Background: A plant photoreceptor “phototropin” is a light-dependent kinase containing the LOV photosensory domains.
Results: Mutations in the N-terminal flanking region of LOV2 elevate kinase activity in darkness.
Conclusion: The N-terminal flanking region is involved in intramolecular signaling from LOV2 to the kinase domain.
Significance: This work provides insights into how the LOV domain can activate the kinase domain intramolecularly.
Keywords: Photobiology, Photoreceptors, Protein Structure, Serine/Threonine Protein Kinase, Yeast, LOV, Phototropin
Abstract
Phototropin is a light-regulated kinase that mediates a variety of photoresponses such as phototropism, chloroplast positioning, and stomata opening in plants to increase the photosynthetic efficiency. Blue light stimulus first induces local conformational changes in the chromophore-bearing light-oxygen and voltage 2 (LOV2) domain of phototropin, which in turn activates the serine/threonine (Ser/Thr) kinase domain in the C terminus. To examine the kinase activity of full-length phototropin conventionally, we employed the budding yeast Saccharomyces cerevisiae. In this organism, Ser/Thr kinases (Fpk1p and Fpk2p) that show high sequence similarity to the kinase domain of phototropins exist. First, we demonstrated that the phototropin from Chlamydomonas reinhardtii (CrPHOT) could complement loss of Fpk1p and Fpk2p to allow cell growth in yeast. Furthermore, this reaction was blue light-dependent, indicating that CrPHOT was indeed light-activated in yeast cells. We applied this system to a large scale screening for amino acid substitutions in CrPHOT that elevated the kinase activity in darkness. Consequently, we identified a cluster of mutations located in the N-terminal flanking region of LOV2 (R199C, L202L, D203N/G/V, L204P, T207I, and R210H). An in vitro phosphorylation assay confirmed that these mutations substantially reduced the repressive activity of LOV2 on the kinase domain in darkness. Furthermore, biochemical analyses of the representative T207I mutant demonstrated that the mutation affected neither spectral nor multimerization properties of CrPHOT. Hence, the N-terminal flanking region of LOV2, as is the case with the C-terminal flanking Jα region, appears to play a crucial role in the regulation of kinase activity in phototropin.
Introduction
To sense and respond to a fluctuating light environment, plants have evolved several classes of photoreceptor molecules that convert visible light stimuli into biological signals. Phototropin (PHOT)2 is a blue light (BL) receptor, which is widely conserved in the plant kingdom (1). Most terrestrial plants possess two isoforms of phototropin designated PHOT1 and PHOT2. In Arabidopsis thaliana (At), PHOT1 and PHOT2 have redundant and distinct functions to mediate responses, such as phototropism (2, 3), chloroplast movement (4, 5), stomatal opening (6), and leaf photomorphogenesis (7, 8) to optimize photosynthesis. On the other hand, the unicellular green alga Chlamydomonas reinhardtii (Cr) possesses a single phototropin homologue (CrPHOT), which has been proposed to regulate sexual differentiation (9, 10) and the expression several photosynthetic genes. Interestingly, CrPHOT is fully functional in Arabidopsis, suggesting that the basic mechanism of phototropin signal transduction is highly conserved (11). Phototropins are BL-dependent Ser/Thr protein kinases that share well conserved structural properties. The kinase domain belongs to the AGC VIII subfamily (protein kinases A, G, and C) (12). Autophosphorylation of a conserved serine residue at the activation loop of the kinase domain is prerequisite for its physiological activity (13, 14). To date, little is known about authentic phosphorylation substrates for phototropins. Recently, however, auxin efflux transporter ATP-binding cassette B19 (ABCB19) has been reported as a putative endogenous substrate in Arabidopsis (15).
In addition to the kinase domain, phototropin contains two N-terminal light-oxygen and voltage (LOV) domains, designated LOV1 and LOV2. The LOV domains, which are members of the Per-ARNT-Sim (PAS) domain superfamily, noncovalently bind flavin mononucleotide (FMN) as chromophores (16). Because the kinase fragment lacking LOV domains displays constitutive activity both in planta (17) and in vitro (18), LOV domains appear to suppress kinase activity in darkness intrinsically.
Upon BL absorption, a covalent adduct is formed between the FMN and a conserved cysteine residue within each LOV domain (19, 20). Consequently, reorganization of the hydrogen bond network takes place in the FMN binding pocket (21, 22). Inactivation of LOV domains by site-directed mutagenesis indicates that LOV2 plays a predominant role in regulating the kinase activity (18, 23, 24), whereas the role of LOV1 is limited to modulate photosensitivity (18). An NMR analysis of Avena PHOT1 has revealed an amphipathic α helix, Jα, which is attached to the C terminus of the LOV2 core domain (25). The Jα helix changes its structure in response to a light stimulus. Namely, it detaches from the LOV2 core (25) and unfolds (26, 27). The prominent role of the Jα helix in the regulation of kinase activity has been evidenced by site-directed mutagenesis in Arabidopsis PHOT1. An amino acid substitution that disrupts the LOV2-Jα interaction leads to constitutive activation of the kinase in vitro (28). More recently, the crystallographic analysis of Avena PHOT1 LOV2 has suggested that not only the C-terminal Jα but also the N-terminal flanking region display a light-induced structural change (22). These structural changes are presumed to disrupt the inhibitory interaction of LOV2 with the kinase domains. Accordingly, photoreversible contact/separation between the LOV2 and the kinase domains has been suggested by small angle x-ray scattering in a LOV2 linker kinase fragment of Arabidopsis PHOT2 (29).
Besides the site-directed mutagenesis approach based on structural information, a random-mutagenesis approach utilizing Escherichia coli as a host has been applied successfully to uncover amino acid residues involved in the photochemistry of LOV2 in Avena PHOT1 (30). Although this method is quite effective for identifying spectral mutants in the LOV domains, it is not applicable for screening mutants that are altered in the photoregulation of the kinase activity. This is because no convenient way to assess the kinase activity of phototropin exists in E. coli.
In this study, we developed a new yeast experimental system in which CrPHOT triggered cell growth depending on its kinase activity. We applied this method to screen for mutations that disrupt repression of the kinase domain. Consequently, we revealed that the N-terminal flanking region of LOV2 is important for the suppression of kinase activity in darkness.
EXPERIMENTAL PROCEDURES
Media and Growth Conditions for Yeast
Yeast strains were cultured in YPDA-rich medium (1% yeast extract, 2% bacto-peptone, 2% glucose, and 0.01% adenine). Strains carrying plasmids were selected in synthetic medium (SD) containing the required nutritional supplements (31). When appropriate, 0.5% casamino acids were added to SD medium without uracil (SDA-Ura). For induction of the GAL1 promoter, 3% galactose and 0.2% sucrose were added as carbon sources instead of glucose (YPGA and SGA-Ura). Growth sensitivity to duramycin was examined on YPDA plates containing 10 μm duramycin (Sigma). Blue (peak at 470 nm) and red (peak at 660 nm) light-emitting diode panels (ISL-150X150 series; CCS, Tokyo, Japan) were used as light sources.
