Background: Alba is a dimeric, highly basic archaeal chromatin protein.
Results: The crystal structure of the Alba2-dsDNA complex was determined.
Conclusion: Upon dsDNA binding, Alba undergoes a significant conformational change that is required for oligomerization.
Significance: This study provides the first structural insights into how the Alba dimer binds and packs the dsDNA.
Keywords: Archaea, Chromatin, DNA-binding Protein, Nucleic Acid Structure, Structural Biology, Alba, Ape1823, Archaeal Chromatin, Sso10b
Abstract
All thermophilic and hyperthermophilic archaea encode homologs of dimeric Alba (Sac10b) proteins that bind cooperatively at high density to DNA. Here, we report the 2.0 Å resolution crystal structure of an Alba2 (Ape10b2)-dsDNA complex from Aeropyrum pernix K1. A rectangular tube-like structure encompassing duplex DNA reveals the positively charged residues in the monomer-monomer interface of each dimer packing on either side of the bound dsDNA in successive minor grooves. The extended hairpin loop connecting strands β3 and β4 undergoes significant conformational changes upon DNA binding to accommodate the other Alba2 dimer during oligomerization. Mutational analysis of key interacting residues confirmed the specificity of Alba2-dsDNA interactions.
Introduction
DNA-binding proteins that compact and regulate the availability of genetic material are essential in all organisms. In eukaryotes, the nucleosome is the fundamental chromatin unit, composed of 150 bp of DNA wrapped around two copies of each of four histones (H2a, H2b, H3, and H4) (1). Unlike the universal chromatin proteins in eukaryotes, prokaryotes and archaea tend to utilize two or more DNA-binding proteins to package DNA (2, 3). Interestingly, archaeal DNA replication and transcription pathways are strikingly similar to those of eukaryotes but quite different from the prokaryotic processes (4). Most euryarchaea have true histone proteins lacking the N- and C-terminal tail extensions, which are organized into tetramers that wrap ∼90 bp of DNA (2, 5) and are not post-translationally modified (6).
Thermophilic and hyperthermophilic archaea exclusively contain one or more copies of the gene encoding Alba (Sac10b) (3), a widely distributed chromatin protein accounting for ∼4% of total soluble proteins in Sulfolobus shibatae (7). Alba is a dimeric, highly basic protein, and each ∼10-kDa subunit adopts a mixed α/β-fold with extended β3-β4 hairpins (8). The crystal structures of various Alba family proteins (Alba1 and Alba2) have been determined (8–12). DNA binding by Alba is nonspecific and cooperative, with a final stoichiometry of one Alba dimer per 6 bp of dsDNA bound (7, 8). Recent studies using EMSAs showed that Alba binds more tightly to dsDNA than to either ssDNA or RNA (13). It is post-translationally modified specifically at Lys-16 by the acetyltransferase (Pat) in Sulfolobus solfataricus (Sso10b) (14). A homolog of Sir2 histone deacetylase was shown to associate with and deacetylate Sso10b, which results in transcriptional repression in a reconstituted in vitro transcriptional system (15).
DNA binding by Alba has been studied using various techniques. Electron microscopy of purified Alba proteins from Sulfolobus acidocaldarius complexed with plasmid DNA has shown two modes of interaction: 1) two DNA duplexes interwound by Alba at subsaturating protein concentrations and 2) a single DNA duplex complexed under saturating protein concentrations with little DNA compaction. Based on this, a model for the Alba-DNA interaction was proposed with two helical protein fibers wound around one or two DNA duplexes (16). Based on structural and functional analyses, several Alba-DNA binding models have been proposed (8, 10, 12, 17). One model predicts that Alba spans ∼ 15 bp of duplex DNA, allowing each β3-β4 hairpin to interact with an equivalent part of the DNA duplex, presumably in the minor groove. The central “belly” of the Alba dimer would interact with the major groove (8). Recent NMR studies of Alba-DNA interactions partially supported the above binding models. They further suggested that the basic surface at the dimer interface is important for DNA binding and that the extended loop region (residues 78–84) connecting β3-β4 hairpins remains flexible and is not required, as assumed previously (13).
Here, we report the first high-resolution structure of the Alba2 (Ape10b2)-dsDNA complex from Aeropyrum pernix K1. The overall complex structure reveals a discrete mode of DNA binding, with the positively charged residues on the monomer-monomer interface of each dimer packing on either side of the bound dsDNA in successive minor grooves. Alba-DNA interactions were also clarified by alanine scanning mutagenesis. Analysis of this structure gives insights into how Alba maintains the integrity of genetic material, which is crucial for life at high temperatures.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification
The Alba2 (Ape10b2) gene from A. pernix K1 was cloned, expressed, and purified as described previously (12). The mutation of the indicated residues to alanine was achieved using the QuikChange II site-directed mutagenesis kit (Stratagene), and mutant proteins were purified as described for the wild-type protein.
