Background: Several new ATP-competitive mTOR inhibitors have been described, but their kinome-wide selectivity profiles have not been disclosed.
Results: Four different profiling technologies revealed a different spectrum of targets for four recently described mTOR inhibitors.
Conclusion: Diverse heterocyclic mTOR inhibitors have unique pharmacology.
Significance: Profiling data guide choices of mTOR inhibitors for particular applications and provide new potential targets for medicinal chemistry efforts.
Keywords: Enzyme Kinetics, mTOR, PI3K, Serine/Threonine-protein Kinase, Tyrosine-protein Kinase (Tyrosine Kinase), KU63794, PP242, Slow Off-rate, Torin1, WYE-354
Abstract
An intensive recent effort to develop ATP-competitive mTOR inhibitors has resulted in several potent and selective molecules such as Torin1, PP242, KU63794, and WYE354. These inhibitors are being widely used as pharmacological probes of mTOR-dependent biology. To determine the potency and specificity of these agents, we have undertaken a systematic kinome-wide effort to profile their selectivity and potency using chemical proteomics and assays for enzymatic activity, protein binding, and disruption of cellular signaling. Enzymatic and cellular assays revealed that all four compounds are potent inhibitors of mTORC1 and mTORC2, with Torin1 exhibiting ∼20-fold greater potency for inhibition of Thr-389 phosphorylation on S6 kinases (EC50 = 2 nm) relative to other inhibitors. In vitro biochemical profiling at 10 μm revealed binding of PP242 to numerous kinases, although WYE354 and KU63794 bound only to p38 kinases and PI3K isoforms and Torin1 to ataxia telangiectasia mutated, ATM and Rad3-related protein, and DNA-PK. Analysis of these protein targets in cellular assays did not reveal any off-target activities for Torin1, WYE354, and KU63794 at concentrations below 1 μm but did show that PP242 efficiently inhibited the RET receptor (EC50, 42 nm) and JAK1/2/3 kinases (EC50, 780 nm). In addition, Torin1 displayed unusually slow kinetics for inhibition of the mTORC1/2 complex, a property likely to contribute to the pharmacology of this inhibitor. Our results demonstrated that, with the exception of PP242, available ATP-competitive compounds are highly selective mTOR inhibitors when applied to cells at concentrations below 1 μm and that the compounds may represent a starting point for medicinal chemistry efforts aimed at developing inhibitors of other PI3K kinase-related kinases.
Introduction
Mammalian target of rapamycin (mTOR)4 is a serine/threonine kinase that is highly conserved across eukaryotic cells and controls several fundamental cellular functions (1). mTOR is a master regulator of cellular growth and proliferation and functions as a component of the PI3K/Akt/TSC/mTOR/S6K (4EBP) signal transduction pathway. mTOR is found in at least two distinct multiprotein complexes, mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2). mTORC1 is composed of the mTOR kinase and four associated proteins, Raptor, mLST8, PRAS40, and DEPTOR. mTORC2 also contains mTOR, mLST8, and DEPTOR, but instead of Raptor and PRAS40, it contains Rictor, mSin1, and Protor. mTORC1 controls cell growth in part by phosphorylating S6 kinase 1 (S6K1) and eIF-4E-binding protein 1 (4E-BP1), key regulators of protein synthesis (2). mTORC2 promotes cell survival and proliferation in response to growth factors by phosphorylating its downstream effector Akt/PKB (3). mTOR shares high sequence similarity in its catalytic domain with PI3K and belongs to a family of PI3K-like kinases (PIKK) that also includes ATR, ATM, DNA-PK, and SMG-1 (4). Deregulation of the PI3K/Akt/TSC/mTOR pathway is common in human tumors, and this has provided the impetus to develop mTOR inhibitors as a new class of anti-cancer drugs (5).
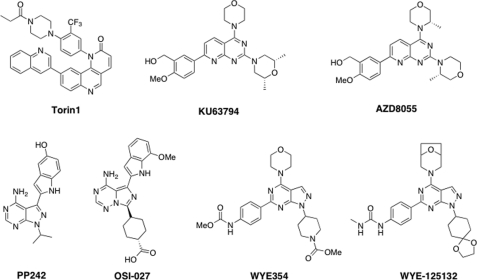
Rapamycin, originally discovered as an antifungal agent with immunosuppressant properties, is an allosteric inhibitor of mTORC1 that acts by recruiting an accessory protein named FKBP-12 to the FRB domain of mTOR; this down-regulates mTOR kinase activity through an as-yet unknown mechanism (6). Rapamycin has demonstrated clinical efficacy in the treatment of renal cell carcinoma and mantle cell lymphoma (7), but it does not have broad anti-tumor activity. This might reflect the fact that treatment of cells with rapamycin generally causes partial inhibition of mTORC1 (8, 9), no inhibition of mTORC2 (10), and activation of PI3Ks as a consequence of inhibiting a negative feedback loop (11). In contrast, ATP-competitive mTOR inhibitors are expected to inhibit both mTORC1 and mTORC2 activities, although mTORC1 inhibition may still lead to PI3K hyperactivation upstream of mTORC2. A number of ATP-competitive heterocyclic inhibitors have been described, including Torin1 (12), PP242 (9), OSI027 (13), KU63794 (14), AZD8055 (15), WYE354 (16), WYE125132 (17), and others (Fig. 1) (18). Several of these compounds have been advanced to phase I clinical investigation, including OSI027, AZD8055, WYE125132, and INK128. To better understand the pharmacological effects of treating cells with mTOR inhibitors, we selected four structurally diverse compounds (Torin1, PP242, KU63794, and WYE354) for kinome-wide selectivity profiling using three complementary approaches as follows: radio-enzymatic assays of 97 recombinant kinases (at the Medical Research Council Phosphorylation Unit), binding to 442 recombinant kinases (using DiscoveRx KinomeScanTM), and chemical proteomics in cellular lysate (involving assays for 121 kinases, using KiNativ methods developed by ActiveX BioSciences). Potential targets other than mTOR identified by these methods were further investigated using cellular assays.
FIGURE 1.
Structures of ATP-competitive mTOR inhibitors.
EXPERIMENTAL PROCEDURES
Materials
Torin1, WYE354, KU63794, and PP242 were prepared following published procedures. MK2206, ZSTK474, PD0325901, and CI-1033 were from Haoyuan Chemexpress Co. The Jak inhibitor (Jak inhibitor I) was from EMD Chemicals. Antibodies to phospho-Thr-389S6K, phospho-Ser-473Akt, phospho-Thr-308Akt, and pan-Akt were from Cell Signaling Technology. Antibody to S6K was from Santa Cruz Biotechnology. ATP was from Sigma, and GFP-4EBP1 was from Invitrogen. Purification of the mTORC1 complex (19) and cell lysis protocols have been described previously (8).
Selectivity Profiling
MRCPPU 97 protein kinase selectivity profiling was performed at the Medical Research Council Phosphorylation Unit in Dundee, Scotland, UK. DiscoveRx 442 kinome-wide selectivity profiling was conducted by DiscoveRx Bioscience with KinomeScanTM Technology. KiNativ® selectivity profiling was performed at ActivX using the Kinativ® platform. Invitrogen SelectScreen® PIKK family selectivity profiling was conducted at Invitrogen.
Binding Modes Modeling Study
mTOR protein structure was obtained from Swiss-Model by homology modeling (20). Docking results were optimized using TINKER 4.2 and AMBER force field. In the process of optimization, the protein conformation was fixed, and the inhibitor was optimized until the root mean square (r.m.s.) energy gradient fell below 0.1 kcal·mol−1·Å−1 (21, 22).