Strains and Plasmids
Saccharomyces cerevisiae strains KKT330 (MATa ura3Δ his3Δ1 leu2Δ0 HIS3MX6::PGAL1-CDC50 fpk1Δ::HphMX4 fpk2Δ::KanMX6, designated hereafter as PGAL1-CDC50 fpk1Δ fpk2Δ) and YKT1638 (MATa ura3Δ his3Δ1 leu2Δ0 fpk1Δ::HphMX4 fpk2Δ::KanMX6, designated hereafter as fpk1Δ fpk2Δ) were used for growth assays (32). The E. coli strains DH5α and BL21 were used for plasmid construction and expression of recombinant proteins, respectively.
For yeast assays, pRS416-AtPHOT1, pRS416-AtPHOT2, and pRS416-CrPHOT were constructed as follows. The coding regions of PHOT1 and PHOT2 from Arabidopsis and PHOT from Chlamydomonas were amplified by PCR and cloned into the BamI/SalI site of pRS416 (33) to allow constitutive expression of phototropins tagged with N-terminal two tandem repeats of the influenza virus hemagglutinin epitope (2HA) under the control of the TPI1 promoter. Site-directed mutations were introduced by using the QuikChange site-directed mutagenesis kit (Stratagene) in accordance with the instructions of the supplier. All amino acid changes were verified by DNA sequencing. For overexpression of CrPHOT protein in E. coli, pET28a-CrPHOT was constructed as follows. Wild-type (WT) and mutant PHOT were amplified from pRS416-CrPHOT and cloned into NdeI/SalI sites of pET28a plasmid vector (Novagen) to allow expression of CrPHOT with an N-terminal histidine tag, which was composed of 20 residues including six histidine residues.
Immunoblot Analysis
Protein extraction from yeast was performed as described (34). Immunoblot analysis was performed essentially as described previously (11). The same amounts of protein extracts were run on 10.0% SDS-PAGE and blotted onto nitrocellulose membrane. Rat anti-HA monoclonal antibody (Roche Applied Science) and alkaline phosphatase-conjugated anti-rat IgG antibody (Promega) were used.
Yeast Growth Assay
The cells were grown in liquid SGA-Ura medium to A600 of 0.6–0.8, harvested by centrifugation, and resuspended into sterile water, resulting in an A600 of 0.1. Serial 5-time dilutions were then prepared for loading 10 μl each of the diluted cell suspension onto a plate with SGA-Ura or SDA-Ura medium. The plates were then placed under different light conditions.
PCR-based Random Mutagenesis and Mutant Screening for Pseudolit Phenotype
Error-prone PCR libraries of the N-terminal (1–1399) and C-terminal (1179–2136) moieties of CrPHOT flanked by sequences from the pRS416 vector (∼150 bp) were generated from the pRS416-CrPHOT using the Diversity PCR random mutagenesis kit (Clontech). The primer sequences are as follows: pRS416 forward for the N-terminal library, 5′-GTGAACTTGCAACATTTACTATTTTCCC; CrPHOT-1399 reverse for the N-terminal library, 5′-CGGTCTGGATGGTGCAGTACAG; CrPHOT-1179 forward for the C-terminal library, 5′-CTACCAGGCGCTGCTGCAGCTG; and pRS416 reverse for the C-terminal library, 5′-AAAGCGGGCAGTGAGCGCAACGC. Final concentrations of 320 μm MnSO4 and 16 μm dGTP were chosen for the PCR to adjust the mutagenesis frequency to 1.5 mutations/kb.
The pRS416-CrPHOT plasmid was digested with BamHI/NruI and SalI/NruI to remove the sequence encoding the N-terminal and C-terminal moieties, respectively. The N- or C-terminal PCR fragments and a pRS416-CrPHOT-derived vector lacking the corresponding part of CrPHOT were mixed and transformed into the PGAL1-CDC50 fpk1Δ fpk2Δ yeast strain, so that homologous recombination generated a vector containing mutagenized PHOT. The yeast strain was then plated on SDA-Ura agar and incubated in the dark at 28 °C for 60 h. The growing colonies were isolated. Among isolated colonies, candidates were randomly selected, and plasmids were rescued using Wizard prep (Promega) according to the instructions in the DUALhybrid kit (Dualsystems Biotech). The candidate plasmids were sequenced in the mutagenized moieties. The second growth assay was performed by repeating the assays using the purified plasmids.
Expression and Purification of Recombinant Proteins
His6-tagged full-length CrPHOT and N-terminal fragment of Arabidopsis PHOT1 (AtP1-Nt) were prepared from E. coli according to the protocols used for LOV2 linker kinase fragment preparation (35) with minor modifications. The E. coli cells were lysed with a French pressure cell (Thermo Fisher Scientific) in a buffer containing 20 mm HEPES-NaOH (pH 7.8), 500 mm NaCl, and 1 mm PMSF. The purification was performed at 0–4 °C under a dim red light. The centrifugation supernatant of the lysate was subjected to Ni2+-ion affinity chromatography (Qiagen). The AtP1-Nt were then desalted by using HiTrap Desalting (GE Healthcare) and stored at −80 °C.
The CrPHOT protein was further purified via size-exclusion column chromatography (Sephacryl HR 200; GE Healthcare) in a buffer containing 100 mm NaCl, 1 mm EGTA, and 20 mm Tris-HCl (pH 7.8). Glycerol was added to the eluted CrPHOT to a final concentration of 10% (v/v). The phosphorylation levels of the products after purification were examined by Pro-Q Diamond phosphoprotein gel stain (Invitrogen) according to the manufacturer's instructions if necessary.
For the spectral analysis, the above sample was further purified using an anion-exchange Resource Q column (GE Healthcare) equilibrated with a buffer containing 10% (v/v) glycerol, 1 mm EGTA, and 20 mm Tris-HCl (pH 7.8). The CrPHOT fraction that did not bind to the Resource Q column was collected and concentrated by ultrafiltration using the Amicon Ultra K15 (Millipore). The purity of CrPHOT in each step was examined by SDS-PAGE.
In Vitro Phosphorylation Assays
The phosphorylation assay was performed under dim green light (18, 35). CrPHOT and AtP1-Nt polypeptides were incubated at 30 °C in a kinase reaction mixture containing 20 mm Tris-HCl (pH 7.8), 100 mm NaCl, 10% glycerol, 10 mm MgCl, 500 μm ATP, and 60 kBq of [γ-32P]ATP. The reaction was terminated by the addition of concentrated SDS-PAGE sample buffer followed by boiling for 3 min. Then, the samples were run on SDS-PAGE to visualize phosphorylation bands using an imaging plate and a bioimaging analyzer (FLA2000; Fuji, Tokyo, Japan). The molecular masses of phosphorylated proteins were estimated by Coomassie Brilliant Blue (CBB) staining with reference to molecular mass standard samples (Sigma).
