Background: ATP is required to generate the pre-chylomicron transport vesicle from intestinal ER.
Results: FABP1, part of a four protein complex, is split from the complex by phosphorylation of Sar1b enabling ER binding.
Conclusion: Phosphorylation of Sar1b enables FABP1 to bind to the ER.
Significance: Control of Sar1b phosphorylation controls chylomicron exit from the ER.
Keywords: Lipid Absorption, Lipid-binding Protein, Protein Kinase C (PKC), Triglyceride, Vesicles, Chylomicrons, Liver Fatty Acid-binding Protein, Protein Phosphorylation
Abstract
Native cytosol requires ATP to initiate the budding of the pre-chylomicron transport vesicle from intestinal endoplasmic reticulum (ER). When FABP1 alone is used, no ATP is needed. Here, we test the hypothesis that in native cytosol FABP1 is present in a multiprotein complex that prevents FABP1 binding to the ER unless the complex is phosphorylated. We found on chromatography of native intestinal cytosol over a Sephacryl S-100 HR column that FABP1 (14 kDa) eluted in a volume suggesting a 75-kDa protein complex that contained four proteins on an anti-FABP1 antibody pulldown. The FABP1-containing column fractions were chromatographed over an anti-FABP1 antibody adsorption column. Proteins co-eluted from the column were identified as FABP1, Sar1b, Sec13, and small VCP/p97-interactive protein by immunoblot, LC-MS/MS, and MALDI-TOF. The four proteins of the complex had a total mass of 77 kDa and migrated on native PAGE at 75 kDa. When the complex was incubated with intestinal ER, there was no increase in FABP1-ER binding. However, when the complex member Sar1b was phosphorylated by PKCζ and ATP, the complex completely disassembled into its component proteins that migrated at their monomer molecular weight on native PAGE. FABP1, freed from the complex, was now able to bind to intestinal ER and generate the pre-chylomicron transport vesicle (PCTV). No increase in ER binding or PCTV generation was observed in the absence of PKCζ or ATP. We conclude that phosphorylation of Sar1b disrupts the FABP1-containing four-membered 75-kDa protein complex in cytosol enabling it to bind to the ER and generate PCTV.
Introduction
The intestinal absorptive cell performs many complex operations in the process of absorbing dietary nutrients. Of these, the absorption of dietary lipid is among the most multifaceted. Most dietary lipid is made available for absorption as fatty acids (FA)2 and sn-2-monoacylglycerols, the hydrolytic products of triacylglycerols (TAG), in the intestinal lumen. The enterocyte has little control over the amount of FA and sn-2-monoacylglycerols presented for absorption and no well defined control over the rate at which these lipids are absorbed. It must rapidly convert them to the more physicochemically inert TAG, however, or risk cell membrane disruption (1).
Accordingly, the re-synthesis of the split lipids to TAG occurs extremely rapidly (2) at the level of the ER. Diet-derived TAG in the ER is then incorporated into the intestine's unique TAG-rich transport lipoprotein, the chylomicron, in a two-step process (3–5). Because of the size of chylomicrons, 250 nm average (6), and the presence of lipase in intestinal cytosol (7), we postulated that a cargo-specific, sealed transport vesicle would be required to transport the chylomicrons from the ER to the Golgi. In support of this hypothesis, we isolated and characterized the pre-chylomicron transport vesicle (PCTV) (8) that accomplishes this task. The formation of PCTV is highlighted by our finding that the ER budding step for PCTV is rate-limiting in the transit of dietary TAG from the ER to the lymph (2). These data led us to concentrate on the mechanisms by which PCTV are budded from the ER.
