Background: HIF-1α can be regulated by both VHL-dependent and VHL-independent mechanisms.
Results: ErbB4 directly interacts with HIF-1α and promotes its stability and signaling in vitro and in vivo.
Conclusion: The interaction between HIF-1α and ErbB4 is a biologically significant mechanism to promote HIF-1α activity.
Significance: A novel VHL-independent mechanism promoting HIF-1α signaling is described.
Keywords: Epidermal Growth Factor Receptor (EGFR), Growth Factors, Hypoxia-inducible Factor (HIF), Receptor-tyrosine Kinase, Signal Transduction
Abstract
The receptor-tyrosine kinase ErbB4 was identified as a direct regulator of hypoxia-inducible factor-1α (HIF-1α) signaling. Cleaved intracellular domain of ErbB4 directly interacted with HIF-1α in the nucleus, and stabilized HIF-1α protein in both normoxic and hypoxic conditions by blocking its proteasomal degradation. The mechanism of HIF stabilization was independent of VHL and proline hydroxylation but dependent on RACK1. ErbB4 activity was necessary for efficient HRE-driven promoter activity, transcription of known HIF-1α target genes, and survival of mammary carcinoma cells in vitro. In addition, mammary epithelial specific targeting of Erbb4 in the mouse significantly reduced the amount of HIF-1α protein in vivo. ERBB4 expression also correlated with the expression of HIF-regulated genes in a series of 4552 human normal and cancer tissue samples. These data demonstrate that soluble ErbB4 intracellular domain promotes HIF-1α stability and signaling via a novel mechanism.
Introduction
Hypoxia-inducible factor-1α (HIF-1α)2 is a basic helix-loop-helix transcription factor that binds DNA at specific hypoxia response elements (HRE) as a heterodimeric complex with HIF-1β (1). Transcriptional activity of HIF-1α regulates expression of genes involved in angiogenesis, pH regulation, glycolysis, and metastasis (2). Expression of HIF-1α is increased in diseases such as neoplasias and ischemia (2, 3), and HIF-1α regulation has potential as a therapeutic approach (2). HIF-1α also has indispensable roles during normal development of multiple tissues such as the heart (4, 5), the central nervous system (4, 5), and the mammary gland (6).
HIF-1α signaling is regulated at the level of protein degradation by a family of oxygen-dependent prolyl hydroxylases (PHD or EGLN), which hydroxylate conserved proline residues of HIF-1α, thereby promoting HIF-1α ubiquitination by the von Hippel-Lindau gene product (VHL) (7). There are, however, also less well-characterized VHL- and oxygen-independent mechanisms of regulation of HIF-1α stability (8). These include regulation of HIF-1α ubiquitination by receptor for activated protein kinase C (RACK1) (9) and hypoxia-associated factor (HAF) (10).
ErbB4 (HER4) is a receptor tyrosine kinase that serves as a receptor for the neuregulins (NRG) and other epidermal growth factor (EGF)-like ligands. ErbB4 belongs to the ErbB/HER subfamily of receptor tyrosine kinases (RTK) that also includes the well-characterized human cancer drug targets EGF receptor (EGFR = ErbB1 = HER1), ErbB2 (HER2), and ErbB3 (HER3) (11). Dysregulated ErbB4 has been associated with several diseases including malignancies (12), cardiovascular diseases (13), and schizophrenia (14). ErbB4 is also necessary for normal development of several tissues, such as heart (15), kidney (16), central nervous system (17, 18), and mammary gland (19, 20).
In addition to signaling via classical RTK-activated cascadical signaling pathways, specific alternatively spliced isoforms of ErbB4 signal via proteolytically released intracellular domain (ErbB4 ICD) that may translocate into the nucleus and regulate transcription (17, 21, 22). Only isoforms of the juxtamembrane-a (JM-a)-type are susceptible to a two-step proteolytic process involving tumor necrosis factor-α converting enzyme (TACE) and γ-secretase activity that releases the soluble ICD. The alternative JM isoform JM-b is resistant to cleavage by TACE and does not release a soluble ICD (22, 23). The ERBB4 gene is further spliced to generate variation in the cytoplasmic (CYT) domain. The CYT-1 isoform includes a stretch of amino acids that are lacking from the alternative CYT-2 isoform and that provide unique interaction sites for proteins such as phosphoinositide 3-kinase (PI3-K) (24).
Here, we describe a novel mechanism by which the proteolytically released ErbB4 ICD regulates HIF-1α stability and signaling both in vitro and in vivo. The mechanism is based on an interaction between the two proteins in the nucleus and leads to RACK1-dependent but VHL-independent suppression of HIF-1α degradation.
EXPERIMENTAL PROCEDURES
Cell Culture
MCF-7 breast cancer cell line was maintained in RPMI supplemented with 10% FCS. 1 nm estrogen (Sigma-Aldrich) was included in MCF-7 culture medium. COS-7 and BT-20 cells were maintained in DMEM supplemented with 10% FCS. RCC4- and RCC4+VHL cells (European Collection of Cell Cultures) were maintained in DMEM, 10% FCS, and 250 μg/ml G418. Cell culture in hypoxic atmosphere was carried out using a hypoxic work station (In vivo2, Ruskinn Technology Ltd.).
Ligands and Inhibitors
Cells were treated for the indicated periods of time with 50 ng/ml NRG-1 (R&D Systems), 200 μm CoCl2 (Sigma-Aldrich), 10 μm AG 1478, 20–50 μm GSI IX, or 20 μm LY294002 (all from Calbiochem) in the absence of FCS.
Expression Plasmids and Retroviruses
ErbB4 deletion mutants with C-terminal HA-tags were cloned by PCR using pcDNA3.1ErbB4ICD2-HA (25) as template and primers 5′-taatacgactcactatagggagacc-3′ and 5′-tattatctagattatgcgtaatcgggtacatcgtatgg-gtagggaagcttcata-3′ for ICD2-ΔC and primers 5′-tagaaggcacagtcgagg-3′ and 5′-ttagctagcaccatgagtccaaatgacagcaagttctttcagaat-3′ for ICD2-ΔN. PCR products were ligated into NheI-XbaI and AflII-XbaI restriction sites in pcDNA3.1(+)hygro vector (Invitrogen), respectively. Other pcDNA3.1ErbB4 constructs (22, 26) have been described earlier. HIF-1α deletion constructs with C-terminal 6×His tags were cloned by PCR using forward primers 5′-ctgggatccaccaatggagggcgccggcggc-3′ for wild-type HIF-1α, 5′-ctgggatccaccaatgactagccgaggaaga-3′ for Δ1–174 deletion, 5′-ctgggatccacca-atgattattcagcacga-3′ for Δ1–343 deletion, 5′-ctgggatccaccaatgttcaagttggaatt-3′ for Δ1–529 deletion, and 5′-ctggga-tccaccaatgtctcatccaagaagc-3′ for Δ1–681 deletion. A common reverse primer 5′-aattgcgccgcttaatggtgatggtgatggtggttaacttgatccaaag-3′ was used for all constructs. PCR products were ligated into BamHI and NotI restriction sites of pcDNA3.1(+)hygro. Wild-type HIF-1α and HIF-1αP402A,P564G expression plasmids were a kind gift from Dr. Peter Ratcliffe (Oxford University, UK).
Cells were transfected with Fugene 6 (Roche) following the manufacturer's recommendations. For retroviral expression, pBABE-puro constructs encoding ErbB4 JM-a CYT-2, ErbB4 JM-b CYT-2, or empty vector (27) were expressed in Phoenix-packaging cell line. Twenty-four hours after transfection, medium was collected and used to infect RCC cells. Stable cell pools were selected using puromycin (Sigma-Adrich).
siRNA Knock-down
One day after plating, MCF-7 cells were treated with siRNAs specifically targeting ErbB4 JM-a (ErbB4 siRNA #1; 5′-gucaugacuagugggaccgtt-3′ and 5′-guauugaagacugcaucggtt-3′) or against all ErbB4 isoforms (ErbB4 siRNA #2) (22). RACK1 targeting siRNA #1 (5′-aucauguccgggaacugcggg-3′) and siRNA #2 (5′-uaaacuucuagcgugugccuu-3′) were purchased from Qiagen. Universal negative control siRNA (Eurogentech) and siRNA targeting ErbB4 JM-b (22), which is not expressed in MCF-7 cells, were used as negative controls. All siRNAs were introduced to cells using Lipofectamine 2000 (Invitrogen) following manufacturer's recommendations. When both siRNA and plasmid DNA were transfected, siRNAs were transfected 4 h after plasmid transfection.