Cellular Selectivity Confirmation Using High Throughput Microscopy
SKBR3 or MCF-7 cells were plated at 7500 cells/well in 96-well microscopy plates (Corning Glass) in McCoy's and DMEM supplemented with 10% fetal bovine serum for 24 h and then starved in media lacking serum for 16 h (23). Cells were pretreated for 10 min with 10-fold stock solutions of inhibitors and treated with 10-fold stock solutions of epidermal growth factor, heregulin, glial cell line-derived neurotrophic factor, or interleukin 6 (all PeproTech) for 10 min. Cells were fixed in 2% paraformaldehyde for 10 min at room temperature and washed with PBS-T (phosphate-buffered saline, 0.1% Tween 20). Cells were then permeabilized in methanol for 10 min at room temperature, washed with PBS-T, and blocked in Odyssey blocking buffer (LI-COR Biosciences) for 1 h at room temperature. Next, cells were incubated overnight at 4 °C with rabbit antibody specific for Ser(P)-473 AKT, Thr(P)-308 AKT, Thr(P)-202/Tyr(P)-204 ERK1/2, or Tyr(P)-701 STAT1 (Cell Signaling Technology) diluted 1:400 in Odyssey blocking buffer (Licor). Cells were then washed three times in PBS-T and incubated with rabbit-specific secondary antibody labeled with Alexa Fluor 647 (Invitrogen) diluted 1:2000 in Odyssey blocking buffer. Cells were washed once in PBS-T and once in PBS and incubated in 250 ng/ml Hoechst 33342 (Invitrogen) and 1:1000 Whole Cell Stain (blue; Thermo Scientific) solution for 15 min. Cells were then washed two times with PBS and imaged in an imageWoRx high throughput microscope (Applied Precision). The average data and standard deviations of six experiments were plotted using DataPflex (24).
ATR, ATM, and DNA-PK Cellular Activity Assays
HeLa cells were seeded in 6-well plates (0.5 × 106/well) and grown overnight. After 1 h of pretreatment with appropriate compounds at 37 °C, the culture media were removed and saved. For the ATR assay, cells were treated with 50 mJ of UV radiation energy using a Stratalinker (10 gray ionizing radiation for the ATM and DNA-PK assays). The culture media were added back to cells, and they were incubated at 37 °C. After 1 h, cells rinsed once with ice-cold PBS were lysed in ice-cold lysis buffer (40 mm HEPES (pH 7.4), 2 mm EDTA, 10 mm pyrophosphate, 10 mm glycerophosphate, 1% Triton X-100, and 1 tablet of EDTA-free protease inhibitors per 25 ml). The soluble fractions of cellular lysates were then separated from cellular debris by centrifugation at 13,000 rpm for 10 min in a microcentrifuge. After the lysates from all plates were collected, the concentration of the protein was normalized by using Bradford assays. 50 μl of sample buffer was added to the normalized lysates and boiled for 5 min. The samples were subsequently analyzed by SDS-PAGE and immunoblotting.
Cellular Viability Assay with JAK-transformed Cells
Ba/F3 derivatives expressing various oncogenic fusion kinases, namely TEL-JAK1, TEL-JAK2, TEL-JAK3, and TEL-ABL, were described previously (25). The cells were maintained in RPMI 1640 medium supplemented with 10% FBS, l-glutamine, and penicillin/streptomycin (Sigma). For cell viability assays, the cells were incubated (density of 5000 cells per well in 96-well plates) in the presence of graded doses of small molecule inhibitors for 72 h. The number of viable cells was determined with the CellTiter Glo assay (Promega, Madison, WI). The 50% growth inhibition (GI50) values with an inhibitor for each cell line were calculated by nonlinear regression using GraphPad Prism software (La Jolla, CA).
Mouse Liver Microsome Stability Study
Mouse liver half-life was evaluated at Scripps Research Institute, Jupiter, FL. Microsome stability was determined by incubating 1 μm test compound with 1 mg/ml mouse hepatic microsomes in 100 mm KPi at pH 7.4. The reaction was initiated by adding NADPH (1 mm final concentration). Aliquots were removed at 0, 5, 10, 20, 40, and 60 min and added to acetonitrile (5× v/v) to stop the reaction and precipitate proteins. The NADPH dependence of the reaction was evaluated using NADPH-free samples. At the end of the assay, the samples were centrifuged through a Millipore Multiscreen Solvinter 0.45-micron low binding PTFE hydrophilic filter plate and analyzed by LC-MS/MS. The data were log transformed, and regression analysis was used to calculate the half-life values.
Time-resolved mTORC1 Enzymatic Activity Study
mTORC1 was incubated with inhibitors (0.5 μm, 1% DMSO) in 5 μl of 1× reaction buffer (25 mm HEPES (pH 7.4), 10 mm MgCl2, and 4 mm MnCl2) for 40 min at room temperature. Then drug-ATP competition was induced by the addition of 245 μl of 1× reaction buffer containing 500 μm ATP and 0.4 μm GFP-4EBP1 (Invitrogen). The reaction mixture was dispensed (10 μl, in triplicate) into a low volume black plate (Corning Glass), and the kinase reaction was stopped at various times with 5 μl of stop solution (Invitrogen) containing 30 mm EDTA. Stop solution (5 μl) containing 4 nm terbium-labeled p-4EBP1 (T46) antibody (Invitrogen) was added, and the FRET signal was read using Envision (PerkinElmer Life Sciences) after 30 min of incubation.
Cell Washout Experiment
HeLa or PC3-S473D cells were seeded in 6-well plates (0.25 × 106/well) and grown overnight. Cells were then treated with the appropriate compounds for 1 h. After 1 h, except for the 0-h plate, the plates were washed of inhibitors with 1 ml/well culture media. Cells were then lysed in regular time intervals of 1, 2, 4, 6, and 16 h. The cells were rinsed once with ice-cold PBS and lysed in ice-cold lysis buffer (40 mm HEPES (pH 7.4), 2 mm EDTA, 10 mm pyrophosphate, 10 mm glycerophosphate, 1% Triton X-100, and 1 tablet of EDTA-free protease inhibitors per 25 ml). The soluble fractions of cell lysates were isolated by centrifugation at 13,000 rpm for 10 min in a microcentrifuge. After lysates from all of the plates were collected, the concentration of the protein was normalized using the Bradford assay. 50 μl of sample buffer was added to the normalized lysates, which were then boiled for 5 min. Samples were subsequently analyzed by SDS-PAGE and immunoblotting.
RESULTS
Potency Profiling of mTOR Inhibitors
To directly compare the relative potency of the four inhibitors, we determined biochemical IC50 values using both a recombinant mTOR kinase domain and an intact mTORC1 complex purified from mammalian cells. All four compounds exhibited highly potent and similar IC50 values against the recombinant mTOR kinase domain. However, using the purified mTORC1 complex 1, Torin1 was the most potent compound by a factor of ∼20–70-fold (Table 1 and supplemental Fig. 1). To establish relative cellular potency, we examined the EC50 values for inhibition of S6 kinase Thr-389 phosphorylation, a residue well established to be modified directly by mTORC1. In this assay, Torin1 exhibited an EC50 of 2 nm, ∼10–20-fold more potent than the other inhibitors. These results demonstrate that assays using purified mTORC1 complexes more accurately reflect the cellular potency of mTOR inhibitors as compared with assays using the recombinant mTOR kinase domain.
TABLE 1.