There are multiple differences between PCTV generation and that of vesicles that transport newly synthesized proteins (protein vesicles) from the ER to the Golgi. These include the cargo selection process and the machinery for budding the vesicles from ER membranes. In contrast to protein vesicles that require COPII proteins for their initiation (9–11), PCTV require only the small molecular weight FA-binding protein, the liver fatty acid-binding protein (FABP1) (12). Also, in contrast to protein vesicles that require ATP and GTP to bud (13), FABP1, in the absence of both ATP and GTP, can bind to the ER (12) and organize the PCTV budding complex (14). This finding is confusing because our original studies, using whole cytosol, identified ATP (not GTP) as being necessary for PCTV budding to occur (8). These conflicting data created a conundrum in which ATP was found to be necessary for PCTV budding when whole cytosol is used, but ATP was not required when FABP1 was utilized to initiate budding. Furthermore, native cytosol contains 2% of total protein as FABP1 (15) suggesting that adequate amounts of FABP1 would be present in the cytosol to bud PCTV. These data could be rationalized if FABP1 in cytosol were sequestered from binding to the ER until ATP was provided.
The treatment of native cytosol with ATP enabling PCTV budding to occur suggests the involvement of a kinase. We have identified the putative kinase as the atypical protein kinase C, PKCζ. Immunodepletion of this kinase greatly attenuates PCTV budding activity that is restored by adding recombinant PKCζ to the depleted cytosol (16). In support of this finding, ER membranes incubated with PKCζ-depleted cytosol do not form the PCTV budding complex despite the addition of ATP (14).
This study tests the hypothesis that FABP1 is a member of a cytosolic multiprotein complex that blocks FABP1 from binding to the ER unless it is treated with PKCζ and ATP. We further identify the substrate for PKCζ as Sar1b, mutations of which result in chylomicron retention disease (17) in which chylomicrons are retained in the ER lumen (18).
EXPERIMENTAL PROCEDURES
Materials
[3H]Oleic acid (9.2 Ci/mm) was obtained from PerkinElmer Life Sciences. Immunoblot reagents were purchased from Bio-Rad. ECL reagents were procured from GE Healthcare. Protease inhibitor mixture tablets were obtained from Roche Applied Science. Albumin was purchased from Sigma. PKCζ was purchased from StressGen Biotechnologies (Victoria, Canada). Other biochemicals used were of analytical grade and were purchased from local companies. Rats, 150–200 g, were purchased from Harlan (Indianapolis, IN).
Antibodies
Polyclonal antibodies against FABP1 were a generous gift from Dr. Judith Storch (Rutgers University). Polyclonal anti-small VCP/p97-interactive protein (SVIP) antibodies were generated commercially (Alpha Diagnostic, Inc. San Antonio, TX), and the results were compared with anti-SVIP antibodies generously provided by Dr. Mitsuo Tagaya (School of Life Science, Tokyo University of Pharmacy and Life Science, Tokyo, Japan). The protein identified by the antibody was confirmed as SVIP by MALDI-TOF with a Z score of 1.9. Polyclonal antibodies to Sar1b were also generated by Alpha Diagnostic using an 18-mer from 102 and an 18-mer from 133. Both peptides were injected into the same rabbit. The antibodies generated could not distinguish Sar1a from Sar1b (Fig. 5B). The anti-Sar1 and anti-SVIP antibodies were generated under the direction of Dr. Shadab A. Siddiqi (University of Central Florida, Orlando) while a member of the laboratory. Polyclonal antibodies to Sec13 were a kind gift of Dr. Chris Kaiser (Massachusetts Institute of Technology, Cambridge, MA). Goat anti-rabbit IgG conjugated with agarose beads was purchased from Sigma. Goat anti-rabbit IgG, goat anti-mouse IgG, and goat anti-rabbit IgG conjugated with horseradish peroxidase were purchased from Santa Cruz Biotechnology (Santa Cruz, CA).
Fig. 5.