Conditional Knock-out Mice and Immunohistochemistry
Mice with mammary gland specific targeting of Erbb4 (Erbb4Flox/FloxWAP-Cre) have been described earlier (28). Paraffin sections of mammary glands of pregnancy day 18 (P18) or lactating day 1 (L1) mice were immunostained with anti-ErbB4 (sc-283; Santa Cruz Biotechnology) or anti-GLUT-1 (ab14683; Abcam) rabbit polyclonal antibodies. Immunohistochemical analysis of HIF-1α expression was carried out with anti-HIF-1α mouse monoclonal antibody (clone H1α67; Abcam) or nonspecific mouse monoclonal IgG (clone 3g6; kind gift of Dr. Sirpa Jalkanen) using a HistomouseMax mouse-on-mouse IHC staining kit (Invitrogen) following manufacturer's protocol. Images were taken using Olympus BX60 microscope equipped UPlanFl 20×/0.50 Ph1 objective and Olympus DP71 digital color camera. Quantification of immunohistochemical stainings was carried out with NIH ImageJ v1.43o using Color Deconvolution plug-in (29).
Real-time RT-PCR
Quantitative real-time RT-PCR was performed, as previously described (30). Shortly, total RNA was extracted from cell cultures using Trizol (Invitrogen) RNA extraction reagent. One microgram of total RNA was subjected to DNase I treatment and cDNA synthesis using random hexamer primers. Primers and Universal Probe library probes (Roche Applied Science) were: EPO (left 5′-tcccagacaccaaagttaatttcta-3′, right 5′-ccctgccagacttctacgg-3′, probe #58), PGK1 (left 5′-tgcaaaggccttggagag-3′, right 5′-tggatcttgtctgcaactttagc-3′, probe #72), and EEF1A (left 5′-ccccaggacacagagacttt-3′, right 5′-gcccattcttggagatacca-3′, probe #56). GLUT1 was detected using: left 5′-gtgggcatgtgcttccagtc, right 5′-aagaacagaaccaggagcacagt-3′, probe aactgtgtggtccctacgtcttcatcatct. Primers and probes for ERBB4 (30) and VEGFA (31) have been described earlier.
Western Blotting and Co-immunoprecipitation
Western analyses were carried out, as previously described (32), using the following antibodies: anti-HIF-1α (Clone 54; BD Biosciences), anti-actin (sc-1616; Santa Cruz Biotechnology), anti-ErbB4 (sc-283 from Santa Cruz Biotechnology or E200 from Abcam), anti-pAkt (Cell Signaling Technology), anti-Akt (sc-1618; Santa Cruz Biotechnology), anti-HA (3F10; Roche Applied Sciences), anti-RACK1 (ab62735; Abcam), and anti-GST (Amersham Biosciences). For immunoprecipitation, HA-tagged ErbB4 was precipitated with anti-HA (3F10; Roche Applied Sciences) and HIF-1α with anti-HIF-1α (Clone 54; BD Biosciences) using protein G-Sepharose beads (Amersham Biosciences). Immunocomplexes were washed four times with co-IP buffer (100 mm NaCl, 50 mm Tris-HCl, pH 7.5, 1% Triton X-100).
GST Pull-down Assay
Inserts encoding HIF-1α or ErbB4-ICD2 (ICD with CYT-2-type of cytoplasmic domain) were cloned using PCR into pGEX-6P1 GST fusion vector (Amersham Biosciences). The GST fusion products were expressed in BL-21 DE3 strain of Escherichia coli (Invitrogen). For experiments testing interaction between recombinant HIF-1α and ErbB4 ICD2-GST fusion, the GST domain of HIF-1α-GST fusion was cleaved using Precission protease (Amersham Biosciences), and free GST removed with glutathione beads.
GST fusion proteins were affinity-purified using glutathione Sepharose 4B beads (Amersham Biosciences), and used directly in pull-down experiments or eluted with 20 mm glutathione, 100 mm NaCl, 0.5% Triton X-100, and 1 mm DTT. In GST pull-down experiments, 1 μg of GST-fusion protein was incubated together with 10 μl of in vitro translation reaction for 2 h at room temperature or overnight at 4 °C with 25 μl of glutathione-Sepharose 4B beads in a total volume of 200 μl of binding buffer (150 mm NaCl, 50 mm Tris-HCl, pH 7.5, 0.5% Triton X-100). Nonspecific binding was removed with at least four washes with 500 μl of binding buffer. Beads were boiled in Laemmli sample buffer, separated with SDS-PAGE and subjected to Western blotting. In vitro translation reactions were performed with pcDNA 3.1+Hygro vectors containing either ERBB4 or HIF1A inserts using TNT T7 Coupled Reticulocyte Lysate System (Promega) according to manufacturer's protocol, except that reactions were performed in the presence of 100 μm ALLN and 1 mm DMOG to prevent degradation of HIF-1α.
Luciferase Assays
MCF-7 cells were plated on 24-well plates at density of 4 × 104 cells/cm2. On the following day, the cells were transfected with 100 ng of pGL3-HRE-luc, 50 ng of pTK-Rluc and 50 ng of other plasmids per well. One day after transfection, cells were subjected to experimental treatments. Luciferase activity was measured using Dual-luciferase assay (Promega) following manufacturer's protocol. pGL3-HRE-luc (kindly provided by Dr. Peter Ratcliffe) activity was normalized with luminescence signal from co-transfected Renilla luciferase (pTK-Rluc; Promega).
Immunofluorescence Staining
COS-7 cells were plated on coverslips at density of 10,000 cells/24-well a day prior to transfection. Twenty-four hours after transfection, the cells were washed with PBS and fixed with −20 °C methanol for 30 min. HA-tagged ErbB4 and HIF-1α were detected using anti-HA (Sigma) and anti-HIF (BD Biosciences) primary antibodies and anti-rat-Alexa-568 or anti-mouse-Alexa-488 (Molecular Probes) secondary antibodies, respectively. Nuclei were visualized with DNA-binding dye 4,6-diamidino-2-phenylindole (DAPI; 0.5 μg/ml; Sigma-Aldrich). Signals were analyzed with Zeiss LSM510 META confocal microscope equipped with Plan-Apochromat 63×/1.4 Oil DIC objective. Images were processed with median 2 × 2 filter using NIH ImageJ v1.36b software before printing to reduce background noise. Quantitative colocalization analysis was performed with NIH ImageJ v1.36b software with the colocalization threshold plug-in (written by Tony Collins, McMaster University, Hamilton, Canada).
Proximity Ligation Assay
In situ proximity ligation assay (PLA) was performed using Duolink in situ PLA kit (Olink Biosciences) following manufacturer's protocol. MCF-7 cells (100,000 cells/24-well) were plated onto glass coverslips. The cells were treated overnight with 200 μm CoCl2, fixed in 4% paraformaldehyde and permeabilized with 0.2% Triton X-100. After blocking nonspecific binding with 10% normal goat serum in PBS for 1 h, the cells were incubated with the primary antibodies anti-ErbB4 (HFR-1; Neomarkers) and/or anti-HIF1α (HPA001275; Atlas Antibodies), or anti-RACK1 ((ab62735; Abcam) and/or anti-HIF-1α (Clone 54; BD Biosciences) overnight at 4 °C in blocking solution. Anti-mouse Minus and anti-rabbit Plus (Olink Biosciences) were used as secondary antibodies. Proximity of the two antibodies was detected using Duolink Brightfield Detection kit and Zeiss AxioImager A1 microscope equipped with Plan-Neofluar 40×/0.75 Ph2 objective and Axiocam ICc3 digital color camera, or with fluorescent Duolink Orange Detection kit (Olink Biosciences) and Zeiss LSM510 META confocal microscope using Plan-Apochromat 63x/1.4 Oil DIC objective. Confocal images were processed with median 2 × 2 filter (NIH ImageJ v1.36b) to reduce background noise.
Apoptosis Assay
Wild-type MCF-7 cells were plated at a density of 10,000 cells/96-well. The following day, the wild-type cells were treated with siRNAs. Cells were transferred into either normoxic (21% O2) or hypoxic (1% O2) conditions 24 h after transfection. After 48 h of incubation, cells were fixed with 1% paraformaldehyde and nuclei were stained with DAPI. Nuclei demonstrating normal or apoptotic morphology were counted from microscopic images (Plan 10×/0.25 Ph1 objective, Zeiss Axiovert 200 m). On average 400 nuclei were counted for each sample and apoptotic indices (percent of apoptotic nuclei per all nuclei) were determined.