Spectral Analyses and Size-exclusion Chromatography
The absorption spectra of CrPHOT under BL illumination and the time course of absorption changes during thermal decay from the cysteinyl adduct state to the dark-adapted state were measured essentially as described previously (35) at 20 °C and pH 7.8. The molecular mass of CrPHOT was estimated by size-exclusion chromatography using a Superdex 200 pg Hi-Load column (GE Healthcare) and the kinase reaction buffer as previously described (35).
RESULTS
Complementation of fpk1Δ fpk2Δ and cdc50Δ Synthetic Lethality in S. cerevisiae by Chlamydomonas PHOT in a BL-dependent Manner
To establish an experimental system that can easily and quickly assess the light-dependent kinase activity of phototropin, we employed S. cerevisiae, in which the protein kinases named flippase-kinase 1 (Fpk1p) and Fpk2p were recently identified (32). Among the yeast kinases, Fpk1p and Fpk2p exhibit the highest sequence homology to the AGCVIII kinase domain of phototropins (32) (Fig. 1A and supplemental Fig. S1). Hence, we reasoned that phototropins might complement the loss of FPKs in yeast in a light-dependent manner.
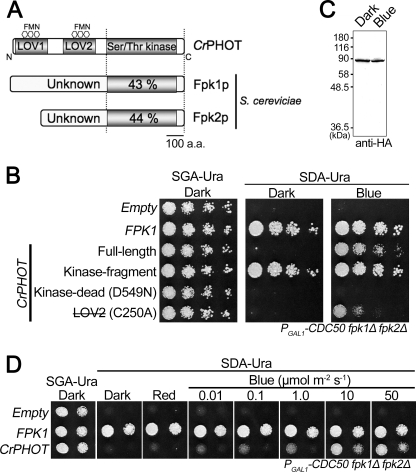
FIGURE 1.
Photoregulation of yeast cell growth by CrPHOT. The yeast conditional mutant PGAL1-CDC50 fpk1Δ fpk2Δ was transformed with a vector containing the FPK1 gene or CrPHOT derivatives fused to the constitutive TPI1 promoter. A, schematic illustration of the phototropin from CrPHOT and Fpk1p and Fpk2p from S. cerevisiae. The numbers within the Ser/Thr kinase domain show the percentage sequence identity with the CrPHOT kinase domain. B, growth of the yeast conditional mutant PGAL1-CDC50 fpk1Δ fpk2Δ expressing the full-length CrPHOT and its derivatives. LOV2 (with a line through it) CrPHOT with a mutation that decreases the extent of light activation. Yeast cells were serially diluted and spotted onto plates containing galactose (SGA-Ura) or glucose (SDA-Ura), followed by incubation in darkness (Dark) or under BL irradiation (50 μmol m−2 s−1; Blue) at 28 °C for 2–3 days. Empty, yeast cells transformed with a control plasmid. C, immunoblot detection of the full-length CrPHOT with an anti-HA monoclonal antibody in yeast grown as for B. Protein extracts from 1 mg of wet yeast cells were separated by 10.0% SDS-PAGE. D, effects of monochromatic light irradiation on growth of yeast expressing CrPHOT. Red light at 50 μmol m−2 s−1 or blue light at different intensities was applied to yeast cells as for B.
FPK1 and FPK2 exhibit synthetic growth defects with the null mutation of CDC50 (36, 37). Their products, Fpk1p and Fpk2p, regulate the Lem3p-Dnf1p and Lem3p-Dnf2p flippases, which act as inward-directed phospholipid translocases (38). Nakano et al. have established a conditional mutant in which Cdc50p is expressed under the control of the glucose-repressible GAL1 promoter in the fpk1Δ fpk2Δ background (PGAL1-CDC50 fpk1Δ fpk2Δ) (32). The resultant PGAL1-CDC50 fpk1Δ fpk2Δ mutant grows normally in galactose-containing medium (SGA-Ura) but exhibits severe growth defect in glucose-containing medium (SDA-Ura) (Fig. 1B) (32). To examine whether phototropins could rescue the loss of FPKs in yeast, we introduced the Arabidopsis PHOT1 (AtPHOT1) and PHOT2 (AtPHOT2), and Chlamydomonas PHOT (CrPHOT) genes into the PGAL1-CDC50 fpk1Δ fpk2Δ strain. Consequently, AtPHOT2 partly suppressed the growth defect in glucose-containing medium regardless of the light condition, whereas AtPHOT1 completely failed to allow growth (supplemental Fig. S2). Remarkably, CrPHOT, albeit partially, suppressed the growth defect under BL but not in darkness (Fig. 1B) or under red light (Fig. 1D). Thus, CrPHOT complemented the loss of Fpk activity in a BL-dependent manner in yeast.
CrPHOT Acts as a Light-dependent Flippase-Kinase in Yeast
We genetically examined whether the kinase activity of CrPHOT was indeed required for the complementation. The kinase domain fragments of phototropins exhibit constitutive kinase activity without light stimulus (17, 18). As expected, the kinase domain fragment of CrPHOT restored yeast growth regardless of the light condition (Fig. 1B). Conversely, the D549N mutant of CrPHOT, which corresponded to the kinase-dead mutation in AtPHOT1 and AtPHOT2 (17, 23), failed to restore growth. The substitution of conserved Cys39 to alanine in the LOV2 domain is known to decrease the extent of light activation in phototropins (18, 23). The corresponding C250A mutant of CrPHOT restored growth but very weakly (Fig. 1B). Taken together, CrPHOT complemented the loss of Fpks in a kinase-like manner.
We then examined whether the complementation by CrPHOT indeed depended on its light activation. First, we confirmed that the level of CrPHOT in yeast was not altered under BL with an anti-HA tag antibody (Fig. 1C). This was because phototropin is known to be degraded more rapidly under BL in some cases (7, 39). It should be noted that electric mobility shift due to BL-induced autophosphorylation was not observed. This was probably because such activity is low in CrPHOT (11).
We determined the dependence of complementation by CrPHOT on BL intensity (Fig. 1D). The degree of complementation increased as BL was increased from 0.01 to 10 μmol m−2 s−1. This result matched well with those from a previous analysis on the biological activity of CrPHOT expressed in Arabidopsis (11).
BL-dependent Regulation of Phospholipid Uptake by CrPHOT
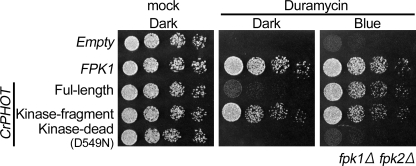
We further examined whether CrPHOT activated flippases in yeast as Fpks do. Phosphatidylethanolamine is enriched in the outer leaflet of the membrane in fpk1Δ fpk2Δ mutant due to reduced flippase activity. This leads to growth defect in the presence of duramycin, a cytotoxic small tetracyclic peptide that binds phosphatidylethanolamine on the outer leaflet. As expected, CrPHOT rescued the duramycin-sensitive growth defect in a BL-dependent manner (Fig. 2). In addition, the kinase fragment rescued the phenotype regardless of the light condition, whereas the putative kinase-dead mutant (D549N) of CrPHOT failed to restore the growth. Hence, CrPHOT was suggested to regulate the flippase activity in yeast.