A, single protein (Sar1) is phosphorylated by PKCζ. Blot A, proteins (150 μg) eluted from the anti-FABP1 antibody adsorption column were incubated with PKCζ and [γ-32P]ATP. The proteins (30 μg) were separated by one-dimensional SDS-PAGE, and an autoradiograph was performed. Only one band at 35 kDa was identified on the gel. Blot B, eluted proteins from the anti-FABP1 adsorption column (150 μg) were incubated with PKCζ and [γ-32P]ATP and separated on a two-dimensional gel (see under “Experimental Procedures”). An autoradiogram was performed. Blot C, eluted proteins from the adsorption column (150 μg) were incubated with PKCζ and ATP, and the proteins were separated on a two-dimensional gel. An immunoblot for Sar1 was performed. Detection was by ECL. Blot D, same experiment was performed as in blot C, but the proteins were immunoblotted using anti-Thr(P) antibodies. The pI of the bands on the two-dimensional gels was calculated to be 5.8. The molecular mass of the bands on the one- and two-dimensional gels is marked. The whole two-dimensional gel is shown for each experiment (blots B–D). B, Sar1 has two paralogs in cytosol as identified by anti-Sar1 antibody immunoblot. Cytosolic proteins (250 μg) were separated by two-dimensional gel electrophoresis, and an anti-Sar1 antibody immunoblot was performed. Because of the minor difference in pI between the paralogs, a narrow pI IPG strip was selected (5.3 to 6.5 as indicated at the top of the gel). The migration in the gel of Sar1a and Sar1b is shown at the bottom of the gel. Detection was by ECL.
Antibody Specificity
Immunogenic peptides used to generate antibodies to SVIP and Sar1 and the recombinant protein, FABP1, were incubated with their respective antibodies to determine whether the signal on immunoblot generated by the antibody could be extinguished. In each case, the antibody gave a strong signal for the protein against which it was directed in intestinal cytosol. This signal was completely blocked by prior incubation of the antibody with its immunogenic peptide (Sar1, SVIP) or recombinant protein (FABP1). These data suggest the monospecificity of the antibodies employed. Antibody inhibition was not done for Sec13 because the immunogenic peptide was not available, although the antibody has been characterized previously (19).
Preparation of Cytosol and Labeling of Enterocytes
Enterocytes from the proximal half of male Sprague-Dawley rat small intestine were isolated and radiolabeled with [3H]oleate as described previously (8). In brief, the isolated enterocytes were incubated with albumin-bound [3H]oleate for 30 min at 35 °C and washed twice with PBS containing 2% BSA to remove the excess [3H]oleate. The labeled enterocytes were homogenized using a Parr bomb, and the cytosol was isolated. The cytosol was dialyzed against Buffer A (0.25 m sucrose, 30 mm HEPES (pH 7.2), 30 mm KCl, 5 mm MgCl2, 5 mm CaCl2, 2 mm DTT) overnight at 4 °C and concentrated 5-fold using a 50-ml Amicon filter with a YM10 membrane (Amicon, Beverly, MA). This cytosol was further concentrated on a Centricon filter (Amicon) with a 10-kDa cutoff to 20 mg of protein/ml.
Preparation of [3H]TAG-loaded ER
Enterocytes from the proximal half of male Sprague-Dawley rat small intestines were isolated and radiolabeled with [3H]oleate as described (20). In brief, enterocytes were isolated from intestinal villi, collected, incubated with albumin-bound [3H]oleate for 30 min at 35 °C, and washed with 2% BSA to remove the excess [3H]oleate. The labeled enterocytes were homogenized using a Parr bomb, and the ER was isolated using a sucrose step gradient, which was repeated to purify the ER (21).
Isolation of a 75-kDa Protein Complex by Gel Filtration Chromatography
1 mg of cytosol was applied to a Sephacryl S-100 HR column (1.5 cm × 45 cm) previously equilibrated with PBS (pH 7.2). The flow rate was 0.5 ml/min, and the cytosol was eluted with PBS (pH 7.2) at 4 °C. 1-ml fractions were collected. 3H disintegrations/min radioactivity was determined by liquid scintillation spectroscopy for each fraction. For immunoblot, proteins in each fraction were concentrated using a Millipore centrifugal filter unit (Millipore Corp., Billerica, MA) and suspended in Laemmli's buffer. The presence of FABP1 in each fraction was analyzed by immunoblot using anti-FABP1 antibodies as indicated.