In Silico Transcriptomics
IST-database (version 2) covering gene expression data from 70 different normal tissue types, 50 tumor types and 176 different functional experiments on cell lines was used (33). All samples were manually annotated including information about anatomical location, cell type, disease, and treatment data. Co-expression in normal (n = 975) and cancer (n = 3577) tissue samples was analyzed. Heat-map of correlations was produced with program JavaTreeView (version 1.1.4) (34).
Statistical Analyses
In vitro experiments were analyzed with two-sided Student's t test or with one-way ANOVA associated with Tukey's post-hoc test. For statistical analyses of data from the in silico data base, Pearson's correlation analysis was performed for each of the cancer or tissue types separately. Only tissues or cancer types showing statistically significant Pearsons's correlations (p < 0.05) between ERBB4 or NRG1 and EPO, CAIX, or GLUT1 were used in further analyses. Statistically significant Pearson's correlations were scored as either positive or negative. Distribution of these scores was analyzed statistically using binomial test (sign test), and was based on assumption that if there was no interaction between the test parameters, there would be an equal probability of 0.5 for both negative and positive values.
RESULTS
Mice with Targeted Erbb4 or Hif1a Genes Exhibit Overlapping Phenotypes
When surveying for novel cancer-associated signaling pathways by comparing reported phenotypes of mice with targeted cancer-associated genes, we made a previously unrecognized observation that targeted gene deletion of either Hif1a (6) or Erbb4 (19, 28) in the mammary gland resulted in strikingly similar phenotypes. Both targeted Hif1a and Erbb4 mice demonstrate impaired expansion of the lobuloalveoli and production of milk proteins during pregnancy and ability to produce milk during lactation. This observation suggests that HIF-1α and ErbB4 may function in the same pathway regulating lactation.
ErbB4 Is Necessary for HIF-1α-regulated Responses in Vitro and in Vivo
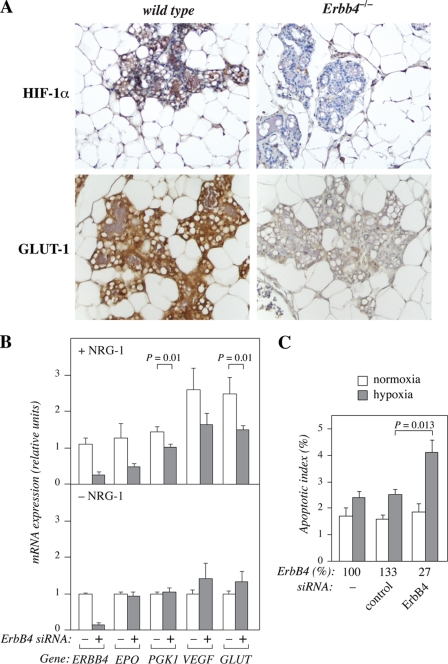
To more directly address the potential effect of ErbB4 on HIF-1α protein in vivo, lactating mouse mammary gland tissues from mice with mammary gland-specific targeting of Erbb4 under the WAP promoter were immunohistochemically analyzed for HIF-1α expression. The Erbb4 null lobuloalveoli demonstrated strongly reduced immunoreactivity for HIF-1α when compared with wild-type controls (Fig. 1A). The reduction was 68% when the immunosignal was quantified by ImageJ software (n = 5, p < 0.001). Erbb4 targeting also significantly reduced (by 47%; n = 5, p < 0.001) expression of the HIF-1α target gene product GLUT-1 in the same tissue (Fig. 1A), indicating that the down-regulation of HIF-1α was of functional significance. As controls, it was demonstrated that Erbb4-targeting resulted in total loss of ErbB4 immunoreactivity (n = 5, p < 0.001), but did not affect staining for a non-sequence-specific anti-phosphotyrosine antibody 4G10 (n = 5, p = 0.36) or the negative control antibody 3g6 (n = 5, p = 0.83) (supplemental Fig. S1).
FIGURE 1.
ErbB4 is necessary for HIF-1α-regulated cellular responses in vitro and in vivo. A, immunohistochemical analysis of HIF-1α and GLUT-1 expression in paraffin sections of lactating (L1) wild-type and Erbb4Flox/FloxWAP-Cre+/− (Erbb4−/−) mice. B, MCF-7 cells were transfected with no siRNA, nonspecific control siRNA or ErbB4 siRNA (#1). Twenty-four hours after transfection, cells were switched into serum-free medium with or without 50 ng/ml NRG-1 and cultured for 20 h in 1% oxygen. Expression of the indicated mRNAs of known HIF-regulated genes and ERBB4 were measured by real-time RT-PCR. For presentation of the data as columns indicating mRNA expression in −siRNA and +siRNA groups, samples demonstrating ERBB4 mRNA expression at levels >75% of control siRNA levels were pooled into −siRNA group (no siRNA and control siRNA), and samples demonstrating ERBB4 mRNA expression at levels <50% of control siRNA levels were pooled into +siRNA group (ErbB4 siRNA #1 and ErbB4 siRNA #2). The data represent normalized values of mean ± S.E. of at least four samples for each gene. In the absence of ErbB4 down-regulation and NRG-1 treatment ERBB4 expression corresponded to 0.02% of the expression of the reference gene EEF1A. The respective expression level for EPO was 0.03%, for PGK1 5.2%, for VEGFA 2.7%, and for GLUT1 3.7% of EEF1A mRNA expression. C, MCF-7 cells transfected with no siRNA, nonspecific control siRNA or ErbB4 siRNA (#1) were cultured in hypoxia (1% O2) or normoxia (21% O2) for 48 h and condensed apoptotic nuclei were visualized by staining with the nuclear stain DAPI. Each column represents the mean apoptotic index ± S.E. from two independent experiments (n = 11). The effects of the treatments on ErbB4 protein levels, as determined by Western analysis and densitometry, are shown relative to no siRNA treatment (ErbB4%).
To receive more information about the biological significance of ErbB4 in regulating HIF-dependent activities, the effect of endogenous ErbB4 knock-down by siRNAs on the transcription of well-characterized HIF-regulated genes was analyzed by real-time RT-PCR. Reducing endogenous ErbB4 levels in MCF-7 cells resulted in a significant suppression of the levels of transcripts encoding phosphoglycerate kinase-1 (PGK-1) and GLUT-1 when the cells were stimulated with the ErbB4 ligand NRG-1 (Fig. 1B). A tendency for down-regulation of transcripts encoding erythropoietin (EPO) and VEGF-A was also observed.
To address the contribution of ErbB4 to cellular responses known to be regulated by HIF-1α, MCF-7 transfectants were cultured in hypoxia (1% O2) or normoxia (21% O2) and the fraction of cells undergoing apoptosis was measured. In accordance with ErbB4 supporting HIF-regulated survival, knock-down of endogenous ErbB4 by siRNA increased the apoptotic effect of hypoxia (Fig. 1C).
ErbB4 Expression Is Associated with HIF-1α Activity in Human Tissues
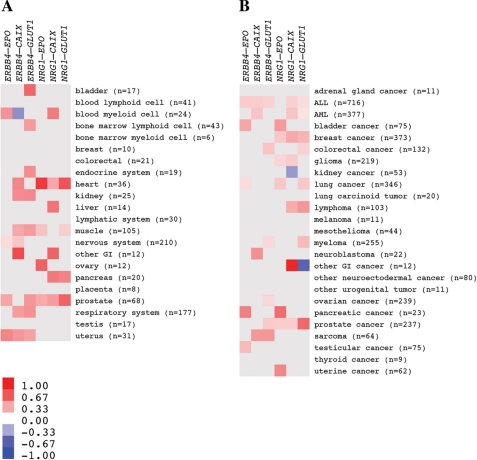
To address the significance of the interplay between ErbB4 and HIF-1α signaling pathways in human tissues in vivo, the association of ERBB4 and NRG1 expression with the expression of known HIF-1α-regulated genes was analyzed from 975 normal and 3577 cancer tissues samples, using human microarray expression data available as an in silico transcriptomics (IST) data base (33). EPO, carbonic anhydrase IX (CAIX) and GLUT1 were chosen as surrogate markers of HIF-1α activity, as they have previously been associated with hypoxia in clinical samples, and are known to be directly regulated by HIF-1α (35).
In normal tissues, a statistically significant positive association was found 32 times between expression of ERBB4 or NRG1 and any of the HIF-1α target genes whereas a significant negative association was only found once (p < 0.001) (Fig. 2A). The respective numbers for the cancer tissues were 37 positive and 2 negative significant associations (p < 0.001) (Fig. 2B).