Crystallization and Data Collection
The 16-bp synthetic oligonucleotides containing the putative promoter sequence 5′-CCCGGCGTGCGGCCCG-3′ and its complementary oligonucleotide 5′-CGGGCCGCACGCCGGG-3′ were annealed completely to form the DNA duplex and complexed with the Ape10b2 protein at an equimolar ratio. The initial crystals of the complex between Ape10b2 and 16-bp DNA were produced at 20 °C by the sitting drop vapor diffusion method (18) by adding 1 μl of protein-DNA complex solution to 1 μl of well solution containing 20% (w/v) PEG 3350 and 0.2 m ammonium dihydrogen phosphate (pH 4.6). The mother liquor with 20% ethylene glycol was used as a cryoprotectant. The complete data sets were collected at RIKEN structural genomics beamline I (BL26B1) at SPring-8. The crystal belongs to the orthorhombic space group P212121 and diffracts up to 2.0 Å resolution. The data set was processed using the HKL2000 suite (see Table 1) (19).
TABLE 1.
Data collection and refinement statistics of the Ape10b2-DNA (16 bp) complex
The r.m.s.d. in bond angles and bond lengths varied from 1.2° to 1.6° and from 0.006 to 0.008 Å, respectively. Solvent content was ∼55%.
| Data collection | |
| Wavelength | 1.0000 |
| Space group | P212121 |
| Cell dimensions (Å) | a = 33.55, b = 74.29, c = 88.19 |
| No. of molecules/asymmetric unit | 2 |
| Resolution range (Å) | 40–2.00 (2.07–2.0)a |
| Unique reflections | 15.617 |
| Redundancy | 14.1 (14.3) |
| Completeness (%) | 99.9 (100) |
| Rmerge (%)b | 0.069 (0.470) |
| Refinement statistics | |
| Resolution range (Å) | 20–2.0 |
| Reflections used in refinement | 15,507 |
| Total no. of reflections used for working set | 14,401 |
| R-factor (%)c | 0.244 |
| Total no. of reflections used for Rfree | 1106 |
| Rfree (%)d | 0.282 |
| No. of protein atoms | 1548 |
| No. of nucleic acid atoms | 199 |
| No. of water molecules | 183 |
| No. of PO4 ions | 1 |
| Average B-factor (Å2) | |
| Protein molecules | 41.2 |
| Nucleic acids | 57.4 |
| Waters | 52.5 |
| Overall | 43 |
| Ramachandran statistics | |
| Most favored regions (%) | 87.1 |
| Allowed regions (%) | 12.9 |
| Protein Data Bank code | 3U6Y |
a Values in parentheses are for the highest resolution shell.
b Rmerge = ΣhΣi|I(h,i) − 〈I(h)〉|/ΣhΣiI(h,i), where I(h,i) is the intensity value of the ith measurement of h, and 〈I(h)〉 is the corresponding mean value of I(h) for all i measurements.
c R-factor = Σ‖Fo| − |Fc‖/|Fo|, where |Fo| and |Fc| are the observed and calculated structure factor amplitudes, respectively.
d Rfree is the same as the R-factor but for a 5–7% subset of all reflections.
Structure Determination and Refinement
The Ape10b2-DNA complex structure with 16-bp nucleotides was determined by the molecular replacement method using our previous Ape10b2 structure (Protein Data Bank code 2H9U) as a search model. The solution was found by automated MOLREP, within the CCP4 program suite (20), and the refinement was carried out using CNS (21). We observed unambiguous density for the DNA bases and built the fragment of the DNA model using the program Quanta (22). The final model was refined and manually fitted using CNS, Coot (23), and Quanta. The final model with 198 protein residues and 10 nucleic acid bases, except for three residues in the C terminus of the A and B chains, respectively, was refined to a crystallographic R-factor of 0.246 (Rfree = 0.286) at 2.0 Å resolution. Figures were prepared with the program PyMOL (24). The coordinates and structure factors for the complex between Ape10b2 and 16-bp DNA have been deposited in the Protein Data Bank under accession code 3U6Y.
EMSA
The purified wild-type and mutant Ape10b2 proteins (0.27–2.1 μm) were incubated with 16 nm 84-bp dsDNA in EMSA buffer (20 mm Tris-HCl (pH 8.0) and 300 mm NaCl) for ∼30 min at room temperature and then run on 5–20% gradient native polyacrylamide gels in 0.5× Tris borate/EDTA buffer (90 V for 2 h at 4 °C). The 84-bp dsDNA fragment from human α-satellite DNA (25) was chosen as the substrate for binding multiple Alba dimers for convenient analysis by gel electrophoresis. Gels were stained with SYBR Green (Invitrogen) and visualized on a phosphor imager (Fujifilm) using MultiGauge image analysis software (Fujifilm).
