Background: Protease-activated receptor 4 (PAR4) mediates thrombin signaling on platelets and other cells.
Results: Disruption of the PAR4 homodimer interface interferes with signaling.
Conclusion: Dimerization of PAR4 is critical for signaling.
Significance: The interactions of PAR4 at the plasma membrane may have important functions that regulate signaling.
Keywords: Bioluminescence Resonance Energy Transfer (BRET), G Protein-coupled Receptors (GPCR), Hemostasis, Membrane Proteins, Mutagenesis Site-specific, Protein-Protein Interactions, Receptor Structure-Function, Protease-activated Receptor, Protease-activated Receptor 4
Abstract
Thrombin activates platelets by binding and cleaving protease-activated receptors 1 and 4 (PAR1 and PAR4). Because of the importance of PAR4 activation on platelets in humans and mice and emerging roles for PAR4 in other tissues, experiments were done to characterize the interaction between PAR4 homodimers. Bimolecular fluorescence complementation and bioluminescence resonance energy transfer (BRET) were used to examine the PAR4 homodimer interface. In bimolecular fluorescence complementation experiments, PAR4 formed homodimers that were disrupted by unlabeled PAR4 in a concentration-dependent manner, but not by rhodopsin. In BRET experiments, the PAR4 homodimers showed a specific interaction as indicated by a hyperbolic BRET signal in response to increasing PAR4-GFP expression. PAR4 did not interact with rhodopsin in BRET assays. The threshold maximum BRET signal was disrupted in a concentration-dependent manner by unlabeled PAR4. In contrast, rhodopsin was unable to disrupt the BRET signal, indicating that the disruption of the PAR4 homodimer is not due to nonspecific interactions. A panel of rho-PAR4 chimeras and PAR4 point mutants has mapped the dimer interface to hydrophobic residues in transmembrane helix 4. Finally, mutations that disrupted dimer formation had reduced calcium mobilization in response to the PAR4 agonist peptide. These results link the loss of dimer formation to a loss of PAR4 signaling.
Introduction
The protease-activated receptors (PAR)2 are a class of G protein-coupled receptors (GPCR) that are activated by the proteolysis of the N-terminal exodomain (1). Upon proteolysis, the newly formed N terminus acts as a tethered ligand that activates the receptor and initiates signaling cascades through multiple G proteins (Gαq, Gαi, and Gα12/13) (2, 3). There are four members of this receptor family (PAR1–4) that are expressed on a variety of cells including platelets, endothelial cells, vascular smooth muscle cells, and inflammatory cells. PAR1, 3, and 4 are activated by thrombin, whereas PAR2 is activated by trypsin or tryptase. Other serine proteases like FVIIa, FXa, plasmin, activated protein C, MMP1, MASP-1, and cathepsin G can activate PARs as well (4–10). Unlike conventional G protein-coupled receptors, PARs are irreversibly activated by cleavage of the exodomain; therefore, specific mechanisms are required to turn off PAR signaling (11).
PAR4 has primarily been studied in the context of platelet activation. PAR1 and PAR4 are expressed on human platelets and mediate thrombin activation of platelets. However, on mouse platelets, PAR4 is the signaling thrombin receptor (12, 13). On cells, PAR1 is activated by low concentrations of thrombin (1 nm), whereas PAR4 is activated by higher concentrations of thrombin. However, PAR1 lowers the amount of thrombin required for PAR4 activation when PAR1 and PAR4 are co-expressed on the same cell, as is the case for human platelets (14, 15). PAR1 and PAR4 signaling first appeared to be redundant, but recent data suggest that each has unique pathways for platelet activation (16–18). A major difference in PAR1 and PAR4 signaling is that, upon activation, PAR1 is phosphorylated on key serine residues, which results in rapid internalization and degradation of the receptor (11). In contrast, PAR4 is not phosphorylated and does not internalize as rapidly, resulting in a prolonged stimulus (19, 20). The sustained signaling from PAR4 may be required for stable clot formation (21, 22). In addition to its role in activation of platelets by thrombin, there is increasing evidence of roles for PAR4 in other physiological processes including nociception, colon cancer cell proliferation, cardiac response to ischemia/reperfusion injury, and diabetic arterial plaques (23–26).
In addition to the interaction between PAR1 and PAR4, PAR4 also interacts with the P2Y12 receptor on platelets to alter signaling (27). Recent studies also suggest that other PAR subtypes can interact with one another to alter signaling from these receptors. Kaneider et al. (28) demonstrated that the PAR1-PAR2 heterodimers signaled through a different set of G proteins than PAR1, which changed the physiological response of the endothelial cells from barrier disruptive to barrier protective. PAR1 and PAR2 also form heterodimers on smooth muscles cells and regulate their response to arterial injury (29). McLaughlin et al. (30) have also demonstrated that PAR1 homodimers couple to a different set of G proteins than PAR1-PAR3 heterodimers in transfected HEK293 cells. In summary, PARs, like other GPCRs, may form dimers to mediate multiple effects on cells depending on the context.
PAR4 has roles in multiple physiological processes. In addition, PAR4 may be implementing cross-talk between other receptors. To fully understand how PAR4 is signaling to cells, it is imperative to determine the molecular details of how PAR4 is arranged in the membrane of cells. The present study uses independent methods to demonstrate that PAR4 forms homodimers: bimolecular fluorescence complementation (BiFC) and bioluminescence resonance energy transfer (BRET). The present study has also used a panel of chimeric proteins and PAR4 point mutants to map the region on PAR4 required for homodimers to a hydrophobic interface within transmembrane helix 4.
EXPERIMENTAL PROCEDURES
Reagents
Unless otherwise stated, all of the reagents were from Sigma-Aldrich.
Cell Culture
HEK293 cells were from American Type Culture Collection (Bethesda, MD) and cultured in DMEM supplemented with 10% fetal bovine serum (Hyclone, Logan, UT). Cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
Molecular Cloning
The human PAR4 cDNA was purchased from UMR cDNA Resource Center. The mouse rhodopsin cDNA (accession number P15409) was provided by Dr. Krzysztof Palczewski (Case Western Reserve University). The restriction enzyme sites HindIII and KpnI were added to PAR4 by PCR to facilitate ligation into the vectors V1 and V2 for BiFC or pLUC-N2 and pGFP2-N2 for BRET. The vectors D2-V1 and D2-V2 were purchased from Addgene (31), and the D2 sequence was replaced with that of PAR4. The vectors pLUC-N2 and pGFP-N2 were purchased from PerkinElmer Life Sciences. The rhodopsin/PAR4 chimeras were generated using overlapping PCR as previously described (32). A detailed list of primers is available by request. Briefly, primers were designed such that the sequence would switch from rhodopsin to PAR4 at the beginning of transmembrane domain (TM) 2 (rho-PAR4-TM2), TM3 (rho-PAR4-TM3), TM4 (rho-PAR4-TM4), TM5 (rho-PAR4-TM5), TM6 (rho-PAR4-TM6), or TM7 (rho-PAR4-TM7) (see Table 1). The resulting chimeras were ligated into the vector pGFP2-N2 (PerkinElmer Life Sciences) for BRET studies. The transmembrane 4 domain swap mutant (rho-PAR4-rho) was made in a similar fashion. Alanine substitution mutants were made using primers that altered the relevant codon(s) to change L2094.58 and L2134.62 (PAR4–2A); L1924.41, L1944.43, M1984.47, and L2024.51 (PAR4–4A); or individual alanine substitutions. The nomenclature is that of Ballesteros and Weinstein (33). For expression without GFP, PAR4 or rhodopsin was ligated into pCDNA3.1. Human PAR4 with an N-terminal T7 epitope (MASMTGGQQMG) has been described (14); T7-rhodopsin was made in a similar fashion. The HA tag (YPYDVPDYA) or V5 tag (GKPIPMPLLGLDST) was added to the N terminus of PAR4 with PCR at amino acid 18 such that the PAR4 signal sequence was removed.