FIGURE 2.
Expression of ErbB4 and an ErbB4 ligand correlates with expression of HIF-1α-regulated genes in human. Messenger RNA expression levels of ERBB4, NRG1, and the HIF-1α-regulated genes EPO, CAIX and GLUT1 were analyzed using an in silico transcriptomic database. Pearson's correlations were separately determined for correlation of ERBB4 or NRG1 mRNA with each of the three HIF-regulated transcripts in different human normal (A) and cancer (B) tissues. Each statistically significant (p < 0.05) positive Pearson's correlation is indicated with red color and each significant negative correlation with blue color. Color intensity reflects Pearson's correlation coefficient (r) as depicted in the scale bar. Numbers of samples available for each tissue (n) are indicated. Only tissues of which 5 or more samples were available are included in the presentation. The gene pair analyzed for each correlation is depicted on top of the heat maps.
Furthermore, a global analysis of all statistically significant associations using binomial test across all normal and cancer samples revealed that ERBB4 mRNA expression was significantly associated with higher mRNA expression of EPO (p = 0.002), CAIX (p = 0.003), and GLUT1 (p < 0.001). Similarly, mRNA expression of NRG1 was significantly associated with higher mRNA expression of EPO (p < 0.001), CAIX (p = 0.002), and GLUT1 (p = 0.002). Data about the sample numbers, correlation coefficients, and p values for each individual normal and cancer tissue type are given for associations with ERBB4 and NRG1 in supplemental Tables S1 and S2, respectively. These data indicate that expression of ErbB4 as well as expression of the ErbB4-stimulating NRG-1 is significantly associated with expression of HIF-1α-responsive genes in human tissues in vivo.
ErbB4 Promotes Accumulation of HIF-1α Protein
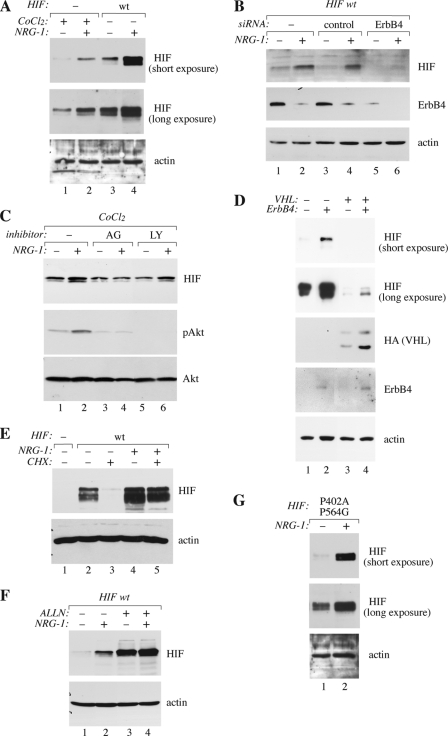
To experimentally address a possible interaction between ErbB4 and HIF-1α signaling pathways at the molecular level, we analyzed the effect of ErbB4 activation on HIF-1α protein levels in human MCF-7 breast cancer cells. Stimulation of serum-starved MCF-7 cells with the ErbB4 ligand NRG-1 increased the steady-state levels of endogenous HIF-1α protein beyond the levels observed in the presence of a well-characterized HIF-inducing agent, CoCl2 (Fig. 3A, lanes 1 and 2), which blocks PHD activity (35). The increase was observed already 15 min after NRG-1 treatment and reached a maximum after 4 h (supplemental Fig. S2A). NRG-1 also promoted accumulation of ectopic wild-type HIF-1α introduced via transfection under the control of a cytomegalovirus (CMV) promoter (Fig. 3A, lanes 3 and 4). Furthermore, NRG-1-stimulated accumulation of both endogenous and overexpressed HIF-1α was specifically blocked by siRNAs targeting endogenous ErbB4 both in the presence and absence of CoCl2 (Fig. 3B; supplemental Fig. S2B).
FIGURE 3.
Accumulation of HIF-1α by ErbB4. Western analyses of the indicated proteins are shown. A, MCF-7 cells were transfected with a vector control or a plasmid encoding wild-type (wt) HIF-1α. Transfectants were treated with or without 50 ng/ml NRG-1 and 200 μm CoCl2 for 20 h. Two different exposures of the same HIF-1α blot are shown. B, MCF-7 cells were transfected with no siRNA, nonspecific control siRNA or ErbB4 siRNA (#2), and a plasmid encoding HIF-1α, and treated with or without NRG-1 for 24 h. C, wild-type MCF-7 cells were treated with CoCl2 with or without NRG-1, AG 1478 or LY2940002 for 20 h. D, RCC4 cells carrying a mutation in the VHL gene (VHL−) or rescued to re-express VHL (VHL+) were infected with an empty retrovirus or a retrovirus encoding ErbB4 JM-a CYT-2. The VHL construct included a HA-tag. Two different exposures of the same HIF-1α blot are shown. E, MCF-7 cells were transfected with a vector control or a plasmid encoding wild-type HIF-1α and treated with or without NRG-1 for 20 h. Cycloheximide (CHX) was added for the last two hours of the experiment to block protein synthesis. F, MCF-7 transfectants overexpressing HIF-1α were treated with or without NRG-1 and the proteasomal inhibitor ALLN for 20 h to analyze the effect of proteasomal activity on NRG-1-stimulated accumulation of HIF-1α. G, MCF-7 transfectants overexpressing a proline hydroxylation resistant HIF-1αP402A,P564G were treated with or without NRG-1 for 20 h. Two different exposures of the same HIF-1α blot are shown.
The effect of NRG-1 on HIF-1α expression was inhibited by an ErbB tyrosine kinase inhibitor, AG 1478 (36) (Fig. 3C, lanes 3 and 4). In contrast, the PI3-K inhibitor LY294002, previously shown to block translational up-regulation of HIF-1α expression by EGFR (37) and ErbB2 (38), did not block the relative effect of NRG-1 (Fig. 3C, lanes 5 and 6). LY294002 did, however, reduce the baseline level of HIF-1α protein both in the absence and presence of NRG-1. Moreover, retroviral overexpression of a constitutively active (22) cleavable ErbB4 JM-a CYT-2 isoform was sufficient to enhance the expression of endogenous HIF-1α protein in naturally ErbB4-negative renal cell carcinoma (RCC4) cells (Fig. 3D). Taken together, these findings demonstrate that ErbB4 activity promotes HIF-1α accumulation and that the regulation takes place at a post-transcriptional level.
ErbB4 Blocks Proteasomal Degradation of HIF-1α by a Mechanism Independent on VHL and Proline Hydroxylation
The observation that regulatory sequences of the endogenous HIF1A locus were not needed, suggested that the increased HIF-1α level induced by ErbB4 activation was due to reduced protein degradation. This hypothesis was tested by blocking translational activity with cycloheximide and following the levels of HIF-1α protein in MCF-7 cells. Indeed, the degradation of HIF-1α was significantly reduced in the presence of NRG-1 (Fig. 3E). In the presence of the proteasomal inhibitor ALLN, the levels of HIF-1α were similar in the presence and absence of NRG-1 (Fig. 3F), indicating that ErbB4 activation suppressed proteasome-dependent HIF-1α degradation.
Previous work has demonstrated a central role for proline hydroxylation and VHL-regulated proteasomal degradation of HIF-1α in the regulation of HIF-1α stability in response to hypoxia (39, 40). Overexpression of the cleavable ErbB4 JM-a CYT-2 increased HIF-1α levels both in a VHL-negative RCC4 line and the corresponding RCC4 line rescued to re-express VHL independently of the VHL status (Fig. 3D). Moreover, mutation of the proline residues in HIF-1α (double mutation P402A/P564G) needed for HIF hydroxylation and HIF-1α/VHL interaction (40, 41), did not affect the ability of NRG-1 to stimulate HIF-1α accumulation in MCF-7 cells (Fig. 3G). Together with the observation that ErbB4-stimulated HIF-1α accumulation was not sensitive to PHD inhibition by CoCl2, these data imply that regulation of HIF-1α stability by ErbB4 activation does not involve the oxygen-dependent PHD/VHL pathway.
