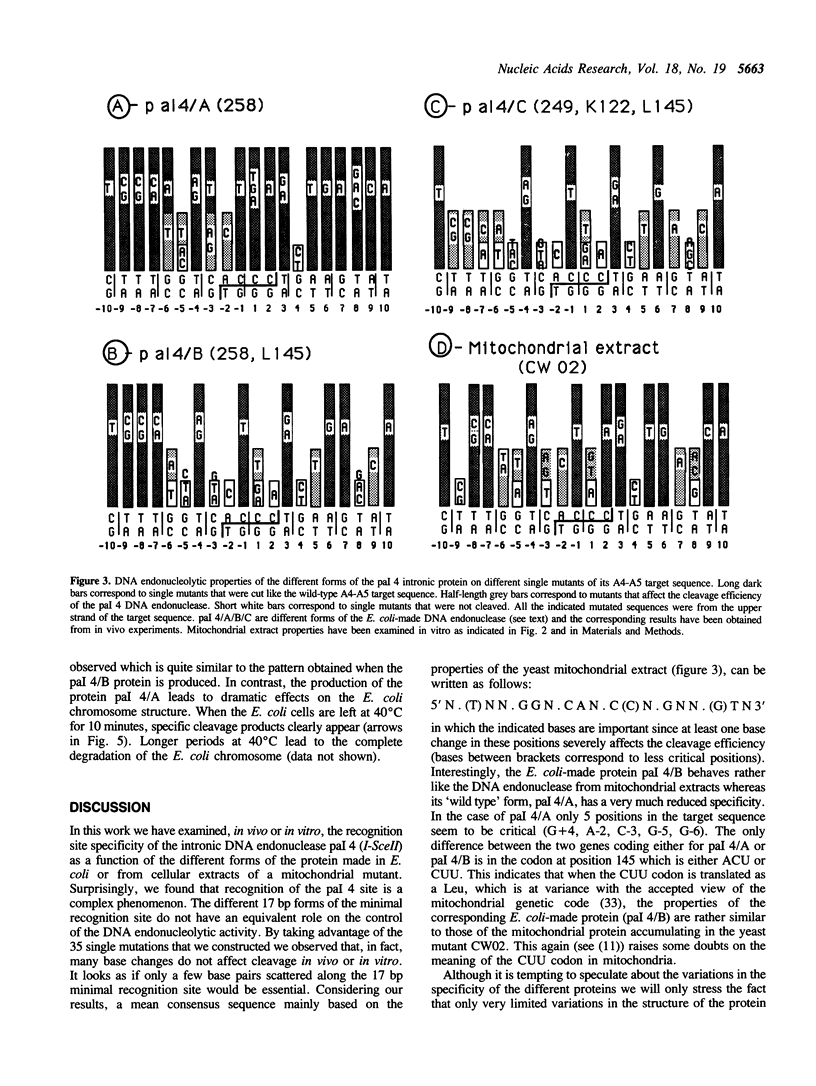
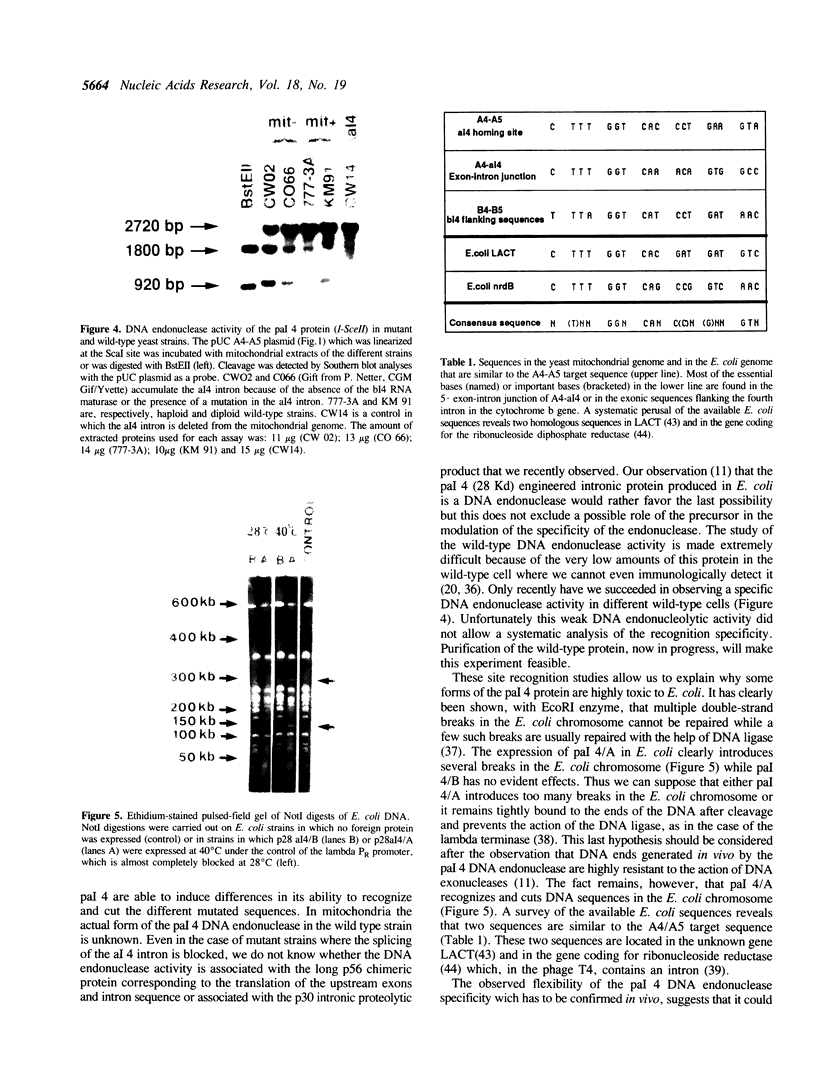
Abstract
The pal 4 nuclease (termed I-Sce II) is encoded in the group I al 4 intron of the COX I gene of Saccharomyces cerevisiae. It introduces a specific double-strand break at the junction of the two exons A4-A5 and thus mediates the insertion of the intron into an intronless strain. To define the sequence recognized by pal 4 we introduced 35 single mutations in its target sequence and examined their cleavage properties either in vivo in E. coli (when different forms of the pal 4 proteins were artificially produced) or in vitro with mitochondrial extracts of a mutant yeast strain blocked in the splicing of the al 4 intron. We also detected the pal 4 DNA endonuclease activity in extracts of the wild type strain. The results suggest that 6 to 9 noncontiguous bases in the 17 base-pair region examined are necessary for pal 4 nuclease to bind and cleave its recognition site. We observed that the pal 4 nuclease specificity can be significantly different with the different forms of the protein thus explaining why only some forms are highly toxic in E. coli. This study shows that pal 4 recognition site is a complex phenomenon and this might have evolutionary implications on the transfer properties of the intron.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Banroques J., Delahodde A., Jacq C. A mitochondrial RNA maturase gene transferred to the yeast nucleus can control mitochondrial mRNA splicing. Cell. 1986 Sep 12;46(6):837–844. doi: 10.1016/0092-8674(86)90065-6. [DOI] [PubMed] [Google Scholar]
- Banroques J., Perea J., Jacq C. Efficient splicing of two yeast mitochondrial introns controlled by a nuclear-encoded maturase. EMBO J. 1987 Apr;6(4):1085–1091. doi: 10.1002/j.1460-2075.1987.tb04862.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belfort M. Bacteriophage introns: parasites within parasites? Trends Genet. 1989 Jul;5(7):209–213. doi: 10.1016/0168-9525(89)90083-8. [DOI] [PubMed] [Google Scholar]
- Bonitz S. G., Berlani R., Coruzzi G., Li M., Macino G., Nobrega F. G., Nobrega M. P., Thalenfeld B. E., Tzagoloff A. Codon recognition rules in yeast mitochondria. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3167–3170. doi: 10.1073/pnas.77.6.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buvinger W. E., Lampel K. A., Bojanowski R. J., Riley M. Location and analysis of nucleotide sequences at one end of a putative lac transposon in the Escherichia coli chromosome. J Bacteriol. 