Background: HIV-1 integration is promoted by viral integrase and its cellular cofactors.
Results: Nucleoporin 62 interacts with HIV-1 integrase in chromatin, and shRNA knockdown of nucleoporin 62 was able to impair integrase chromatin association and viral replication.
Conclusion: Interaction of nucleoporin 62 and HIV-1 integrase contributes to viral DNA integration.
Significance: A new nucleoporin was identified as an integrase-binding cofactor required for HIV-1 integration and replication.
Keywords: Chromatin, Integrase, Nuclear Pore, Retrovirus, RNA Interference (RNAi), HIV-1, Chromatin Association, Integration, Nucleoporin 62, Protein-Protein Interaction
Abstract
HIV-1 integration is promoted by viral integrase (IN) and its cellular cofactors. The lens epithelium-derived growth factor (LEDGF/p75), an IN interacting cellular cofactor, has been shown to play an important role in HIV-1 chromatin targeting and integration. However, whether other cellular cofactors are also involved in viral replication steps is still elusive. Here, we show that nucleoporin 62 (Nup62) is a chromatin-bound protein and can specifically interact with HIV-1 IN in both soluble nuclear extract and chromatin-bound fractions. The knockdown of Nup62 by shRNA reduced the association of IN with host chromatin and significantly impaired viral integration and replication in HIV-1-susceptible cells. Furthermore, the expression of the IN-binding region of Nup62 in CD4+ T cells significantly inhibited HIV-1 infection. Taken together, these results indicate that the cellular Nup62 is specifically recruited by HIV-1 IN and contribute to an efficient viral DNA integration.
Introduction
The integration of viral DNA into the host genome is an absolute requirement for the completion of the HIV-1 life cycle and progeny virus production. To achieve this integration, the reverse-transcribed HIV-1 DNA, as a component of a large preintegration complex (PIC),6 must translocate into the nucleus through nuclear pore complexes (NPC), attach to chromatin, and integrate to specific sites of the host genome. Several viral proteins, especially integrase (IN), and a number of cellular cofactors are involved in this long and complex journey.
HIV-1 IN is a critical enzymatic molecule that catalyzes the integration process, including 3′-processing of viral DNA in the cytoplasm and the strand transfer reaction at the integration site. In addition, IN also plays an important role during HIV-1 nuclear import (1–5), although the exact nuclear import pathway(s) undertaken by IN has yet to be defined. Accumulated evidence clearly indicates that the nuclear import function of IN is essential for successful HIV-1 infection, not only in nondividing cells but also in dividing cells, suggesting that the proper transport of the HIV-1 PIC through nuclear pore may facilitate post-nuclear entry steps such as chromatin targeting and integration. Following nuclear translocation, the PIC-associated IN is well documented to achieve a secure and cell cycle-persistent chromatin attachment to support HIV-1 DNA integration. It is also evident that the chromosomal site of viral integration is not random. The lentiviruses favor insertion of their genomes into active transcription units of the host chromosome (6, 7), whereas in murine leukemia virus the viral integration favors chromatin loci near transcription start sites and CpG islands. The integration model of tethering has been proposed to explain integration site selection, and any cellular protein that is able to bind both chromatin and PIC-associated IN may represent a candidate tethering factor (8). To date, numerous cellular proteins have been proposed, but only LEDGF/p75 is proved to be a bona fide tethering protein (9–11). LEDGF/p75 not only provides a means for attachment of viral cDNA to chromatin, but it is also required for directing HIV PICs to active transcription sites in the chromatin (10, 12–16). However, other studies have indicated that the specific knockdown or knock-out of LEDGF does not completely abolish HIV-1 cDNA integration (12, 16). In addition, HIV-1 IN was shown to be able to bind to chromatin in yeast cells, which have no LEDGF homolog, and induce a lethal phenotype (17). All of these observations suggest that there may be other unidentified cellular cofactors of IN that also contribute to IN chromatin targeting and integration, either together with LEDGF/p75 or by a LEDGF/p75-independent pathway.
Nucleoporins (Nups) are the major components of the nuclear pore complex (NPC), a macromolecular structure that form channels spanning the double lipid bilayer of the nuclear envelope (18, 19). The Nups not only contribute to NPC assembly, overall architecture, and permeability barriers but also play an important role in the active translocation of macromolecules through the NPC. In addition, the regulation of gene expression was recently identified as an important function of Nup. The first evidence for the transcriptional modulation role of Nup is the finding that the oncogenic phenylalanine-glycine (FG) repeat of Nup214 or Nup98 fuses to homeodomain transcription factor, resulting in the transactivation or transrepression of gene expression (20–24). Second, chromatin interaction with the nuclear envelope can be observed both at the nuclear lamina and at the NPCs (25), and studies in yeast have revealed that Nups can bind to transcriptionally active genes (26, 27). Interestingly, several studies have also shown that some nucleoporins (e.g. Nup98, Nup50, and Nup153) are mobile and localize to the nucleoplasm (28–32), suggesting that Nup-chromatin interaction may also occur in the nucleoplasm. The most recent evidence has come from reports that demonstrated the specific interaction of Nups, such as Sec13, Nup98, Nup88, Nup62, and Nup50, with transcriptionally active genes of chromatin at the NPC or within the nucleoplasm of embryonic Drosophila cells (33, 34). Importantly, several observations suggest that Nup153 specifically participates in the nuclear entry of HIV-1 cDNA (35, 36), and Nup98 was also implicated in HIV nuclear import (35) or integration (37). Recently, Ocwieja et al. (38) showed that the HIV-1 nuclear import mediated by transportin-3 and RanBP2/Nup358 facilitates proper site selection in chromatin during integration, suggesting that Nups are required for HIV-1 nuclear import and subsequent integration step.
Nup62, with its homolog Nsp1p in Saccharomyces cerevisiae, is a glycosylated FG-Nup that is located near the central channel of the NPC together as a complex with Nup58, Nup45, and Nup54 (39–41). Nup62 consist of two domains, the N-terminal FG-rich region (amino acids 1–327) and the C-terminal α-helical coiled-coil domain (amino acids 328–522). The C-terminal domain of Nup62 contributes to nucleocytoplasmic transport by direct interaction with nuclear transport receptors or with cargo and facilitates the anchoring of Nup62 to the NPC (42–46). Although the importance of Nup62 in nuclear transport has been intensively investigated, Han et al. (47) demonstrated that Nup62 interacts with transcription factor Sp1, suggesting that Nup62 might be involved in the transcription regulation. A recent study revealed that Nup62 is able to associate directly with active genes inside the nucleoplasm (33). In this study, we provided evidence that supports that HIV-1 IN interacts with Nup62 for an efficient viral DNA integration and replication.
EXPERIMENTAL PROCEDURES
Construction of Expression Plasmids
To construct SVCMV-T7-Nup62(1–325) or T7-Nup62(328–522) plasmid, the cDNAs encoding Homo sapiens Nup62(1–325) and Nup62(328–522) were PCR-amplified with the corresponding primers from pCMV6-entry-Nup62-myc (OriGene Technologies) and cloned into the SVCMVin-T7 vector at the 3′ end of a T7 tag using BamHI/HindIII or BglII/HindIII restriction sites. To construct the full-length T7-Nup62(1–522), a cDNA fragment encoding the C-terminal region of Nup62 (amino acids 300–522) was digested from the pCMV6-entry-Nup62-myc using BamHI and NotI and cloned into the T7-Nup62(1–327) plasmid by using the same restriction enzymes. To generate a lentiviral vector pEF1-T7-Nup62(328–522)-puro, the corresponding cDNA from SVCMVin-T7-Nup62–328-522 was digested and subcloned into the pEF1-T7-Puro vector (48) through the BamHI and ClaI sites.
To generate ProLabel (PL)-IN, the cDNA encoding full-length IN was digested from AcGFP-IN (49) using BamHI and cloned into the ProLabel-C vector (Clontech). A ProLabel-tagged HA-APOBEC3G (A3G) expression plasmid (PL-HA-A3G) was also constructed by PCR amplifying the HA-A3G cDNA from CMVin-HA-A3G (48) and subcloned into the ProLabel-C vector at the KpnI and SmaI restriction sites. To generate GST-IN construct, an IN cDNA from pNL-4.3 HIV-1 provirus was PCR-amplified and cloned into pGEX4-GST vector at the BamHI/SmaI sites. AcGFP-IN, MA-YFP, Vpr-GFP, CMV-Vpr-RT-IN-PL, the HIV-1 proviral clones, including pNL4.3-Nef+/GFP+ (pNL4.3-GFP), HxBruΔRI, and pNL-BruΔBgl/Luc, have been described previously (49, 50).
Antibodies and Chemicals
Nup98 and Nup88 mouse monoclonal antibodies, and rabbit anti-Nup50 and anti-TPR polyclonal antibodies were purchased from Santa Cruz Biotechnology. The mouse anti-histone H1 monoclonal antibody was obtained from Abcam. The rabbit anti-GFP polyclonal antibody, mouse anti-T7 monoclonal antibody, rabbit anti-HA antibody, and horseradish peroxidase (HRP)-conjugated anti-GFP antibody have been described previously (50). The rat and rabbit anti-Nup62, monoclonal anti-α-tubulin and anti-γ-actin antibodies were obtained from Sigma. The ProLabel detection kit II was obtained from Clontech.