In vitro characterization of mTOR inhibitors
mTOR enzymatic assay was performed using SelectScreen® technology (Invitrogen), and the mTORC1 assay employed purified Raptor-tagged mTORC1 complex obtained from HEK293T cells. The EC50 was calculated based upon the ability to suppress phosphorylation of pS6K-Thr-389.
| Entry | mTOR-recombinant, IC50 | mTORC1 complex, IC50 | mTOR (HeLa), EC50 |
|---|---|---|---|
| nm | nm | nm | |
| Torin1 | 4.3 | 1 | 2 |
| PP242 | 13.7 | 24.7 | 40 |
| KU63794 | 8.57 | 35.4 | 30 |
| WYE354 | 15.8 | 77.1 | 20 |
Selectivity Profiling of mTOR Inhibitors
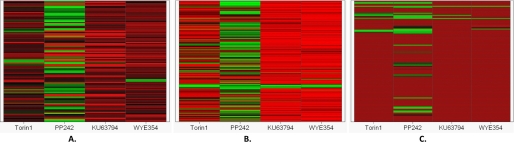
Next we evaluated kinase selectivity against a panel of 97 recombinant protein kinases (Fig. 2A, Table 2, and supplemental Table 1). At a concentration of 10 μm, Torin1, KU63794, and WYE354 were not observed to inhibit any protein kinase in the panel (at using a threshold of 50% inhibition relative to a DMSO-only control), whereas 10 μm PP242 exhibited strong (90%) inhibition of a number of kinases in the tyrosine kinase family/TK (including BTK, Eph, FGF receptor, VEGF receptor, Src, LCK, and YES), calcium/calmodulin-dependent protein kinase family/CAMK (BRSK2, CHK2, and MLCK), CMGC family (DYK2/3, HIPK2 CDK, MAPK, GSK3, CLK, and ERK8), casein kinase family (CK1δ), and AGC family (PKCα). The relative lack of selectivity of PP242 can be rationalized based on the fact that its binding mode and scaffold are most similar to ATP and its structural similarity to PP1, a relatively nonselective inhibitor of Src family kinases.
FIGURE 2.
Selectivity profiling of Torin1, PP242, KU63794, and WYE354 at 10 μm. A, Medical Research Council protein phosphorylation unit 97 protein kinase panel; B, KinomeScan (DiscoveRx) 442 kinome-wide kinase panel; C, KiNativ (ActivX) 121 Hela cell kinase panel. In general, green indicates strong inhibition, and red indicates weak inhibition. The lighter the color, the stronger the inhibition (green). The deeper the color, the weaker the inhibition (red).
TABLE 2.
mTOR inhibitors in the Medical Research Council protein kinase panel
Inhibitors were screened at 10 μm, and numbers represent the percent of kinase activity remaining relative to the control. Only kinases with remaining activity of 10% or less are shown. The full dataset is available in the supplemental material.
| Family | Kinase | Torin1 | PP242 | KU63694 | WYE354 |
|---|---|---|---|---|---|
| TK | BTK | 56 ± 0 | 5 ± 2 | 81 ± 21 | 99 ± 6 |
| EPHA2 | 87 ± 0 | 3 ± 0 | 96 ± 2 | 104 ± 2 | |
| EPHB3 | 94 ± 6 | 5 ± 4 | 78 ± 4 | 116 ± 11 | |
| FGF-R1 | 79 ± 8 | 2 ± 0 | 87 ± 10 | 109 ± 3 | |
| LCK | 78 ± 4 | 4 ± 0 | 91 ± 2 | 91 ± 2 | |
| Src | 52 ± 5 | 2 ± 1 | 86 ± 5 | 67 ± 2 | |
| VEG-FR | 84 ± 6 | 2 ± 1 | 92 ± 8 | 88 ± 10 | |
| YES1 | 78 ± 3 | −8 ± 15 | 90 ± 1 | 113 ± 3 | |
| CAMK | BRSK2 | 104 ± 7 | 4 ± 2 | 72 ± 7 | 88 ± 8 |
| CHK2 | 83 ± 4 | 2 ± 0 | 96 ± 9 | 88 ± 3 | |
| MLCK | N.D. | 6 ± 1 | 104 ± 13 | 91 ± 2 | |
| CMGC | DYRK2 | 99 ± 16 | 7 ± 2 | 96 ± 12 | 96 ± 8 |
| DYRK3 | 52 ± 3 | 2 ± 1 | 84 ± 1 | 77 ± 1 | |
| HIPK2 | 88 ± 6 | 7 ± 1 | 89 ± 1 | 94 ± 6 | |
| ERK8 | 46 ± 7 | 6 ± 0 | 92 ± 5 | 98 ± 1 | |
| CK1 | CK1δ | 104 ± 0 | 9 ± 1 | 109 ± 12 | 92 ± 6 |
| AGC | PKCα | 68 ± 3 | 9 ± 2 | 86 ± 18 | 75 ± 1 |
The four mTOR inhibitors were also subjected to binding assays at a screening concentration of 10 μm using the KinomeScanTM approach, which tests for association with 442 distinct kinases (Fig. 2B, Table 3, and supplemental Table 2). The results were consistent with the biochemical profiling (Fig. 2A and Table 2). PP242 strongly inhibited a number of TK family kinases (ABL, FLT, JAK, KIT, LCK, PDGF receptor, and RET), TKL family kinases (ACVR1/2 and BMPR), CAMK family kinases (BRSK2, MLCK, and PIM2), CMGC family kinases (HIPKs), STE family kinases (LOK, GCK, MEK1/2/5, SLK, TAO1, and YSK4), AGC family kinases (DMPK, MRCKα, PKCϵ, MSK2, and RSK2), PI3K family kinases (PI3Kβ/δ/γ), and CK1 family kinases (CSNK1E). In contrast, Torin1, KU63794, and WYE354 were much more selective. Torin1 exhibited strong off-target binding to MRCKa in the AGC family and PI3Kα in the PIKK family. KU63794 had the most selective profile and did not appear to bind any protein kinases other than mTOR. The only strong off-target binding by KU63794 involved the I800L mutant form of PI3Kα. WYE354 also bound to PI3K-I800L as well as p38 β/γ. Off-target binding of mTOR inhibitors to members of the PI3K family was expected because the mTOR shares a high level of sequence identity to PI3K family members in the kinase catalytic domain. Off-target effects are most easily visualized with respect to a kinome dendrogram (Fig. 3). All four mTOR inhibitors exhibited greater potency against the PI3Kα-I800L as compared with wild-type PI3Kα (Table 4) by KinomeScan profiling, but follow-on determination of Kd values did not confirm this result. For example, cellular assays examining the inhibition of Akt phosphorylation by mTOR in human mammary endothelial cells (HMECs) expressing PI3Kα-I800L did not reveal significant activity against this target at 1 μm drug (data not shown).
TABLE 3.
mTOR inhibitors in KinomeScanTM kinase panel
Inhibitors were screened at a single concentration of 10 μm. Scores are related to the probability of a hit and are not strictly an affinity measurement. At a screening concentration of 10 μm, a score of less than 10% implies that the false positive probability is less than 20% and that the Kd value is most likely less than 1 μm. A score between 1 and 10% implies that the false positive probability is less than 10%, although it is difficult to assign a quantitative affinity from a single-point primary screen. A score of less than 1% implies that the false positive probability is less than 5% and that the Kd value is most likely less than 1 μm. Hits of less than 1% are shown in the list, and the full list is shown in the supplemental material.