1984 Aug;159(2):618–623. doi: 10.1128/jb.159.2.618-623.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson J., Fuchs J. A., Messing J. Primary structure of the Escherichia coli ribonucleoside diphosphate reductase operon. Proc Natl Acad Sci U S A. 1984 Jul;81(14):4294–4297. doi: 10.1073/pnas.81.14.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter P., Bedouelle H., Winter G. Improved oligonucleotide site-directed mutagenesis using M13 vectors. Nucleic Acids Res. 1985 Jun 25;13(12):4431–4443. doi: 10.1093/nar/13.12.4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu F. K., Maley G., Pedersen-Lane J., Wang A. M., Maley F. Characterization of the restriction site of a prokaryotic intron-encoded endonuclease. Proc Natl Acad Sci U S A. 1990 May;87(9):3574–3578. doi: 10.1073/pnas.87.9.3574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colleaux L., D'Auriol L., Galibert F., Dujon B. Recognition and cleavage site of the intron-encoded omega transposase. Proc Natl Acad Sci U S A. 1988 Aug;85(16):6022–6026. doi: 10.1073/pnas.85.16.6022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colleaux L., d'Auriol L., Betermier M., Cottarel G., Jacquier A., Galibert F., Dujon B. Universal code equivalent of a yeast mitochondrial intron reading frame is expressed into E. coli as a specific double strand endonuclease. Cell. 1986 Feb 28;44(4):521–533. doi: 10.1016/0092-8674(86)90262-x. [DOI] [PubMed] [Google Scholar]
- Del Sal G., Manfioletti G., Schneider C. A one-tube plasmid DNA mini-preparation suitable for sequencing. Nucleic Acids Res. 1988 Oct 25;16(20):9878–9878. doi: 10.1093/nar/16.20.9878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delahodde A., Goguel V., Becam A. M., Creusot F., Perea J., Banroques J., Jacq C. Site-specific DNA endonuclease and RNA maturase activities of two homologous intron-encoded proteins from yeast mitochondria. Cell. 1989 Feb 10;56(3):431–441. doi: 10.1016/0092-8674(89)90246-8. [DOI] [PubMed] [Google Scholar]
- Dujardin G., Jacq C., Slonimski P. P. Single base substitution in an intron of oxidase gene compensates splicing defects of the cytochrome b gene. Nature. 1982 Aug 12;298(5875):628–632. doi: 10.1038/298628a0. [DOI] [PubMed] [Google Scholar]
- Dujon B. Group I introns as mobile genetic elements: facts and mechanistic speculations--a review. Gene. 1989 Oct 15;82(1):91–114. doi: 10.1016/0378-1119(89)90034-6. [DOI] [PubMed] [Google Scholar]