Cell Culture and Transfection
Human embryonic kidney 293T cells and CD4+ C8166 T lymphocyte cells were maintained as described previously (50). Human monocytes (purchased from Advanced Biotechnologies, Inc) were allowed to differentiate to macrophages in DMEM containing 20% FBS and 10 ng/ml macrophage colony-stimulating factor (M-CSF; R&D Systems) for 1 week. The DNA transfection experiments in 293T cells were using the standard calcium phosphate DNA precipitation method.
Generation of the Nup62 Knockdown (KD) Cell Line
The pLKO1 lentiviral vector plasmids containing the Nup62 siRNA hairpin, which consists of a 21-base stem and 6-base loop, were obtained from Open Biosystems. The sense oligonucleotide sequence was 5′-GCAGATCTGCAAGATCCTCAA-3′, which targeted nucleotides 1415–1436 of Nup62. The Nup62 shRNA pseudotyped lentiviral vector particle) produced in 293T cells were used to establish cell lines stably expressing shNup62 in 293T or C8166 T cells, as described previously (50). To obtain Nup62-KD and empty vector-treated monocyte-derived macrophages (MDMs), the MDMs were treated with empty or shNup62 lentiviral vector particles, followed by Nup62 siRNA treatment 24 h later. The Nup62-specific stealth siRNA duplex (Invitrogen) targeted Nup62 mRNA nucleotides 1635–1659 (5′UCAAGGACAUCAUCGAGCACCUGAA3′) and was transfected into MDMs using LipofectamineTM RNAiMAX reagent (Invitrogen). The knockdown level of Nup62 in each cell type was evaluated by Western blot (WB) using an anti-Nup62 antibody.
To generate the CD4+ T cell line stably expressing T7-Nup62–328-522 peptide, the C8166 T cells were transduced by a VSV-G pseudotyped lentiviral vectors carrying the T7-Nup62-(328–522) gene. Then the transduced cells were selected using puromycin for 1 week and were ready for the infection experiment.
In Vitro Binding Assays and Cell-based Coimmunoprecipitation (Co-IP) Experiments
In Vitro Binding Studies
GST and GST-IN fusion protein were purified from Escherichia coli BL21 cell lysates on glutathione-agarose beads, and the interactions between GST-IN and endogenous Nups (TPR, Nup98, Nup88, Nup62, and Nup50) were assessed as described previously (50, 51).
Co-IP Experiments
The Co-IP assay was carried out as described previously (50, 51). Briefly, GFP, GFP-IN, MA-YFP, or Vpr-GFP was expressed in 293T cells. The cells were then lysed in 0.5% Nonidet P-40 lysis buffer and immunoprecipitated with either the rabbit anti-GFP or anti-Nup62 antibody to pull down the bound proteins that were subsequently detected by WB using mouse anti-Nup62 or anti-GFP antibody. To detect the interaction between viral IN and the endogenous Nup62 in HIV-1-infected CD4+ C8166 T cells, HxBru or HxBru-IN-HA virus-infected C8166 T cells were lysed and immunoprecipitated with anti-HA or -Nup62 antibodies. The coprecipitated Nup62 or IN-HA was detected by WB with the corresponding antibody. The analyses for the interaction between GFP-IN and different T7-Nup62 deletion mutants were performed with immunoprecipitated anti-GFP antibody followed by WB with anti-T7 antibody.
To quantitatively analyze the Nup62-IN interaction, a chemiluminescent co-IP system was employed and carried out following the manufacturer's manual (Clontech). Briefly, 48 h after the transfection of 293T cells with PL-IN, cells were harvested, fractionated, and followed by IP with rabbit anti-Nup62 antibody. The immunoprecipitates were resuspended with lysis/complementation buffer, provided with the ProLabel detection kit II, and transferred to a 96-well plate. The substrate mix was then added, and the chemiluminescent signal was measured with a PolarStar Optima microplate reader (BMG Labtech).
Subcellular Protein Fractionation and Detection
A Thermo Scientific subcellular protein fractionation kit (Thermo Scientific, catalog no. 78840) was used to prepare cytoplasmic, nuclear soluble, and chromatin-bound protein extracts from cells. Briefly, 293T cells (transfected or nontransfected) or HIV-infected C8166 T cells were first treated with cytoplasmic extraction (CE) buffer to release of soluble cytoplasmic contents. The pellets were treated with membrane extraction buffer to dissolve plasma, mitochondria, and endoplasmic reticulum/Golgi membranes but not nuclear membranes. The intact nuclei were recovered by centrifugation, and a nuclear extraction buffer was added to yield the soluble nuclear extract (NE). The chromatin-bound nuclear proteins (CE) were released by treated nuclear pellet again with micrococcal nuclease. The final insoluble pellet was then extracted with the final reagent to obtain cytoskeletal proteins. Each subcellular fraction was subjected to WB to detect proteins by using specific antibodies or was subjected to the measurement of IN-related ProLab activity.
Virus Production and Infection
HIV-1 pNL4.3-GFP and VSV-G pseudotyped pNL-BruΔBgl/Luc/R− viruses were produced in 293T cells and used to infect Nup62-KD or the control C8166 T cell lines, as described previously (50). 48 h later, HIV replication levels were monitored by measuring HIV-1 Gag-p24 antigen or luciferase (Luc) activity. To test the effect of Nup62-KD on HIV infection in macrophages, the Nup62-KD and control MDMs were infected with macrophage-tropic pNL4.3-Bal virus as described previously (52). At different time points after infection, the viral production in the supernatant was monitored using an anti-p24 ELISA.
HIV-1 single cycle IN-PL virus was generated by cotransfecting 293T cells with the HxBruR−ΔRI provirus, in which RT and IN genes were deleted (53) and the Vpr-RT-IN-PL plasmid. An equal amount of viruses was used to infect Nup62-KD or empty vector-transduced C8166T cells for 3–4 h. Cells were then washed three times and kept in fresh medium. At different time points, the cells were collected and fractionated, and the IN-PL activity in the different subcellular fractions was measured using a ProLabelTM detection kit II.
To test the effect of the C-terminal domain polypeptide of Nup62 on HIV-1 replication, the VSV-G pseudotyped pNL4.3-Luc+ virus or Ad5-CMV-luciferase (Vector Biolabs) was used to infect 293T cells that had been transfected with different amounts of CMV-T7-Nup62(328–522) plasmid for 24 h. HIV or adenovirus replication levels were monitored by measuring Luc activity after 48 h of infection. Also, the C8166 T cells stably expressing Nup62(328–522) peptide were challenged with pNL4.3 GFP+, and HIV-p24 levels in the supernatants were detected at different time points after infection.
Real Time Quantitative PCR Analysis
The Nup62 knockdown or control C8166 cells were infected with the pNL4.3-GFP+ virus and subjected to RQ-PCR analysis. Heat-inactivated virus (pretreated at 70 °C for 45 min) was used as a negative control. Twenty hours post-infection, the viral DNA was isolated by using a QIAmp blood DNA minikit (Qiagen). Total HIV-1 DNA, two-LTR circle, and integrated DNA levels were quantified using a Mx3000PTM real time PCR system (Stratagene, CA) with the protocols described previously (50).
RESULTS
Nup62 Specifically Interacts with HIV-1 IN
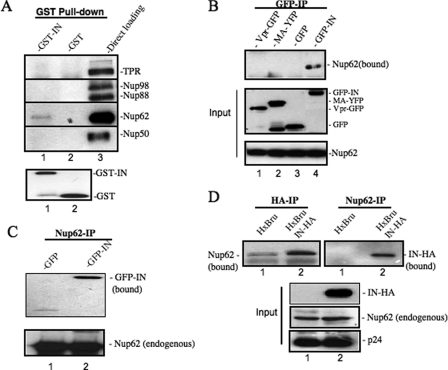
IN plays a critical role in ensuring that nuclear import and integration steps proceed. To investigate whether Nups could participate in the above viral replication steps, we first examined the potential interaction between HIV-1 IN and various nucleoporins. The interaction of HIV-1 IN with several previously described chromatin-bound nucleoporins, including Nup98, Nup88, Nup62, Nup50, and an intranuclear filamentous nucleoporin TPR, were tested by using a GST pulldown assay. Briefly, the purified recombinant GST-IN fusion protein was incubated with 293T cell lysate, and the IN-bound endogenous Nups after GST pulldown were detected by WB using specific antibodies. The in vitro binding data revealed that GST-IN, but not GST, was able to pull down Nup62 (Fig. 1A). To confirm this viral-cellular protein interaction and test its specificity, we expressed GFP-IN, Vpr-GFP, or MA-YFP in 293T cells and tested the interaction between these HIV-1 proteins and the endogenous Nup62 by using anti-GFP coimmunoprecipitation (50) followed by WB with anti-Nup62 antibody (Fig. 1B). Results clearly showed that the endogenous Nup62 was only coprecipitated with GFP-IN and not with Vpr-GFP or MA-YFP (Fig. 1B, upper panel). This result suggests a specific interaction between Nup62 and HIV-1 IN. Consistently, the interaction between IN and Nup62 was also detected by anti-Nup62 coimmunoprecipitation followed by an anti-GFP WB (Fig. 1C).