TABLE 1.
Rhodopsin-PAR4 chimeras
Sequences are shown with standard one-letter abbreviations. Rhodopsin sequences are shown in plain font. PAR4 sequences are shown in bold font.
| Chimera | Sequence |
|---|---|
| Rho-PAR4-TM2 | … VTVQHKKLRT70S111TMLLMNLA … |
| Rho-PAR4-TM3 | … TSLHGYFVFG106L151ATAALLYG … |
| Rho-PAR4-TM4 | … KPMSNFRFGE150L192ALGLCMAA … |
| Rho-PAR4-TM5 | … DYYTLKPEVN199C247LALLGCFL … |
| Rho-PAR4-TM6 | … QQESATTQKA246L284TAVVLASA … |
| Rho-PAR4-TM7 | … FTHQGSNFGP285G320AYVPSLAL … |
| Rho-PAR4-Rho | … MSNFRFGE150L192ALGLCMAAWLMAAALALPLTL213G174WSRYIPE … |
Bimolecular Fluorescence Complementation
HEK293 cells were transfected with HA-PAR4-V1 (0.1 μg) and V5-PAR4-V2 (0.01–0.5 μg). For competition experiments, T7-PAR4-pCDNA3.1 or T7-rhodopsin-pCDNA3.1 (0–1 μg) was co-transfected with HA-PAR4-V1 or V5-PAR4-V2 (0.1 μg each). Forty-eight hours following transfection, the cells were removed from plates by rinsing in PBS containing 1% glucose and immediately analyzed for fluorescence complementation or stained with anti-HA or anti-V5 antibodies to determine expression levels. Fluorescence complementation was determined by measuring the mean fluorescence of 10,000 cells using a BD Biosciences LSRII flow cytometer.
Bioluminescence Resonance Energy Transfer
For BRET experiments, HEK293 cells (1 × 105) were plated on 6-well dishes. The cells were transfected with 0.5 μg of donor plasmid (PAR4-Luc) and increasing amounts (0–5 μg) of acceptor plasmid (PAR4-GFP, rho-GFP, or rho-PAR4-GFP chimeras) (30, 34). For BRET studies with PAR4 point mutants, 0.03 μg of donor plasmid and 0–0.24 μg of acceptor plasmid was used. Emission was detected using a PerkinElmer Life Sciences Victor 3 plate reader equipped with the appropriate BRET2 filter set (410 nm with 80-nm bandpass and 515 nm with 30-nm bandpass; PerkinElmer Life Sciences). Emission at 410 and 515 nm was collected immediately after the addition of 5 μm luciferase substrate (coelenterazine 400a; Biotium Inc., Hayward, CA). BRET signal was calculated by the ratio of emission at 515 nm to emission at 410 nm minus the BRET in the absence of GFP (35). In BRET studies, specific interactions are detected by a hyperbolic increase in BRET signal (ratio of emission at 510/410 nm) as the ratio of GFP-receptor/RLuc-receptor increases (34). In contrast, nonspecific interactions increase linearly. The graphing software Prism was used to choose the best model (hyperbolic versus linear) for each experiment. GFP expression was determined by excitation at 495 nm and emission at 515 nm. Luciferase expression was determined by adding 5 μm coelenterazine H (Invitrogen) and reading total light emission without a filter. The BRET data from three or four independent experiments are pooled and analyzed by global fit to all of the data.
Flow Cytometry
Cell surface expression of PAR4, rhodopsin, or rho-PAR4 chimeras was determined by flow cytometry. HEK293 cells in 6-well plates were transfected with 0.5 μg of plasmid and were removed from plates 48 h post-transfection by rinsing with PBS. The cells were incubated with antibodies to PAR4 (32) or rhodopsin (monoclonal antibody B6–30N (36), kindly provided by Dr. Krzysztof Palczewski (Case Western Reserve University)) followed by incubation with species specific antibodies conjugated to Alexa Fluor 647 (Invitrogen) to detect surface expression distinct from GFP expression. Mean fluorescence was determined by counting 10,000 cells on an LSRII (BD Biosciences) in the Case Comprehensive Cancer Center flow cytometry core. Surface detection of HA-, V5-, or T7-tagged proteins were detected with an HA tag antibody conjugated to Alexa Fluor 647 with 1:50 dilution (6E2; Cell Signaling Technology, Danvers, MA), V5 tag antibody conjugated to DyLight 649 at 1 μg/ml (ab82889; Abcam, Cambridge, MA), or T7 tag antibody conjugated to SureLight allophycocyanin at 1 μg/ml (ab72565, Abcam).
Quantitative Flow Cytometry
To quantitate the cell surface expression of HA- or V5-tagged PAR4, quantitative flow cytometry was performed using QuantumTM Simply Cellular® anti-mouse IgG (Bangs Laboratories, Inc., Fishers, IN) according to the manufacturer's protocol. Beads coated with 0, 7675, 50411, 127964, or 357217 antibody binding sites were incubated with anti-HA or anti-V5 antibodies, washed, and analyzed by flow cytometry for mean fluorescence. These values were used to generate a standard curve with the manufacturer's QuickCal® analysis template v2.3. The expression levels of HA-PAR4 or V5-PAR4 were analyzed by flow cytometry as described above and converted to antibody-binding sites using this template and standard curve.
Calcium Mobilization
HEK 293 cells were transfected with V5-PAR4-wt, V5-PAR4–2A, or V5-PAR4–4A (1 μg) and removed from plates 48 h post-transfection. Calcium mobilization was performed as previously described (37). Briefly, the cells were removed from plates with Versene, washed, and loaded with 5 μm Fura2-AM (Invitrogen) in Hepes-Tyrode's buffer supplemented with magnesium and calcium. Cells (2.5 × 105) were placed into 96-well plates, stimulated with PAR4 agonist peptide (AYPGKF-NH2, 1 mm; Polypeptide Group, Strasbourg, France), and read in a NOVOstar plate reader (BMG Labtech, Durham, NC). Fluorescence measurements were converted to intracellular calcium concentration by the formula of Grynkiewicz et al. (38).
RESULTS
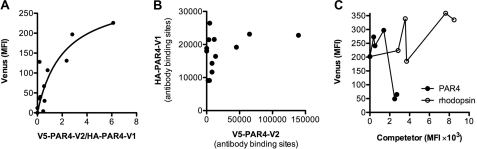
PAR4 participates in multiple physiologic processes. However, the molecular assembly of PAR4 in the membrane of cells is not known. To investigate whether PAR4 forms homodimers on cell surfaces, we used BiFC. HEK293 cells were co-transfected with PAR4 fused with the N-terminal region of Venus (V1) or the C-terminal region of Venus (V2) to test for complementation (Fig. 1A). An HA epitope (HA-PAR4-V1) or V5 epitope (V5-PAR4-V2) was added to the N terminus to monitor the expression level of each of the PAR4 constructs. For these experiments, the cells were co-transfected with HA-PAR4-V1 (0.1 μg) and V5-PAR4-V2 (0.01–0.5 μg) (Fig. 1). The cells were washed from plates in PBS with 1% glucose and immediately analyzed by flow cytometry to determine the mean fluorescence resulting from the complementation or stained with anti-HA or V5 antibodies to analyze expression levels. The mean fluorescence intensity from each antibody was converted to antibody binding sites using a quantitative flow cytometry assay (Bangs Laboratories), allowing the direct comparison of the expression of the HA- and V5-tagged PAR4 proteins. These values were used to calculate the V2/V1 ratio in Fig. 1A. The individual split Venus constructs did not have a fluorescence signal above the background of mock transfected cells (data not shown). However, as V5-PAR4-V2 expression increased, the mean fluorescence fit a hyperbolic curve with a plateau at 302 fluorescence units with a 95% confidence interval of 113–491 (Fig. 1A). Importantly, the expression of HA-PAR4-V1 was not altered as the expression of V5-PAR4-V2 increased (Fig. 1B). To examine the specificity of the interaction that was detected with BiFC, unlabeled T7-PAR4 or T7-rhodopsin (0–1.0 μg) was co-transfected with HA-PAR4-V1 and V5-PAR4-V2 (0.1 μg each) (Fig. 1C). The expression levels of PAR4 and rhodopsin were directly compared with flow cytometry using an antibody to the T7 epitope conjugated to SureLight allophycocyanin. PAR4 was able to disrupt the BiFC in a concentration-dependent manner. In contrast, rhodopsin was unable disrupt the PAR4-PAR4 interaction, even though it was expressed 5-fold higher. It is of note that rhodopsin consistently had a higher expression than PAR4 when equivalent amounts of plasmid were used in the transfection.