RESULTS AND DISCUSSION
Overall Structure of Alba2-DNA Complex
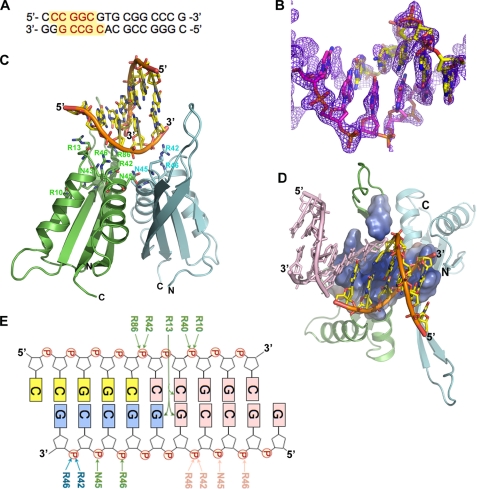
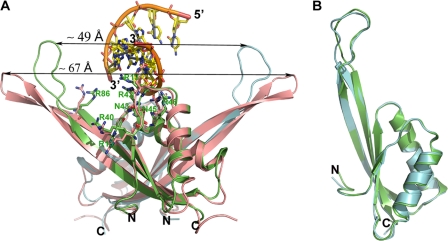
On the basis of our earlier work (12), we crystallized Alba2 with 16-bp duplex DNA and collected a data set to 2.0 Å resolution under space group P212121. The Alba2-DNA complex structure was refined to a final R-value of 24% and an Rfree value of 28% (Table 1). Here, a high-quality electron density map enabled us to build the protein and nucleic acids unambiguously (Fig. 1, A and B). The Alba2 complex crystallized as a dimer with 4-bp dsDNA with a one-nucleotide overhang at the 5′-ends in the asymmetric unit, with overall dimensions of 58 × 57 × 35 Å (Fig. 1C). Using the 21 symmetry operation, we could generate continuous DNA duplexes stabilized by stacking interactions with those of the symmetry-related molecules (Fig. 2). The bound duplex DNA adopted a B-form right-handed structure.
FIGURE 1.
Crystal structure of Alba2-dsDNA complex. A, 16-mer dsDNA sequence used for crystallization. The indicated red residues are modeled in the electron density. B, final 1Fo − Fc omit electron density map, with nucleic acids contoured at the 2σ level shown in blue mesh. The template strand (magenta) and its complementary strands (yellow) are shown in stick models. C, ribbon diagram of Alba2, with distal (green) and proximal (cyan) subunits and stick model of the bound DNA (yellow). The above color code is uniformly used in all of the figures. D, Alba2-dsDNA complex viewed from the “top,” with electrostatic surface representation of the interacting residues. The dsDNA from the adjacent asymmetric unit (light pink) was included for a complete view of the “tripartite clamp.” E, schematic representation showing Alba2-dsDNA contacts. The distal (green) and proximal (cyan) interacting residues are marked by arrows. The dsDNA from the adjacent asymmetric unit (light pink) was included to show the symmetric interaction.
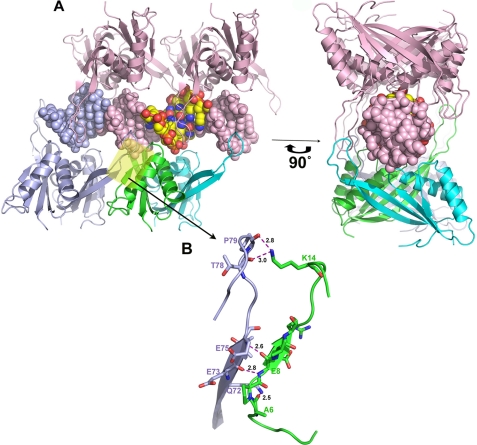
FIGURE 2.
Oligomerization of Alba2-DNA complex. A, individual asymmetric unit of the complex showing the distal (green) and proximal (cyan) subunits and bound dsDNA as a spherical model (yellow). The adjacent asymmetric units arranged on either side of the dsDNA are marked (top, light pink; bottom, light blue). The same figure is rotated −90° along the y axis. The Alba2 dimer-dimer interface is marked with a transparent yellow box. B, in this close-up view, the residues involved in dimer-dimer interactions (indicated in transparent yellow) are indicated by their respective chain color, and hydrogen bonds are represented by dashed magenta lines.