FIGURE 1.
Nup62 interacts with HIV-1 integrase in vitro, in cotransfected 293T cells, and in HIV-infected C8166 T cells. A, cell lysates from 293T cells were incubated with GST-IN (lane 1) or GST (lane 2) recombinant proteins. After GST pulldown, the bound proteins were analyzed by WB with a variety of nucleoporin antibodies, including anti-TPR and anti-Nup98, -88, -62, and -50. Lane 3, whole cell lysate from 293T cells. B, endogenous Nup62 interacts with HIV-1 IN and not MA or Vpr. 293T cells were transfected with GFP, GFP-IN, MA-YFP, or Vpr-GFP plasmid; 48 h later, transfected cells were lysed in 0.5% Nonidet P-40 buffer and immunoprecipitated with rabbit anti-GFP antibody. The bound Nup62 was detected by WB with anti-Nup62 antibody (upper panel). Middle and lower panels, overexpressed GFP fusion proteins or endogenous Nup62 in 1/10 of transfected cells. C, GFP or GFP-IN was transfected into 293T cells, and the cell lysates were immunoprecipitated with rabbit anti-Nup62. The bound GFP fusion protein was detected by WB with an anti-GFP antibody (upper panel). D, Nup62 interacts with HIV-1 IN in virus-infected C8166 T cells. C8166 T cells were infected with HxBru or HxBru-IN-HA viruses for 48 h, and the cell lysates were immunoprecipitated with rabbit anti-HA antibody (left upper panel) or rabbit anti-Nup62 antibody (right upper panel). The bound endogenous Nup62 or HIV-1 IN was detected by WB with specific antibody. The immunoprecipitated HA-IN was also detected by WB using a mouse anti-HA antibody (middle, panel 1). 1/20 infected C8166 cell lysates were used to detect endogenous Nup62 and HIV Gag-p24 protein by WB (middle, panel 2, and lower panel).
To further test whether this virus-host protein interaction occurs during HIV infection, we infected a CD4+ C8166 T cell line with HxBru-IN-HA (50) or HxBru virus for 48 h. The infected cells were then lysed and immunoprecipitated with anti-HA followed by WB using an anti-Nup62 antibody (Fig. 1D, left panel) or immunoprecipitated with anti-Nup62 followed by WB using an anti-HA antibody (Fig. 1D, right panel). Results clearly showed an interaction of HIV-1 IN with Nup62 in HIV-1-infected T cells (Fig. 1D, upper panel). We also observed a similar level of HIV p24 and endogenous Nup62 expression in both HxBru-IN-HA and HxBru virus-infected cells (Fig. 1D, middle and lower panels), indicating that the detected bands were not the result of increased infection or cell number in the HxBru-IN-HA sample. Therefore, we conclude that HIV-1 IN is able to specifically interact with Nup62 during HIV-1 infection.
Nup62 Knockdown Significantly Inhibits HIV-1 Infection by Affecting Viral cDNA Integration
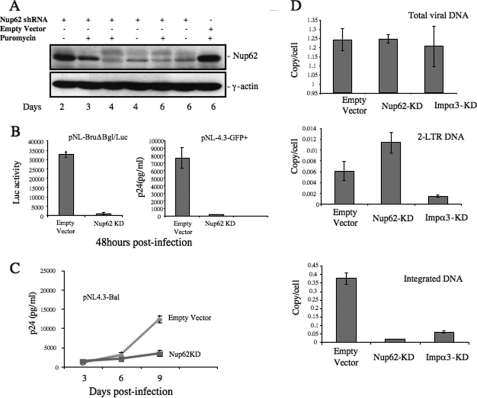
Because Nup62 is found to interact with HIV IN, we next asked whether and how Nup62 contributes to HIV-1 replication. Nup62 expression in CD4+ C8166 T cells was knocked down by using a specific Nup62-shRNA lentiviral vector, as described under “Experimental Procedures.” The down-regulation of Nup62 was verified by WB with an anti-Nup62 antibody. Results indicated that the expression of Nup62 started to decrease at day 3, and ∼80–90% of the Nup62 in transduced T cells was knocked down at days 4 and 6. Because the pLKO1-shRNA lentiviral vector contains a puromycin resistance gene, the transduced cells were selected with puromycin (0.5 mg/ml). However, the knockdown levels were not significantly affected by the presence of puromycin (Fig. 2A), possibly due to the high efficiency of transduction. We also noticed that the down-regulation of Nup62 indeed affected C8166 T cell growth, and after 10 days of Nup62-shRNA treatment, low cell viability was observed (data not shown). Therefore, at day 4 after transduction, we infected Nup62-KD T cells with equal amounts of VSV-G pseudotyped luciferase-expressing HIV-1 (pNL-BruΔBgl/Luc) (Fig. 2B, left panel) or the wild-type HIV-1 (pNL-4.3-GFP+) (Fig. 2B, right panel). After 48 h, HIV-1 infection levels were monitored by measuring the Luc activity from an equal amount of infected cells or measuring HIV Gag-p24 production in the supernatant via HIV-p24 ELISA. Results showed that viral infection was significantly inhibited, and no virus-associated luciferase activity (Fig. 2B, left panel) or viral Gag-p24 (Fig. 2B, right panel) was detected in Nup62-KD T cells after 48 h of infection. We then investigated whether Nup62 is also necessary for HIV-1 infection in nondividing primary macrophages. The MDMs from healthy donors were transduced with Nup62-shRNA lentiviral vectors followed by a Nup62 siRNA treatment 24 h later, as described under “Experimental Procedures.” Meanwhile, MDMs were treated with empty lentiviral vector and scramble siRNA as control. WB showed that about 50% of Nup62 protein expression was knocked down at 6 days post-transduction (data not shown). Different than the Nup62-shRNA treated C8166 T cells, the Nup62-KD MDMs showed a similar cell viability as compared with the empty vector-treated MDMs (data not shown), probably due to a less sufficient down-regulation of Nup62. After 4 days of transduction, MDMs were infected with the pNL4.3-Bal virus strain, and viral replication was monitored by measuring the levels of the HIV-1 Gag-p24 antigen from the supernatants at different post-infection time points (Fig. 2C). Results showed that in Nup62-KD MDMs, HIV-1 could not mediate a productive infection. After 9 days post-infection, supernatants from Nup62-KD MDMs showed a 3-fold lower Gag-p24 content compared with control cells. These data indicate that the Nup62 knockdown significantly inhibits HIV-1 infection in dividing and nondividing susceptible cells.
FIGURE 2.
Nup62 knockdown significantly inhibited HIV-1 infection by reducing viral DNA integration. A, C8166 cells were transduced with lentiviral vectors harboring Nup62 shRNA or empty vector and were subsequently analyzed for Nup62 expression by WB at different time points. γ-Actin was included as an internal control. B, inhibitory effect of Nup62-KD on HIV-1 replication in CD4+ C8166 T cells and macrophages. Nup62-KD and empty C8166 cells were infected with VSV-G pseudotyped luc-expressing HIV-1 (pNL-BruΔBgl/Luc) (left panel) or HIV-1 pNL 4.3-GFP (right panel). The Luc activity from cells or HIV Gag-p24 levels of supernatants was measured to monitor viral replication after 48 h. C, monocyte-derived macrophages from healthy donors were transduced with lentiviral vectors encoding Nup62-shRNA and Nup62-siRNA, as described under “Experimental Procedures.” At day 4 post-transduction, MDMs were infected with the pNL4.3-Bal virus strain. Viral replication was monitored by measuring the levels of the HIV-1 Gag-p24 antigen in supernatants at different time points after infection. D, Nup62-KD, importin α3-kDa (Impα3-kDa), and ScRNA-C8166 T cells were infected with HIV-1 pNL4.3 Env+/R−/Luc virus, and 20 h later, DNA was extracted from infected cells, and HIV-1 late reverse transcription products (upper panel), HIV-1 two-LTR circles (middle panel), and the integrated DNA levels (lower panel) were analyzed by RQ-PCR using corresponding primers, as described under “Experimental Procedures.” Means and standard errors are representative of the results for duplicate samples from a typical experiment and were confirmed in two other independent experiments.