FIGURE 1.
PAR4 homodimers are detected with BiFC. HEK293 cells were transiently transfected with HA-PAR4-V1 (0.1 μg) and V5-PAR4-V2 (0.01–0.5 μg). Cells were removed from plates 48 h post-transfection and analyzed immediately by flow cytometry to determine fluorescence from BiFC or stained with anti-HA or anti-V5 antibodies. A, the mean fluorescence intensity (MFI) from BiFC is plotted against the ratio of V5-PAR4-V2 to HA-PAR4-V1 expression determined by quantitative flow cytometry. B, the expression level of HA-PAR4-V1 plotted against the expression level of V5-PAR4-V2. C, for competition experiments, unlabeled T7-PAR4 (closed circles) or T7-rhodopsin (open circles) (0–1.0 μg) was co-transfected with HA-PAR4-V1 (0.1 μg) and V5-PAR4-V2 (0.1 μg) and analyzed for BiFC as in A. The expression level of T7-PAR4 and T7-rhodopsin was determined by flow cytometry with an anti-T7 antibody. The data are a global fit to three independent experiments.
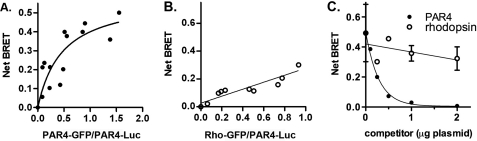
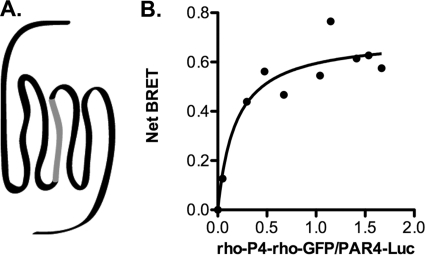
We have demonstrated that PAR4 forms homodimers by BiFC. To further validate these studies, we performed BRET. The PAR4 homodimers show a specific interaction as indicated by a hyperbolic BRET signal in response to an increase in the ratio of PAR4-GFP:PAR4-Luc expression (Fig. 2A). In contrast, a nonspecific BRET signal increases linearly with increasing GFP expression. Importantly, PAR4 did not form dimers with rhodopsin (Fig. 2B), indicating the specificity of the PAR4-PAR4 interactions. Rhodopsin was used as an unrelated GPCR to verify that the interactions were not due merely to overexpression of the receptors. Because we were able to detect a strong BRET signal from the PAR4-Luc and PAR4-GFP interaction, we sought to disrupt this interaction by competing with unlabeled PAR4 (Fig. 2C). Increasing amounts of PAR4 in pCDNA3.1 (0–2 μg) were co-transfected with the ratio of the PAR4 BRET constructs that gave the threshold maximum BRET signal (0.5 μg of PAR4-Luc and 2.5 μg of PAR4-GFP). Unlabeled PAR4 was able to disrupt the BRET signal in a concentration-dependent manner (Fig. 2C, closed circles). In contrast, rhodopsin was unable to disrupt the BRET signal from the PAR4 homodimer (Fig. 2C, open circles), indicating that the disruption of the PAR4 homodimer with PAR4 is not due to nonspecific interactions.
FIGURE 2.
PAR4 homodimers are detected with BRET. A and B, HEK293 cells were transfected with PAR4-Luc (0.5 μg) and PAR4-GFP (0–5.0 μg) (A) or rho-GFP (0–5.0 μg) (B). Forty-eight hours post-transfection, the cells were analyzed for GFP expression, luciferase expression, and BRET. The data are from three or four independent experiments, in which all points were analyzed by global fit to a hyperbolic or linear curve. C, PAR4 homodimers are disrupted by expression of unlabeled PAR4. PAR4-Luc (0.5 μg) and PAR4-GFP (2.5 μg) were transfected in to HEK293 cells for BRET assays as described in A with increasing amounts of unlabeled PAR4 (closed circles) or rhodopsin (open circles). The data are from three independent experiments, and the error bars represent the standard deviation; the apparent absence of error bars indicates a small standard deviation.
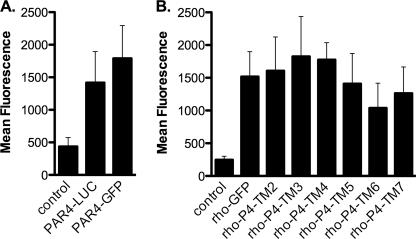
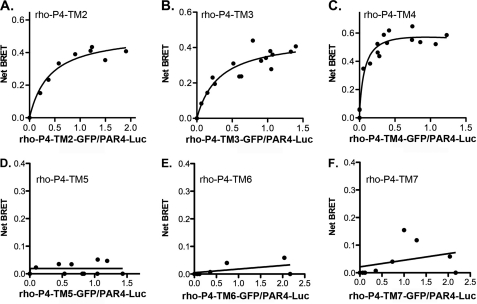
To fully understand the interaction between PAR4 homodimers, we initiated investigations to map the homodimer interface. We have demonstrated that PAR4 does not interact with rhodopsin using BRET studies (Fig. 2B). To take advantage of this observation, we generated a series of rhodopsin-PAR4 chimeras that have the junction at the beginning of transmembrane (TM) helix 2, 3, 4, 5, 6, or 7 (Table 1). The boundaries for the transmembrane helices for PAR4 were determined by comparing the amino acid sequence with that of rhodopsin using the program T-coffee (39, 40). Because we are able to detect a strong interaction using BRET for PAR4 homodimers but not with heterodimers between PAR4 and rhodopsin, the strategy for these experiments was to use the panel of chimeras to determine when dimer formation is restored. Because these chimeras are not naturally occurring molecules, initial experiments examined the surface expression of these chimeras by flow cytometry (Fig. 3). PAR4-Luc and PAR4-GFP were detected with a goat polyclonal antibody directed to PAR4 (29) and a secondary antibody conjugated to Alexa Fluor 647, which allowed specific detection of PAR4 on the surface without interference from the GFP signal (Fig. 3A). Rhodopsin-GFP and the rhodopsin-PAR4-GFP chimeras were detected with a monoclonal antibody directed to the N terminus of rhodopsin (Fig. 3B). Each of these chimeras was expressed on the cell surface at comparable levels. To examine the interaction of the rho-PAR4 chimeras with PAR4-wt, HEK293 cells were transfected with PAR4-Luc (0.5 μg) and rho-PAR4-chimera-GFP (0–5 μg) (Fig. 4, A–F). The chimeras that have junctions at TM2, 3, or 4 were able to interact with PAR4-wt in a manner similar to the PAR4 homodimers (Fig. 4, A–C). In contrast, when the junction was moved to the beginning of TM5, 6, or 7, the BRET signal was linear, indicating that these molecules were unable to interact with PAR4-wt (Fig. 4, D–F). These results indicate that the region on PAR4 required for homodimer formation is between the start of TM4 and TM5.