FIGURE 2.
Growth sensitivity to duramycin of the fpk1Δ fpk2Δ mutant expressing CrPHOT and its derivatives. The yeast cells were serially diluted and spotted onto YPDA plates with or without 10 μm duramycin. The plates were then incubated in darkness (Dark) or under BL irradiation (50 μmol m−2 s−1; Blue) at 28 °C for 1.5 days.
Search for Novel CrPHOT Variants Showing Pseudolit Activity
The above experimental system should be potentially useful in identifying amino acid residues important for phototropin functions. To explore this possibility, mutations were introduced separately into the N- and C-terminal moieties of CrPHOT by a PCR-based method at the rate of 1.5 mutations/kb (Fig. 3A). Pools of transformed cells corresponding to 5.0 × 105 each of successful recombination events were screened for mutations that allowed growth in darkness on SDA-Ura plates. Consequently, 1504 and 107 growth-positive colonies were obtained for the N- and C-terminal mutant pools, respectively.
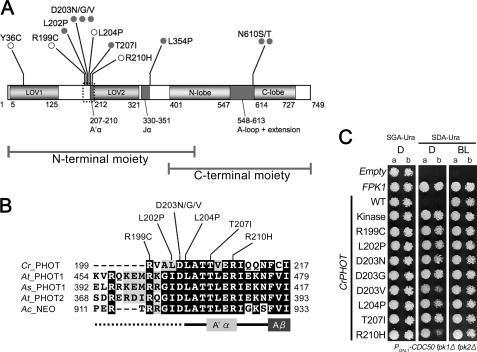
FIGURE 3.
CrPHOT mutants that restored yeast growth in darkness. A, schematic drawing of the domain structure of CrPHOT and locations of amino acid substitutions found in the present studies. Gray filled circles indicate mutations identified in clones with a single mutation. Open circles indicate mutations found in more than three independent clones with multiple mutations. B, sequence alignment of N-terminal flanking regions of LOV2 among different phototropin species. Cr_PHOT, C. reinhardtii PHOT; At_PHOT1, A. thaliana PHOT1; As_PHOT1, Avena sativa PHOT1; and At_PHOT2, A. thaliana PHOT2; Ac_NEO, Adiantum capillus-veneris NEOCHROME1. Sequences were aligned using the ClustalW and BOXSHADE program. C, growth of the PGAL1-CDC50 fpk1Δ fpk2Δ mutant expressing CrPHOT with single amino acid substitutions. Cells were treated as for Fig. 1B. Two concentrations of yeast cells (a, A600 = 0.004; b, A600 = 0.0008) were spotted.
We then sequenced 96 randomly selected clones for the N-terminal mutations and all 107 clones for the C-terminal mutations. Consequently, we found that many of them carried multiple amino acid substitutions. In addition, a large deletion, probably due to irregular homologous recombination, was found in some clones. Nonetheless, eight clones with single amino acid substitutions were identified (Fig. 3A). Additionally, four substitutions were shared in more than three independent clones with multiple mutations. Reintroduction of the latter mutations into the wild-type CrPHOT confirmed that they indeed conferred the pseudolit phenotype (Fig. 3C and supplemental Fig. S3). We also confirmed that the expression levels of these mutant proteins were not elevated in yeast cells (supplemental Fig. S4).
Kinase Activities of Full-length CrPHOT Carrying Mutations in the N-terminal Flanking Region of LOV2
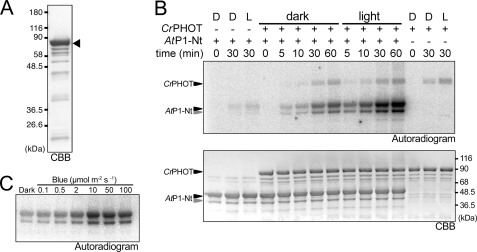
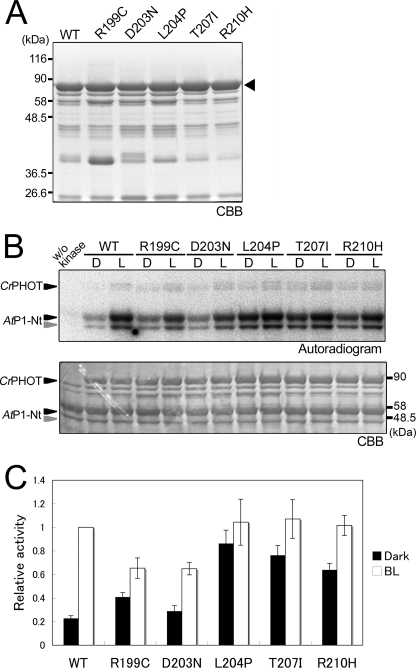
Interestingly, 8 of the above 12 mutations were clustered in a well conserved N-terminal LOV2-flanking region (residues Arg199-Arg210) (Fig. 3, A and B). Hence, their kinase activity was examined in vitro. For this purpose, we attempted to express the recombinant full-length CrPHOT protein tagged with an N-terminal His6 tag in E. coli. Full-length CrPHOT protein was recovered as a yellow-colored, soluble protein from the E. coli extract. For the kinase assay, the CrPHOT protein was purified from the extract by Ni2+-affinity chromatography and size-exclusion chromatography. SDS-PAGE revealed that the resulting sample consisted of a main band of 90 kDa, which corresponded to the theoretical mass of the protein, 85.7 kDa, with several minor bands of lower molecular masses. The purity was then estimated to be ∼75% (Fig. 4A), which was comparable with those of samples used for the previous kinase assays of LOV kinase fragments (18, 35).
FIGURE 4.
In vitro phosphorylation assay of the full-length CrPHOT. A, 10.0% SDS-polyacrylamide gel pattern of full-length CrPHOT stained with CBB. The sample with an N-terminal His6 tag was successively purified by Ni2+-ion affinity chromatography and size-exclusion chromatography. The triangle indicates the position of the intact CrPHOT. B, in vitro phosphorylation assay for CrPHOT under BL (100 μmol m−2 s−1) or in the dark. CrPHOT and/or AtP1-Nt was incubated with radioactive ATP for the indicated durations. The upper and lower panels show the 10.0% SDS-PAGE gel patterns visualized by autoradiography and CBB staining, respectively. + or −, the presence or absence of the kinase and/or the substrate, respectively; D, dark condition; L, BL irradiation. C, dependence of the kinase activation on the light intensity. In vitro phosphorylation assay of CrPHOT was performed as described for B. The samples were incubated for 10 min under BL at the indicated light intensities. Phosphorylated AtP1-Nt was visualized by autoradiography.