To identify the step(s) affected by Nup62 knockdown during HIV-1 replication, we infected the Nup62-KD with pNL4.3-GFP+ virus and examined HIV-1 reverse transcription, two-LTR circles, and integrated DNA levels by RQ-PCR analysis, as described previously (50). In parallel, an importin α3-knockdown (Impα3-KD) cell line was used as the control cells because Impα3 has recently been demonstrated to be able to interact with HIV IN and specifically contributes to viral nuclear import (50). By RQ-PCR analysis, we found that similar levels of viral cDNA synthesis were detected in the control cells, Nup62-KD, and Impα3-KD cells after 20 h of infection (Fig. 2D, upper panel), indicating that Nup62 is not involved in the reverse transcription step. However, for the nucleus-associated two-LTR DNA circles, an approximate 1-fold increase was detected in Nup62-KD cells (Fig. 2D, middle panel), whereas the integrated viral DNA was almost undetectable as compared with control cells (Fig. 2D, lower panel). In contrast, in Impα3-KD cells, HIV-1 displayed a severe reduction of two-LTR DNA circles (4-fold) and a similar decrease of integrated DNA levels (Fig. 2D, middle and lower panels). These data suggest that the Nup62 knockdown significantly impaired viral integration. However, compared with the IN integration-deficient mutant D64E virus that generated significantly excessive two-LTR circular forms (data not shown), the nearly 1-fold increase of two-LTR circles in Nup62-KD cells also implies that the down-regulation of Nup62 may at the same time affect the nuclear translocation of viral cDNA, but to a much lesser extent compared with its effect on viral integration. In summary, these results have provided evidence that Nup62-KD in CD4+ T cells disrupts the events contributing to HIV integration.
Nup62 Binds to Chromatin and Interacts with HIV-1 IN Both in the Nuclear and Chromatin-bound Extracts
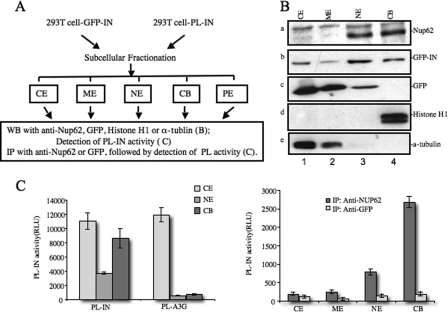
The question of how Nup62 is involved in HIV-1 integration remains. Nup62 was first characterized as a GlcNac-containing nucleoporin, which forms a subcomplex with Nup54, Nup58, and Nup45 that localizes to the NPC, either at the cytoplasmic face or both sides of the NPC (40, 54–57). Interestingly, by using the in vivo mapping technique, DamID, a recent study reported that Nup62 interacts with chromatin inside the nucleoplasm of embryonic Drosophila cells (33). Because our above data indicate that Nup62 interacts with HIV-1 IN and Nup62-KD was shown to affect HIV integration, we then aimed to test whether Nup62 and HIV IN are both associated with cellular chromatin. GFP-IN-transfected or mock-transfected 293T cells were fractionated into cytoplasmic (CE), membrane (ME), nuclear soluble (NE), chromatin-bound (CB), and pellet extract fractions by using the Thermo Scientific subcellular protein fractionation kit (Fig. 3A). It is worth noticing that, after separating the CE and ME fractions, the remaining nucleus was treated with a detergent-free high-salt nuclear extraction buffer. After centrifugation, the soluble nucleoplasmic components, as well as some hydrophilic nuclear envelope proteins, were present in the supernatant, forming what is termed the nuclear soluble (NE) fraction. The pellet containing the chromatin and CB proteins was treated with micrococcal nuclease to release chromatin-associated proteins (CB), and the final nuclear pellet extract contained cytoskeletal and hydrophobic envelope proteins. Each subcellular fraction was Western-blotted to detect endogenous Nup62 proteins (Fig. 3B, panel a). Meanwhile, the presence of α-tubulin and histone H1 in each fraction was monitored as a quality control of the experiment (Fig. 3B, panels d and e). Our data clearly show that both Nup62 and GFP-IN was primarily present in the NE and CB fractions, whereas GFP alone was not associated with chromatin (Fig. 3B, panels a–c). Because NE contains both soluble nucleoplasmic components and hydrophilic nuclear envelope proteins, we could not rule out the possibility of the Nup62 in the NE portion also came from nucleoplasma. In fact, researchers have already found that some nucleoporins are mobile and localize inside the nucleoplasm (31, 32).
FIGURE 3.
Nup62 binds to chromatin and interacts with HIV-1 IN in both nuclear and chromatin-bound extracts. A, experimental setup. B, subcellular distribution patterns of endogenous Nup62, α-tubulin, and histone H1, and exogenous GFP and GFP-IN were determined by the subcellular fractionation method and WB in 293T cells transfected with the GFP or GFP-IN plasmid. CE, cytoplasmic extracts; ME, membrane extracts; NE, nuclear extracts; CB, chromatin-bound extract. C, 293T cells were transfected with the PL-IN or PL-A3G plasmid for 48 h, fractionated, and then assessed for PL activity (left panel), as described under “Experimental Procedures.” To test Nup62-IN interaction in different fractions, 293T cells were fractionated 48 h after transfection with PL-IN, and each fraction was immunoprecipitated (IP) with anti-Nup62 or anti-GFP (as the control). The coimmunoprecipitated PL-IN in each fraction was evaluated by measuring PL activity (right panel).
Because HIV-1 IN and Nup62 interact with each other and both were bound to the host chromatin, it was of interest to examine whether this viral-cellular protein complex could be associated with the chromatin. A ProLabel-tagged IN (PL-IN) expression plasmid was used for quantitatively detecting the association of IN with chromatin or Nup62. The subcellular fractionation experiments in PL-IN-transfected 293T cells clearly confirmed that PL-IN was associated with chromatin, whereas another PL-tagged cellular protein PL-APOBEC3G (PL-A3G) was only found in the cytoplasmic fraction (Fig. 3C, left panel). Then we examined the interaction of IN and Nup62 in each subcellular fraction by anti-Nup62 immunoprecipitation followed by the detection of the ProLabel activity of each immunoprecipitate (Fig. 3C, right panel). The anti-GFP immunoprecipitation was used as a negative control. Interestingly, the results showed that the Nup62/IN association was mainly found in the chromatin-bound fraction and to a lesser extent in the nuclear soluble fraction, suggesting the Nup62-IN interaction may take place at chromatin-bound sites, NPC, or in nucleoplasm. Moreover, the data also indicated that the Nup62-IN interaction does not require the presence of DNA because a large amount of Nup62-IN complexes was still detected after the chromatin-bound proteins (CE) were treated with micrococcal nuclease.
Down-regulation of Nup62 Affects the HIV-1 IN/Chromatin Association
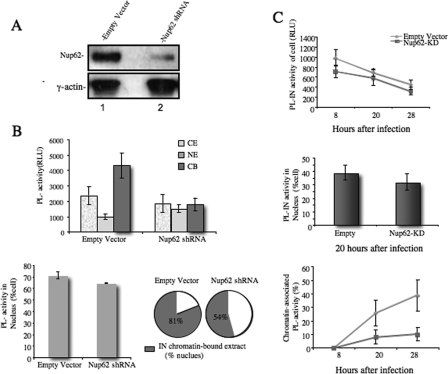
The above studies showed that Nup62 interacts with HIV-1 IN inside the nucleus, especially in the chromatin-bound fraction, and the knockdown of Nup62 significantly inhibited viral DNA integration. Therefore, the participation of Nup62 in HIV IN/chromatin tethering and/or chromatin association consequently affecting HIV DNA integration may be speculated. To further test this hypothesis, we first expressed the PL-IN fusion protein in Nup62-KD or empty vector-treated 293T cells (Fig. 4A) and analyzed PL-IN activity in different subcellular fractions (Fig. 4B). Consistent with our previous finding, the majority of the PL-IN was found associated with the host chromatin in the empty vector-treated 293T cells. However, in Nup62-KD cells, the total PL-IN activity in the cells was reduced to 67% that of the control cells (Fig. 4B, upper panel). The nuclear translocation of PL-IN also slightly decreased (from 71 to 64% of the total cells), but the difference is not statistically significant (p = 0.0883) (Fig. 4B, lower left panel). These findings suggest that Nup62-KD did not significantly affect IN nuclear translocation. In contrast, the chromatin-associated PL-IN in Nup62-KD cells was significantly reduced; the percentage of chromatin-bound PL-IN activity per nucleus was only 54%, whereas the percentage reached 81% in empty vector-treated cells (Fig. 4B, lower right panel). These results indicate that Nup62 is required for the association of HIV-1 IN with the host chromatin.
FIGURE 4.
Effect of Nup62 on the chromatin association and nuclear localization of HIV-1 IN in 293T cells and HIV-1-infected T cells. A, 293T cells were transduced with a different panel of lentiviral vectors harboring Nup62 shRNA (lane 2) or empty vector (lane 1). Nup62 levels were measured by WB using anti-Nup62 antibody. γ-Actin was included as an internal control. B, at 4 days post-transduction, cells were transfected with PL-IN, fractionated, and then analyzed for PL activity in each subcellular fraction (upper panel). The PL-IN activity in the nucleus is presented as the percentage of the total cellular PL-IN activity (lower left panel). The PL-IN activity of the chromatin-bound fraction is presented as the percentage of total nuclear PL-IN activity (lower right panel). C, RT/IN-deleted HIV-1 provirus (BruΔRI) and the Vpr-RT-IN-PL fusion protein were cotransfected into 293T cells to generate the HIV-1 single cycle IN-PL virus. The Nup62-KD- or empty vector-transduced C8166 cells were infected with IN-PL virus, fractionated at different time points, and measured for PL activity. Upper panel, the PL activity of the total cell lysates. Middle panel, The PL-IN activity in the nucleus is presented as the percentage of the total cellular PL-IN activity. Lower panel, PL-IN activity in the chromatin-bound is presented as the percentage of the total cell lysate PL-IN activity. RLU, relative luminescence units.