FIGURE 3.
Rhodopsin-PAR4 chimeras were expressed on the cell surface. HEK293 cells were transfected with 0.5 μg of plasmid, and cells were prepared for flow cytometry. A, PAR4 was detected with a polyclonal antibody (5 μg/ml) and anti-goat secondary antibody conjugated to Alexa Fluor647 (2 μg/ml). B, rhodopsin and rhodopsin-PAR4 chimeras were detected with a monoclonal antibody to rhodopsin (B6–30N) 1:10 dilution and anti-mouse secondary antibody conjugated to Alexa Fluor 647 (2 μg/ml). Control experiments used mock transfected cells followed by staining with anti-PAR4 or anti-rhodopsin. The data are from three or four independent experiments, and error bars represent the standard deviation.
FIGURE 4.
Mapping the domain on PAR4 for homodimer formation using rhodopsin-PAR4 chimeras. HEK293 cells were transfected with PAR4-Luc (0.5 μg) and rho-P4-TM2-GFP (0–5.0 μg) (A), rho-P4-TM3-GFP (0–5.0 μg) (B), rho-P4-TM4-GFP (0–5.0 μg) (C), rho-P4-TM5-GFP (0–5.0 μg) (D), rho-P4-TM6-GFP (0–5.0 μg) (E), or rho-P4-TM7-GFP (0–5.0 μg) (F). Forty-eight hours post-transfection, the cells were analyzed for GFP expression, luciferase expression, and BRET. The data are from three or four independent experiments in which all points were analyzed by global fit to a hyperbolic or linear curve.
We have narrowed the PAR4 homodimer interface to a 56-amino acid region between the start of TM4 and TM5. To determine whether TM4 was sufficient to interact with PAR4, we generated a chimera in which the TM4 of rhodopsin was replaced with that of PAR4 (Fig. 5A). The rho-PAR4-rho chimera was tested in BRET experiments, and it showed a specific interaction with PAR4 (Fig. 5B).
FIGURE 5.
Transmembrane helix 4 is sufficient for PAR4 homodimer formation. A, diagrammatic representation of rho-PAR4-rho. TM4 of rhodopsin was replaced with that of PAR4. B, HEK293 cells were transfected with PAR4-Luc (0.5 μg) and rho-PAR4-rho-GFP (0–5.0 μg). Forty-eight hours post-transfection, the cells were analyzed for GFP expression, luciferase expression, and BRET. The data are from three independent experiments in which all points were analyzed by global fit to a hyperbolic or linear curve.
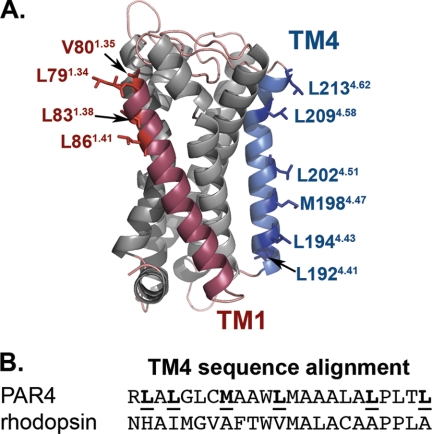
We next wanted to understand the structural basis for the homodimer interface. Because the structure of PAR4 is not known, we generated models using Swiss Modeler (Fig. 6A) (41–43). The models are in good agreement with our experimental data in that transmembrane helix 4 has residues that are directed away from the other helices, making them accessible for potential interactions with adjacent PAR4 molecules. An interesting feature of TM4 is a series of leucine residues that are directed outward. The TM4 between PAR4 and rhodopsin were aligned using the alignment program T-coffee (Fig. 6B). This alignment is based on the arrangement of all seven transmembrane helices.
FIGURE 6.
Molecular models of PAR4. A, structural model of PAR4 based on squid rhodopsin generated with Swiss modeler. TM1 is indicated in red, and TM4 is indicated in blue. Residues that are directed away from the other TM helices are shown as stick diagrams and are labeled using the nomenclature of Ballesteros and Weinstein (33). B, alignment of TM4 sequence from PAR4 and rhodopsin using the alignment program T-coffee.
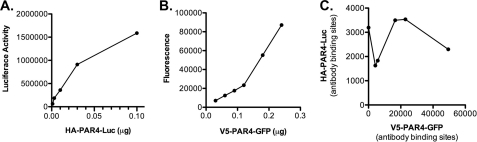
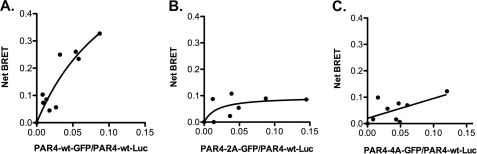
Our data using the rhodopsin/PAR4 chimeras and the PAR4 model strongly suggest that transmembrane helix 4 is the homodimer interface. Studies next directly examined the influence of the hydrophobic residues identified in the PAR4 model as a potential interface (L1924.41, L1944.43, M1984.47, L2024.51, L2094.58, and L2134.62). In addition, we wanted to minimize the influence of overexpression, which may force nonphysiologic interactions to occur. Therefore, an HA epitope (HA-PAR4-Luc) or V5 epitope (V5-PAR4-GFP) was added to allow us to determine the expression levels of each PAR4 on the surface of cells. Initial studies determined the minimum amount of plasmid required to achieve sufficient expression for the BRET assays (Fig. 7). The expression levels were determined using antibodies to HA or V5 tags and analyzed by flow cytometry (Fig. 7). The minimum amount of HA-PAR4-Luc plasmid required for detection was 0.03 μg. Similar to the BiFC experiments (Fig. 1B), increasing V5-PAR4-GFP did not alter HA-PAR4-Luc expression (Fig. 7C). The BRET experiments with PAR4 wild type were repeated using the minimum amount of plasmid (0.03 μg for HA-PAR4-Luc). The V5-PAR4-GFP concentrations were chosen to be 0.3–6 times that of HA-PAR4-Luc to achieve a wide range of expression for the saturation BRET studies (Fig. 8A). The specific interactions for PAR4 homodimers were also observed when we used the reduced amounts of plasmid, confirming the result from the previous experiments (compare Fig. 2A and Fig. 8A). Using these conditions, studies next examined the residues in transmembrane helix 4 that were suggested to be an interface in the PAR4 model (Fig. 6). Initial studies mutated L2094.58 and L2134.62 to alanine (PAR4–2A) or L1924.41, L1944.43, M1984.47, and L2024.51 to alanine (PAR4–4A) (Fig. 6). The mutants were labeled with a V5 epitope, ligated into the GFP vector and examined with BRET assays with PAR4-wt (Fig. 8). V5-PAR4–2A interacted with HA-PAR4-Luc as indicated by a hyperbolic binding curve (Fig. 8B); however, the binding was significantly reduced compared with PAR4-wt. In contrast, V5-PAR4–4A-GFP was unable to interact with HA-PAR4-Luc as indicated by a linear relationship with the GFP:Luc ratio (Fig. 8C). These studies suggested that several residues may be forming the interaction interface. To investigate whether these residues are acting in concert or whether individual residues were supporting dimerization, each of the four residues were mutated individually. For each of the point mutations, the binding curve and the maximum BRET signal were unchanged from wild type PAR4 (Table 2). These data indicate that the hydrophobic residues identified in TM4 act as a hydrophobic interface to mediate homodimers and that mutating a single residue is insufficient to disrupt dimerization.