| Family | Kinase | Torin1 | PP242 | KU63794 | WYE354 |
|---|---|---|---|---|---|
| TK | ABL1(E255K) | 85 | 0.1 | 100 | 100 |
| ABL1(H396P)np | 68 | 0 | 100 | 91 | |
| ABL1(H396P)p | 81 | 0.05 | 100 | 100 | |
| ABL1(Q252H)np | 89 | 0.05 | 83 | 100 | |
| ABL1(Q252H)p | 99 | 0.2 | 71 | 94 | |
| ABL1(T315I)p | 100 | 0.95 | 100 | 100 | |
| ABL1(Y253F)p | 83 | 0.1 | 100 | 90 | |
| ABL1np | 60 | 0.75 | 98 | 84 | |
| ABL1p | 77 | 0.05 | 99 | 100 | |
| ERBB3 | 44 | 0 | 85 | 100 | |
| FLT4 | 74 | 0.3 | 100 | 92 | |
| JAK1 | 76 | 0.2 | 100 | 74 | |
| JAK2 | 65 | 0.95 | 71 | 73 | |
| JAK3 | 100 | 0 | 100 | 100 | |
| KIT | 59 | 0.8 | 87 | 100 | |
| KIT(L576P) | 48 | 0.8 | 93 | 100 | |
| KIT(V559D) | 50 | 0.3 | 90 | 96 | |
| LCK | 77 | 0.95 | 98 | 88 | |
| PDGFRB | 24 | 0 | 95 | 100 | |
| RET | 44 | 0 | 91 | 93 | |
| RET(M918T) | 51 | 0 | 100 | 87 | |
| TKL | ACVR1 | 28 | 0.1 | 100 | 100 |
| ACVR2A | 47 | 0.5 | 100 | 100 | |
| BMPR1B | 100 | 0.1 | 99 | 78 | |
| BMPR2 | 85 | 0.1 | 99 | 78 | |
| CAMK | BRSK2 | 76 | 0.95 | 98 | 100 |
| MYLK | 87 | 0.9 | 57 | 81 | |
| PIM2 | 62 | 0.6 | 100 | 100 | |
| CMGC | HIPK2 | 58 | 0.9 | 58 | 65 |
| HIPK3 | 86 | 0.9 | 76 | 72 | |
| P38δ | 39 | 97 | 71 | 0 | |
| P38γ | 71 | 59 | 100 | 0 | |
| STE | LOK | 79 | 0.5 | 92 | 100 |
| MAP4K2 | 35 | 0 | 86 | 100 | |
| MEK1 | 100 | 0.25 | 100 | 100 | |
| MEK2 | 100 | 0.4 | 92 | 100 | |
| MEK5 | 100 | 0.35 | 100 | 100 | |
| SLK | 94 | 0.2 | 98 | 70 | |
| TAOK1 | 58 | 0.2 | 83 | 83 | |
| YSK4 | 38 | 0 | 100 | 75 | |
| AGC | DMPK | 77 | 0.55 | 86 | 100 |
| MRCKA | 0.65 | 74 | 92 | 100 | |
| PRKCE | 86 | 0 | 82 | 94 | |
| RPS6KA4 | 100 | 0 | 95 | 88 | |
| RSK2 | 38 | 0.35 | 99 | 76 | |
| PI3K | mTOR | 0 | 0 | 0 | 0 |
| PIK3CA(C420R) | 0.7 | 30 | 2.3 | 3 | |
| PIK3CA(E545K) | 0.6 | 21 | 2 | 4.9 | |
| PIK3CA(I800L) | 0 | 1.6 | 0 | 0.4 | |
| PIK3CB | 65 | 0 | 30 | 42 | |
| PIK3CD | 51 | 0 | 0.95 | 40 | |
| PIK3CG | 1.2 | 0.55 | 23 | 78 | |
| CK1 | CSNK1E | 59 | 0.1 | 87 | 72 |
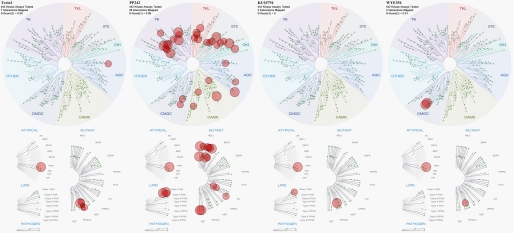
FIGURE 3.
Kinome tree depiction of the mTOR inhibitor off-targets in protein kinases. Figures were generated with DiscoveRx TREEspotTM Version 4. The original results were shown as percent control to DMSO, and targets exhibiting less than 1% remaining activity were selected in the figures. S score indicated the relative selectivity properties of the drugs with smaller S values signifying a more selective compound. The sizes of the red circle are proportional to the strength of the binding; the larger circles imply higher affinity.
TABLE 4.
Comparison of mTOR inhibitor sensitivity to PI3Kα versus PI3Kα (I800L)
| Entry | PI3Kα (Ambit Score) | PI3Kα (I800L) (Ambit score) | PI3Kα (Kd, nm) | PI3Kα (I800L) (Kd, nm) |
|---|---|---|---|---|
| Torin1 | 1.3 | 0 | 23 | 14 |
| PP242 | 32 | 1.6 | 200 | 230 |
| KU63794 | 2.5 | 0 | 71 | 57 |
| WYE354 | 5.4 | 0.4 | 140 | 190 |
Given the structural similarities among PIKK family members, we subjected mTOR inhibitors to ActivX KiNativTM kinase target profiling, a method that has the most extensive coverage of PIKK family members (Fig. 2C, Table 5, and supplemental Table 3). The KiNativ assay measures the ability of small molecules to protect kinases present in cell extract from binding to and forming an adduct with a lysine-reactive ATP-biotin (26). PP242 exhibited potent protection of all PIKK family members (ATM, ATR, PI3Ks, DNA-PK, and SMG1), lipid kinase (PIP5K3), as well as some protein kinases in the CMGC, AGC, and STE families, confirming earlier results with recombinant kinase assays. Torin1 strongly inhibited ATM, ATR, PI3Kα, and DNA-PK in the PIKK family. KU63794 targeted both PI3Kα and -δ, whereas WYE354 exhibited the greatest selectivity and only inhibited PI3Kδ.
TABLE 5.
mTOR inhibitors in KiNativTM kinase panel
Inhibitors were screened at 0.01, 0.1, 1, and 10 μm, and IC50 values were determined (μm) in Jurkat cells. The values are Kdapp if the Kdapp is different from the IC50 value.
| Family | Kinase | Torin1 IC50 (μm) | IC50 (μm) | KU63794 IC50 (μm) | WYE354 IC50 (μm) |
|---|---|---|---|---|---|
| PIKK(L) | ATM | 0.64 | 0.79 | 10 | >10 |
| ATR | <0.01 | <0.01 | >10 | >10 | |
| mTOR | <0.01 | <0.01 | <0.01 | 0.022 | |
| PIK3C2B | 3.6 | 0.6 | >10 | >10 | |
| PIK3CA | 0.26 | 0.95 | 1 | 1.2 | |
| PIK3CB | 4.9 | 0.17 | 10 | 10 | |
| PIK3CD | 1.6 | 0.1 | 0.096 | 0.98 | |
| PIP5K3 | >10 | 0.061 | >10 | 2 | |
| DNA-PK | <0.01 | 0.061 | 10 | >10 | |
| SMG1 | 4.4 | 0.38 | >10 | >10 | |
| CMGC | CHED | >10 | 0.3 | >10 | >10 |
| AGC | PKCa/b | >10 | 0.15 | >10 | >10 |
| RSK1/2/3 | >10 | 0.38 | >10 | >10 | |
| RSK2 | >10 | 0.046 | >10 | >10 | |
| STE | SLK | >10 | 0.1 | >10 | >10 |
To further investigate the activity of mTOR inhibitors against PIKK family kinases, we examined 10 kinases present in the Invitrogen SelectScreen® PIKK panel (Table 6). SelectScreen® is a FRET-based technology using fluorescence signal change as readout to detect inhibition of small molecules against protein kinases. PP242 exhibited a low IC50 value against PI3K-C2β, PI3Kδ, and DNA-PK (IC50 <100 nm) and moderate IC50 values for PI3K-C2α, PI3Kα, PI3Kβ, and PI3Kγ (IC50 ∼100–1000 nm). Torin1 was a potent inhibitor of DNA-PK (IC50 ∼6.3 nm) and moderate inhibitor of PI3K-C2α, PI3K-C2β, hVPS34, PI3Kα, PI3Kδ, and PI3Kγ. Both KU63794 and WYE354 were very selective against the PIKKs, and exhibited no activity against PI4K, group II PI3K, and group III PI3K. They only weakly inhibited PI3Kα and PI3Kδ in the group I PI3K subfamily(IC50, 100–1000 nm), and neither of them had apparent activity against PI3Kβ, PI3Kγ, or DNA-PK.