The effect of Nup62 on the IN/chromatin association was also observed in HIV-1-infected cells. A previously described HIV-1 single cycle replicating viruses (HIV-IN-PL), in which a PL tag was fused with IN at its C terminus, was used to infect the Nup62-KD C8166 T cells (49, 53). The intracellular localization of IN-PL in HIV-infected cells was traced by detecting the PL activity in different subcellular fractions. Briefly, the Nup62-KD C8166 T cells were generated using the Nup62 shRNA lentiviral vector and infected by the HIV-IN-PL single cycle replicating virus for 2 h. At 8, 20, and 28 h post-infection, infected cells were subjected to subcellular fractionation, and the PL activity of each fraction was measured. Compared with the control cells, the IN-PL activity in HIV-infected Nup62-KD cells was slightly reduced (15–20%) throughout all time points (Fig. 4C, upper panel). Also, an ∼20% reduction of IN-PL activity was found in the nuclei of HIV-infected Nup62-KD cells (Fig. 4C, middle panel). Strikingly, the chromatin-bound HIV IN was significantly decreased in Nup62-KD cells, and at 28 h, the amount of IN associated with chromatin in the control cells was 3–4-fold higher than that of Nup62-KD cells (Fig. 4C, lower panel). In summary, these results provide further evidence that Nup62 is required for an efficient HIV-1 IN/chromatin association.
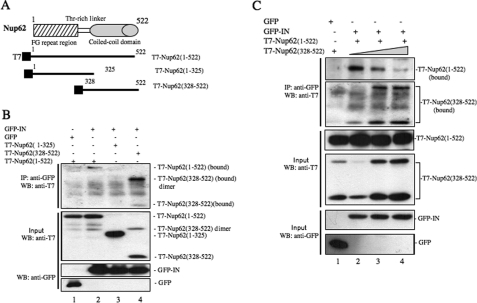
C-terminal Domain of Nup62 Sufficiently Competes with Nup62 for Binding to HIV-1 IN
The above studies have shown the requirement of Nup62 for the HIV-1 IN/chromatin association and viral cDNA integration. To further understand the mechanism of the Nup62-IN interaction, we constructed two Nup62-truncated protein expression plasmids, a T7-tagged Nup62 N-terminal domain (residues 1–325) and the C-terminal coiled-coil domain (residues 328–522) (Fig. 5A). These T7-Nup62-truncated mutants were expressed in 293T cells and detected by WB with an anti-T7 antibody (Fig. 5B, middle panel). As expected, the T7-Nup62(1–325) and T7-Nup62(328–522) bands corresponded to a molecular mass of 40 and 22 kDa, respectively. In the T7-Nup62(328–522) sample, we detected another band at ∼44 kDa, which appears to be a dimer form of T7-Nup62(328–522). To test the IN-binding ability of these two Nup62-truncated mutants, each of the mutants or the full-length Nup62 (T7-Nup62(1–522)) were cotransfected with GFP-IN in 293T cells (Fig. 5B, lanes 2–4). In parallel, GFP and T7-Nup62(1–522) cotransfected cells were included as the control (Fig. 5B, lane 1). After 48 h of transfection, cells were collected, and the interactions between GFP-IN and T7-Nup62WT/del were revealed by coimmunoprecipitation with rabbit anti-GFP antibody followed by WB using an anti-T7 mouse monoclonal antibody. The result showed that, like the full-length Nup62, the T7-Nup62(328–522) was able to bind to GFP-IN, whereas the N-terminal domain of Nup62 (T7-Nup62(1–325)) did not (Fig. 5B, upper panel). Interestingly, the association level of the T7-Nup62(328–522) with IN was significantly higher than that of T7-Nup62(1–522). We therefore consider the possibility that the C-terminal fragment of Nup62 was able to compete with the full-length Nup62 for binding to HIV-1 IN. To test this possibility, we cotransfected GFP-IN with T7-Nup62(1–522) (2 μg) and an increased amount of T7-Nup62(328–522) (2, 4, and 6 μg) into 293T cells (Fig. 5C, lanes 2–4). As a control, a GFP plasmid was cotransfected with T7-Nup62(1–522) and T7-Nup62(328–522) (Fig. 5C, lane 1). The interactions between IN and T7-Nup62(1–522) or T7-Nup62(328–522) are depicted in Fig. 5C (top two panels). Results showed that the association of T7-Nup62(1–522) with IN was significantly decreased when T7-Nup62(328–522) expression was increased (Fig. 5C, upper 1st and 2nd panels). These data strongly suggest that the C-terminal coiled-coil domain of Nup62 is responsible for binding to HIV-1 IN, and the overexpression of T7-Nup62(328–522) in 293T cells is able to compete with wild-type Nup62 for IN binding.
FIGURE 5.
C-terminal coiled-coil domain of Nup62 is required for the IN-Nup62 interaction. A, schematic representation of Nup62 and the constructs of the T7-Nup62 fragments. B, C-terminal coiled-coil domain of Nup62 is required for its interaction with IN. Each of the T7-tagged plasmids encoding full-length Nup62 (amino acids 1–522), the FG-repeated fragment of Nup62 (amino acids 1–325), or the C-terminal domain (amino acids 328–522) were cotransfected with the GFP or GFP-IN expressors in 293T cells, and their interactions with IN were analyzed by anti-GFP immunoprecipitation followed by WB with an anti-T7 antibody. Upper panel, bound full-length or truncated T7-Nup62. Middle and lower panels, expression of GFP, GFP-IN, T7-Nup62 in 1/10 transfected 293T cells. C, C-terminal domain of Nup62 competes with Nup62 for binding to HIV-1 IN. 293T cells were cotransfected with GFP (lane 1), GFP-IN (lanes 2–4), T7-Nup62(1–522) (lanes 1–4), and an increased amount of T7-Nup62(328–522) (lanes 1–4). The interactions of IN with full-length Nup62 and the C-terminal domain of Nup62 were analyzed by anti-GFP immunoprecipitation (IP) followed by WB with an anti-T7 antibody. Upper two panels, bound T7-Nup62(1–522) and T7-Nup62(328–522). Middle and lower panels, expression of GFP, GFP-IN, T7-Nup62(1–522) and T7-Nup62(328–522) in 1/10-transfected 293T cells.
Expression of the Nup62 C-terminal Domain Reduces the HIV-1 IN/Chromatin Association and Inhibits HIV-1 Infection
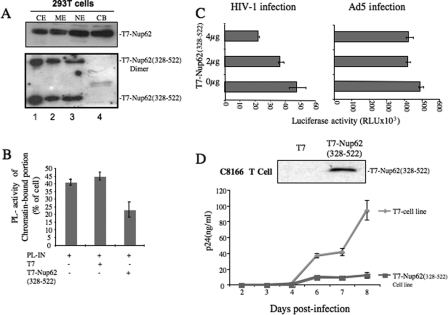
After the C-terminal domain of Nup62 was identified to be critical for binding to HIV-1 IN, we further examined the intracellular localization of T7-Nup62(328–522) by the subcellular fractionation method. Markedly, the T7-Nup62(328–522) failed to be associated with chromatin, even though it was located in the nucleus (Fig. 6A). These data suggest that the IN binding region of Nup62 might be distinct from that required for chromatin association. It might also be predicted that the presence of the C-terminal region of Nup62 may affect HIV-1 IN chromatin association and consequently disrupt HIV-1 infection.
FIGURE 6.
Impact of Nup62 C-terminal domain (residues 328–522) expression on HIV-1 infection. A, 293T cells were transfected with either T7-Nup62 or T7-Nup62(328–522) and subsequently fractionated and analyzed by WB using an anti-T7 antibody. B, 293T cells were cotransfected with T7-Nup62(328–522) and PL-IN plasmids for 48 h, followed by fractionation and analysis for PL activity in each subcellular fraction. The PL-IN activity of the chromatin-bound fraction is presented as the percentage of total cellular PL-IN activity. C, 293T cells were transfected with the T7 vector or a different dose of the T7-Nup62(328–522) plasmid. After 24 h, equal numbers of transfected cells were infected with p24-normalized VSV-G pseudotyped pNL 4.3 ΔBgl/Luc+ or Ad5-CMV-luciferase. At 48 h post-infection, equal amounts of infected cells were collected, and the luciferase activities were measured. D, C8166 cells were transduced with the lentiviral vector pEF1-Nup62(328–522)-puro or control vector pEF1-T7-puro and were then infected by pNL4.3 GFP+ viruses. Upper panel, expression of T7-Nup62(328–522) in a different panel of transduced cells. Lower panel, HIV-Gag-p24 levels in supernatants were measured at different time points to monitor virus replication.