Alba2 Dimer Interacts with Minor Groove of dsDNA
The overall structure of the Alba2 subunit in complex with the DNA is similar to the apo-Alba2 structure (12), consisting of two α-helices and four β-strands arranged in the order β1-α1-β2-α2-β3-β4 in the primary structure. In the double helical B-DNA, the major and minor grooves lay 180° opposite each other, spiraling along the axis of the molecule. The Alba2-DNA complex structure reveals how the Alba2 dimer binds the extended duplex DNA; the positively charged residues in the monomer-monomer interface of each dimer pack on either side of the bound DNA in the successive minor grooves. The residues in the loops connect to the C-terminal ends of helices α1 and α3 from each subunit, like a helix-turn-helix motif, making a tripartite clamp that binds diagonally across the minor groove (Fig. 1D). Each Alba2 subunit within a given asymmetric unit recognizes distinct bases and docks differently onto the DNA, with the distal (with respect to the bound DNA) subunit (green) making the majority of the base-specific and phosphate contacts and the proximal subunit (cyan) making only phosphate interactions (Fig. 1, C and E). The total solvent-accessible surface area buried between the Alba2-DNA interfaces is 1453 Å2 (using a 1.4 Å probe), including the dsDNA in the adjacent asymmetric unit, which interacts with distal subunits. The Alba2 dimer binding of the minor groove of the duplex DNA is further supported by displacement of DAPI from the minor groove (8).
As shown in Fig. 1C, part of the DNA fragment (4-bp dsDNA with a one-nucleotide overhang at the 5′-end) bound to the dimeric molecules of Alba2. The DNA fragments observed in three adjacent asymmetric units of the dimeric molecules probably form one full-length (16-bp) dsDNA of the cassette/duplex used for analysis of this complex (Fig. 2A). The full-length DNA duplex in the Alba2-DNA complex was also confirmed by dissolving the crystals in water, after washing them several times in reservoir solution. Analysis of these dissolved samples by agarose gel electrophoresis indicated that the DNA duplex present in the crystals was indeed the full-length DNA (data not shown). It is noteworthy to mention that the 16-bp dsDNA has no sequence repeats (Fig. 1A); however, the DNA is intrinsically disordered and only partly bound to the protein molecules. Although we built the DNA model based on the observed density, it can be modeled with other nucleotides because the densities may be disordered and averaged out. In the distal monomer, Arg-13, Arg-42, Asn-43, Asn-45, and Arg-46 form the center of the clamp, in which Arg-13 and Arg-42 are buried within the minor groove. Arg-13 forms three hydrogen bonds with three bases (one base within the asymmetric unit and two bases through symmetric interactions) (Fig. 1E). Arg-42 interacts with the backbone phosphate (O1P) and also with O4′ of the sugar moiety with the symmetry-related molecule. Arg-46 interacts with a phosphate in the DNA backbone, and Asn-43 symmetrically interacts with the sugar. In addition, the side chains of Arg-46 NH2 and Asn-43 Nδ2 form hydrogen bonds with the Arg-13 and Gly-12 main chain CO groups at distances of 3.29 and 3.25 Å, respectively. These interactions shift the position of the loop connecting β1 and α1 by 2.5 Å compared with the apo-Alba structure to place Arg-13 optimally within the minor groove. Arg-10, Arg-40, and Arg-86 from the distal monomer are placed on the major groove side of the DNA backbone, and the side chains of these residues symmetrically interact with phosphate groups. Interestingly, Asn-45 is in the center of the monomer-monomer interface (Fig. 1C). The side chains of Asn-45 Nδ2 and Oδ1 form strong hydrogen bonds with Asn-45 Oδ1 and a main chain NH group in the proximal subunit, as well as a phosphate group of the DNA backbone. In the proximal monomer, Arg-42 and Arg-46 on the major groove side of the DNA backbone interact with the phosphate group.
The extended hairpin loop connecting strands β3 and β4 does not contact the bound DNA (Fig. 1D). The average B-factor of the loop region (residues 74–86, 76 Å2) was higher compared with that of other regions of the structure (38 Å2), and it was evident that the loop remains mobile. Recent NMR studies mapping the amide backbone chemical shift changes in Sso10b support our structural data. No changes are observed in chemical shifts, especially in this region, when Alba binds to dsDNA, ssDNA, and RNA, and the loop remains flexible (13). Alba oligomerizes through a dimer-dimer interface on both sides of the DNA (180° opposite to each other), and the extended hairpin loops connecting strands β3 and β4 are arranged in a zip-like structure. Thus, the overall extended structure of the Alba2-DNA complex forms a rectangular tube-like structure consisting of two faces of the zip-like structure with the duplex DNA at the center, to which the positively charged residues in the Alba monomer-monomer interface are anchored in the minor groove (Fig. 2A). The width and height of the rectangular pipe are ∼84 and 47 Å, respectively, comparable with reported electron microscopic studies on an Alba-DNA complex of high binding density showing a diameter of ∼10–11 nm (16). Thus, the Alba2-DNA structure shows a stoichiometric ratio of one 4-bp duplex DNA fragment with a one-nucleotide overhang at the 5′-end per dimer, consistent with the existing ratio of 6 bp/dimer at the high-density level based on biochemical analyses (7, 8).