Cleaved ErbB4 Promotes HIF-1α Activity
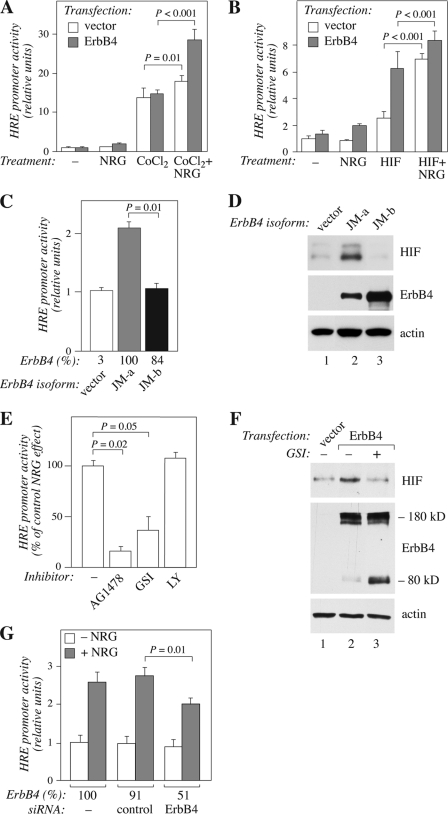
To analyze the functional significance of ErbB4-stimulated HIF-1α stability, promoter assays measuring hypoxia response element (HRE)-mediated transcription of a luciferase gene were carried out in the presence and absence of ErbB4 activation. HREs have previously been shown to function as genomic regulatory elements mediating the transcriptional activity of HIF-1α (1). While CoCl2 treatment substantially increased the basal level of HRE-luciferace activity, ErbB4 activation further increased the activity (Fig. 4A). Similar findings were observed when HIF-1α was overexpressed by transfection (Fig. 4B), providing more direct evidence that the effect of ErbB4 activation on the HRE element involved the HIF-1α protein.
FIGURE 4.
Stimulation of hypoxia response element-mediated transcription by a cleavable ErbB4 isoform. A, MCF-7 cells were transfected with plasmids constructs encoding firefly luciferase under the control of HIF-1α response element (HRE-luc), control Renilla luciferase under thymidine kinase promoter (TK-Rluc), and ErbB4 JM-a CYT-2 or an empty vector control. Transfectants were treated with or without NRG-1 and CoCl2 for 18 h. HRE-regulated luciferase activity was normalized with luminescence signal from co-transfected Renilla luciferase. Columns represent mean ± S.E. from three independent experiments (n = 12). B, HRE-mediated luciferase activity was measured as in A except that CoCl2-treatment was replaced by overexpression of HIF-1α by transfection. Data represent mean ± S.E. from three independent experiments (n = 6). C, MCF-7 cells were transfected with a plasmid encoding HIF-1α, and with vector control or plasmids encoding either ErbB4 JM-a CYT-2 or ErbB4 JM-b CYT-2. HRE-mediated luciferase activity was measured as in A. ErbB4 expression levels in transfectants, as determined by Western analysis and densitometry, are indicated relative to ErbB4 JM-a CYT-2 expression (ErbB4%). D, Western analysis of RCC4 cells transfected with plasmids encoding ErbB4 JM-a CYT-2 or JM-b CYT-2 or with vector control. E, MCF-7 transfectants overexpressing HIF-1α were treated for 18 h with AG 1478, GSI IX, or LY294002 in the presence or absence of NRG-1. HRE-mediated luciferase activity was measured as in A. Data are presented as the relative effect of NRG stimulation obtained in the presence of the inhibitor as compared with the relative NRG effect in the absence of inhibitors. Data represent mean ± S.E. from three independent experiments (n = 8). F, Western analysis of BT-20 cells transfected with a plasmid encoding ErbB4 JM-a CYT-2 or an empty vector. The cells were treated for 6 h with GSI IX. Full-length ErbB4 migrates at 180 kDa. The accumulation of 80 kDa ErbB4 product demonstrates the effect of GSI (22). G, MCF-7 cells transfected with plasmids encoding HRE-luc, TK-Rluc and HIF-1α, and with no siRNA, nonspecific control siRNA or ErbB4 siRNA (#1). The transfectants were treated with or without NRG-1 for 18 h. HRE-mediated luciferase activity was measured as in A. Data represent mean ± S.E. from three independent experiments performed in triplicates (n = 9). The effects of the treatments on ErbB4 protein levels, as determined by Western analysis and densitometry, are shown relative to no siRNA treatment (ErbB4%).
Interestingly, when the cleavable JM-a CYT-2 ErbB4 isoform was compared with the non-cleavable JM-b CYT-2 for its potential to up-regulate HRE-mediated transcription in the presence of HIF-1α overexpression, only JM-a CYT-2 demonstrated activity (Fig. 4C). Similarly, overexpression of cleavable JM-a isoforms in MCF-7 cells selectively promoted transcription of the HRE-containing VEGFA gene in the absence of HIF-1α overexpression under normoxic conditions (supplemental Fig. S3A). To address whether the selective stimulation of transcriptional activity by the cleavable ErbB4 isoforms was associated with selective accumulation of HIF-1α protein, constructs encoding ErbB4 JM-a CYT-2 or JM-b CYT-2 were introduced into the naturally ErbB4-negative RCC4 cells. As expected based on the transcriptional analyses, only the cleavable JM-a efficiently accumulated HIF-1α (Fig. 4D). Further consistent with an activity selective for the cleavable ErbB4 isoforms, γ-secretase inhibitor GSI IX known to block the cleavage of ErbB4 JM-a isoforms and subsequent release of soluble ErbB4 ICDs (21, 22) suppressed the JM-a isoform-promoted HRE activity (Fig. 4E), VEGFA mRNA expression (supplemental Fig. S3B), as well as HIF-1α protein accumulation (Fig. 4F).
The ability of NRG-1 to promote HRE-mediated activity was also blocked by the ErbB kinase inhibitor AG 1478 but not by the PI3-K inhibitor LY294002 (Fig. 4E; supplemental Fig. S3B). Moreover, siRNA targeting of endogenous ErbB4 (Fig. 4G) significantly down-regulated the NRG-1-specific effect on the HRE activity. These data indicate that cleavable ErbB4 isoforms, but not their non-cleavable counterparts, can promote transcriptional activity of HIF-1α.
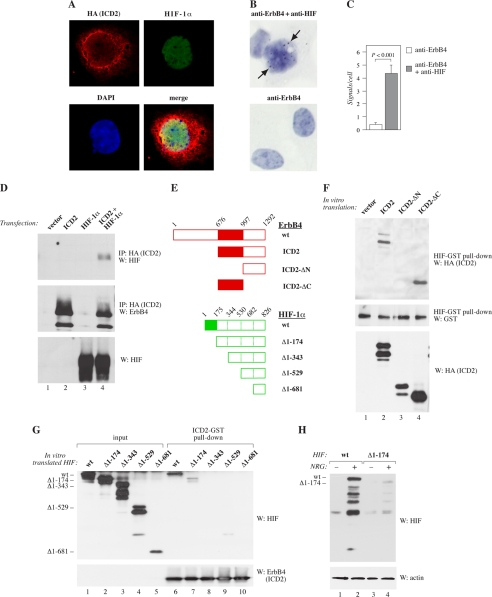
Direct Interaction between HIF-1α and ErbB4 Intracellular Domain in the Nucleus
The findings that ErbB4 cleavage was necessary for its effect on HIF-1α activity raised the possibility that the γ-secretase-released soluble ErbB4 ICD and HIF-1α could physically associate. To experimentally address this, both co-localization and co-precipitation studies were carried out. Immunofluorescence staining detected by confocal microscopy demonstrated clear co-localization of the two proteins in the nuclei of COS-7 transfectants expressing HIF-1α and ErbB4 ICD constructs (Fig. 5A). In quantitative image analysis of confocal data, 27 ± 10% of pixels positive for ErbB4 were also positive for HIF-1α, and 72 ± 19% of pixels positive for HIF-1α were also positive for ErbB4 (n = 16).
FIGURE 5.