TABLE 6.
mTOR inhibitors in the Invitrogen SelectScreen® PIKK family panel
| Kinases | Torin1 IC50 (nm) | PP242 IC50 (nm) | KU63794 IC50 (nm) | WYE354 IC50 (nm) |
|---|---|---|---|---|
| PIK4CA | >10,000 | >10,000 | >10,000 | >3330 |
| PIK4CB | 6680 | 2130 | >10,000 | >10,000 |
| PIK3C2A | 176 | 866 | >10,000 | >10,000 |
| PIK3C2B | 549 | 82.9 | >10,000 | 5150 |
| PIK3C3 | 533 | >10,000 | >10,000 | >10,000 |
| PIK3CA | 250 | 201 | 654 | 886 |
| PIK3CD | 564 | 34.2 | 173 | 1290 |
| PIK3CB | >10,000 | 750 | >10,000 | >12,000 |
| PIK3CG | 171 | 289 | 2130 | 1870 |
| DNA-PK | 6.34 | 92.1 | 9120 | 9270 |
Binding Modes Modeling Study
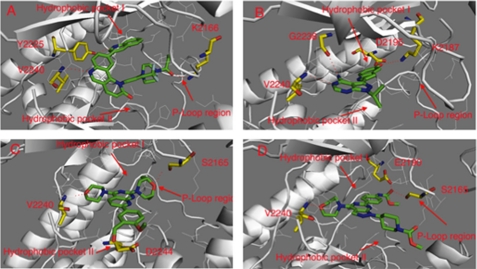
The distinct selectivity profiles of the four mTOR inhibitors in our study prompted us to investigate their potential binding conformations using molecular modeling. As the crystallographic structure of mTOR is unknown, we used the reported structure of PI3Kγ (Protein Data Bank code 3DBS) to create a homology model. Docking studies suggest that KU63794 and WYE354 share a similar binding mode, with compounds exploiting the morpholine oxygen to form a hydrogen bond with Val-2240 located in the “hinge region” of the kinase (Fig. 4). Modeling predicts that KU63794 forms two more hydrogen bonds, one with Asp-2244 near the solvent-exposed area (hydrophobic pocket II) and another with Ser-2165 located in a loop structurally analogous to the P-loop of protein kinases (P-loop-like region). WYE354 is predicted to form two hydrogen bonds between the carbamate moiety and Ser-2165 in the P-loop-like position and Glu-2190 in the C-helix region. Torin1 is predicted to form a hinge-binding hydrogen bond with Val-2240 via the nitrogen atom of the core tricyclic quinoline moiety. Lys-2166 in the P-loop-like area forms a hydrogen bond with the amide moiety in the piperazine side chain. The quinolone side chain occupies an inner hydrophobic pocket (hydrophobic pocket I) that is not utilized by ATP and forms a hydrogen bond in this region with Tyr-2225. Confirming this, mutation the of tyrosine (Tyr-2166) in Saccharomyces cerevisiae TOR2 that is analogous to human mTOR-Tyr-2225 resulted in reduced affinity for Torin1-like compound.5 PP242 is predicted to form two hydrogen bonds in the hinge area with Val-2240 and Gly-2238. The phenol moiety forms two hydrogen bonds, one in the hydrophobic pocket I region with Asp-2195 in the C-helix and the other with Lys-2195. In comparison, all of the inhibitors are class I kinase inhibitors and occupy the ATP adenine binding area to bind the hinge (27). PP242 explores the adjacent hydrophobic pocket (I), whereas KU63794 and WYE354 explore the hydrophobic pocket in the hinge area (II) and P-loop region. Torin1 utilizes hydrophobic pocket (I) and the P-loop region. The molecular modeling provides ideas about how to modify the chemical structures to exploit different regions of the ATP-binding pocket to modulate potency and selectivity.
FIGURE 4.
Molecular modeling of binding modes. A, binding modes of Torin1 in mTOR; B, binding mode of PP242 in mTOR; C, binding mode of KU63794 in mTOR; D, binding mode of WYE354 in mTOR.
Selectivity Profiles in Cellular Assays
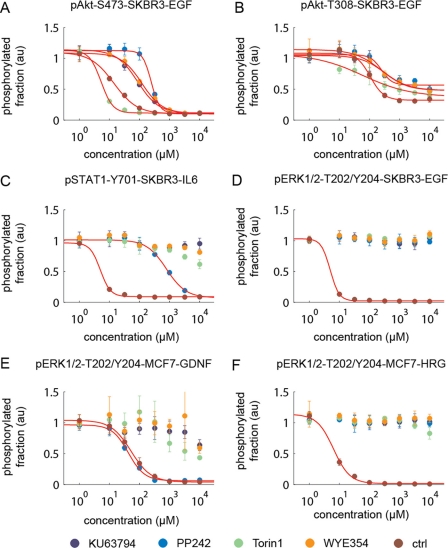
Biochemical and proteomic assays performed in vitro show that Torin1, KU63794, and WYE354 are more selective kinase inhibitors than PP242. However, targets identified by these approaches are simply candidates until they can be validated using an appropriate cellular assay. By looking at individual signal transduction pathways activated through growth factors and cytokines, we determined the efficacy of specific kinases in intact cells. We first investigated the selectivity for mTOR inhibition relative to PI3K inhibition by examining the levels of phosphorylation of Akt-Thr-308, which lies downstream of PI3K, and Akt-Ser-473, a substrate of mTORC2, in cells treated with epidermal growth factor (EGF). It is important to note that this assay does not fully differentiate between cellular mTOR and PI3K inhibition because the inhibition of Ser-473 of Akt in turn affects the phosphorylation of Thr-308. The potency for inhibition of Akt-Thr-308 phosphorylation was Torin1 (IC50 = 5 nm) > KU63794 (IC50 = 86 nm) > WYE354 (IC50 = 120 nm) > PP242 (IC50 = 245 nm). For Torin1, the IC50 for inhibition of Akt-Ser-473 was significantly lower than for Thr-308 (12-fold), marginally lower than KU63794 (4-fold) and WYE354 (2-fold), and identical to that of PP242 (Fig. 5, A and B, and Table 7).
FIGURE 5.
mTOR inhibitor selectivity profiles in cellular assays in SKBR cells stimulated with various ligands. A, EGF-induced pAkt-Ser-473 monitors mTORC2 inhibition; B, EGF-induced pAkt-Thr-308 monitors PI3K inhibition; C, IL6-induced pSTAT1-Tyr-702 monitors JAK1/2/3 inhibition; D, EGF-induced pErk1/2-Thr-202/Tyr-204 monitors MEK1/2; E, GDNF-induced pErk1/2-Thr-202/Tyr-204 monitors RET inhibition in MCF7 cells; F, HRG-induced pErk1/2-Thr-202/Tyr-204 monitors MEK1/2 and ErbB3 inhibition in MCF7 cells. Control compounds are as follows: A, MK2206; B, ZSTK474; C, JAK inhibitor I; D, PD0325901; E, ALW-II-41–27; F, CI-1033.
TABLE 7.
Cellular mTOR inhibitor selectivity profile
NA means not applicable.
| Kinase | mTORC2 (EC50, nm) | PI3K (EC50, nm) | RET (EC50, nm) | JAK1,2,3 (EC50, nm) | ATR (EC50, nm) | ATM (EC50, nm) |
|---|---|---|---|---|---|---|
| Torin1 | 5 | 60 | NA | NA | >1000 | >1000 |
| PP242 | 245 | 250 | 42 | 780 | >1000 | >1000 |
| KU63794 | 86 | 320 | NA | NA | NA | NA |
| WYE354 | 120 | 245 | NA | NA | NA | NA |
To investigate whether the binding of PP242 to MEK1/2 observed in the KinomeScanTM assay was evident in a cellular context, we asked whether phosphorylation of ERK1/2, the immediate downstream target of MEK1/2, was inhibited in SKBR3 cells treated with EGF (Fig. 5D). No inhibition of ERK1/2 phosphorylation was observed, suggesting that PP242 is not a functional MEK1/2 inhibitor. However, PP242 was capable of inhibiting JAK1–3 (IC50 = 780 nm), as measured by the inhibition of STAT1 phosphorylation in SKBR3 cells stimulated with interleukin 6, and RET kinase (IC50 = 42 nm), as measured by inhibition of ERK1/2 phosphorylation in MCF-7 cell lines stimulated with glial cell line-derived neurotrophic factor treatment. Further testing of PP242 on TEL-transformed JAK-dependent BaF3 cells showed that PP242 had a moderate inhibitory effect on JAK3 (0.91 μm) and a mutant form JAK4 (0.98 μm) (Table 8).