FIGURE 7.
The minimal expression of PAR4 required for BRET assays. A and B, HEK293 cells were transfected with HA-PAR4-Luc (0–0.1 μg) (A) or V5-PAR4-GFP (0–0.25 μg) (B) and assayed for luciferase activity with colenterazine H (5 μm) or GFP expression to determine the minimal amount of plasmid required for BRET assays. C, HEK293 cells were transfected with HA-PAR4-Luc (0.03 μg) and V5-PAR4-GFP (0–0.24 μg). Expression levels were determined by quantitative flow cytometry with anti-HA or anti-V5 antibodies. HA-PAR4-Luc is plotted against increasing expression of V5-PAR4-GFP.
FIGURE 8.
Mapping the residues within transmembrane helix 4 that are required for PAR4 homodimers. HEK293 cells were transfected with HA-PAR4-Luc (0.03 μg) and V5-PAR4-GFP (0–0.24 μg) (A), V5-PAR4–2A-GFP (0–0.24 μg) (B), or V5-PAR4–4A-GFP (0–0.24 μg) (C). PAR4–2A is PAR4-L209A/L213A, and PAR4–4A is PAR4-L192A/L194A/M198A/L202A. Forty-eight hours post-transfection, the cells were analyzed for GFP expression, luciferase expression, and BRET. The data are from three or four independent experiments in which all points were analyzed by global fit to a hyperbolic or linear curve.
TABLE 2.
BRETmax values for PAR4 point mutants
PAR4 variants were tagged with GFP at the C terminus and examined in BRET assays with PAR4-wt-Luc. The data were fit to a hyperbolic curve, and each PAR4 variant was compared with PAR4-wt.
| PAR4 variant | BRETmax | 95% confidence interval |
|---|---|---|
| PAR4-wt | 0.46 | 0.23–0.70 |
| PAR4–2A | 0.07 | 0.02–0.12 |
| PAR4–4A | Linear | |
| PAR4-L192A | 0.55 | 0.44-.066 |
| PAR4-L194A | 0.39 | 0.25–0.53 |
| PAR4-M198A | 0.31 | 0.23–0.40 |
| PAR4-L202A | 0.31 | 0.05–0.57 |
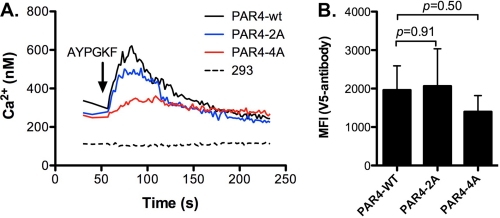
Finally, experiments were designed to determine whether disrupting the PAR4 homodimers had a functional consequence. For these studies, HEK293 cells were transfected with V5-PAR4-wt, V5-PAR4–2A, or V5-PAR4–4A in pCNDA3.1. The expression level was verified by flow cytometry. The mean fluorescence was similar for PAR4-wt (1960 ± 629) and PAR4–2A (2064 ± 560, p = 0.91). The expression of PAR4–4A (1397 ± 422, p = 0.50) was also similar to PAR4-wt. These data agree with the GFP tagged PAR4 mutants in which no differences in expression were observed. Studies next analyzed the ability of each PAR4 construct to mobilize intracellular calcium in response to the PAR4-activating peptide (AYPGKF) (Fig. 9). V5-PAR4–2A had slightly reduced Ca2+ mobilization compared with V5-PAR4-wt. However, V5-PAR4–4A had drastically reduced Ca2+ mobilization. These data correlate the loss of dimer formation to a loss of signaling function for PAR4.
FIGURE 9.
Disrupting PAR4 homodimers influences calcium mobilization in response to PAR4 agonist peptide. HEK293 cells were transfected with V5-PAR4-wt, V5-PAR4–2A, or V5-PAR4–4A in pCDNA3.1(1 μg). Forty-eight hours post-transfection the cells were removed from plates and loaded with Fura2-AM (A) or stained for flow cytometry (B) with an anti-V5 antibody. A, calcium mobilization was measured over time in response to the PAR4 agonist peptide (AYPGKF-NH2, 1 mm). Calcium concentration was determined as described under “Experimental Procedures.” B, cell surface expression of PAR4-wt, PAR4–2A, or PAR4–4A by flow cytometry. The mean fluorescence is from three independent experiments. The error bars indicate the standard deviation.
DISCUSSION
The current report demonstrates that PAR4 forms homodimers and maps the PAR4 homodimer interface to a series of hydrophobic residues in transmembrane helix 4. The dimerization of PAR4 was determined by independent techniques: BiFC and BRET. The dimerization was specific in that PAR4 did not heterodimerize with rhodopsin, another GPCR from the same class of GPCRs. Further, the current report shows that point mutations that disrupt PAR4 homodimers also impair signaling as measured by calcium mobilization.
G protein-coupled receptors have seven transmembrane domains that, when stimulated, signal via heterotrimeric G proteins, which are linked to many intracellular signaling pathways (44). Because of the complex arrangement of GPCRs in the membranes of cells, there has been much debate regarding the molecular assembly of GPCRs in vivo (45, 46). The traditional views were that the signaling GPCR occurred with the 1:1 complex with the intracellular G protein. In support of this view, recent studies with a reconstituted system demonstrate that a monomeric β2-adrenergic receptor was able to bind agonists with high affinity (47). Importantly, in this system, the binding of agonists was able to mediate rapid exchange of nucleotide from the Gs heterotrimer, demonstrating that monomers of the β2-adrenergic receptor are functional. Other studies have used the same high-density lipoprotein (HDL) disks to demonstrate that bovine rhodopsin is also capable of signaling as a monomer (48). Therefore, on a biophysical level, two prototypical GPCRs (β2-adrenergic receptor and rhodopsin) are functional as monomers in a minimal system.
There is also a growing body of evidence suggesting that GPCRs function as oligomeric structures in the plasma membrane of cells (49, 50). The most compelling evidence that GPCRs form oligomers in vivo is the high resolution crystal structure of one of the most well studied GPCRs, rhodopsin (45, 50). When the structure of rhodopsin is compared with models of Gtα subunits that are based on several lines of biochemical analysis, the oligomeric structure of rhodopsin can be modeled to interact with oligomers of Gtα (45). Recently, a heteropentameric complex consisting of two rhodopsin molecules and one Gt complex purified from native bovine retinas was characterized biochemically and by scanning transmission electron microscopy (51). These studies provide the most compelling data to date regarding the functionality of GPCR dimers in native tissues. As more is learned about the arrangement of PAR4 in the membrane, it will be important to consider how dimer or oligomer formation may alter PAR4 activity. The current report provides a key starting point for analyzing PAR4 in native tissues.