To test the effect of the Nup62(328–522) on HIV-1 IN chromatin association, we cotransfected PL-IN plasmid with either the T7-Nup62(328–522) or T7 vector plasmid into 293T cells, and the ratio of PL-IN activity of the chromatin-bound portion compared with the total cell PL-IN activity was examined (Fig. 6B). Results showed that in the presence of Nup62(328–522), the levels of chromatin-associated PL-IN were reduced by 50% when compared with levels in the control cells.
Given that Nup62(328–522) is able to inhibit the IN associated to chromatin, we next investigated whether the expression of Nup62(328–522) could affect HIV-1 replication. The 293T cells were first transfected with different amounts of the T7-Nup62(328–522) plasmid, and after 24 h, cells were infected with VSV-G pseudotyped luciferase-expressing HIV-1 (pNL-BruΔBgl/Luc). After 48 h of infection, cells were collected, and the virus-associated luciferase activity was measured from an equal number of cells. The results showed that the HIV-1 infection was clearly impaired in the presence of Nup62(328–522) in a dose-dependent manner (Fig. 6C, left panel). To exclude the possibility that the inhibition of HIV infection mediated by Nup62(328–522) could be due to a nonspecific effect, we also infected Nup62(328–522)-expressing 293T cells with Ad5-CMV-luciferase, which carried the luciferase gene under a CMV promoter, and the results showed no significant difference of luciferase activity in either T7-Nup62(328–522) or control cells (Fig. 6C, right panel). Moreover, we have generated a CD4+ C8166 T cell line stably expressing T7-Nup62(328–522) through a lentiviral vector system, as described previously (48). The expression levels of T7-Nup62(328–522) were much lower than those in the 293T cells (Fig. 6D, upper panel), and neither abnormal nuclei nor cell growth defects were observed in this cell lines. Then we assessed HIV-1 infection in the T7-Nup62(328–522) or T7 tag-expressing T cell line by challenging with pNL4.3 GFP+ virus (multiplicity of infection of 0.01). At different time points, the supernatants were collected, and the levels of p24 antigen in the supernatants were measured. Compared with the control cell line, an ∼3–5-fold reduction of viral replication was steadily observed in the T7-Nup62(328–522) cell line (Fig. 6D, lower panel). These observations, together with the observed interaction between Nup62 and HIV-1 IN and the inhibitory effect of the Nup62 knockdown on IN chromatin targeting, demonstrate that Nup62 is a new IN cellular cofactor that is required for efficient HIV-1 integration.
DISCUSSION
Retroviral integration, which occurs between the viral preintegration complex and the host chromatin, is catalyzed by viral IN, whereas cellular cofactors also play important roles in this process. Recent studies have highlighted that LEDGF/p75, a ubiquitous chromatin-bound protein and HIV-1 IN binding partner, acts as a chromatin docking factor for the lentiviral PIC. Depleting LEDGF/p75 from cells or overexpressing its integrase-binding domain inhibits HIV-1 replication (15, 58). However, it appears that HIV-1 does not entirely rely on this host factor. Even in a genetic knock-out model, LEDGF-null MEFs still supported ∼10% of the level of HIV-1 integration and replication (16), suggesting the possibility that other unrecognized cellular factor(s) may also contribute to HIV-1 integration. Here, we report that another cellular protein, Nup62, binds to chromatin and interacts with HIV-1 IN in the nucleus, especially in the chromatin-bound fraction of cells. We show that the shRNA-mediated knockdown of Nup62 significantly reduced the association of IN with chromatin and led to impaired HIV-1 integration and viral replication in CD4+ T cells. Moreover, the expression of the C-terminal domain of Nup62 in CD4+ T cells was shown to reduce the association of IN with chromatin and inhibit HIV-1 infection.
It is well known that the nuclear import of viral reverse-transcribed DNA and its integration are two necessary steps for HIV-1 productive replication, and viral IN plays a critical role in ensuring that these replication steps proceed. Recently, a nuclear import-coupled integration model has been proposed, which suggests that the HIV-1 PIC may require necessary cellular cofactor(s) or pathways during nuclear translocation that facilitate both HIV-1 chromatin targeting and integration (37). Nups, which were originally thought to be involved in macromolecular transport between the nucleus and cytoplasm (35–38), have recently been reported to be able to interact with chromatin at transcriptionally active genes at the NPC or in the nuclear interior and are involved in regulating the expression of genes (33, 34). All of these observations raised the question of whether Nups could participate in the steps of HIV replication, especially as an important cofactor for HIV-1 nuclear import and integration. To investigate this possibility, we examined the potential interaction between HIV-1 IN and various nucleoporins. From a GST pulldown assay, we identified Nup62 as a potential IN-binding cofactor (Fig. 1A). Their interaction was further confirmed through coimmunoprecipitation analysis in 293T cells and in HIV-1-infected T cells. Interestingly, the results clearly showed that Nup62 is only associated with HIV-1 IN, but not with HIV-1 Vpr and MA proteins. In addition, the IN-Nup62 interaction was independent of the presence of DNA. In an attempt to delineate the region in Nup62 necessary for its IN-binding, we have also found that the C-terminal coiled-coil domain of Nup62 is sufficient for binding to HIV IN. Interestingly, our results indicated that both monomer and a possible dimer form of T7-Nup62(328–522) could interact with HIV-1 IN (Fig. 5, B and C), suggesting that the dimerization of the C-terminal domain of Nup62 may also contribute to its binding to HIV-1 IN. However, more detailed studies, including mutagenic analysis, are required to draw a clear conclusion. Overall, these data demonstrate a specific interaction between Nup62 and HIV-1 IN.
Even though our data provided evidence for the Nup62-IN interaction, we still did not know whether and how Nup62 could impact HIV-1 replication. To address this question, we knocked down Nup62 in CD4+ T cells with an shRNA vector and investigated its impact on HIV-1 infection. Our data revealed no significant inhibitory effect on either HIV-1 reverse transcription or viral DNA nuclear import in Nup62-KD cells, suggesting that Nup62 is not directly involved in these viral replication steps. Interestingly, the level of virally integrated DNA in Nup62-KD cells was drastically decreased to only 4% that in the HIV-infected control cells (Fig. 2D), indicating that the major effect of Nup62-KD on viral replication occurs at the integration step. As previous studies have demonstrated that Nups are able to bind to transcriptionally active genes at the NPC or in the nuclear interior (33, 34), we speculate that by interacting with HIV IN, Nup62 offers the HIV PIC a direct link from the NPC to the chromosomal DNA, thus enabling efficient integration. The evidence that supports this hypothesis includes the following. 1) Nup62 is also present in the chromatin-bound fraction (Fig. 3B). 2) The Nup62-IN complexes were detected in both the chromatin-bound fraction and in soluble nuclear extract (Fig. 3C). Because the soluble nuclear extract contains proteins from both the soluble nucleoplasm and the nuclear envelope and because we could not distinguish them from each other, their interaction may occur at either the NPC, in the nucleoplasm, or in both compartments. Afterward, the Nup62-IN complex may shuttle to the chromatin for viral DNA integration. Indeed, some studies have indicated that communication between intranuclear transcription sites and the nuclear pore may be conducted by some nucleoporins, including Nup98 and Nup153, as they serve as shuttling nucleoporins (31, 32). 3) The above mentioned speculations are further supported by the finding that Nup62 was required for an efficient IN/chromatin association. In Nup62-KD 293T cells, the IN/chromatin association rate dropped from 81 to 54% (Fig. 4B), and in HIV-infected Nup62-KD cells, the chromatin-associated viral IN-PL activity was reduced 3–4-fold (Fig. 4C). 4) The expression of the IN-binding domain of Nup62 reduced the IN/chromatin association and inhibited HIV-1 infection. Although more studies are required regarding the role of Nup62 in HIV-1 integration site selection and its direct effect on integration reaction, the above data strongly suggest that during HIV replication, IN recruits Nup62 to facilitate HIV cDNA chromatin targeting and integration. In addition, this study provides further evidence for the scenario of HIV nuclear import-coupled integration.
We also noticed in this study that the knockdown of Nup62 did not eliminate the HIV IN/chromatin association in either the IN overexpression system or in HIV-infected cells, supporting that other host factor(s) is also involved in IN chromatin targeting. In fact, the functional role of LEDGF/p75 in HIV chromatin targeting and integration has been well documented (12–16, 59). At this point, we still do not know whether Nup62 and LEDGF/p75 could cooperate for some cellular activities. It would also be interesting to investigate how HIV IN coordinates with LEDGF and Nup62 to facilitate viral DNA chromatin targeting and integration. Notably, our preliminary study has suggested an association of Nup62 and LEDGF/p757; however, its biological relevance and impact on HIV-1 replication are still open questions. Potentially, HIV IN may form a functional complex with both Nup62 and LEDGF or either of them individually in the nucleus to contribute to HIV cDNA chromatin targeting and integration.