Intermolecular Interactions between Dimer-Dimer Interfaces
The dimer-dimer interactions were mediated by docking of β3 with the β1 strand from the neighboring symmetry-related Alba protein through two main chain hydrogen bonds, resulting in the extended structure. In addition, the side chain Nϵ2 of Gln-72 in the β3 strand forms a hydrogen bond with the carbonyl group of Ala-6 in the symmetry-related molecule at a distance of 2.5 Å. Intriguingly, Lys-14 of the Alba2 molecule forms two hydrogen bonds with the main chain CO groups of Pro-79 and Glu-80 in the extended loop at distances of 2.8 and 3.0 Å, respectively (Fig. 2B). Lys-14 in this dimer-dimer interface is highly conserved within this family of proteins. It might play an important role in positioning the extended loop. The mutational analysis of equivalent residues in Sso10b (15) and Archaeoglobus fulgidus Alba (10) showed reduced DNA-binding affinity. Moreover, the crystal packing shows that DNA is surrounded by the positive electrostatic potential of the oligomerized Alba dimer. The other side of the dimer-dimer interface shows no interaction, and the closest distance between two subunits is 11.3 Å. The total buried surface area in the dimer-dimer interface is 931 Å2.
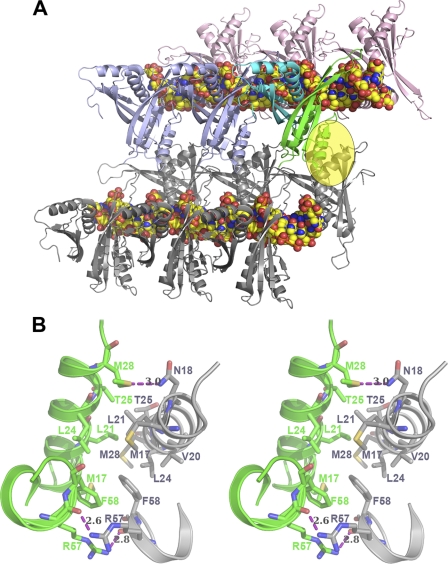
We also noticed the dimer-dimer contacts between the two fibers as observed by Jelinska et al. (13) in our structure (Fig. 3). This dimer-dimer interface consists of two distinct regions. First, the α1 helices (Met-17–Met-28) generate an antiparallel interaction between hydrophobic residues with the adjacent dimer subunit. Most of the residues in the α1 helix are highly conserved within this family of proteins (Fig. 4). In addition, one hydrogen bond contact was observed between Asn-18 Nδ2 and Met-28 sulfur δ with a distance of 3.0 Å. Second, Arg-57 NH2 forms a symmetric hydrogen bond with Arg-57 CO in the adjacent subunit at distances of 2.6 and 2.8 Å, respectively. Those two observed hydrogen bonds further stabilize the hydrophobic stacking interactions between symmetric Phe-58 residues. Recent NMR analysis of Arg-59 and Phe-60 in Sso10b, which correspond to Arg-57 and Phe-58 in Ape10b2, showed that these residues are important for oligomerization and protein-DNA interaction (13). Based on biochemical as well as our current structural studies, the above dimer-dimer contact might play a key role in bringing several extended fibers of Alba-DNA complex chains together during the DNA compaction. However, the B-form DNA packed by Alba proteins in this way remains straight. Previous studies have shown that a small chromosomal protein, such as Sac7d, can bind nonspecifically to the DNA minor groove and sharply kink duplex DNA (∼60°) via the intercalation of both Val-26 and Met-29 (26). Thus, we suggest that these Sac7d families of proteins may also be involved in bending/kinking the DNA in such a way as to pack the chromosomal DNA more tightly in the cell.
FIGURE 3.
Compaction of Alba2-DNA complex. A, extended Alba2-dsDNA complex (fiber) made similarly to Fig. 2. The adjacent Alba2-dsDNA extended fiber is shown in gray. The Alba2 dimer-dimer interface between the fibers is marked with a transparent yellow oval. B, stereo view of the dimer-dimer interface. In this close-up view, the residues involved in dimer-dimer interactions (transparent yellow oval) are indicated by their respective chain color, and hydrogen bonds are represented by dashed magenta lines.
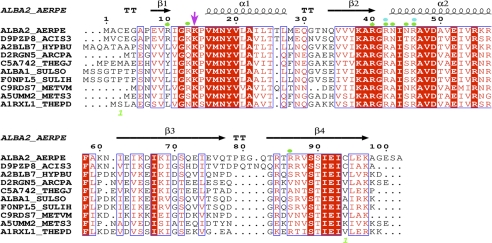
FIGURE 4.
Sequence alignment of archaeal Alba homologs: A. pernix (Q9YAX2), Acidilobus saccharovorans (D9PZP8), Hyperthermus butylicus (A2BLB7), Archaeoglobus profundus (D2RGN5), Thermococcus gammatolerans (C5A742), S. solfataricus (P60849), Sulfolobus islandicus (F0NPL5), Methanocaldococcus vulcanius (C9RDS7), Methanobrevibacter smithii (A5UMM2), and Thermofilum pendens (A1RXL1). The secondary structure of the apo-Ape10b2 structure (Protein Data Bank code 2H9U) is shown above the alignment. Residues involved in DNA contacts are indicated by green and cyan dots, and the conserved Lys-17 residue (Sso10b2) is indicated by the magenta arrow.