Although there has been recent progress in methodologies to study the structural properties of GPCRs (52), there is little or no structural data for most GPCRs. Therefore, we must rely on biochemical data and molecular models to define interaction partners and their interaction surfaces. The current report maps the homodimer interface of PAR4 to transmembrane helix 4. In molecular models, transmembrane helix 4 of PAR4 has several leucine residues that are directed away from the other helices that may allow these side chains to mediate dimerization (Fig. 8A). The membrane leucine zipper has been described for other membrane proteins such as E-cadherin, erythropoietin, and DDR1 (53–56). However, one must be cautious in drawing general conclusions based on results from single membrane domain-containing proteins to GPCRs, which have seven transmembrane domains. In other GPCRs, such as the dopamine D2 receptor, rhodopsin, and α-factor receptor (Ste2), TM4 is also at the homodimer interface (49, 57, 58). However, GPCRs use other regions in addition to or instead of TM4 for dimer interactions (59). A recent mutagenesis study with the M3 muscarinic receptor demonstrated that multiple interfaces are possible with the M3 receptor dimer/oligomer (60). Molecular models and biochemical data suggest TM4:TM4, TM5:TM5, TM6:TM7, TM4:TM5, and TM1:TM2 interfaces for the M3 receptor. As the authors of this study suggest, these interfaces may be in dynamic equilibrium. Another example, the dopamine D2 receptor forms oligomers with two interfaces, TM1 and TM4. Our data do not support TM1 as a homodimer interface for PAR4. However, in the PAR4 model, TM1 is angled and may have a smaller potential interface (Fig. 8A). The smaller interface may have weaker interactions that play a role in oligomer formation.
PAR1 has been well studied in human platelet activation by thrombin. However, there is a growing appreciation for the role of PAR4 in platelet activation. The sustained intracellular Ca2+ mobilization as a result of PAR4 stimulation may be required for stable clot formation (20–22). Further, PAR4 cooperates with the P2Y12 receptor to mediate platelet aggregation (16). In contrast, PAR1-mediated platelet aggregation is independent of P2Y12 signaling. PAR1 signals through PI3K and phosphatidic acid formed by phospholipase D (17, 61). Inhibitors to PI3K or phospholipase D do not disrupt PAR4 signaling. Further, PAR4 activation is required for full spreading of platelets on fibrinogen in response to α-thrombin in a p38- and ERK1/2-dependent manner (22). These data suggest that PAR1 and PAR4 stimulate different signaling pathways in platelets and likely cooperate to mediate the complete response of the platelets to thrombin. Understanding the arrangement of these receptors in the membrane may shed light on these unique signaling pathways in platelets.
In addition to expression on platelets, PAR4 is also expressed in smooth muscle cells, endothelial cells, cardiac tissue, brain, and other neurological tissues. Dangwal et al. (26) demonstrated that PAR4 expression on carotid artery plaques and saphenous vein smooth muscle cells in diabetic patients was increased compared with normal controls. These observations were supported by in vitro experiments in which human saphenous vein smooth muscle cells up-regulated PAR4 expression when grown in the presence of high glucose (26). In cardiac tissue, inhibition of PAR4 with two independent antagonists was cardioprotective in a rat ischemia/reperfusion model (23). Inhibiting PAR4 elicited cell survival pathways through PI3K/Akt, ERK1/2, nitric-oxide synthase, and KATP channels in a adenosine receptor-mediated pathway (23). In addition to roles in cardiovascular tissues, PAR4 has been implicated in nociception (24, 25). However, the details appear to be tissue-specific. For example, in dorsal root ganglia, PAR4 sensitized TRPV1, resulting in the release of calcitonin gene-related peptide (25). In other studies, PAR4 sensitization was independent of TRPV1 (24). In contrast, PAR4 stimulation inhibited visceral pain induced by PAR2 and TRPV4 agonists, and PAR4−/− mice had increased pain response compared with wild type controls (62). Taken together, these recent studies provide evidence that PAR4 has roles in a number of physiological processes in addition to its well studied role in platelet activation.
PAR1 co-expression with PAR4 enhances thrombin activation of PAR4 on human platelets and transfected cells (14, 15). In addition, the possibility exists that PAR4 interacts with other PARs or other receptors in other tissues. One example is the recent study by Li et al. (27) that describes a signaling interaction between PAR4 and the P2Y12 receptor. It will be important to examine the role of PAR4 dimerization in the context of these new roles ascribed to PAR4. The current report details the PAR4 homodimer interface and provides the methodology to examine the heterodimer interface with PAR1 or other receptors. Future studies will need to examine the formation of homodimers in the context of these potential heterodimer interactions. An important question to be considered in these studies is the relative affinity for homodimers versus heterodimers. In addition to the relative binding affinities, the dynamics of these interactions are also of interest.
The present study provides a detailed analysis of the PAR4 homodimer. Understanding the molecular details of receptor-receptor interactions may provide insights for the development of pharmacological agents that specifically target the receptor dimer, disrupt the dimer formation, or both. These pharmacological agents may be an effective add on therapy for patients who do not respond well to current antiplatelet therapies (63). In addition, as novel roles for PAR4 continue to be uncovered, there may also be new opportunities for pharmacological intervention.
Acknowledgments
We thank Michele Mumaw and Amal Arachiche for helpful suggestions and editing of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant 1R01HL098217-01A1 (to M. N.). This work was also supported by an American Heart Association beginning grant in aid, American Heart Association Scientist Development Grant 10SDG2600021, and an American Society for Hematology Scholar Award. This work was also supported by the Cytometry and Light Microscopy Core Facility of the Comprehensive Cancer Center of Case Western Reserve University and University Hospitals of Cleveland under Grant P30 CA43703.
- PAR
- protease-activated receptor
- BiFC
- bimolecular fluorescence complementation
- BRET
- bioluminescence resonance energy transfer
- TM
- transmembrane helix
- GPCR
- G protein-coupled receptor(s).
REFERENCES
- 1. Coughlin S. R. (2000) Thrombin signalling and protease-activated receptors. Nature 407, 258–264 [DOI] [PubMed] [Google Scholar]
- 2. Swift S., Sheridan P. J., Covic L., Kuliopulos A. (2000) PAR1 thrombin receptor-G protein interactions. Separation of binding and coupling determinants in the Gα subunit. J. Biol. Chem. 275, 2627–2635 [DOI] [PubMed] [Google Scholar]
- 3. Steinhoff M., Buddenkotte J., Shpacovitch V., Rattenholl A., Moormann C., Vergnolle N., Luger T. A., Hollenberg M. D. (2005) Proteinase-activated receptors. Transducers of proteinase-mediated signaling in inflammation and immune response. Endocr. Rev. 26, 1–43 [DOI] [PubMed] [Google Scholar]