In vitro phosphorylation assay for the purified CrPHOT was performed using AtP1-Nt as a substrate (35). As expected, CrPHOT phosphorylated AtP1-Nt under BL, whereas phosphorylation was substantially reduced in darkness (Fig. 4B). The phosphorylation level increased during the incubation period of 60 min. The effects of BL were saturated at ∼10 μmol m−2 s−1 (Fig. 4C). It should be noted that the background activity in darkness was relatively high, which has been reported for other phototropin samples as well (40, 41).
We then examined the kinase activities in 6 of 8 mutants carrying the mutation in the N-terminal flanking region of LOV2 (R199C, D203N, L204P, T207I, and R210H). The full-length proteins were expressed in E. coli and purified as described above. Consequently, protein samples in similar qualities were obtained with the exception of R199C in which a larger amount of impurity at around 36.5 kDa was detected (Fig. 5A).
FIGURE 5.
Effects of CrPHOT mutations on in vitro phosphorylation activity. A, 10.0% SDS-polyacrylamide gel patterns of the mutant CrPHOT proteins stained with CBB. The wild-type (WT) and the mutant CrPHOT proteins were prepared as for Fig. 4A. The triangle indicates the position of the intact CrPHOT. B, in vitro phosphorylation assay for the WT and mutant CrPHOT samples. The reaction proceeded for 30 min under BL (100 μmol m−2 s−1) or in the dark condition. Sample preparation and experimental procedures were the same as those for Fig. 4B. The upper and lower panels show the 10.0% SDS-polyacrylamide gel patterns visualized by autoradiography and CBB staining, respectively. C, quantification of the phosphorylation activity in the WT and the mutant CrPHOT. An in vitro phosphorylation assay was performed as for B. Band intensities were quantified by a bioimaging analyzer and expressed relative to the maximal phosphorylation level in the WT under BL. The data represent the means of three independent experiments. Error bars, S.D.
We examined the kinase activity using AtP1-Nt as a substrate. Consistent with the yeast phenotype (Fig. 3), the above mutants exhibited increased kinase activities in darkness compared with the WT with the exception of D203N (Fig. 5, B and C). Hence, most of the CrPHOT mutants disrupted repression of the kinase activity in vitro.
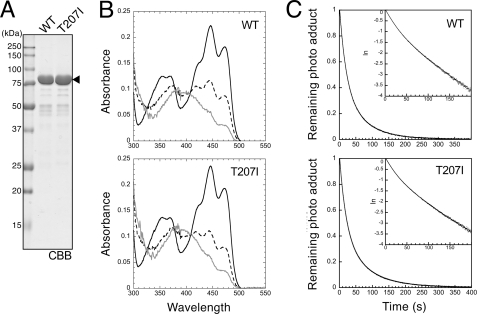
Spectroscopic Analysis of Mutation in N-terminal Flanking Region of LOV2
We examined whether the present mutations affected the spectral nature of CrPHOT. For this purpose, the T207I mutant was selected as a representative mutant and expressed in E. coli. The protein sample conventionally prepared according to the procedure listed for the kinase assay was further purified by negative ion exchange chromatography. Consequently, a sample with >95% purity was obtained (Fig. 6A). The absorption spectroscopy revealed that both WT and T207I mutant exhibited the typical fine structured absorption spectra of protein-bound FMN with absorption maxima at 425, 447, and 472 nm (Fig. 6B). Under continuous BL irradiation, both WT and T207I CrPHOT proteins were converted to cysteinyl adduct state with absorption maxima at 390 nm (Fig. 6B). Those absorption spectra were nearly identical to those of the LOV1-hinge-LOV2 fragment of CrPHOT, which lacked the kinase domain (42).
FIGURE 6.
Effects of the Thr207 mutation on photochemical properties. A, 10.0% SDS-polyacrylamide gel pattern of full-length CrPHOT samples stained with CBB. The sample was prepared using the same procedures as those for Fig. 5 and was further purified by anion-exchange chromatography. The triangle indicates the position of the intact CrPHOT. B, absorption spectra of the WT and the T207I mutant CrPHOT in darkness (solid black lines) and under BL irradiation at 50 μmol m−2 s−1 (dashed black lines) and 270 μmol m−2 s−1 (gray lines). The data represent the means of three measurements. C, kinetics of the reversion of the cysteinyl adduct state to the dark state monitored by the absorption at 450 nm. The samples were equilibrated under BL at 270 μmol m−2 s−1, and the light was then tuned off at time 0. The remaining photoadduct level was calculated as 1 − (At − A0)/(A∞ − A0), where At, A∞, and A0 are absorptions at 450 nm at time t in seconds, in darkness, and under BL, respectively. The logarithm of the remaining photoadduct level was plotted against t in the inset, indicating the fast and slow components of the reversion kinetics.
At a fluence rate of 270 μmol m−2 s−1, adduct formation was saturated within 24 s, whereas the dark reversion was completed within 400 s after the illumination was turned off (data not shown). By exponential fitting analysis, the half-lives for dark decay of two photoproduct components in WT CrPHOT were calculated to be 15 and 50 s, whereas those for the T207I mutant were 15 and 56 s. Hence, the T207I mutant was almost indistinguishable from the WT. Taken together, the T207I mutation in the N-terminal flanking region of LOV2 activated the kinase domain without affecting the spectral nature of CrPHOT.
Estimation of Molecular Mass by Size-exclusion Chromatography
LOV1 domains in phototropins have been suggested to form a dimer (43–45). Hence, we examined the possibility that the T207I mutation affected kinase activity by altering the multimerization state of full-length CrPHOT. To test this possibility, we determined the molecular masses of full-length WT and T207I CrPHOT by size-exclusion chromatography. For this purpose, the samples were purified as for the spectral analysis.
The size-exclusion elution profile demonstrated that both WT and T207I CrPHOT proteins exhibited a major distinct peak at 71.14 ml, corresponding to a molecular mass of 123.7 kDa (supplemental Fig. S5). Hence, the T207I mutation did not alter the multimerization state of full-length CrPHOT. Another minor peak at 78 ml (corresponding to 68 kDa) probably represented a degradation product.
DISCUSSION
CrPHOT Complements Loss of Fpk1p and Fpk2p Kinases in Yeast
A bacterial system has been employed successfully to identify amino acid residues important for the spectral integrity of the LOV domains of phototropins (30). The present study further extended this strategy to assess the signaling activity of phototropin derivatives in a microorganism.
Indeed, CrPHOT successfully rescued the fpk1Δ fpk2Δ phenotype to allow cell growth in a BL-dependent manner in yeast (Fig. 1). We further demonstrated that both photoexcitation of the LOV domain and the kinase activity of CrPHOT are necessary to promote yeast cell growth (Fig. 1B). Hence, we have developed an experimental system suitable for a large scale mutant screening for CrPHOT.