Interaction of HIF-1α with ErbB4 ICD. A, COS-7 cells were transfected with plasmids encoding HA-tagged ErbB4 ICD of CYT-2-type (ICD2) and HIF-1α. Colocalization of the two proteins was analyzed by immunofluorescence staining with antibodies against HA (ErbB4; red) and HIF-1α (green) and confocal microscopy. No specific signals were observed when vector control cells were stained. Nuclei were stained with DAPI (blue). B, proximity ligation assay of endogenous proteins in MCF-7 cells treated for 24 h with CoCl2. Analysis was carried out using antibodies against ErbB4 and HIF-1α, or ErbB4 alone (negative control). Arrows indicate signals positive for proximity of the epitopes in ErbB4 and HIF-1α. Nuclei were stained with DAPI (blue). C, quantification of the proximity ligation assay data (n = 34 for anti-ErbB4 control analysis; n = 92 for anti-ErbB4/anti-HIF-1α analysis). Columns represent the mean number of signals per cell ± S.E. D, co-immunoprecipitation of HIF-1α with ErbB4. Lysates of COS-7 transfectants expressing or not HA-tagged ErbB4 ICD2 and HIF-1α were immunoprecipitated with anti-HA and analyzed by Western blotting with anti-HIF. Membranes were reprobed with anti-ErbB4. HIF-1α expression was controlled from cell lysates with anti-HIF. E, deletion constructs of ErbB4 ICD2 and HIF-1α. Numbers of amino acids are indicated. F, GST pull-down of recombinant ErbB4 with HIF-GST. In vitro translated HA-tagged ErbB4 ICD2 constructs including the whole ICD of CYT-2 type (aa 676–1292), N-terminally truncated ICD2 (aa 997–1292) or C-terminally truncated ICD2 (aa 676–996) were co-precipitated with bacterially produced HIF-1α-GST fusion protein and visualized by Western blotting with anti-HA antibodies. Loading was controlled by anti-HIF and input of ErbB4 proteins by anti-HA. G, GST pull-down of recombinant HIF with ErbB4-GST. In vitro translated HIF-1α constructs including the indicated amino acids were co-precipitated with bacterially produced recombinant ErbB4 ICD2-GST fusion protein, and visualized by Western blotting with anti-HIF antibodies (lanes 6–10). Lanes 1–5 show Western analysis of the input of HIF deletion constructs. Input of the ICD2-GST protein was controlled by Western analysis with anti-ErbB4. H, effect of NRG-1 on the stability of HIF-1α constructs. MCF-7 transfectants expressing the indicated HIF-1α constructs were treated with or without NRG-1 for 20 h. HIF-1α protein levels were analyzed by Western blotting.
In situ proximity ligation assay with antibodies recognizing endogenous ErbB4 and HIF-1α indicated an interaction of the two proteins in the nuclei of CoCl2-treated MCF-7 cells (Fig. 5, B and C). Consistently, overexpressed HIF-1α also co-precipitated with ErbB4 ICD (Fig. 5D). Moreover, bacterially expressed recombinant HIF-1α interacted with a bacterially expressed ErbB4 ICD-GST fusion protein in an in vitro GST pull-down experiment (supplemental Fig. S4A), and endogenous ErbB4 co-precipitated with endogenous HIF-1α in hypoxic (1% O2) MCF-7 cells (supplemental Fig. S4B).
The interaction sites in both ErbB4 and HIF-1α were mapped using in vitro translated deletion constructs of ErbB4 and of HIF-1α (Fig. 5E). In GST pull-down experiments, HIF-1α-GST associated with C-terminally truncated ErbB4 ICD (ICD2-ΔC; consisting of amino acids 676–996) (Fig. 5F, lane 4) but not with N-terminally truncated ICD (ICD2-ΔN; amino acids 997–1292) (Fig. 5F, lane 3). In a reciprocal experiment with ErbB4 ICD-GST and a deletion series of in vitro translated HIF-1α constructs, most HIF-1α binding was lost when amino acids 1–174 were deleted (Fig. 5G, lane 7). The interaction was totally abrogated by additional deletion of amino acids 175–343 (Fig. 5G, lane 8). To test whether the physical interaction of HIF-1α with ErbB4 was associated with the ErbB4-promoted accumulation of HIF-1α, HIF-1α deletion constructs were expressed in MCF-7 cells. Similarly to the HIF-1α domains necessary for ErbB4 binding, deletion of amino acids 1–174 of HIF-1α almost totally prevented NRG-1-stimulated HIF-1α accumulation (Fig. 5H). These findings demonstrate that the ICD of ErbB4 physically interacts with HIF-1α in the nucleus.
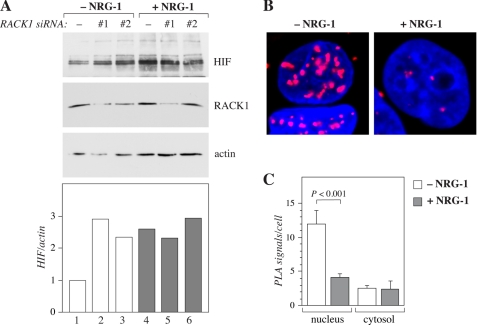
RACK1 in the Regulation of HIF-1α by ErbB4
Interestingly, RACK1 has been shown to promote HIF-1α degradation by interacting with the amino acids 81–200 of HIF-1α (9), partially overlapping with the amino acids 1–174 most critical for the interaction between HIF-1α and ErbB4. RACK1 has also been described to promote proteasomal degradation of HIF-1α by a mechanism independent of proline hydroxylation and VHL (9).
To address the role of RACK1 in HIF-1α accumulation in MCF-7 cells, RACK1 expression was down-regulated by RNA interference. Targeting of endogenous RACK1 led to accumulation of endogenous HIF-1α protein (Fig. 6A, lanes 1–3), indicating that RACK1 indeed promoted HIF-1α degradation in these cells. However, while ErbB4 activation by NRG-1 promoted HIF-1α accumulation to the same level as RACK1 down-regulation, no further accumulation of HIF-1α was observed when NRG-1 treatment and RACK1 siRNAs were combined (Fig. 6A, lanes 4–6). These finding are consistent with a model in which ErbB4 activation interferes with RACK1-mediated degradation of HIF-1α. Indeed, ErbB4 stimulation by NRG-1 reduced the interaction of endogenous HIF-1α with endogenous RACK1 in the nuclei of CoCl2-treated MCF-7 cells in an in situ proximity ligation assay (Fig. 6, B and C). These observations indicate that the mechanism by which ErbB4 activity suppresses HIF-1α degradation involves RACK1 and that ErbB4 may promote HIF-1α stability by disrupting an interaction between HIF-1α and RACK1.
FIGURE 6.
Role of RACK1 in the regulation of HIF-1α by ErbB4. A, MCF-7 cells were transfected with a plasmid encoding HIF-1〈 and with a nonspecific control siRNA (−) or with two different RACK1-targeted siRNAs (#1 and #2). The cells were stimulated with or without NRG-1 for 18 h, and analyzed by Western blotting. B, proximity ligation assay of endogenous proteins in MCF-7 cells treated for 24 h with CoCl2 and for 4 h with or without NRG-1. Analysis was carried out using antibodies against RACK1 and HIF-1α. Proximity of the epitopes is indicated by red fluorescent signals. Nuclei were stained with DAPI (blue). C, quantification of the proximity ligation assay data (n = 40 for -NRG-1; n = 35 for +NRG-1). Columns represent the mean number of signals per cell ± S.E.
DISCUSSION
Our findings indicate that an interaction between ErbB4 and HIF-1α is a biologically significant mechanism to promote HIF-regulated responses. These observations represent the first description of a direct physical interaction between a RTK and HIF-1α. They also provide a novel mechanism of cross-talk between the ErbB and HIF signaling systems, two protein families with major significance in developmental and tumor biology and in clinical applications for cancer therapy.
A well-characterized mechanism to regulate HIF-1α level is the oxygen-induced proline hydroxylation of HIF-1α leading to VHL-dependent ubiquitination and proteasomal degradation of HIF-1α (40). There are, however, indications that HIF-1α level is additionally regulated by oxygen-independent mechanisms (42). Here, we identified ErbB4 activation as a novel oxygen-independent HIF-regulatory mechanism. ErbB4 activation promoted HIF-1α accumulation in normoxia but also resulted in an additive increase in HIF-1α protein levels when combined to hypoxia, or when the PHD/VHL pathway was either chemically or genetically inactivated. To quantitatively address the relative contributions of the two pathways, a densitometric analysis of the Western analyses of RCC4 cells with and without VHL and/or ErbB4 was carried out (Fig. 3D). Inactivation of VHL resulted in a 15-fold and overexpression of ErbB4 in a 3-fold increase in the HIF-1α level, respectively. These observations suggest that the VHL pathway is quantitatively more significant although the minor effect achieved by ErbB4 was still shown to be sufficient for several biological effects such as enhancement of HIF-regulated transcription. The quantitative estimation about the relative significance of ErbB4 for HIF-1α accumulation is also consistent with the ∼3-fold increase in the amount of immunohistochemically detected HIF-1α in vivo in wild-type mouse mammary glands when compared with mammary glands with targeted Erbb4 alleles.