Affinity Purification of the 75-kDa Complex
500 μg of protein from the gel filtration column was purified using a SulfoLink coupling gel kit according to the manufacturer's instructions (Pierce) in three steps. In brief, 10 μl of anti-FABP1 antibody was incubated with 6 mg of 2-methylamine at 37 °C for 1.5 h. This reduced sample was first incubated with a SulfoLink coupling gel column for 15 min at room temperature (RT) by rocking and another 30 min without rocking. Second, the nonspecific binding sites on the column gel were blocked by first incubating 15.8 mg of l-cysteine for 15 min at RT by rocking and then for 30 min without rocking. The column was washed with PBS. Third, 1 ml (500 μg of protein) of the sample was applied to the SulfoLink coupling column and incubated for 2 h at RT; the column was washed with 15 ml of sample buffer, and the bound proteins were eluted by applying 8 ml of 100 mm glycine buffer (pH 2.5). 1-ml fractions were collected, immediately neutralized by 1 m Tris (pH 9.0), and kept at 4 °C. The fractions were pooled and concentrated using a Millipore centrifugal filter unit.
Native-PAGE Electrophoresis, Immunoblots, and Autoradiography
For immunoblots, proteins were separated by SDS-PAGE and then transblotted to nitrocellulose membranes (Bio-Rad). After blocking the membrane with 5% Blotto, the membranes were incubated with specific primary antibodies and then peroxidase-conjugated secondary antibodies. Labeled proteins were detected using ECL (GE Healthcare) and Biomax film (Eastman Kodak Co.). For native gels, samples were not boiled, and SDS or DTT was not used in the sample preparation or running gel buffer. Otherwise, the procedure was the same as with the SDS-polyacrylamide gels. For autoradiography, proteins were separated by SDS-PAGE, 2D gels, or native PAGE. The gels were rinsed three times with distilled water, dried, and autoradiographed using Kodak Biomax film at −70 °C for 10 days.
Two-dimensional Gel Electrophoresis
Cytosol (250 μg of protein) or the 75-kDa protein complex (150 μg of protein) were solubilized in 500 μl of sample buffer (7 m urea, 2 m thiourea, 4% CHAPS, 2 mm tri-butyl phosphine, 0.5% carrier ampholyte, 40 mm Tris) for 1 h at 37 °C. The solubilized protein sample was loaded onto immobilized pI gradient gel (IPG) strips (pH 3–11 or pH 5.3–6.5) (GE Healthcare) by incubation (RT) overnight. Isoelectric focusing was performed using a Multiphor II apparatus (GE Healthcare). A total of 50,500 V-h was applied to each IPG strip. After isoelectric focusing, the IPG strip was first equilibrated in buffer I (0.5 m Tris-HCl (pH 6.8), 6 m urea, 30% glycerol (v/v), 2% SDS, 25 mm DTT) for 10 min and then in buffer II (0.5 m Tris-HCl (pH 6.8), 6 m urea, 30% glycerol (v/v), 2% SDS, 12 mm iodoacetamide) for 10 min. For the second dimension, the proteins were separated on an 8–18% SDS-polyacrylamide gel, transblotted onto nitrocellulose membranes, and probed with specific antibodies.
Immunoprecipitation of the FABP1 from the Sephacryl S-100 HR Column Eluates
Immunoprecipitation of proteins from fractions 3 to 5 of the Sephacryl S-100 HR column was accomplished by incubating the eluted fractions (250 μg of protein) with 10 μl of anti-FABP1 antibody at 4 °C for 4 h. 40 μl of anti-rabbit IgG bound to agarose beads (Sigma) were added and incubated for another 4 h at 4 °C. The beads were collected by centrifugation, washed 10 times with cold PBS, and resuspended in Laemmli's buffer. The proteins were separated by native or SDS-PAGE, transblotted to a nitrocellulose membrane, and immunoblotted using anti-SVIP, Sec13, or Sar1 antibodies. Detection was by ECL.
Phosphorylation of the 75-kDa Complex
The 75-kDa complex (150 μg) was incubated with 5 μg of recombinant PKCζ, an ATP-generating system (1 mm ATP, 5 mm phosphocreatine, 5 units of creatine phosphokinase), 50 μCi of [γ-32P]ATP, 2 mm NaF, 5 mm Mg2+, 2 mm DTT, and 2.5 mm Ca2+ for 30 min at 37 °C or as indicated. Postincubation, the reaction was stopped by placing the tubes on ice and by adding cold HEPES buffer (10 mm) supplemented with 10 units of apyrase.