It remains unclear why the Arabiodopsis PHOT1 and PHOT2 did not function properly in yeast. To gain further insights, the complementation activities of respective kinase fragments were examined. Consequently, the PHOT1 kinase fragment failed to promote the cell growth, whereas the PHOT2 fragment did (supplemental Fig. S2). Hence, intrinsic kinase activities of Arabiodopsis PHOT1 and PHOT2 appeared to be quite different in yeast for an unknown reason.
Kinase Activity of Full-length CrPHOT
It had long been believed that expression of recombinant full-length phototropins in E. coli for biochemical analyses was very difficult. Recently, however, Pfeifer et al. successfully purified full-length CrPHOT in E. coli to conduct FTIR spectroscopy (46). Similarly, we were able to prepare full-length WT and mutated CrPHOT proteins pure enough for the in vitro kinase assay, photochemical kinetic analysis, and multimerization analysis.
The BL-dependent kinase activity of the purified CrPHOT was successfully shown in vitro using AtP1-Nt as a substrate (Fig. 4B). As has been shown already for CrPHOT expressed in insect cells (11), autophosphorylation activity was relatively low. This was not because the purified CrPHOT was already phosphorylated in E. coli (supplemental Fig. S6). We suppose that fewer phosphorylation sites exist in CrPHOT (11).
Spectral and Multimerization Properties of Full-length CrPHOT
Fast and slow components observed in the dark decay kinetics (Fig. 6) presumably represented LOV2 and LOV1 (t½ = 15 and 50 s, respectively). Compared with the CrPHOT LOV1+LOV2 fragment (t½ = 17 and 200 s) (47), the slower component was four times faster in the full-length CrPHOT. Hence, the presence of the C-terminal kinase domain might alter the photochemical kinetics of LOV1 as previously reported for Arabidopsis PHOT1 and PHOT2 (47).
The LOV1 domains of Arabidopsis PHOT1 and PHOT2 (43–45) and CrPHOT (48) have been suggested to form a dimer and higher oligomers. However, size-exclusion chromatography in this study hinted that full-length CrPHOT may exist as a monomer (apparent molecular mass of 123.7 kDa versus theoretical mass of 85.7 kDa) (supplemental Fig. S5). This possibility should be investigated further by other methods such as small angle x-ray scattering or crystallization analysis.
Mutations Outside LOV2 N-terminal Flanking Region
Prior to the present work, several amino acid substitutions in the N-terminal region of phototropins had been known to cause the pseudolit activation. Those include the ones in the Jα helix (V601E, A605E, and I608E in Arabidopsis PHOT1) (28) and the LOV2 core region (G513N in Avena PHOT1) (49). By contrast, such a mutation had not been known within the C-terminal region. In the present study, four novel mutations were found in addition to those clustered in the N-terminal LOV2 flanking region (see below).
Among them, L354P resides in the vicinity of Jα (Fig. 3A and supplemental Fig. S3). This is not surprising because several mutations are already known in this region to cause the pseudolit phenotype (see above). By contrast, Y39C in the LOV1 core region (Fig. 3A and supplemental Fig. S3) was somewhat unexpected because LOV1 is less important than LOV2 to regulate the kinase activity (18, 23, 24). However, LOV1 modifies the light sensitivity of PHOT2 (18). Hence, this mutation may affect the presumed LOV1/LOV2 or LOV1/kinase interaction to substantially increase the light sensitivity.
The N610S/T mutations in the activation loop of the kinase domain conferred the pseudolit phenotype (Fig. 3A and supplemental Fig. S3). Interestingly, the Asn610 residue is adjacent to Ser611, which corresponds to the autophosphorylation site required for the activation of both PHOT1 and PHOT2 in Arabidopsis (13, 14) (supplemental Fig. S1). We constructed the N610A CrPHOT to show that Ser610 and Thr610 but not Ala610 caused the pseudolit activation (supplemental Fig. S3). Hence, the newly introduced Ser/Thr610 might have acted as a secondary phosphorylation site to increase the kinase activity.
Involvement of N-terminal Flanking Region of LOV2 in Light-induced Signal Transduction
In our screening, we identified a novel mutational hot spot in the N-terminal flanking region of LOV2 (Arg199-Arg210 in Fig. 3A). These mutations substantially reduced repression of the kinase activity by the N-terminal moiety in darkness (Figs. 3 and 5) without affecting the photochemical (Fig. 6) or multimerization properties of CrPHOT (supplemental Fig. S5).
The crystal structure of LOV2 with N- and C-terminal flanking regions from Avena PHOT1 (residues Leu404-Ala559 in Avena PHOT1) indicates that the N-terminal region (residues Leu404-Arg414 in Avena PHOT1) is in the turn-helix-turn structure, in which the helix has been denoted as A′α (residues Thr407-Arg410 in Avena PHOT1) (22). In this structure, A′α is packed against the surface of the β sheet of the LOV2 core domain to interact with the Jα helix (residues Asp522-Ala543 in Avena PHOT1) in the C-terminal flanking region. Importantly, the light-induced structural rearrangements within the LOV2 core are associated with displacement of residues surrounding A′α (22). Hence, not only the C-terminal Jα helix but also the N-terminal flanking region are proposed to act as interfaces relaying the signal from the LOV2 core to the outside domain.
Our mutational analysis sheds new light on the biochemical function of the N-terminal flaking region of LOV2. The sequences in this region (Arg199-Arg210) are strikingly conserved among different phototropins (Fig. 3B). The alignment suggests that T207I and R210H reside within A′α whereas L204P is in the loop upstream of A′α. Although the other three mutational sites, R199C, L202P, and D203N/G/V, are excluded from the above structural study, they are in close proximity on the primary structure. It should be noted here that a high degree of homology does not warrant similar phenotype in corresponding mutants of other phototropins. Indeed, Arabidopsis PHOT1 carrying T469I (corresponding to T207I in CrPHOT) failed to show pseudolit activation (data not shown).
Hypothetical Extended A′α Helix May Play an Important Role for Intramolecular Signaling
Interestingly, a secondary structure prediction suggests that the A′α helix could be extended toward the N terminus to form a longer α helix (Arg199-Arg210), within which all the six mutational sites are mapped (supplemental Fig. S7A). Furthermore, the presumed α helix shows amphipathic character (supplemental Fig. S7B). The hydrophobic surface of this helix might then be packed against the hydrophobic LOV2 core. If this prediction is correct, the mutated residues residing in the polar surface of the helix (Arg199, Asp203, Thr207, and Arg210) might be directly involved in the physical interaction with other functional domains, such as the Jα helix and the C-terminal kinase. The existence of and a structural role for the predicted helix must be investigated in future studies.
Supplementary Material
Acknowledgments
We thank Prof. Masato Umeda and Dr. Utako Kato from Kyoto University for helpful discussions.