Conformational Change
Similar topologies allowed superposition of the apo- and DNA-bound Alba2 structures, giving a root mean standard deviation (r.m.s.d.)3 of 0.76 Å for the Cα atoms. Interestingly, large conformational changes are observed in the extended loop connecting the β3-β4 hairpins upon DNA binding (Fig. 5A). In the apo form, the β3-β4 hairpins (residues 61–96) form 16-residue-long β-sheets connected by a short turn with four residues. In the DNA-bound complex, the length of the β-sheets is reduced to 12 residues, and that of the extended loop is increased to 13 residues. Superposition of the β3-β4 hairpins (residues 61–96) between the apo- and DNA-bound structures gave an r.m.s.d. of 1.28 Å. This structural change may play an important role in oligomerization during DNA binding to fit the incoming Alba2 dimer cooperatively without steric clashing. In comparing both structures, it is evident that the key residues involved in Alba2-DNA interactions, such as Arg-10, Arg-13, Arg-42, Arg-46, and Arg-86, changed their side chain orientation to establish contacts with the bound DNA. Superposition of Alba2 subunits within the asymmetric unit of the DNA-bound complex shows an r.m.s.d. of 0.55 Å. Intriguingly, the length of the β-sheets in the β3-β4 hairpins differs by one residue, with little change in the conformation of the extended loop region. Superposition of the β3-β4 hairpins (residues 61–96) between the subunits gives an r.m.s.d. of 0.64 Å, slightly higher than with the full-length molecule (Fig. 5B). The distances between the extended loops in the apo- and DNA-bound complexes are 67 and 49 Å, respectively. The shortened distance between the extended loops is due to the conformational changes in the β3-β4 hairpins and translocation of the extended loops by 12 Å toward the monomer-monomer interface.
FIGURE 5.
Conformational changes observed upon DNA binding. A, superposition of the apo-Alba2 structure (light red; Protein Data Bank 2H9U) onto the Alba2-dsDNA complex. Distances between the extended loops connecting the β3-β4 hairpins are marked. Key residues interacting with DNA are shown as a stick model. B, superposition of Alba2 subunits within the asymmetric unit of the DNA-bound complex.
Transcription factors use cooperativity for binding specificity even at low protein concentrations. In effect, cooperativity leads to synergistic responses, so small changes in activator concentration can dramatically alter binding and promoter activity (27). Accordingly, the Alba2-DNA complex exhibits cooperative binding through protein-protein interactions resulting from conformational changes in the β3-β4 hairpins. This suggests that the interaction of the first Alba2 protein with DNA induces an allosteric change in the protein, which in turn increases its affinity for an incoming one. Thus, the interaction between two Alba2 proteins is enhanced and stabilized by DNA.
Alba is subject to post-translational acetylation of Lys-16 in Sso10b, which appears to play a key role in chromatin regulation in archaea (14, 15). Interestingly, the acetylated Lys-16 residue of Sso10b is not conserved among thermophiles, whereas the adjacent Lys-17 residue is highly conserved within this protein family; these residues are equivalent to Arg-13 and Lys-14 in Ape10b2 (Fig. 4). In the Alba2-DNA complex, Arg-13 is important for DNA binding, whereas Lys-14 forms intermolecular interactions with the extended loop in the β3-β4 hairpins, thereby fixing the loop position and helping to stack the incoming Alba protein along the DNA axis. Because the acetylated Lys-16 residue is not conserved within the Alba protein family, potential acetylated residues in other archaea will need to be identified to understand its role in chromatin regulation.