- Fisher R. P., Parisi M. A., Clayton D. A. Flexible recognition of rapidly evolving promoter sequences by mitochondrial transcription factor 1. Genes Dev. 1989 Dec;3(12B):2202–2217. doi: 10.1101/gad.3.12b.2202. [DOI] [PubMed] [Google Scholar]
- Flamm E. L., Weisberg R. A. Primary structure of the hip gene of Escherichia coli and of its product, the beta subunit of integration host factor. J Mol Biol. 1985 May 25;183(2):117–128. doi: 10.1016/0022-2836(85)90206-2. [DOI] [PubMed] [Google Scholar]
- Goguel V., Perea J., Jacq C. Synthesis and function of the mitochondrial intron--encoded bI4 RNA maturase from Saccharomyces cerevisiae. Effects of upstream frame-shift mutations. Curr Genet. 1989 Oct;16(4):241–246. doi: 10.1007/BF00422109. [DOI] [PubMed] [Google Scholar]
- Gott J. M., Shub D. A., Belfort M. Multiple self-splicing introns in bacteriophage T4: evidence from autocatalytic GTP labeling of RNA in vitro. Cell. 1986 Oct 10;47(1):81–87. doi: 10.1016/0092-8674(86)90368-5. [DOI] [PubMed] [Google Scholar]
- Heitman J., Zinder N. D., Model P. Repair of the Escherichia coli chromosome after in vivo scission by the EcoRI endonuclease. Proc Natl Acad Sci U S A. 1989 Apr;86(7):2281–2285. doi: 10.1073/pnas.86.7.2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudspeth M. E., Shumard D. S., Tatti K. M., Grossman L. I. Rapid purification of yeast mitochondrial DNA in high yield. Biochim Biophys Acta. 1980 Dec 11;610(2):221–228. doi: 10.1016/0005-2787(80)90003-9. [DOI] [PubMed] [Google Scholar]
- Jacq C., Banroques J., Becam A. M., Slonimski P. P., Guiso N., Danchin A. Antibodies against a fused 'lacZ-yeast mitochondrial intron' gene product allow identification of the mRNA maturase encoded by the fourth intron of the yeast cob-box gene. EMBO J. 1984 Jul;3(7):1567–1572. doi: 10.1002/j.1460-2075.1984.tb02012.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacq C., Lazowska J., Slonimski P. P. Sur un nouveau mécanisme de la régulation de l'expression génétique. C R Seances Acad Sci D. 1980 Jan 14;290(2):89–92. [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Labouesse M., Netter P., Schroeder R. Molecular basis of the 'box effect', A maturase deficiency leading to the absence of splicing of two introns located in two split genes of yeast mitochondrial DNA. Eur J Biochem. 1984 Oct 1;144(1):85–93. doi: 10.1111/j.1432-1033.1984.tb08434.x. [DOI] [PubMed] [Google Scholar]