This work was supported Grant-in-aid for Scientific Research on Priority Areas 17084002 (to A. N.), Grant-in-aid for Scientific Research on Innovative Areas 22120002 (to A. N.), a Grant-in-aid for the Global COE (Center of Excellence) Program “Formation of a Strategic Base for Biodiversity and Evolutionary Research: From Genome to Ecosystem” (to A. N.) from the Ministry of Education, Culture, Sports, Science, and Technology, Japan, Grant-in-aid for Scientific Research (B) 21370020 (to A. N.), and Research Fellowship 22.1094 from the Japan Society for the Promotion of Science (to Y. A.).

This article contains supplemental Figs. S1–S7.
- PHOT
- phototropin
- AtP1-Nt
- N-terminal fragment of Arabidopsis PHOT1
- BL
- blue light
- CBB
- Coomassie Brilliant Blue
- CrPHOT
- Chlamydomonas phototropin
- Fpk1p
- flippase-kinase 1
- LOV
- light-oxygen and voltage.
REFERENCES
- 1. Christie J. M. (2007) Phototropin blue light receptors. Annu. Rev. Plant Biol. 58, 21–45 [DOI] [PubMed] [Google Scholar]
- 2. Huala E., Oeller P. W., Liscum E., Han I. S., Larsen E., Briggs W. R. (1997) Arabidopsis NPH1: a protein kinase with a putative redox-sensing domain. Science 278, 2120–2123 [DOI] [PubMed] [Google Scholar]
- 3. Sakai T., Kagawa T., Kasahara M., Swartz T. E., Christie J. M., Briggs W. R., Wada M., Okada K. (2001) Arabidopsis nph1 and npl1: blue light receptors that mediate both phototropism and chloroplast relocation. Proc. Natl. Acad. Sci. U.S.A. 98, 6969–6974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jarillo J. A., Gabrys H., Capel J., Alonso J. M., Ecker J. R., Cashmore A. R. (2001) Phototropin-related NPL1 controls chloroplast relocation induced by blue light. Nature 410, 952–954 [DOI] [PubMed] [Google Scholar]
- 5. Kagawa T., Sakai T., Suetsugu N., Oikawa K., Ishiguro S., Kato T., Tabata S., Okada K., Wada M. (2001) Arabidopsis NPL1: a phototropin homolog controlling the chloroplast high-light avoidance response. Science 291, 2138–2141 [DOI] [PubMed] [Google Scholar]
- 6. Kinoshita T., Doi M., Suetsugu N., Kagawa T., Wada M., Shimazaki K. (2001) Phot1 and phot2 mediate blue light regulation of stomatal opening. Nature 414, 656–660 [DOI] [PubMed] [Google Scholar]
- 7. Sakamoto K., Briggs W. R. (2002) Cellular and subcellular localization of phototropin 1. Plant Cell 14, 1723–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kozuka T., Kong S. G., Doi M., Shimazaki K., Nagatani A. (2011) Tissue-autonomous promotion of palisade cell development by phototropin 2 in Arabidopsis. Plant Cell 23, 3684–3695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huang K., Beck C. F. (2003) Phototropin is the blue light receptor that controls multiple steps in the sexual life cycle of the green alga Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. U.S.A. 100, 6269–6274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Im C. S., Eberhard S., Huang K., Beck C. F., Grossman A. R. (2006) Phototropin involvement in the expression of genes encoding chlorophyll and carotenoid biosynthesis enzymes and LHC apoproteins in Chlamydomonas reinhardtii. Plant J. 48, 1–16 [DOI] [PubMed] [Google Scholar]
- 11. Onodera A., Kong S. G., Doi M., Shimazaki K., Christie J., Mochizuki N., Nagatani A. (2005) Phototropin from Chlamydomonas reinhardtii is functional in Arabidopsis thaliana. Plant Cell Physiol. 46, 367–374 [DOI] [PubMed] [Google Scholar]
- 12. Galván-Ampudia C. S., Offringa R. (2007) Plant evolution: AGC kinases tell the auxin tale. Trends Plant Sci. 12, 541–547 [DOI] [PubMed] [Google Scholar]
- 13. Inoue S., Kinoshita T., Matsumoto M., Nakayama K. I., Doi M., Shimazaki K. (2008) Blue light-induced autophosphorylation of phototropin is a primary step for signaling. Proc. Natl. Acad. Sci. U.S.A. 105, 5626–5631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Inoue S., Matsushita T., Tomokiyo Y., Matsumoto M., Nakayama K. I., Kinoshita T., Shimazaki K. (2011) Functional analyses of the activation loop of phototropin2 in Arabidopsis. Plant Physiol. 156, 117–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Christie J. M., Yang H., Richter G. L., Sullivan S., Thomson C. E., Lin J., Titapiwatanakun B., Ennis M., Kaiserli E., Lee O. R., Adamec J., Peer W. A., Murphy A. S. (2011) phot1 inhibition of ABCB19 primes lateral auxin fluxes in the shoot apex required for phototropism. PLoS Biol. 9, e1001076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Crosson S., Moffat K. (2001) Structure of a flavin-binding plant photoreceptor domain: insights into light-mediated signal transduction. Proc. Natl. Acad. Sci. U.S.A. 98, 2995–3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kong S. G., Kinoshita T., Shimazaki K., Mochizuki N., Suzuki T., Nagatani A. (2007) The C-terminal kinase fragment of Arabidopsis phototropin 2 triggers constitutive phototropin responses. Plant J. 51, 862–873 [DOI] [PubMed] [Google Scholar]
- 18. Matsuoka D., Tokutomi S. (2005) Blue light-regulated molecular switch of Ser/Thr kinase in phototropin. Proc. Natl. Acad. Sci. U.S.A. 102, 13337–13342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Salomon M., Christie J. M., Knieb E., Lempert U., Briggs W. R. (2000) Photochemical and mutational analysis of the FMN-binding domains of the plant blue light receptor, phototropin. Biochemistry 39, 9401–9410 [DOI] [PubMed] [Google Scholar]
- 20. Swartz T. E., Corchnoy S. B., Christie J. M., Lewis J. W., Szundi I., Briggs W. R., Bogomolni R. A. (2001) The photocycle of a flavin-binding domain of the blue light photoreceptor phototropin. J. Biol. Chem. 276, 36493–36500 [DOI] [PubMed] [Google Scholar]
- 21. Crosson S., Moffat K. (2002) Photoexcited structure of a plant photoreceptor domain reveals a light-driven molecular switch. Plant Cell 14, 1067–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Halavaty A. S., Moffat K. (2007) N- and C-terminal flanking regions modulate light-induced signal transduction in the LOV2 domain of the blue light sensor phototropin 1 from Avena sativa. Biochemistry 46, 14001–14009 [DOI] [PubMed] [Google Scholar]
- 23. Christie J. M., Swartz T. E., Bogomolni R. A., Briggs W. R. (2002) Phototropin LOV domains exhibit distinct roles in regulating photoreceptor function. Plant J. 32, 205–219 [DOI] [PubMed] [Google Scholar]