An interaction between ErbB4 and HIF-1α was necessary for the ErbB4-stimulated accumulation of HIF-1α as experiments with HIF-1α deletion series demonstrated that similar regions of HIF-1α were required both for mutual interaction and for ErbB4-stimulated HIF-1α accumulation. The interaction was likely to be direct as it was also observed when bacterially expressed recombinant proteins were analyzed in GST-pull-down experiments. The domains needed for the interaction were mapped to the kinase domain of ErbB4 ICD (amino acids 676–996) and to the N-terminal part of HIF-1α (amino acids 1–174). As no model or structure of N-terminal region of HIF-1α has been published, the actual nature and the potential residues involved in the interaction remain elusive. The kinase domain of ErbB4 is relatively well-conserved within the ErbB family of RTKs (43), and the other ErbB receptors have also been found in the nucleus as full-length receptors (44), suggesting that the role of the other ErbB receptors as direct regulators of nuclear HIF-1α should also be considered in future experiments.
Interestingly, it has recently been reported that RACK1 interacts with HIF-1α at amino acids 81–200 partially overlapping with the amino acids 1–174 mediating binding to ErbB4, and that this interaction with RACK1 promotes oxygen-independent proteasomal degradation of HIF-1α (9). In support for a role of RACK1 in ErbB4-dependent regulation of HIF-1α, the siRNA-mediated RACK1 knock-down blocked NRG-1-induced HIF-1α accumulation. Moreover, NRG-1 reduced the amount of RACK1/HIF-1α complex suggesting that ErbB4 regulates HIF-1α stability by interfering with the interaction between RACK1 and HIF-1α.
HIF-1α accumulation and transcriptional activity was only promoted by the cleavable JM-a-type of ErbB4 isoforms, suggesting that ErbB4 cleavage by TACE-like sheddases that process the JM-a isoforms, but not the JM-b isoforms (22, 23, 45), was needed. Indeed, a chemical inhibitor of γ-secretase down-regulated HIF-1α accumulation and activity stimulated by the ErbB4 activation. In addition, ErbB4 ICD co-localized and interacted with HIF-1α in the nuclei. These observations suggest that the mechanism by which ErbB4 activity enhances HIF signaling involves release of a soluble ErbB4 ICD, translocation to the nucleus, and direct interaction with nuclear HIF-1α leading to suppression of RACK1-dependent proteasomal degradation of HIF-1α.
Targeted Erbb4 and Hif1a null mice had similar phenotypes in lactating mammary glands (6, 19, 28), strongly indicating that the two signaling systems also interact in vivo. Analysis of conditionally targeted Erbb4-null mice demonstrated that expression of ErbB4 is indeed necessary to maintain normal HIF-1α protein levels in the lactating mammary gland. Our in silico analysis further indicated that expression of ErbB4 or its ligand NRG-1 correlated with expression of HIF-1α-regulated genes in a number of normal and malignant human tissues. While the specific contribution of the ErbB4/HIF interaction for the biology of each of these tissues needs to be experimentally addressed, the interaction may e.g. augment cellular adaptation to hypoxia, as well as enhance expression of HIF-regulated genes in normoxic conditions. Our unpublished observations indicate that cleavable ErbB4 overexpressed in breast cancer cells is sufficient to promote typical HIF-regulated responses, such as induced VEGF-A expression, endothelial chemotaxis and tumor angiogenesis, and increased cancer cell survival in hypoxic conditions in vitro and in xenografts in vivo. Taken together, these findings suggest that the interaction between ErbB4 HIF-1α is biologically significant in vivo.
Supplementary Material
Acknowledgments
We thank Maria Tuominen, Minna Santanen, and Mika Savisalo for excellent technical assistance and John Eriksson and Johanna Ivaska for valuable comments.
This work was supported by Academy of Finland, Finnish Cancer Organizations, Foundation of the Finnish Cancer Institute, Marie Curie Canceromics (MEXT-CT-2003-2728), Sigrid Juselius Foundation, Turku University Foundation, and Turku University Central Hospital.

This article contains supplemental Figs. S1–S4 and Tables S1–S2.
- HIF-1α
- hypoxia-inducible factor-1α
- HRE
- hypoxia response elements
- CYT
- cytoplasmic
- PI3-K
- phosphoinositide 3-kinase
- PLA
- proximity ligation assay.
REFERENCES
- 1. Semenza G. L., Wang G. (1992) A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol. Cell Biol. 12, 5447–5454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pouysségur J., Dayan F., Mazure N. (2006) Hypoxia signaling in cancer and approaches to enforce tumour regression. Nature 441, 437–443 [DOI] [PubMed] [Google Scholar]
- 3. Semenza G. (2001) Hypoxia-inducible factor 1: oxygen homeostasis and disease pathophysiology. Trends Mol. Med. 7, 345–350 [DOI] [PubMed] [Google Scholar]
- 4. Iyer N. V., Kotch L. E., Agani F., Leung S. W., Laughner E., Wenger R. H., Gassmann M., Gearhart J. D., Lawler A. M., Yu A. Y., Semenza G. L. (1998) Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1α. Genes Dev. 12, 149–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ryan H. E., Lo J., Johnson R. S. (1998) HIF-1α is required for solid tumor formation and embryonic vascularization. EMBO J. 17, 3005–3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Seagroves T. N., Hadsell D., McManaman J., Palmer C., Liao D., McNulty W., Welm B., Wagner K. U., Neville M., Johnson R. S. (2003) HIF1α is a critical regulator of secretory differentiation and activation, but not vascular expansion, in the mouse mammary gland. Development 130, 1713–1724 [DOI] [PubMed] [Google Scholar]
- 7. Kaelin W. J., Jr., Ratcliffe P. J. (2008) Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol. Cell 30, 393–402 [DOI] [PubMed] [Google Scholar]
- 8. Yee Koh M., Spivak-Kroizman T. R., Powis G. (2008) HIF-1 regulation: not so easy come, easy go. Trends Biochem. Sci. 33, 526–534 [DOI] [PubMed] [Google Scholar]
- 9. Liu Y. V., Baek J. H., Zhang H., Diez R., Cole R. N., Semenza G. L. (2007) RACK1 competes with HSP90 for binding to HIF-1α and is required for O(2)-independent and HSP90 inhibitor-induced degradation of HIF-1α. Mol. Cell 25, 207–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Koh M. Y., Darnay B. G., Powis G. (2008) Hypoxia-associated factor, a novel E3-ubiquitin ligase, binds and ubiquitinates hypoxia-inducible factor 1α, leading to its oxygen-independent degradation. Mol. Cell Biol. 28, 7081–7095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hynes N. E., MacDonald G. (2009) ErbB receptors and signaling pathways in cancer. Curr. Opin. Cell Biol. 21, 177–184 [DOI] [PubMed] [Google Scholar]
- 12. Hollmén M., Elenius K. (2010) Potential of ErbB4 antibodies for cancer therapy. Future Oncol. 6, 37–53 [DOI] [PubMed] [Google Scholar]
- 13. García-Rivello H., Taranda J., Said M., Cabeza-Meckert P., Vila-Petroff M., Scaglione J., Ghio S., Chen J., Lai C., Laguens R. P., Lloyd K. C., Hertig C. M. (2005) Dilated cardiomyopathy in Erb-b4-deficient ventricular muscle. Am. J. Physiol. Heart Circ Physiol. 289, H1153–H1160 [DOI] [PubMed] [Google Scholar]
- 14. Silberberg G., Darvasi A., Pinkas-Kramarski R., Navon R. (2006) The involvement of ErbB4 with schizophrenia: association and expression studies. Am. J. Med. Genet. B Neuropsychiatr. Genet. 141, 142–148 [DOI] [PubMed] [Google Scholar]
- 15. Gassmann M., Casagranda F., Orioli D., Simon H., Lai C., Klein R., Lemke G. (1995) Aberrant neural and cardiac development in mice lacking the ErbB4 neuregulin receptor. Nature 378, 390–394 [DOI] [PubMed] [Google Scholar]