Mutagenesis of DNA-interacting Residues in Alba2
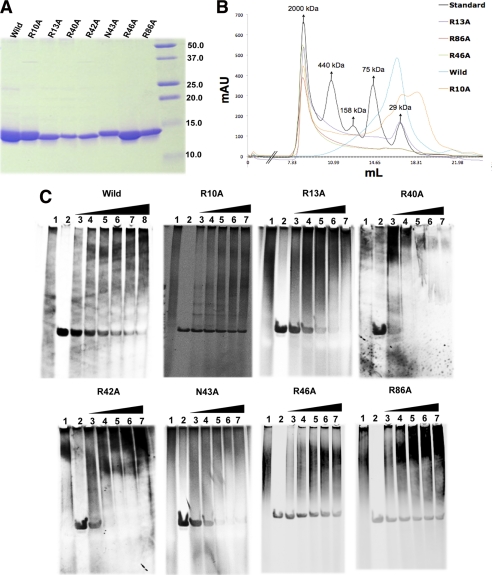
To determine the contribution of key residues, we expressed Alba2 mutants with alanine substituted at Arg-10, Arg-13, Arg-40, Arg-42, Asn-43, Arg-46, or Arg-86 (Fig. 6A). Analytical gel filtration and spectrometric analysis of the purified wild-type protein and seven mutants showed higher order oligomers of various sizes in solution with high absorbance at 260 nm (Fig. 6B). Consistent with these results, the wild-type and mutant proteins also migrated as a smeared band on the native gel stained with SYBR Green and Coomassie blue. Thus, more than one species was present through nonspecific binding to genomic DNA, and these complexes were in equilibrium while migrating through the gel. EMSA was performed with purified proteins incubated with a fixed quantity of unlabeled 84-bp dsDNA. The complexes were resolved on a native polyacrylamide gel and visualized using SYBR Green (Fig. 6C). The results could not be quantified due to high background signals and the lack of discrete bands in the gel. However, decreasing oligonucleotide levels with increasing protein concentrations were observed for both wild-type and mutant proteins. The difference in the affinity for dsDNA, ssDNA, and RNA for Alba (13) may have played a role in the exchange of nonspecifically bound host nucleic acid with the 84-bp dsDNA during the incubation. Intriguingly, the extent of decrease in the 84-bp dsDNA varied between the wild-type and mutant proteins, suggesting differences in DNA-binding affinity. These differences may be due to a weaker interaction of the mutant with genomic DNA compared with the wild-type protein, which resulted in a greater exchange rate and shift in the 84-bp dsDNA compared with the wild-type protein. Notably, we observed a 4–12-fold lower DNA-binding ability for the R13A, R40A, R42A, and N43A mutants. The R86A mutant had a moderate effect, whereas the R10A and R46A mutants showed weak or no contribution to DNA binding. These results are supported by previous alanine substitutions at Lys-16 and Lys-17 in Sso10b (15) and the fact that acetylated and non-acetylated forms of Alba show only a 3–4-fold difference in binding affinity (28). Together, these results support the specificity of the Alba2-DNA interaction.
FIGURE 6.
Alba2 purification and DNA binding analysis. A, Coomassie Blue-stained gel showing purified wild-type and mutant proteins. B, analytical gel filtration (Superdex 200 10/300 GL) showing the oligomerization state of purified wild-type and mutant proteins. mAU, milli-absorbance units. C, EMSAs with increasing concentrations of purified recombinant wild-type and mutant proteins with a fixed concentration of 84-bp dsDNA stained with SYBR Green. Lanes 1 and 2, protein and 84-bp dsDNA control; lanes 3–8, wild-type protein at 270 nm, 526 nm, 800 nm, 1.05 μm, 1.5 μm, and 2.1 μm, respectively; lanes 3–7, mutant proteins at 53, 167, 270, 416, and 526 nm, respectively.
In summary, we have reported here the first high-resolution structure of the Alba2-DNA complex. Consistent with biochemical analyses of the Alba-DNA complex, our structure shows the Alba2 monomer-monomer interface docking on either side of the cognate DNA in successive minor grooves. Alba2 binds to a 4-bp duplex DNA fragment with a one-nucleotide overhang at the 5′-end, supporting the existing stoichiometric ratio of 6 bp/dimer at the high-density level. Upon DNA binding, Alba2 undergoes conformational changes in the extended hairpin loops connecting strands β3 and β4, which are important for packing the other symmetry-related Alba molecule along the DNA axis. Based on our structure and available biochemical data, the dimer-dimer (intra- and inter-fiber) interface might be biologically important for packaging and compaction of DNA. Further studies of Alba and other hyperthermophilic DNA-binding proteins may reveal how these proteins function in chromatin organization and gene regulation.
Acknowledgments
We thank M. Okada for purification of the mutant proteins and Dr. T. Ishikawa for support and encouragement.
This research work was supported by the “Program for RIKEN SPring-8 Center projects to meet society's needs.”
The atomic coordinates and structure factors (code 3U6Y) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- r.m.s.d.
- root mean standard deviation.