- 24. Cho H. Y., Tseng T. S., Kaiserli E., Sullivan S., Christie J. M., Briggs W. R. (2007) Physiological roles of the light, oxygen, or voltage domains of phototropin 1 and phototropin 2 in Arabidopsis. Plant Physiol. 143, 517–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Harper S. M., Neil L. C., Gardner K. H. (2003) Structural basis of a phototropin light switch. Science 301, 1541–1544 [DOI] [PubMed] [Google Scholar]
- 26. Chen E., Swartz T. E., Bogomolni R. A., Kliger D. S. (2007) A LOV story: the signaling state of the phot1 LOV2 photocycle involves chromophore-triggered protein structure relaxation, as probed by far-UV time-resolved optical rotatory dispersion spectroscopy. Biochemistry 46, 4619–4624 [DOI] [PubMed] [Google Scholar]
- 27. Yamamoto A., Iwata T., Sato Y., Matsuoka D., Tokutomi S., Kandori H. (2009) Light signal transduction pathway from flavin chromophore to the Jα helix of Arabidopsis phototropin1. Biophys. J. 96, 2771–2778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Harper S. M., Christie J. M., Gardner K. H. (2004) Disruption of the LOV-Jα helix interaction activates phototropin kinase activity. Biochemistry 43, 16184–16192 [DOI] [PubMed] [Google Scholar]
- 29. Takayama Y., Nakasako M., Okajima K., Iwata A., Kashojiya S., Matsui Y., Tokutomi S. (2011) Light-induced movement of the LOV2 domain in an Asp720Asn mutant LOV2-kinase fragment of Arabidopsis phototropin 2. Biochemistry 50, 1174–1183 [DOI] [PubMed] [Google Scholar]
- 30. Christie J. M., Corchnoy S. B., Swartz T. E., Hokenson M., Han I. S., Briggs W. R., Bogomolni R. A. (2007) Steric interactions stabilize the signaling state of the LOV2 domain of phototropin 1. Biochemistry 46, 9310–9319 [DOI] [PubMed] [Google Scholar]
- 31. Rose M. D., Winston F., Hieter P. (1990) Methods in Yeast Genetics: A Laboratory Course Manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 32. Nakano K., Yamamoto T., Kishimoto T., Noji T., Tanaka K. (2008) Protein kinases Fpk1p and Fpk2p are novel regulators of phospholipid asymmetry. Mol. Biol. Cell 19, 1783–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sikorski R. S., Hieter P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122, 19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kushnirov V. V. (2000) Rapid and reliable protein extraction from yeast. Yeast 16, 857–860 [DOI] [PubMed] [Google Scholar]
- 35. Okajima K., Matsuoka D., Tokutomi S. (2011) LOV2-linker-kinase phosphorylates LOV1-containing N-terminal polypeptide substrate via photoreaction of LOV2 in Arabidopsis phototropin1. FEBS Lett. 585, 3391–3395 [DOI] [PubMed] [Google Scholar]
- 36. Saito K., Fujimura-Kamada K., Furuta N., Kato U., Umeda M., Tanaka K. (2004) Cdc50p, a protein required for polarized growth, associates with the Drs2p P-type ATPase implicated in phospholipid translocation in Saccharomyces cerevisiae. Mol. Biol. Cell 15, 3418–3432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen S., Wang J., Muthusamy B. P., Liu K., Zare S., Andersen R. J., Graham T. R. (2006) Roles for the Drs2p-Cdc50p complex in protein transport and phosphatidylserine asymmetry of the yeast plasma membrane. Traffic 7, 1503–1517 [DOI] [PubMed] [Google Scholar]
- 38. Pomorski T., Lombardi R., Riezman H., Devaux P. F., van Meer G., Holthuis J. C. (2003) Drs2p-related P-type ATPases Dnf1p and Dnf2p are required for phospholipid translocation across the yeast plasma membrane and serve a role in endocytosis. Mol. Biol. Cell 14, 1240–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kong S. G., Suzuki T., Tamura K., Mochizuki N., Hara-Nishimura I., Nagatani A. (2006) Blue light-induced association of phototropin 2 with the Golgi apparatus. Plant J. 45, 994–1005 [DOI] [PubMed] [Google Scholar]
- 40. Jones M. A., Christie J. M. (2008) Phototropin receptor kinase activation by blue light. Plant Signal. Behav. 3, 44–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jones M. A., Feeney K. A., Kelly S. M., Christie J. M. (2007) Mutational analysis of phototropin 1 provides insights into the mechanism underlying LOV2 signal transmission. J. Biol. Chem. 282, 6405–6414 [DOI] [PubMed] [Google Scholar]
- 42. Guo H., Kottke T., Hegemann P., Dick B. (2005) The phot LOV2 domain and its interaction with LOV1. Biophys. J. 89, 402–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nakasako M., Iwata T., Matsuoka D., Tokutomi S. (2004) Light-induced structural changes of LOV domain-containing polypeptides from Arabidopsis phototropin 1 and 2 studied by small-angle x-ray scattering. Biochemistry 43, 14881–14890 [DOI] [PubMed] [Google Scholar]
- 44. Salomon M., Lempert U., Rüdiger W. (2004) Dimerization of the plant photoreceptor phototropin is probably mediated by the LOV1 domain. FEBS Lett. 572, 8–10 [DOI] [PubMed] [Google Scholar]
- 45. Nakasako M., Zikihara K., Matsuoka D., Katsura H., Tokutomi S. (2008) Structural basis of the LOV1 dimerization of Arabidopsis phototropins 1 and 2. J. Mol. Biol. 381, 718–733 [DOI] [PubMed] [Google Scholar]
- 46. Pfeifer A., Mathes T., Lu Y., Hegemann P., Kottke T. (2010) Blue light induces global and localized conformational changes in the kinase domain of full-length phototropin. Biochemistry 49, 1024–1032 [DOI] [PubMed] [Google Scholar]
- 47. Kasahara M., Swartz T. E., Olney M. A., Onodera A., Mochizuki N., Fukuzawa H., Asamizu E., Tabata S., Kanegae H., Takano M., Christie J. M., Nagatani A., Briggs W. R. (2002) Photochemical properties of the flavin mononucleotide-binding domains of the phototropins from Arabidopsis, rice, and Chlamydomonas reinhardtii. Plant Physiol. 129, 762–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kutta R. J., Hofinger E. S., Preuss H., Bernhardt G., Dick B. (2008) Blue light induced interaction of LOV domains from Chlamydomonas reinhardtii. ChemBioChem 9, 1931–1938 [DOI] [PubMed] [Google Scholar]
- 49. Nash A. I., Ko W. H., Harper S. M., Gardner K. H. (2008) A conserved glutamine plays a central role in LOV domain signal transmission and its duration. Biochemistry 47, 13842–13849 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.