TABLE 8.
Activity of PP242s on TEL-transformed BaF3 cells
| Entry | GI50 (μm) | JAK2 GI50 (μm) | JAK3 GI50 (μm) | TYK2 (E957D) GI50 (μm) |
|---|---|---|---|---|
| PP242 | 1.449 | 1.709 | 0.910 | 0.982 |
To determine whether the putative binding of Torin1 and PP242 to ATR and ATM observed in the KinativTM scan would translate into inhibitory activity in cells, we examined the ability of the compounds to inhibit phosphorylation of Chk1-Ser-317 and Chk2-Thr-68 in HeLa cells exposed to UV and ionizing radiation. Surprisingly, neither Torin1 nor PP242 was capable of inhibiting Chk1 and Chk2 phosphorylation up to a concentration of 1 μm, suggesting that these compounds are not potent inhibitors of ATM or ATR in cells (supplemental Fig. 2). In addition, as judged by monitoring a putative DNA-PK auto-phosphorylation site (Ser-2056), neither Torin1 nor PP242 measurably blocked the activity of DNA-PK up to a concentration of 1 μm (8).
Mouse Liver Microsome Stability
To complement the comparisons of kinase selectivity, we also examined the chemical stabilities of the compounds during incubation with mouse liver microsomes (Table 9). Torin1 and PP242 were rapidly metabolized, whereas KU63794 and WYE354 were considerably more resistant (11, 16). These results suggest that further chemical optimization of Torin1 and PP242 needs to be performed to improve metabolic stability.
TABLE 9.
Mouse liver microsome stability evaluation of mTOR inhibitors
In general, a 0–10-min half-life indicates a poor in vivo stability; 10–20 min indicates a moderate stability; 20–40 min indicates a good hepatic stability, and over 40 min indicates that there might be some other mechanism involved instead of typical hepatic enzyme.
| Entry | Torin1 | PP242 | KU63794 | WYE354 |
|---|---|---|---|---|
| T½ (min) | 4 | 2 | 61 | 15 |
Torin1 Has Slow Off-binding Kinetics
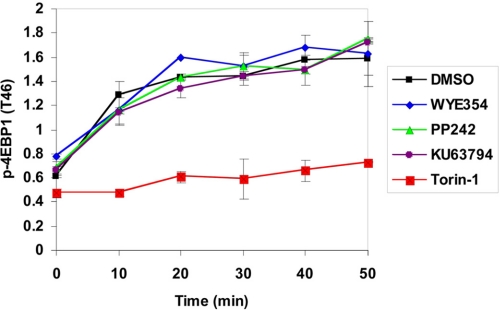
The off-rates of kinase inhibitors are considered to be a primary driver of efficacy in cellular and in vivo models; inhibitors with slow off-kinetics display longer duration and more effective pharmacological inhibition (28). To investigate potential differences in the off-rates of mTOR inhibitors and their targets, we evaluated the time-dependent inhibition of the mTORC1 complex in the presence of a relatively high concentration of ATP (500 nm, apparent Km under assay conditions was 4 μm). Interestingly, Torin1 exhibited a substantially longer lived inhibition of mTORC1 kinase activity relative to the other inhibitors (Fig. 6). This implies that Torin1 has a slow off-rate.
FIGURE 6.
Binding kinetics of mTOR inhibitors with mTORC1 complex; mTORC1 complex with treatment of 500 nm drug in the presence of 500 mm ATP. The activity of the enzyme was monitored using GFP-4EBP1 as a substrate.
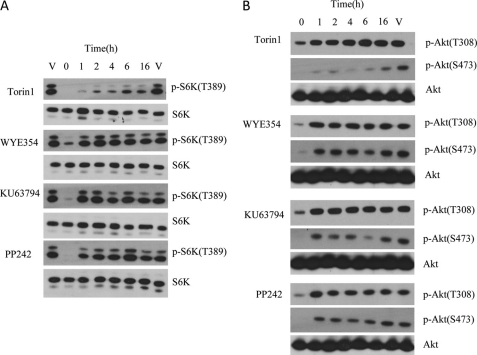
To investigate whether the slow off-rate of Torin1 bound to mTOR would also hold in a cellular context, cellular washout experiments were performed. HeLa cells were treated with inhibitors for 1 h at a concentration of 250 nm, followed by extensive washing to remove excess drug, and the recovery of mTORC1-dependent (pS6K-Thr-389) and PI3K-dependent (p-AKT-Thr-308) phosphorylation was monitored as a function of time (Fig. 7). For these experiments, we employed PC-3 cells stably expressing the S473D mutant of Akt, which results in a decoupling of the Thr-308 and Ser-473 sites, allowing for the PI3K inhibition to be monitored independent of mTORC2 (29). Consistent with results using purified compounds, Torin1 was able to inhibit mTORC1 for up to 16 h after washout, although other compounds only suppressed S6K phosphorylation for 1–2 h.
FIGURE 7.
ATR, ATM, and DNA-PK cellular activity characterization and off-rate assay of mTOR inhibitors. A, cellular mTORC1 assay, HeLa cells were treated with inhibitors at a concentration of 250 nm for 1 h followed by washing off unbound drug. The lysates were then Western-blotted for pS6K (Thr-389) (mTORC1 substrate) and total S6K. B, cellular PI3K assay, PC-3 cells stably expressing the AktS473D mutation were treated with inhibitors at a concentration of 250 nm, and lysates were Western-blotted for pAkt(Thr-308) and total Akt.
DISCUSSION
Achieving a high degree of selectivity is a key challenge when developing ATP-competitive kinase inhibitors because of the highly conserved nature of the kinase ATP-binding site. Most clinically approved and “tool” kinase inhibitors have a number of kinase targets, which seem to grow as profiling technology matures. In this study, we undertook a comprehensive assessment of the kinase selectivity of recently reported ATP-competitive mTOR inhibitors. With the exception of PP242, the inhibitors profiled in this study demonstrate that it is possible to attain a high degree of selectivity for the ATP site of mTOR relative to other PIKK family kinases as well as other serine/threonine and tyrosine kinases. As expected, each heterocyclic template exhibited a unique spectrum of additional targets.
Torin1 was developed based on a quinoline core structure and is presumed to share the same binding mode with the PI3K, mTOR, and other PIKK family inhibitor BEZ-235 (11, 30). PP242 has a pyrazolopyrimidine core structure and was discovered as an mTOR inhibitor based on analoging PP2, a low specificity Src family kinase inhibitor (31). Both KU63794 and WYE354 were derived from PI-103, a morpholino-substituted heterocycle related to the original PI3K inhibitor LY294002. Based upon the binding and enzymatic kinase panel data, Torin1, KU63794, and WYE354 exhibited a high degree of selectivity and did not inhibit most of the protein kinases at 10 μm, except for MRCKA (Torin1) and P38δ/γ (WYE354). At a concentration of 10 μm, PP242 displayed broad cross-inhibition against almost all of the protein kinase subfamilies, including TK, TKL, STE, CAMK, CMGC, AGC, and CK1. Profiling these inhibitors across the more structurally related PIKK family revealed some interesting cross-activities. Although KU63794 and WYE354 maintained high selectivity for mTOR, Torin1 exhibited potent inhibition of the enzymatic activity of ATR, ATM, and DNA-PK. but none of this activity could be observed in a cellular assay at much higher concentrations (1 μm) relative to what would be predicted based upon the biochemical assays. As all three inhibitors are capable of inhibiting mTOR activity at low nanomolar concentrations in cellular assays, cell penetration is likely not the explanation for this discrepancy (8). These results suggest that either the currently available biochemical assays for ATR, ATM, and DNA-PK do not faithfully recapitulate the physiological form of these kinases or that the current cellular readouts of phosphorylation of Chk1, Chk2, and DNA-PK are not solely dependent on ATM, ATR, and DNA-PK, respectively. We favor the second explanation because the KiNativ approach, which monitored kinase complexes generated following cell lysis, also demonstrated potent binding of Torin1 to ATR and DNA-PK.