REFERENCES
- 1. Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, 251–260 [DOI] [PubMed] [Google Scholar]
- 2. Sandman K., Reeve J. N. (2005) Archaeal chromatin proteins: different structures but common function? Curr. Opin. Microbiol. 8, 656–661 [DOI] [PubMed] [Google Scholar]
- 3. White M. F., Bell S. D. (2002) Holding it together: chromatin in the Archaea. Trends Genet. 18, 621–626 [DOI] [PubMed] [Google Scholar]
- 4. Keeling P. J., Doolittle W. F. (1995) Archaea: narrowing the gap between prokaryotes and eukaryotes. Proc. Natl. Acad. Sci. U.S.A. 92, 5761–5764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Marc F., Sandman K., Lurz R., Reeve J. N. (2002) Archaeal histone tetramerization determines DNA affinity and the direction of DNA supercoiling. J. Biol. Chem. 277, 30879–30886 [DOI] [PubMed] [Google Scholar]
- 6. Forbes A. J., Patrie S. M., Taylor G. K., Kim Y. B., Jiang L., Kelleher N. L. (2004) Targeted analysis and discovery of post-translational modifications in proteins from methanogenic archaea by top-down MS. Proc. Natl. Acad. Sci. U.S.A. 101, 2678–2683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xue H., Guo R., Wen Y., Liu D., Huang L. (2000) An abundant DNA-binding protein from the hyperthermophilic archaeon Sulfolobus shibatae affects DNA supercoiling in a temperature-dependent fashion. J. Bacteriol. 182, 3929–3933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wardleworth B. N., Russell R. J., Bell S. D., Taylor G. L., White M. F. (2002) Structure of Alba: an archaeal chromatin protein modulated by acetylation. EMBO J. 21, 4654–4662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jelinska C., Conroy M. J., Craven C. J., Hounslow A. M., Bullough P. A., Waltho J. P., Taylor G. L., White M. F. (2005) Obligate heterodimerization of the archaeal Alba2 protein with Alba1 provides a mechanism for control of DNA packaging. Structure 13, 963–971 [DOI] [PubMed] [Google Scholar]
- 10. Zhao K., Chai X., Marmorstein R. (2003) Structure of a Sir2 substrate, Alba, reveals a mechanism for deacetylation-induced enhancement of DNA binding. J. Biol. Chem. 278, 26071–26077 [DOI] [PubMed] [Google Scholar]
- 11. Wang G., Guo R., Bartlam M., Yang H., Xue H., Liu Y., Huang L., Rao Z. (2003) Crystal structure of a DNA-binding protein from the hyperthermophilic euryarchaeon Methanococcus jannaschii. Protein Sci. 12, 2815–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kumarevel T., Sakamoto K., Gopinath S. C., Shinkai A., Kumar P. K., Yokoyama S. (2008) Crystal structure of an archaeal specific DNA-binding protein (Ape10b2) from Aeropyrum pernix K1. Proteins 71, 1156–1162 [DOI] [PubMed] [Google Scholar]
- 13. Jelinska C., Petrovic-Stojanovska B., Ingledew W. J., White M. F. (2010) Dimer-dimer stacking interactions are important for nucleic acid binding by the archaeal chromatin protein Alba. Biochem. J. 427, 49–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marsh V. L., Peak-Chew S. Y., Bell S. D. (2005) Sir2 and the acetyltransferase, Pat, regulate the archaeal chromatin protein, Alba. J. Biol. Chem. 280, 21122–21128 [DOI] [PubMed] [Google Scholar]
- 15. Bell S. D., Botting C. H., Wardleworth B. N., Jackson S. P., White M. F. (2002) The interaction of Alba, a conserved archaeal chromatin protein, with Sir2 and its regulation by acetylation. Science 296, 148–151 [DOI] [PubMed] [Google Scholar]
- 16. Lurz R., Grote M., Dijk J., Reinhardt R., B D. (1986) Electron microscopic study of DNA complexes with proteins from the archaebacterium Sulfolobus acidocaldarius. EMBO J. 5, 3715–3721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cui Q., Tong Y., Xue H., Huang L., Feng Y., Wang J. (2003) Two conformations of archaeal Ssh10b. The origin of its temperature-dependent interaction with DNA. J. Biol. Chem. 278, 51015–51022 [DOI] [PubMed] [Google Scholar]
- 18. McPherson A. (1990) Current approaches to macromolecular crystallization. Eur. J. Biochem. 189, 1–23 [DOI] [PubMed] [Google Scholar]
- 19. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 20. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 21. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 22. Oldfield T. J. (2001) A number of real-space torsion-angle refinement techniques for proteins, nucleic acids, ligands, and solvent. Acta Crystallogr. D Biol. Crystallogr. 57, 82–94 [DOI] [PubMed] [Google Scholar]
- 23. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 24. DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
- 25. Dyer P. N., Edayathumangalam R. S., White C. L., Bao Y., Chakravarthy S., Muthurajan U. M., Luger K. (2004) Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 375, 23–44 [DOI] [PubMed] [Google Scholar]
- 26. Robinson H., Gao Y. G., McCrary B. S., Edmondson S. P., Shriver J. W., Wang A. H. (1998) The hyperthermophile chromosomal protein Sac7d sharply kinks DNA. Nature 392, 202–205 [DOI] [PubMed] [Google Scholar]
- 27. Georges A. B., Benayoun B. A., Caburet S., Veitia R. A. (2010) Generic binding sites, generic DNA-binding domains: where does specific promoter recognition come from? FASEB J. 24, 346–356 [DOI] [PubMed] [Google Scholar]
- 28. Guo R., Xue H., Huang L. (2003) Ssh10b, a conserved thermophilic archaeal protein, binds RNA in vivo. Mol. Microbiol. 50, 1605–1615 [DOI] [PubMed] [Google Scholar]