- Labouesse M., Slonimski P. P. Construction of novel cytochrome b genes in yeast mitochondria by subtraction or addition of introns. EMBO J. 1983;2(2):269–276. doi: 10.1002/j.1460-2075.1983.tb01416.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambowitz A. M. Infectious introns. Cell. 1989 Feb 10;56(3):323–326. doi: 10.1016/0092-8674(89)90232-8. [DOI] [PubMed] [Google Scholar]
- Michel F., Lang B. F. Mitochondrial class II introns encode proteins related to the reverse transcriptases of retroviruses. Nature. 1985 Aug 15;316(6029):641–643. doi: 10.1038/316641a0. [DOI] [PubMed] [Google Scholar]
- Nickoloff J. A., Singer J. D., Heffron F. In vivo analysis of the Saccharomyces cerevisiae HO nuclease recognition site by site-directed mutagenesis. Mol Cell Biol. 1990 Mar;10(3):1174–1179. doi: 10.1128/mcb.10.3.1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman P. S., Butow R. A. Mobile introns and intron-encoded proteins. Science. 1989 Dec 1;246(4934):1106–1109. doi: 10.1126/science.2479980. [DOI] [PubMed] [Google Scholar]
- Quirk S. M., Bell-Pedersen D., Belfort M. Intron mobility in the T-even phages: high frequency inheritance of group I introns promoted by intron open reading frames. Cell. 1989 Feb 10;56(3):455–465. doi: 10.1016/0092-8674(89)90248-1. [DOI] [PubMed] [Google Scholar]
- Roberts D., Kleckner N. Tn10 transposition promotes RecA-dependent induction of a lambda prophage. Proc Natl Acad Sci U S A. 1988 Aug;85(16):6037–6041. doi: 10.1073/pnas.85.16.6037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scazzocchio C. Group I introns: do they only go home? Trends Genet. 1989 Jun;5(6):168–172. doi: 10.1016/0168-9525(89)90068-1. [DOI] [PubMed] [Google Scholar]
- Wenzlau J. M., Saldanha R. J., Butow R. A., Perlman P. S. A latent intron-encoded maturase is also an endonuclease needed for intron mobility. Cell. 1989 Feb 10;56(3):421–430. doi: 10.1016/0092-8674(89)90245-6. [DOI] [PubMed] [Google Scholar]
- Wolf K., Del Giudice L. The variable mitochondrial genome of ascomycetes: organization, mutational alterations, and expression. Adv Genet. 1988;25:185–308. doi: 10.1016/s0065-2660(08)60460-5. [DOI] [PubMed] [Google Scholar]
- Zinn A. R., Butow R. A. Nonreciprocal exchange between alleles of the yeast mitochondrial 21S rRNA gene: kinetics and the involvement of a double-strand break. Cell. 1985 Apr;40(4):887–895. doi: 10.1016/0092-8674(85)90348-4. [DOI] [PubMed] [Google Scholar]