- 4. Kuliopulos A., Covic L., Seeley S. K., Sheridan P. J., Helin J., Costello C. E. (1999) Plasmin desensitization of the PAR1 thrombin receptor. Kinetics, sites of truncation, and implications for thrombolytic therapy. Biochemistry 38, 4572–4585 [DOI] [PubMed] [Google Scholar]
- 5. Riewald M., Petrovan R. J., Donner A., Ruf W. (2003) Activated protein C signals through the thrombin receptor PAR1 in endothelial cells. J. Endotoxin. Res. 9, 317–321 [DOI] [PubMed] [Google Scholar]
- 6. Ruf W., Dorfleutner A., Riewald M. (2003) Specificity of coagulation factor signaling. J. Thromb. Haemost. 1, 1495–1503 [DOI] [PubMed] [Google Scholar]
- 7. Quinton T. M., Kim S., Derian C. K., Jin J., Kunapuli S. P. (2004) Plasmin-mediated activation of platelets occurs by cleavage of protease-activated receptor 4. J. Biol. Chem. 279, 18434–18439 [DOI] [PubMed] [Google Scholar]
- 8. Sambrano G. R., Huang W., Faruqi T., Mahrus S., Craik C., Coughlin S. R. (2000) Cathepsin G activates protease-activated receptor-4 in human platelets. J. Biol. Chem. 275, 6819–6823 [DOI] [PubMed] [Google Scholar]
- 9. Boire A., Covic L., Agarwal A., Jacques S., Sherifi S., Kuliopulos A. (2005) PAR1 is a matrix metalloprotease-1 receptor that promotes invasion and tumorigenesis of breast cancer cells. Cell 120, 303–313 [DOI] [PubMed] [Google Scholar]
- 10. Megyeri M., Makó V., Beinrohr L., Doleschall Z., Prohászka Z., Cervenak L., Závodszky P., Gál P. (2009) Complement protease MASP-1 activates human endothelial cells. PAR4 activation is a link between complement and endothelial function. J. Immunol. 183, 3409–3416 [DOI] [PubMed] [Google Scholar]
- 11. Arora P., Ricks T. K., Trejo J. (2007) Protease-activated receptor signalling, endocytic sorting and dysregulation in cancer. J. Cell Sci. 120, 921–928 [DOI] [PubMed] [Google Scholar]
- 12. Nakanishi-Matsui M., Zheng Y. W., Sulciner D. J., Weiss E. J., Ludeman M. J., Coughlin S. R. (2000) PAR3 is a cofactor for PAR4 activation by thrombin. Nature 404, 609–613 [DOI] [PubMed] [Google Scholar]
- 13. Sambrano G. R., Weiss E. J., Zheng Y. W., Huang W., Coughlin S. R. (2001) Role of thrombin signalling in platelets in haemostasis and thrombosis. Nature 413, 74–78 [DOI] [PubMed] [Google Scholar]
- 14. Nieman M. T. (2008) Protease-activated receptor 4 uses anionic residues to interact with α-thrombin in the absence or presence of protease-activated receptor 1. Biochemistry 47, 13279–13286 [DOI] [PubMed] [Google Scholar]
- 15. Leger A. J., Jacques S. L., Badar J., Kaneider N. C., Derian C. K., Andrade-Gordon P., Covic L., Kuliopulos A. (2006) Blocking the protease-activated receptor 1–4 heterodimer in platelet-mediated thrombosis. Circulation 113, 1244–1254 [DOI] [PubMed] [Google Scholar]
- 16. Holinstat M., Voss B., Bilodeau M. L., McLaughlin J. N., Cleator J., Hamm H. E. (2006) PAR4, but not PAR1, signals human platelet aggregation via Ca2+ mobilization and synergistic P2Y12 receptor activation. J. Biol. Chem. 281, 26665–26674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Voss B., McLaughlin J. N., Holinstat M., Zent R., Hamm H. E. (2007) PAR1, but not PAR4, activates human platelets through a Gi/o/phosphoinositide-3 kinase signaling axis. Mol. Pharmacol. 71, 1399–1406 [DOI] [PubMed] [Google Scholar]
- 18. Fälker K., Haglund L., Gunnarsson P., Nylander M., Lindahl T. L., Grenegård M. (2011) Protease-activated receptor 1 (PAR1) signalling desensitization is counteracted via PAR4 signalling in human platelets. Biochem. J. 436, 469–480 [DOI] [PubMed] [Google Scholar]
- 19. Kahn M. L., Nakanishi-Matsui M., Shapiro M. J., Ishihara H., Coughlin S. R. (1999) Protease-activated receptors 1 and 4 mediate activation of human platelets by thrombin. J. Clin. Invest. 103, 879–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Covic L., Gresser A. L., Kuliopulos A. (2000) Biphasic kinetics of activation and signaling for PAR1 and PAR4 thrombin receptors in platelets. Biochemistry 39, 5458–5467 [DOI] [PubMed] [Google Scholar]
- 21. Covic L., Singh C., Smith H., Kuliopulos A. (2002) Role of the PAR4 thrombin receptor in stabilizing platelet-platelet aggregates as revealed by a patient with Hermansky-Pudlak syndrome. Thromb. Haemost. 87, 722–727 [PubMed] [Google Scholar]
- 22. Mazharian A., Roger S., Berrou E., Adam F., Kauskot A., Nurden P., Jandrot-Perrus M., Bryckaert M. (2007) Protease-activating receptor-4 induces full platelet spreading on a fibrinogen matrix. Involvement of ERK2 and p38 and Ca2+ mobilization. J. Biol. Chem. 282, 5478–5487 [DOI] [PubMed] [Google Scholar]
- 23. Strande J. L., Hsu A., Su J., Fu X., Gross G. J., Baker J. E. (2008) Inhibiting protease-activated receptor 4 limits myocardial ischemia/reperfusion injury in rat hearts by unmasking adenosine signaling. J. Pharmacol. Exp. Ther. 324, 1045–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Russell F. A., Veldhoen V. E., Tchitchkan D., McDougall J. J. (2010) Proteinase-activated receptor-4 (PAR4) activation leads to sensitization of rat joint primary afferents via a bradykinin B2 receptor-dependent mechanism. J. Neurophysiol. 103, 155–163 [DOI] [PubMed] [Google Scholar]
- 25. Vellani V., Kinsey A. M., Prandini M., Hechtfischer S. C., Reeh P., Magherini P. C., Giacomoni C., McNaughton P. A. (2010) Protease activated receptors 1 and 4 sensitize TRPV1 in nociceptive neurones. Mol. Pain 6, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dangwal S., Rauch B. H., Gensch T., Dai L., Bretschneider E., Vogelaar T., Schror K., Rosenkranz A. C. (2011) High glucose enhances thrombin responses via protease-activated receptor-4 in human vascular smooth muscle cells. Arterioscler. Thromb. Vasc. Biol. 31, 624–633 [DOI] [PubMed] [Google Scholar]
- 27. Li D., D'Angelo L., Chavez M., Woulfe D. S. (2011) Arrestin-2 differentially regulates PAR4 and ADP receptor signaling in platelets. J. Biol. Chem. 286, 3805–3814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kaneider N. C., Leger A. J., Agarwal A., Nguyen N., Perides G., Derian C., Covic L., Kuliopulos A. (2007) 'Role reversal' for the receptor PAR1 in sepsis-induced vascular damage. Nat. Immunol. 8, 1303–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sevigny L. M., Austin K. M., Zhang P., Kasuda S., Koukos G., Sharifi S., Covic L., Kuliopulos A. (2011) Protease-activated receptor-2 modulates protease-activated receptor-1-driven neointimal hyperplasia. Arterioscler. Thromb. Vasc. Biol. 31, e100–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McLaughlin J. N., Patterson M. M., Malik A. B. (2007) Protease-activated receptor-3 (PAR3) regulates PAR1 signaling by receptor dimerization. Proc. Natl. Acad. Sci. U.S.A. 104, 5662–5667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Guo W., Urizar E., Kralikova M., Mobarec J. C., Shi L., Filizola M., Javitch J. A. (2008) Dopamine D2 receptors form higher order oligomers at physiological expression levels. EMBO J. 27, 2293–2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nieman M. T., Pagan-Ramos E., Warnock M., Krijanovski Y., Hasan A. A., Schmaier A. H. (2005) Mapping the interaction of bradykinin 1–5 with the exodomain of human protease activated receptor 4. FEBS Lett. 579, 25–29 [DOI] [PubMed] [Google Scholar]