Molecular modeling provides a means to rationalize the observed selectivity differences. The most selective inhibitors, KU63794 and WYE354, are predicted to exploit diverse areas of the ATP-binding site such as hydrophobic pocket region II and the P-loop region. Torin1 utilized hydrophobic region I and introduced one more selectivity factor via the P-loop region. Compared with ATP, PP242 only introduced one more selectivity factor by occupying hydrophobic pocket region I, which is more conserved in the kinome than the other regions that Torin1, KU63794, and WYE354 exploit. This, combined with the smaller size of PP242, may explain why it exhibits more broad cross-activity toward a variety of kinases. The current results indicated that Torin1 could be used as a highly selective mTOR inhibitor below a concentration of 1 μm. PP242 exhibited moderate selectivity against PIKK family kinases and displayed potent cellular activity against RET and JAK family kinases, a property that has been used to elaborate this scaffold to develop inhibitors of a number of other tyrosine kinases (31).
The I800L PI3Kα mutation was previously identified as a mutation that conferred resistance to PI3K inhibitors such as PI-103 as well as sensitization to PI3K/mTOR dual inhibitor BEZ235 (27). Interestingly, the single point KinomeScanTM data demonstrated that the I800L PI3Kα mutant consistently bound mTOR inhibitors more potently than wild-type PI3Kα. Sequence alignments of PI3Kα and mTOR demonstrate that mTOR possesses a leucine (Leu-2185) in place of the isoleucine of PI3Kα (Ile-800, Protein Data Bank code 2RD0). This would suggest that the ATP-binding site of I800L PI3Kα more closely resembles mTOR relative to wild-type PI3Kα (32). Transfection of HMEC cells with I800L PI3Kα did not demonstrate a differential sensitivity of wild-type versus I800L of PI3K to inhibition by the mTOR inhibitors. However, further work is required to investigate this possibility because inhibition of TORC2 leads to suppression of phosphorylation of the hydrophobic motif (Ser-473) of Akt, which is known to couple to Thr-308 and might thereby mute any differences that may have originated from differential PI3K inhibition. Although the in vivo half-life of Torin1 is short (t½ = 40 min), anti-tumor efficacy has been observed following once daily oral administration of 20 mg/kg with suppression of phosphorylation of Thr-389 of S6K observed at least after 6 h after dosing (12). We discovered a possible explanation for this phenomenon; Torin1 exhibited very sustained inhibition of mTOR in both enzymatic and cellular assays. For example, treatment of cells with Torin1 followed by extensive washing resulted in sustained inhibition of mTOR substrates to 16 h, whereas the other mTOR inhibitors experienced recovery of substrate phosphorylation after 2 h. More sustained inhibition of mTORC1 by Torin1 relative to the other mTOR inhibitors was also observed in biochemical assays, suggesting that it is not simply greater cellular retention that is causing this phenomenon. Further investigation will be required to determine whether Torin1 exhibits slow binding kinetics and what structural features of the inhibitor impart this property.
In summary, we have performed extensive profiling of the recently developed ATP-competitive small molecule mTOR inhibitors Torin1, PP242, KU63794, and WYE354 using four distinct biochemical approaches. In addition, we have performed side-by-side comparisons of the relative potencies of these inhibitors in mTOR-dependent cellular assay as well as cellular assays that report on potential off-targets. We conclude that Torin1, KU63794, and WYE354 are selective inhibitors of mTOR when used at concentrations below 1 μm. At concentrations above 1 μm, other PIKK family kinases such as DNA-PK and possibly others are inhibited by Torin1. The sustained kinetics for inhibition of mTOR by Torin1 versus the other three inhibitors is a unique attribute of this compound. The different selectivity and pharmacokinetic profiles reflect the different intrinsic chemical properties of the respective pharmacophores, which can be further explored in the targeted polypharmacology direction. The selectivity profiling reported herein should serve as a useful guide for use of these inhibitors in basic and clinical research.
Supplementary Material
Acknowledgments
We thank Dr. Michael Cameron (Scripps Research Institute, Jupiter, FL) for the mouse microsome stability study and Lili Zhou for assistance with the high throughput microscopy. We thank Dr. Richard Moriggl for providing us with the TEL fusion constructs used in the Ba/F3 cell experiments. We also thank SelectScreen® Kinase Profiling Service (Invitrogen) for performing enzymatic biochemical kinase profiling, DiscoveRx Bioscience for performing KinomeScanTM profiling, ActivX Biosciences for KiNatix® profiling, and The National Centre for Protein Kinase Profiling or Medical Research Council biochemical profiling. The TREEspot view image was generated using the web-based TREEspotTM software (DiscoveRx Biosciences). The kinase dendrogram was adapted and reproduced with permission from Cell Signaling Technology, Inc.
This work was supported, in whole or in part, by National Institutes of Health Grant HG006097 (to Q. L., M. N., and P. K. S.).

This article contains supplemental Tables 1–3 and Figs. 1 and 2.
N. S. Gray, unpublished results.
- mTOR
- mammalian target of rapamycin
- PIKK
- PI3K-like kinase
- ATM
- ataxia telangiectasia mutated
- ATR
- ATM and Rad3-related protein
- S6K
- S6 kinase.