- 16. Veikkolainen V., Naillat F., Railo A., Chi L., Manninen A., Hohenstein P., Hastie N., Vainio S., Elenius K. (2012) ErbB4 modulates tubular cell polarity and lumen diameter during kidney development. J. Am. Soc. Nephrol. 23, 112–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sardi S. P., Murtie J., Koirala S., Patten B. A., Corfas G. (2006) Presenilin-dependent ErbB4 nuclear signaling regulates the timing of astrogenesis in the developing brain. Cell 127, 185–197 [DOI] [PubMed] [Google Scholar]
- 18. Tidcombe H., Jackson-Fisher A., Mathers K., Stern D., Gassmann M., Golding J. P. (2003) Neural and mammary gland defects in ErbB4 knockout mice genetically rescued from embryonic lethality. Proc. Natl. Acad. Sci. U.S.A. 100, 8281–8286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jones F. E., Welte T., Fu X. Y., Stern D. F. (1999) ErbB4 signaling in the mammary gland is required for lobuloalveolar development and Stat5 activation during lactation. J. Cell Biol. 147, 77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Williams C. C., Allison J. G., Vidal G. A., Burow M. E., Beckman B. S., Marrero L., Jones F. E. (2004) The ERBB4/HER4 receptor tyrosine kinase regulates gene expression by functioning as a STAT5A nuclear chaperone. J. Cell Biol. 167, 469–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ni C. Y., Murphy M. P., Golde T. E., Carpenter G. (2001) γ-Secretase cleavage and nuclear localization of ErbB-4 receptor tyrosine kinase. Science 294, 2179–2181 [DOI] [PubMed] [Google Scholar]
- 22. Määttä J. A., Sundvall M., Junttila T. T., Peri L., Laine V. J., Isola J., Egeblad M., Elenius K. (2006) Proteolytic cleavage and phosphorylation of a tumor-associated ErbB4 isoform promote ligand-independent survival and cancer cell growth. Mol. Biol. Cell 17, 67–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Elenius K., Corfas G., Paul S., Choi C. J., Rio C., Plowman G. D., Klagsbrun M. (1997) A novel juxtamembrane domain isoform of HER4/ErbB4. Isoform-specific tissue distribution and differential processing in response to phorbol ester. J. Biol. Chem. 272, 26761–26768 [DOI] [PubMed] [Google Scholar]
- 24. Elenius K., Choi C. J., Paul S., Santiestevan E., Nishi E., Klagsbrun M. (1999) Characterization of a naturally occurring ErbB4 isoform that does not bind or activate phosphatidyl inositol 3-kinase. Oncogene 18, 2607–2615 [DOI] [PubMed] [Google Scholar]
- 25. Sundvall M., Peri L., Määttä J. A., Tvorogov D., Paatero I., Savisalo M., Silvennoinen O., Yarden Y., Elenius K. (2007) Differential nuclear localization and kinase activity of alternative ErbB4 intracellular domains. Oncogene 26, 6905–6914 [DOI] [PubMed] [Google Scholar]
- 26. Junttila T. T., Sundvall M., Lundin M., Lundin J., Tanner M., Härkönen P., Joensuu H., Isola J., Elenius K. (2005) Cleavable ErbB4 isoform in estrogen receptor-regulated growth of breast cancer cells. Cancer Res. 65, 1384–1393 [DOI] [PubMed] [Google Scholar]
- 27. Tvorogov D., Sundvall M., Kurppa K., Hollmén M., Repo S., Johnson M. S., Elenius K. (2009) Somatic mutations of ErbB4: selective loss-of-function phenotype affecting signal transduction pathways in cancer. J. Biol. Chem. 284, 5582–5591 [DOI] [PubMed] [Google Scholar]
- 28. Long W., Wagner K. U., Lloyd K. C., Binart N., Shillingford J. M., Hennighausen L., Jones F. E. (2003) Impaired differentiation and lactational failure of Erbb4-deficient mammary glands identify ERBB4 as an obligate mediator of STAT5. Development 130, 5257–5268 [DOI] [PubMed] [Google Scholar]
- 29. Ruifrok A. C., Johnston D. A. (2001) Quantification of histochemical staining by color deconvolution. Anal. Quant. Cytol. Histol. 23, 291–299 [PubMed] [Google Scholar]
- 30. Junttila T. T., Laato M., Vahlberg T., Söderström K. O., Visakorpi T., Isola J., Elenius K. (2003) Identification of patients with transitional cell carcinoma of the bladder overexpressing ErbB2, ErbB3, or specific ErbB4 isoforms: real-time reverse transcription-PCR analysis in estimation of ErbB receptor status from cancer patients. Clin. Cancer Res. 9, 5346–5357 [PubMed] [Google Scholar]
- 31. Iivanainen E., Paatero I., Heikkinen S. M., Junttila T. T., Cao R., Klint P., Jaakkola P. M., Cao Y., Elenius K. (2007) Intra- and extracellular signaling by endothelial neuregulin-1. Exp. Cell Res. 313, 2896–2909 [DOI] [PubMed] [Google Scholar]
- 32. Kainulainen V., Sundvall M., Määttä J. A., Santiestevan E., Klagsbrun M., Elenius K. (2000) A natural ErbB4 isoform that does not activate phosphoinositide 3-kinase mediates proliferation but not survival or chemotaxis. J. Biol. Chem. 275, 8641–8649 [DOI] [PubMed] [Google Scholar]
- 33. Kilpinen S., Autio R., Ojala K., Iljin K., Bucher E., Sara H., Pisto T., Saarela M., Skotheim R. I., Björkman M., Mpindi J. P., Haapa-Paananen S., Vainio P., Edgren H., Wolf M., Astola J., Nees M., Hautaniemi S., Kallioniemi O. (2008) Systematic bioinformatic analysis of expression levels of 17,330 human genes across 9,783 samples from 175 types of healthy and pathological tissues. Genome Biol. 9, R139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Saldanha A. (2004) Java Treeview, extensible visualization of microarray data. Bioinformatics 20, 3246–3248 [DOI] [PubMed] [Google Scholar]
- 35. Schofield C. J., Ratcliffe P. J. (2004) Oxygen sensing by HIF hydroxylases. Nat. Rev. Mol. Cell Biol. 5, 343–354 [DOI] [PubMed] [Google Scholar]
- 36. Egeblad M., Mortensen O. H., van Kempen L. C., Jäättelä M. (2001) BIBX1382BS, but not AG1478 or PD153035, inhibits the ErbB kinases at different concentrations in intact cells. Biochem. Biophys. Res. Commun. 281, 25–31 [DOI] [PubMed] [Google Scholar]
- 37. Zhong H., Chiles K., Feldser D., Laughner E., Hanrahan C., Georgescu M. M., Simons J. W., Semenza G. L. (2000) Modulation of hypoxia-inducible factor 1α expression by the epidermal growth factor/phosphatidylinositol 3-kinase/PTEN/AKT/FRAP pathway in human prostate cancer cells: implications for tumor angiogenesis and therapeutics. Cancer Res. 60, 1541–1545 [PubMed] [Google Scholar]
- 38. Laughner E., Taghavi P., Chiles K., Mahon P. C., Semenza G. L. (2001) HER2 (neu) signaling increases the rate of hypoxia-inducible factor 1α (HIF-1α) synthesis: novel mechanism for HIF-1-mediated vascular endothelial growth factor expression. Mol. Cell Biol. 21, 3995–4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ivan M., Kondo K., Yang H., Kim W., Valiando J., Ohh M., Salic A., Asara J. M., Lane W. S., Kaelin W. J., Jr. (2001) HIFα targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292, 464–468 [DOI] [PubMed] [Google Scholar]
- 40. Jaakkola P., Mole D. R., Tian Y. M., Wilson M. I., Gielbert J., Gaskell S. J., Kriegsheim Av, Hebestreit H. F., Mukherji M., Schofield C. J., Maxwell P. H., Pugh C. W., Ratcliffe P. J. (2001) Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292, 468–472 [DOI] [PubMed] [Google Scholar]
- 41. Masson N., Willam C., Maxwell P. H., Pugh C. W., Ratcliffe P. J. (2001) Independent function of two destruction domains in hypoxia-inducible factor-α chains activated by prolyl hydroxylation. EMBO J. 20, 5197–5206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Harris A. (2002) Hypoxia, a key regulatory factor in tumour growth. Nat. Rev. Cancer 2, 38–47 [DOI] [PubMed] [Google Scholar]
- 43. Plowman G. D., Culouscou J. M., Whitney G. S., Green J. M., Carlton G. W., Foy L., Neubauer M. G., Shoyab M. (1993) Ligand-specific activation of HER4/p180erbB4, a fourth member of the epidermal growth factor receptor family. Proc. Natl. Acad. Sci. U.S.A. 90, 1746–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang S. C., Hung M. C. (2009) Nuclear translocation of the epidermal growth factor receptor family membrane tyrosine kinase receptors. Clin. Cancer Res. 15, 6484–6489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rio C., Buxbaum J. D., Peschon J. J., Corfas G. (2000) Tumor necrosis factor-α-converting enzyme is required for cleavage of erbB4/HER4. J. Biol. Chem. 275, 10379–10387 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.