- 33. Ballesteros J. A., Weinstein H. (1995) Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci. 25, 366–428 [Google Scholar]
- 34. Mercier J. F., Salahpour A., Angers S., Breit A., Bouvier M. (2002) Quantitative assessment of β1- and β2-adrenergic receptor homo- and heterodimerization by bioluminescence resonance energy transfer. J. Biol. Chem. 277, 44925–44931 [DOI] [PubMed] [Google Scholar]
- 35. Herrick-Davis K., Grinde E., Mazurkiewicz J. E. (2004) Biochemical and biophysical characterization of serotonin 5-HT2C receptor homodimers on the plasma membrane of living cells. Biochemistry 43, 13963–13971 [DOI] [PubMed] [Google Scholar]
- 36. Adamus G., Zam Z. S., Arendt A., Palczewski K., McDowell J. H., Hargrave P. A. (1991) Anti-rhodopsin monoclonal antibodies of defined specificity. Characterization and application. Vision Res. 31, 17–31 [DOI] [PubMed] [Google Scholar]
- 37. Nieman M. T., Warnock M., Hasan A. A., Mahdi F., Lucchesi B. R., Brown N. J., Murphey L. J., Schmaier A. H. (2004) The preparation and characterization of novel peptide antagonists to thrombin and factor VIIa and activation of protease-activated receptor 1. J. Pharmacol. Exp. Ther. 311, 492–501 [DOI] [PubMed] [Google Scholar]
- 38. Grynkiewicz G., Poenie M., Tsien R. Y. (1985) A new generation of Ca2+ indicators with greatly improved fluorescence properties. J. Biol. Chem. 260, 3440–3450 [PubMed] [Google Scholar]
- 39. Notredame C., Higgins D. G., Heringa J. (2000) T-Coffee. A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302, 205–217 [DOI] [PubMed] [Google Scholar]
- 40. Poirot O., O'Toole E., Notredame C. (2003) Tcoffee@igs. A web server for computing, evaluating and combining multiple sequence alignments. Nucleic Acids Res. 31, 3503–3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Arnold K., Bordoli L., Kopp J., Schwede T. (2006) The SWISS-MODEL workspace. A web-based environment for protein structure homology modelling. Bioinformatics 22, 195–201 [DOI] [PubMed] [Google Scholar]
- 42. Bordoli L., Kiefer F., Arnold K., Benkert P., Battey J., Schwede T. (2009) Protein structure homology modeling using SWISS-MODEL workspace. Nat. Protoc. 4, 1–13 [DOI] [PubMed] [Google Scholar]
- 43. Schwede T., Kopp J., Guex N., Peitsch M. C. (2003) SWISS-MODEL. An automated protein homology-modeling server. Nucleic Acids Res. 31, 3381–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Woulfe D. S. (2005) Platelet G protein-coupled receptors in hemostasis and thrombosis. J. Thromb. Haemost. 3, 2193–2200 [DOI] [PubMed] [Google Scholar]
- 45. Park P. S., Filipek S., Wells J. W., Palczewski K. (2004) Oligomerization of G protein-coupled receptors. Past, present, and future. Biochemistry 43, 15643–15656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pin J. P., Neubig R., Bouvier M., Devi L., Filizola M., Javitch J. A., Lohse M. J., Milligan G., Palczewski K., Parmentier M., Spedding M. (2007) International Union of Basic and Clinical Pharmacology. LXVII. Recommendations for the recognition and nomenclature of G protein-coupled receptor heteromultimers. Pharmacol. Rev. 59, 5–13 [DOI] [PubMed] [Google Scholar]
- 47. Whorton M. R., Bokoch M. P., Rasmussen S. G., Huang B., Zare R. N., Kobilka B., Sunahara R. K. (2007) A monomeric G protein-coupled receptor isolated in a high-density lipoprotein particle efficiently activates its G protein. Proc. Natl. Acad. Sci. U.S.A. 104, 7682–7687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Whorton M. R., Jastrzebska B., Park P. S., Fotiadis D., Engel A., Palczewski K., Sunahara R. K. (2008) Efficient coupling of transducin to monomeric rhodopsin in a phospholipid bilayer. J. Biol. Chem. 283, 4387–4394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fotiadis D., Jastrzebska B., Philippsen A., Müller D. J., Palczewski K., Engel A. (2006) Structure of the rhodopsin dimer. A working model for G-protein-coupled receptors. Curr. Opin. Struct. Biol. 16, 252–259 [DOI] [PubMed] [Google Scholar]
- 50. Palczewski K., Kumasaka T., Hori T., Behnke C. A., Motoshima H., Fox B. A., Le Trong I., Teller D. C., Okada T., Stenkamp R. E., Yamamoto M., Miyano M. (2000) Crystal structure of rhodopsin. A G protein-coupled receptor. Science 289, 739–745 [DOI] [PubMed] [Google Scholar]
- 51. Jastrzebska B., Ringler P., Lodowski D. T., Moiseenkova-Bell V., Golczak M., Müller S. A., Palczewski K., Engel A. (2011) Rhodopsin-transducin heteropentamer. Three-dimensional structure and biochemical characterization. J. Struct. Biol. 176, 387–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cherezov V., Abola E., Stevens R. C. (2010) Recent progress in the structure determination of GPCRs, a membrane protein family with high potential as pharmaceutical targets. Methods Mol. Biol. 654, 141–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Huber O., Kemler R., Langosch D. (1999) Mutations affecting transmembrane segment interactions impair adhesiveness of E-cadherin. J. Cell Sci. 112, 4415–4423 [DOI] [PubMed] [Google Scholar]
- 54. Kubatzky K. F., Ruan W., Gurezka R., Cohen J., Ketteler R., Watowich S. S., Neumann D., Langosch D., Klingmüller U. (2001) Self assembly of the transmembrane domain promotes signal transduction through the erythropoietin receptor. Curr. Biol. 11, 110–115 [DOI] [PubMed] [Google Scholar]
- 55. Ruan W., Becker V., Klingmüller U., Langosch D. (2004) The interface between self-assembling erythropoietin receptor transmembrane segments corresponds to a membrane-spanning leucine zipper. J. Biol. Chem. 279, 3273–3279 [DOI] [PubMed] [Google Scholar]
- 56. Noordeen N. A., Carafoli F., Hohenester E., Horton M. A., Leitinger B. (2006) A transmembrane leucine zipper is required for activation of the dimeric receptor tyrosine kinase DDR1. J. Biol. Chem. 281, 22744–22751 [DOI] [PubMed] [Google Scholar]
- 57. Guo W., Shi L., Filizola M., Weinstein H., Javitch J. A. (2005) Crosstalk in G protein-coupled receptors. Changes at the transmembrane homodimer interface determine activation. Proc. Natl. Acad. Sci. U.S.A. 102, 17495–17500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang H. X., Konopka J. B. (2009) Identification of amino acids at two dimer interface regions of the α-factor receptor (Ste2). Biochemistry 48, 7132–7139 [DOI] [PubMed] [Google Scholar]
- 59. Prinster S. C., Hague C., Hall R. A. (2005) Heterodimerization of G protein-coupled receptors. Specificity and functional significance. Pharmacol. Rev. 57, 289–298 [DOI] [PubMed] [Google Scholar]
- 60. McMillin S. M., Heusel M., Liu T., Costanzi S., Wess J. (2011) Structural basis of M3 muscarinic receptor dimer/oligomer formation. J. Biol. Chem. 286, 28584–28598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Holinstat M., Voss B., Bilodeau M. L., Hamm H. E. (2007) Protease-activated receptors differentially regulate human platelet activation through a phosphatidic acid-dependent pathway. Mol. Pharmacol. 71, 686–694 [DOI] [PubMed] [Google Scholar]
- 62. Augé C., Balz-Hara D., Steinhoff M., Vergnolle N., Cenac N. (2009) Protease-activated receptor-4 (PAR 4). A role as inhibitor of visceral pain and hypersensitivity. Neurogastroenterol. Motil. 21, 1189-e107 [DOI] [PubMed] [Google Scholar]
- 63. Simon D. I., Schmaier A. H. (2007) Sweet and sticky. Diabetic platelets, enhanced reactivity, and cardiovascular risk. J. Am. Coll. Cardiol. 50, 1548–1550 [DOI] [PubMed] [Google Scholar]