Nup62 has been implicated to act at several steps during HIV-1 replication. In addition to its effect on viral DNA integration as presented in this study, Nup62 has also been shown to be involved in HIV vRNA export and to be incorporated into virions (60). Although the functional role of encapsidated Nup62 is still not clear, it might be required for early step(s), such as nuclear import and other steps in the new infection cycle. Even though a significant reduction of viral DNA integration was observed in Nup62-KD T cells when the virus stock was produced from normal 293T cells, we still could not exclude the possibility that the encapsidated Nup62 may also facilitate IN-associated viral PIC translocation into the nucleus and/or to target the chromatin. Furthermore, it would be of interest to investigate the impact of the IN-Nup62 interaction on viral incorporation of Nup62 and to test whether the expression of the IN-binding domain of Nup62 might specifically affect the incorporation of endogenous Nup62 into viruses. Taken together, our study provides evidence for the requirement of Nup62 for HIV-1 integration and replication. More detailed studies on the molecular mechanism(s) underlying the effect of Nup62 on HIV-1 integration should enhance our understanding of how HIV employs multiple cellular factors for its survival and may also provide the opportunity for the development of a new anti-HIV treatment approach.
This work was supported in part by Canadian Institutes of Health Research Grants HOP 81180 and HBF 103212, Canadian Foundation for AIDS Research Grant 023-013, and the Leaders Opportunity Fund Award from the Canadian Foundation of Innovation (to X.-J. Y.).
Z. Ao, unpublished data.
- PIC
- preintegration complex
- NPC
- preintegration complex
- IN
- integrase
- CE
- cytoplasmic extract
- NE
- nuclear extract
- Nup
- nucleoporin
- MDM
- monocyte-derived macrophage
- WB
- Western blot
- Luc
- luciferase
- PL
- ProLabel
- CB
- chromatin-bound
- TPR
- translocated promoter region
- RQ-PCR
- real time quantitative PCR.
REFERENCES
- 1. Ao Z., Fowke K. R., Cohen E. A., Yao X. (2005) Contribution of the C-terminal tri-lysine regions of human immunodeficiency virus type 1 integrase for efficient reverse transcription and viral DNA nuclear import. Retrovirology 2, 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Christ F. (2008) Transportin-SR2 imports HIV into the nucleus. Curr. Biol. 18, 1192–1202 [DOI] [PubMed] [Google Scholar]
- 3. Ikeda T., Nishitsuji H., Zhou X., Nara N., Ohashi T., Kannagi M., Masuda T. (2004) Evaluation of the functional involvement of human immunodeficiency virus type 1 integrase in nuclear import of viral cDNA during acute infection. J. Virol. 78, 11563–11573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bouyac-Bertoia M., Dvorin J. D., Fouchier R. A., Jenkins Y., Meyer B. E., Wu L. I., Emerman M., Malim M. H. (2001) HIV-1 infection requires a functional integrase NLS. Mol. Cell 7, 1025–1035 [DOI] [PubMed] [Google Scholar]
- 5. Gallay P., Hope T., Chin D., Trono D. (1997) HIV-1 infection of nondividing cells through the recognition of integrase by the importin/karyopherin pathway. Proc. Natl. Acad. Sci. U.S.A. 94, 9825–9830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schröder A. R., Shinn P., Chen H., Berry C., Ecker J. R., Bushman F. (2002) HIV-1 integration in the human genome favors active genes and local hot spots. Cell 110, 521–529 [DOI] [PubMed] [Google Scholar]
- 7. Wu X., Li Y., Crise B., Burgess S. M. (2003) Transcription start regions in the human genome are favored targets for MLV integration. Science 300, 1749–1751 [DOI] [PubMed] [Google Scholar]
- 8. Desfarges S., Ciuffi A. (2010) Retroviral integration site selection. Viruses 2, 111–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Llano M., Delgado S., Vanegas M., Poeschla E. M. (2004) Lens epithelium-derived growth factor/p75 prevents proteasomal degradation of HIV-1 integrase. J. Biol. Chem. 279, 55570–55577 [DOI] [PubMed] [Google Scholar]
- 10. Maertens G., Cherepanov P., Pluymers W., Busschots K., De Clercq E., Debyser Z., Engelborghs Y. (2003) LEDGF/p75 is essential for nuclear and chromosomal targeting of HIV-1 integrase in human cells. J. Biol. Chem. 278, 33528–33539 [DOI] [PubMed] [Google Scholar]
- 11. Cherepanov P., Maertens G., Proost P., Devreese B., Van Beeumen J., Engelborghs Y., De Clercq E., Debyser Z. (2003) HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 278, 372–381 [DOI] [PubMed] [Google Scholar]
- 12. Ciuffi A., Llano M., Poeschla E., Hoffmann C., Leipzig J., Shinn P., Ecker J. R., Bushman F. (2005) A role for LEDGF/p75 in targeting HIV DNA integration. Nat. Med. 11, 1287–1289 [DOI] [PubMed] [Google Scholar]
- 13. Emiliani S., Mousnier A., Busschots K., Maroun M., Van Maele B., Tempé D., Vandekerckhove L., Moisant F., Ben-Slama L., Witvrouw M., Christ F., Rain J. C., Dargemont C., Debyser Z., Benarous R. (2005) Integrase mutants defective for interaction with LEDGF/p75 are impaired in chromosome tethering and HIV-1 replication. J. Biol. Chem. 280, 25517–25523 [DOI] [PubMed] [Google Scholar]
- 14. Hombrouck A., De Rijck J., Hendrix J., Vandekerckhove L., Voet A., De Maeyer M., Witvrouw M., Engelborghs Y., Christ F., Gijsbers R., Debyser Z. (2007) Virus evolution reveals an exclusive role for LEDGF/p75 in chromosomal tethering of HIV. PLoS Pathog. 3, e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Llano M., Saenz D. T., Meehan A., Wongthida P., Peretz M., Walker W. H., Teo W., Poeschla E. M. (2006) An essential role for LEDGF/p75 in HIV integration. Science 314, 461–464 [DOI] [PubMed] [Google Scholar]
- 16. Shun M. C., Raghavendra N. K., Vandegraaff N., Daigle J. E., Hughes S., Kellam P., Cherepanov P., Engelman A. (2007) LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 21, 1767–1778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zheng Y., Ao Z., Jayappa K. D., Yao X. (2010) Characterization of the HIV-1 integrase chromatin- and LEDGF/p75-binding abilities by mutagenic analysis within the catalytic core domain of integrase. Virol. J. 7, 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Alber F., Dokudovskaya S., Veenhoff L. M., Zhang W., Kipper J., Devos D., Suprapto A., Karni-Schmidt O., Williams R., Chait B. T., Sali A., Rout M. P. (2007) The molecular architecture of the nuclear pore complex. Nature 450, 695–701 [DOI] [PubMed] [Google Scholar]
- 19. Hetzer M. W., Walther T. C., Mattaj I. W. (2005) Pushing the envelope. Structure, function, and dynamics of the nuclear periphery. Annu. Rev. Cell Dev. Biol. 21, 347–380 [DOI] [PubMed] [Google Scholar]
- 20. Kasper L. H., Brindle P. K., Schnabel C. A., Pritchard C. E., Cleary M. L., van Deursen J. M. (1999) CREB-binding protein interacts with nucleoporin-specific FG repeats that activate transcription and mediate NUP98-HOXA9 oncogenicity. Mol. Cell. Biol. 19, 764–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang G. G., Cai L., Pasillas M. P., Kamps M. P. (2007) NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukemogenesis. Nat. Cell Biol. 9, 804–812 [DOI] [PubMed] [Google Scholar]
- 22. Romana S. P., Radford-Weiss I., Ben Abdelali R., Schluth C., Petit A., Dastugue N., Talmant P., Bilhou-Nabera C., Mugneret F., Lafage-Pochitaloff M., Mozziconacci M. J., Andrieu J., Lai J. L., Terre C., Rack K., Cornillet-Lefebvre P., Luquet I., Nadal N., Nguyen-Khac F., Perot C., Van den Akker J., Fert-Ferrer S., Cabrol C., Charrin C., Tigaud I., Poirel H., Vekemans M., Bernard O. A., Berger R. (2006) NUP98 rearrangements in hematopoietic malignancies. A study of the Groupe Francophone de Cytogénétique Hématologique. Leukemia 20, 696–706 [DOI] [PubMed] [Google Scholar]
- 23. von Lindern M., van Baal S., Wiegant J., Raap A., Hagemeijer A., Grosveld G. (1992) Can, a putative oncogene associated with myeloid leukemogenesis, may be activated by fusion of its 3′-half to different genes. characterization of the set gene. Mol. Cell. Biol. 12, 3346–3355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. von Lindern M., Fornerod M., van Baal S., Jaegle M., de Wit T., Buijs A., Grosveld G. (1992) The translocation (6;9), associated with a specific subtype of acute myeloid leukemia, results in the fusion of two genes, dek and can, and the expression of a chimeric, leukemia-specific dek-can mRNA. Mol. Cell. Biol. 12, 1687–1697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Marshall W. F. (2002) Order and disorder in the nucleus. Curr. Biol. 12, R185–R192 [DOI] [PubMed] [Google Scholar]