In Vitro PCTV Formation
[3H]TAG-pre-loaded intestinal ER was used to generate PCTV containing [3H]TAG (8). In brief, [3H]TAG-loaded ER (500 μg) was incubated at 37 °C for 30 min with cytosol (1 mg) and an ATP-regenerating system in the absence of Golgi acceptor (total volume 500 μl). When the 75-kDa protein complex was used to generate PCTV instead of cytosol, PKCζ (5 μg) and an ATP-generating system were used. The incubation mixture was resolved on a continuous sucrose gradient (0.1–1.15 m sucrose) and PCTV isolated from the light portions of the gradient. The PCTV thus formed were concentrated using a Millipore centrifugal filter unit.
Specificity of Antibodies
30 μg of cytosolic protein was separated by SDS-PAGE, transblotted to nitrocellulose membranes, and immunoblotted using specific antibodies as indicated. To test for the specificity of the antibodies utilized, prior to incubating the transblotted membrane with antibody, the antibodies were first incubated with either their immunogenic peptide or FABP1 as indicated. The antibodies were incubated with 3-fold (by weight) excess of the immunogenic peptide or FABP1 in 500 μl of PBS overnight at 4 °C. Following the incubation, the antibody-peptide or FABP1 mixture was diluted. The treated antibody was used to probe the nitrocellulose membrane after it was treated with 5% Blotto and washed with PBS. Detection was by ECL.
Determination of Protein and Oleate Concentration in the Heterotetrameric Complex
The purified four-membered protein complex (5, 10, and 15 μg of protein) was separated by SDS-PAGE, and the proteins were stained by SimplyBlue SafeStain. The gel was completely destained, and each of the four-protein bands were scanned and their densities determined by using the Gel Doc XR. The three differing protein concentrations gave similar results on a per mg basis indicating that the measured densities were proportional to the amount of protein present. The oleate concentration in the protein complex was determined by the [3H]oleate-specific activity fed to the primary enterocyte culture (5,000 dpm/10−12 mol) and subsequently the [3H]oleate dpm in a known amount of complex-FABP1.
LC-MS/MS and MALDI-TOF
The isolated heterotetramer eluted from the anti-FABP1 adsorption column was separated by SDS-PAGE and stained with SimplySafe Blue (Invitrogen). The bands at 33, 22, 14, and 8.6 kDa were cut from the gel and digested by trypsin. Data were acquired on a Thermo Scientific LTQ Orbitrap (Thermo Fisher Scientific, Waltham, MA) equipped with a Waters nanoAcquity UPLC system using a Waters Symmetry® C18 180-μm × 20-mm trap column and a 1.7-μm, 75-μm × 250-mm nanoAcquityTM UPLCTM column (Waters Corp., Milford, MA) (35 °C) for peptide separation. Trapping was done at 15 μl/min. MS was acquired in the Orbitrap using one microscan over the range 400–2000 m/z at 60,000 resolutions and a maximum injection time of 900 ms, followed by six data-dependent MS/MS acquisitions in the ion trap. The data were searched using The MASCOT Distiller and the MASCOT search algorithm (22). The data were searched against the MASCOT Swiss-Prot data base using partial methionine oxidation and propionamide-modified cysteine, a peptide tolerance of +20 ppm, MS/MS fragment tolerance of +0.6 Da, and peptide charges of +2 or +3. Normal and decoy data base searches were run. The band at 9 kDa of the complex could not be analyzed by LC-MS/MS because of contaminating peptides derived from predominantly FABP1 but also Sar1. For this reason we did a pulldown using our anti-SVIP antibody and analyzed the one bound protein by MALDI-TOF. The MALDI-TOF was performed as described previously (21).
Measurement of TAG Radioactivity
TAG radioactivity was determined by liquid scintillation spectroscopy.
Statistical Analysis
Significant differences between two means were tested by Student's t test using the nonpaired, two-tailed method. When more than two means were compared, analysis of variance was used with post-Bonferroni corrections (Instat, GraphPad, San Diego) in combination with Student's nonpaired two-tailed t test. p < 0.05 was taken as a significant difference between means.