REFERENCES
- 1. Guertin D. A., Sabatini D. M. (2007) Two adenovirus mRNAs have a common 5′-terminal leader sequence encoded at least 10 kb upstream from their main coding regions. Cancer Cell 12, 9–22 [DOI] [PubMed] [Google Scholar]
- 2. Hara K., Maruki Y., Long X., Yoshino K., Oshiro N., Hidayat S., Tokunaga C., Avruch J., Yonezawa K. (2002) Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 110, 177–189 [DOI] [PubMed] [Google Scholar]
- 3. Sarbassov D. D., Ali S. M., Sengupta S., Sheen J. H., Hsu P. P., Bagley A. F., Markhard A. L., Sabatini D. M. (2006) Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol. Cell 22, 159–168 [DOI] [PubMed] [Google Scholar]
- 4. Abraham R. T. (2004) PI 3-kinase related kinases. “Big” players in stress-induced signaling pathways. DNA Repair 3, 883–887 [DOI] [PubMed] [Google Scholar]
- 5. Liu P., Cheng H., Roberts T. M., Zhao J. J. (2009) Targeting the phosphoinositide 3-kinase pathway in cancer. Nat. Rev. Drug Discov. 8, 627–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sabers C. J., Martin M. M., Brunn G. J., Williams J. M., Dumont F. J., Wiederrecht G., Abraham R. T. (1995) Isolation of a protein target of the FKBP12-rapamycin complex in mammalian cells. J. Biol. Chem. 270, 815–822 [DOI] [PubMed] [Google Scholar]
- 7. Lane H. A., Breuleux M. (2009) Optimal targeting of the mTORC1 kinase in human cancer. Curr. Opin. Cell Biol. 21, 219–229 [DOI] [PubMed] [Google Scholar]
- 8. Thoreen C. C., Kang S. A., Chang J. W., Liu Q., Zhang J., Gao Y., Reichling L. J., Sim T., Sabatini D. M., Gray N. S. (2009) An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem. 284, 8023–8032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Feldman M. E., Apsel B., Uotila A., Loewith R., Knight Z. A., Ruggero D., Shokat K. M. (2009) Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 7, e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jacinto E., Loewith R., Schmidt A., Lin S., Rüegg M. A., Hall A., Hall M. N. (2004) Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat. Cell Biol. 6, 1122–1128 [DOI] [PubMed] [Google Scholar]
- 11. Wan X., Harkavy B., Shen N., Grohar P., Helman L. J. (2007) Rapamycin induces feedback activation of Akt signaling through an IGF-1R-dependent mechanism. Oncogene 26, 1932–1940 [DOI] [PubMed] [Google Scholar]
- 12. Liu Q., Chang J. W., Wang J., Kang S. A., Thoreen C. C., Markhard A., Hur W., Zhang J., Sim T., Sabatini D. M., Gray N. S. (2010) Discovery of 1-(4-(4-propionylpiperazin-1-yl)-3-(trifluoromethyl)phenyl)-9-(quinolin-3-yl)benzo[h][1,6]naphthyridin-2(1H)-one as a highly potent, selective mammalian target of rapamycin (mTOR) inhibitor for the treatment of cancer. J. Med. Chem. 53, 7146–7155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bhagwat V. S., Gokhale C. P., Crew P. A., Cooke A., Yao Y., Mantis C., Kahler J., Workman J., Bittner M., Dudkin L., Epstein M. D., Gibson W. N., Wild R., Arnold D. L., Houghton J. P., Pachter A. J. (2011) Preclinical characterization of OSI-027, a potent and selective inhibitor of mTORC1 and mTORC2: distinct from rapamycin. Mol. Cancer. Ther. 10, 1394–1406 [DOI] [PubMed] [Google Scholar]
- 14. García-Martínez J. M., Moran J., Clarke R. G., Gray A., Cosulich S. C., Chresta C. M., Alessi D. R. (2009) Ku-0063794 is a specific inhibitor of the mammalian target of rapamycin (mTOR). Biochem. J. 421, 29–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chresta C. M., Davies B. R., Hickson I., Harding T., Cosulich S., Critchlow S. E., Vincent J. P., Ellston R., Jones D., Sini P., James D., Howard Z., Dudley P., Hughes G., Smith L., Maguire S., Hummersone M., Malagu K., Menear K., Jenkins R., Jacobsen M., Smith G. C., Guichard S., Pass M. (2010) AZD8055 is a potent, selective, and orally bioavailable ATP-competitive mammalian target of rapamycin kinase inhibitor with in vitro and in vivo antitumor activity. Cancer Res. 70, 288–298 [DOI] [PubMed] [Google Scholar]
- 16. Yu K., Toral-Barza L., Shi C., Zhang W. G., Lucas J., Shor B., Kim J., Verheijen J., Curran K., Malwitz D. J., Cole D. C., Ellingboe J., Ayral-Kaloustian S., Mansour T. S., Gibbons J. J., Abraham R. T., Nowak P., Zask A. (2009) Biochemical, cellular, and in vivo activity of novel ATP-competitive and selective inhibitors of the mammalian target of rapamycin. Cancer Res. 69, 6232–6240 [DOI] [PubMed] [Google Scholar]
- 17. Yu K., Shi C., Toral-Barza L., Lucas J., Shor B., Kim J. E., Zhang W. G., Mahoney R., Gaydos C., Tardio L., Kim S. K., Conant R., Curran K., Kaplan J., Verheijen J., Ayral-Kaloustian S., Mansour T. S., Abraham R. T., Zask A., Gibbons J. J. (2010) Beyond rapalog therapy. Preclinical pharmacology and antitumor activity of WYE-125132, an ATP-competitive and -specific inhibitor of mTORC1 and mTORC2. Cancer Res. 70, 621–631 [DOI] [PubMed] [Google Scholar]
- 18. Liu Q., Thoreen C., Wang J., Sabatini D., Gray N. S. (2009) mTOR-mediated anti-cancer drug discovery. Drug Discov. Today Ther. Strateg. 6, 47–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yip C. K., Murata K., Walz T., Sabatini D. M., Kang S. A. (2010) Structure of the human mTOR complex I and its implications for rapamycin inhibition. Mol. Cell 38, 768–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schwede T., Kopp J., Guex N., Peitsch M. C. (2003) SWISS-MODEL. An automated protein homology-modeling server. Nucleic Acids Res. 31, 3381–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ren P., Ponder W. J. (2003) Polarizable atomic multipole water model for molecular mechanics simulation. J. Phys. Chem. B 107, 5933–5947 [Google Scholar]
- 22. Cornell W. D., Cieplak P., Bayly C. I., Gould I. R., Merz K. M., Ferguson D. M., Spellmeyer D. C., Fox T., Caldwell J. W., Kollman P. A. (1995) A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 117, 5179–5197 [Google Scholar]
- 23. Millard B. L., Niepel M., Menden M. P., Muhlich J. L., Sorger P. K. (2011) Adaptive informatics for multifactorial and high content biological data. Nat. Methods 8, 487–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hendriks B. S., Espelin C. W. (2010) DataPflex. A MATLAB-based tool for the manipulation and visualization of multidimensional datasets. Bioinformatics 26, 432–433 [DOI] [PubMed] [Google Scholar]
- 25. Lacronique V., Boureux A., Monni R., Dumon S., Mauchauffé M., Mayeux P., Gouilleux F., Berger R., Gisselbrecht S., Ghysdael J., Bernard O. A. (2000) Transforming properties of chimeric TEL-JAK proteins in Ba/F3 cells. Blood 95, 2076–2083 [PubMed] [Google Scholar]
- 26. Patricelli M. P., Szardenings A. K., Liyanage M., Nomanbhoy T. K., Wu M., Weissig H., Aban A., Chun D., Tanner S., Kozarich J. W. (2007) Functional interrogation of the kinome using nucleotide acyl phosphates. Biochemistry 46, 350–358 [DOI] [PubMed] [Google Scholar]
- 27. Liu Y., Gray N. S. (2006) Rational design of inhibitors that bind to inactive kinase conformations. Nat. Chem. Biol. 2, 358–364 [DOI] [PubMed] [Google Scholar]
- 28. Bellacosa A., Chan T. O., Ahmed N. N., Datta K., Malstrom S., Stokoe D., McCormick F., Feng J., Tsichlis P. (1998) Akt activation by growth factors is a multiple-step process. The role of the PH domain. Oncogene 17, 313–325 [DOI] [PubMed] [Google Scholar]
- 29. Copeland R. A., Pompliano D. L., Meek T. D. (2006) Drug-target residence time and its implications for lead optimization. Nat. Rev. Drug Discov. 5, 730–739 [DOI] [PubMed] [Google Scholar]
- 30. Toledo L. I., Murga M., Zur R., Soria R., Rodriguez A., Martinez S., Oyarzabal J., Pastor J., Bischoff J. R., Fernandez-Capetillo O. (2011) A cell-based screen identifies ATR inhibitors with synthetic lethal properties for cancer-associated mutations. Nat. Struct. Mol. Biol. 18, 721–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Apsel B., Blair J. A., Gonzalez B., Nazif T. M., Feldman M. E., Aizenstein B., Hoffman R., Williams R. L., Shokat K. M., Knight Z. A. (2008) Targeted polypharmacology. Discovery of dual inhibitors of tyrosine and phosphoinositide kinases. Nat. Chem. Biol. 4, 691–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zunder E. R., Knight Z. A., Houseman B. T., Apsel B., Shokat K. M. (2008) Discovery of drug-resistant and drug-sensitizing mutations in the oncogenic PI3K isoform p110α. Cancer Cell 14, 180–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.