- 26. Casolari J. M., Brown C. R., Komili S., West J., Hieronymus H., Silver P. A. (2004) Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 117, 427–439 [DOI] [PubMed] [Google Scholar]
- 27. Schmid M., Arib G., Laemmli C., Nishikawa J., Durussel T., Laemmli U. K. (2006) Nup-PI. The nucleopore-promoter interaction of genes in yeast. Mol. Cell 21, 379–391 [DOI] [PubMed] [Google Scholar]
- 28. Daigle N., Beaudouin J., Hartnell L., Imreh G., Hallberg E., Lippincott-Schwartz J., Ellenberg J. (2001) Nuclear pore complexes form immobile networks and have a very low turnover in live mammalian cells. J. Cell Biol. 154, 71–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rabut G., Doye V., Ellenberg J. (2004) Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat. Cell Biol. 6, 1114–1121 [DOI] [PubMed] [Google Scholar]
- 30. Lindsay M. E., Plafker K., Smith A. E., Clurman B. E., Macara I. G. (2002) Npap60/Nup50 is a tri-stable switch that stimulates importin-α:β-mediated nuclear protein import. Cell 110, 349–360 [DOI] [PubMed] [Google Scholar]
- 31. Griffis E. R., Craige B., Dimaano C., Ullman K. S., Powers M. A. (2004) Distinct functional domains within nucleoporins Nup153 and Nup98 mediate transcription-dependent mobility. Mol. Biol. Cell 15, 1991–2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Griffis E. R., Altan N., Lippincott-Schwartz J., Powers M. A. (2002) Nup98 is a mobile nucleoporin with transcription-dependent dynamics. Mol. Biol. Cell 13, 1282–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kalverda B., Pickersgill H., Shloma V. V., Fornerod M. (2010) Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell 140, 360–371 [DOI] [PubMed] [Google Scholar]
- 34. Capelson M., Liang Y., Schulte R., Mair W., Wagner U., Hetzer M. W. (2010) Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell 140, 372–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ebina H., Aoki J., Hatta S., Yoshida T., Koyanagi Y. (2004) Role of Nup98 in nuclear entry of human immunodeficiency virus type 1 cDNA. Microbes Infect. 6, 715–724 [DOI] [PubMed] [Google Scholar]
- 36. Woodward C. L., Prakobwanakit S., Mosessian S., Chow S. A. (2009) Integrase interacts with nucleoporin NUP153 to mediate the nuclear import of human immunodeficiency virus type 1. J. Virol. 83, 6522–6533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. König R., Zhou Y., Elleder D., Diamond T. L., Bonamy G. M., Irelan J. T., Chiang C. Y., Tu B. P., De Jesus P. D., Lilley C. E., Seidel S., Opaluch A. M., Caldwell J. S., Weitzman M. D., Kuhen K. L., Bandyopadhyay S., Ideker T., Orth A. P., Miraglia L. J., Bushman F. D., Young J. A., Chanda S. K. (2008) Global analysis of host-pathogen interactions that regulate early stage HIV-1 replication. Cell 135, 49–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ocwieja K. E., Brady T. L., Ronen K., Huegel A., Roth S. L., Schaller T., James L. C., Towers G. J., Young J. A., Chanda S. K., König R., Malani N., Berry C. C., Bushman F. D. (2011) HIV integration targeting: a pathway involving Transportin-3 and the nuclear pore protein RanBP2. PLoS Pathog. 7, e1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Carmo-Fonseca M., Kern H., Hurt E. C. (1991) Human nucleoporin p62 and the essential yeast nuclear pore protein NSP1 show sequence homology and a similar domain organization. Eur. J. Cell Biol. 55, 17–30 [PubMed] [Google Scholar]
- 40. Guan T., Müller S., Klier G., Panté N., Blevitt J. M., Haner M., Paschal B., Aebi U., Gerace L. (1995) Structural analysis of the p62 complex, an assembly of O-linked glycoproteins that localize near the central gated channel of the nuclear pore complex. Mol. Biol. Cell 6, 1591–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hu T., Guan T., Gerace L. (1996) Molecular and functional characterization of the p62 complex, an assembly of nuclear pore complex glycoproteins. J. Cell Biol. 134, 589–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Frey S., Görlich D. (2007) A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell 130, 512–523 [DOI] [PubMed] [Google Scholar]
- 43. Van Impe K., Hubert T., De Corte V., Vanloo B., Boucherie C., Vandekerckhove J., Gettemans J. (2008) A new role for nuclear transport factor 2 and Ran. Nuclear import of CapG. Traffic 9, 695–707 [DOI] [PubMed] [Google Scholar]
- 44. Buss F., Kent H., Stewart M., Bailer S. M., Hanover J. A. (1994) Role of different domains in the self-association of rat nucleoporin p62. J. Cell Sci. 107, 631–638 [DOI] [PubMed] [Google Scholar]
- 45. Melcák I., Hoelz A., Blobel G. (2007) Structure of Nup58/45 suggests flexible nuclear pore diameter by intermolecular sliding. Science 315, 1729–1732 [DOI] [PubMed] [Google Scholar]
- 46. Percipalle P., Clarkson W. D., Kent H. M., Rhodes D., Stewart M. (1997) Molecular interactions between the importin α/β heterodimer and proteins involved in vertebrate nuclear protein import. J. Mol. Biol. 266, 722–732 [DOI] [PubMed] [Google Scholar]
- 47. Han I., Roos M. D., Kudlow J. E. (1998) Interaction of the transcription factor Sp1 with the nuclear pore protein p62 requires the C-terminal domain of p62. J. Cell. Biochem. 68, 50–61 [DOI] [PubMed] [Google Scholar]
- 48. Ao Z., Yu Z., Wang L., Zheng Y., Yao X. (2008) Vpr14–88-Apobec3G fusion protein is efficiently incorporated into Vif-positive HIV-1 particles and inhibits viral infection. PLoS One 3, e1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zheng Y., Ao Z., Wang B., Jayappa K. D., Yao X. (2011) Host protein Ku70 binds and protects HIV-1 integrase from proteasomal degradation and is required for HIV replication. J. Biol. Chem. 286, 17722–17735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ao Z., Danappa Jayappa K., Wang B., Zheng Y., Kung S., Rassart E., Depping R., Kohler M., Cohen E. A., Yao X. (2010) Importin α3 interacts with HIV-1 integrase and contributes to HIV-1 nuclear import and replication. J. Virol. 84, 8650–8663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ao Z., Huang G., Yao H., Xu Z., Labine M., Cochrane A. W., Yao X. (2007) Interaction of human immunodeficiency virus type 1 integrase with cellular nuclear import receptor importin 7 and its impact on viral replication. J. Biol. Chem. 282, 13456–13467 [DOI] [PubMed] [Google Scholar]
- 52. Ao Z., Wang X., Bello A. J., Danappa Jayappa K., Yu Z., Fowke K., He X., Chen X., Li J., Kobinger G. P., Yao X. (2011) Characterization of anti-HIV activity mediated by R88-APOBEC3G mutant fusion proteins in CD4+ T Cells, peripheral blood mononuclear cells, and macrophages. Hum. Gene Ther. 22, 1225–1237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ao Z., Yao X., Cohen E. A. (2004) Assessment of the role of the central DNA flap in human immunodeficiency virus type 1 replication by using a single-cycle replication system. J. Virol. 78, 3170–3177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Davis L. I., Blobel G. (1986) Identification and characterization of a nuclear pore complex protein. Cell 45, 699–709 [DOI] [PubMed] [Google Scholar]
- 55. Davis L. I., Blobel G. (1987) Nuclear pore complex contains a family of glycoproteins that includes p62. Glycosylation through a previously unidentified cellular pathway. Proc. Natl. Acad. Sci. U.S.A. 84, 7552–7556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cordes V. C., Krohne G. (1993) Sequential O-glycosylation of nuclear pore complex protein gp62 in vitro. Eur. J. Cell Biol. 60, 185–195 [PubMed] [Google Scholar]
- 57. Schwarz-Herion K., Maco B., Sauder U., Fahrenkrog B. (2007) Domain topology of the p62 complex within the three-dimensional architecture of the nuclear pore complex. J. Mol. Biol. 370, 796–806 [DOI] [PubMed] [Google Scholar]
- 58. De Rijck J., Vandekerckhove L., Gijsbers R., Hombrouck A., Hendrix J., Vercammen J., Engelborghs Y., Christ F., Debyser Z. (2006) Overexpression of the lens epithelium-derived growth factor/p75 integrase binding domain inhibits human immunodeficiency virus replication. J. Virol. 80, 11498–11509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Morerio C., Acquila M., Rosanda C., Rapella A., Tassano E., Micalizzi C., Panarello C. (2005) t(9;11)(p22;p15) with NUP98-LEDGF fusion gene in pediatric acute myeloid leukemia. Leuk. Res. 29, 467–470 [DOI] [PubMed] [Google Scholar]
- 60. Monette A., Panté N., Mouland A. J. (2011) HIV-1 remodels the nuclear pore complex. J. Cell Biol. 193, 619–631 [DOI] [PMC free article] [PubMed] [Google Scholar]