RESULTS
FABP1 Is a Member of a Four-component 75-kDa Protein Complex in Cytosol
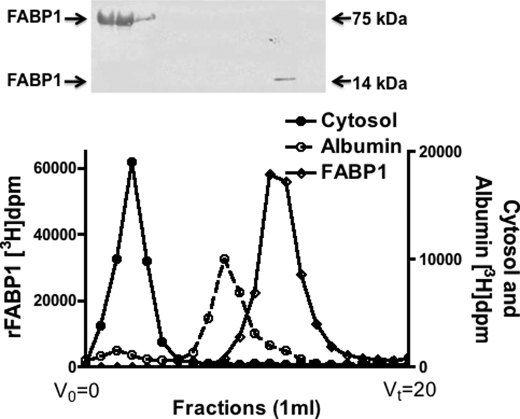
To test if absorbed oleate were present in intestinal cytosol bound to FABP1 as a monomer or as part of a multiprotein complex, we incubated primary cultures of rat intestinal cells with [3H]oleate, disrupted the cells, and isolated the cytosol. The cytosol was passed over a Sephacryl S-100 HR column; the eluted fractions were collected, and the oleate disintegrations/min were determined for each fraction. The oleate eluted in a single peak coincident with the elution of conalbumin (76 kDa), clearly different from the elution volumes of albumin ( 66.5 kDa) or FABP1 (14 kDa) (Fig. 1). We suspected that the [3H]oleate marked the elution volume of FABP1. This was confirmed by immunoblot as shown in the inset of Fig. 1. Only minor amounts of FABP1 (Mr ≈14,000) eluted at its monomer molecular weight.
Fig. 1.
FABP1 elutes in a volume suggesting a 75-kDa protein from a Sephacryl S-100 HR column. Primary cultures of proximal intestinal enterocytes were incubated with [3H]oleate. The cells were homogenized, and cytosol was prepared (“Experimental Procedures”). 1 mg of cytosol was applied to a Sephacryl S-100 HR column, and 1-ml fractions were collected. FABP1 and albumin were incubated with [3H]oleate and were separately applied to the column. The disintegrations/min from each fraction are shown on the ordinate. The disintegrations/min for cytosol are shown as filled circles, for albumin as open circles, and for FABP1 as filled diamonds. Fractions 3–16 were immunoblotted for FABP1 as shown above the column eluent (ECL was used for identification). The kDa of the bands on native PAGE (no SDS, no DTT, no boiling) is shown. V0 is the void volume, and Vt is the total volume of the column.
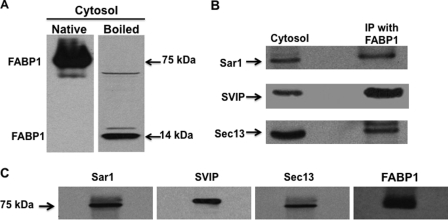
Consistent with the data in Fig. 1, cytosol, whose proteins had been separated by native PAGE (no boiling, no SDS, and no DTT) and immunoblotted for FABP1, showed that FABP1 migrated as a 75-kDa protein (Fig. 2A, Native). However, on SDS-PAGE, FABP1 migrated at its expected monomer mass of 14 kDa (Fig. 2A, Boiled).
Fig. 2.
In intestinal cytosol, FABP1 is in a 75-kDa multiprotein complex. A, intestinal cytosol (30 μg) was separated using native PAGE (A, Native) or by SDS-PAGE (A, Boiled). The proteins from the gel were transblotted onto a nitocellulose membrane and immunoblotted for FABP1. Bands at 75 and 14 kDa are indicated as is FABP1. B, cytosol, proteins from native cytosol (30 μg) were separated by SDS-PAGE, transblotted to a nitocellulose membrane, and immunobloted for Sar1, SVIP, and Sec13. The membrane was washed between blots. Immunoprecipitation (IP) was with FABP1. Anti-FABP1 antibodies were bound to beads and incubated with fractions 3–5 from the Sephacryl S-100 HR column. The beads were washed, and the proteins eluted from the beads and separated by SDS-PAGE. The gel was transblotted to a nitocellulose membrane, and sequential immunoblots for Sar1, SVIP, and Sec13 were perfomed after washing the membrane between blots. C, proteins (30 μg) from fractions 3–5 from the Sephacryl S-100 HR column were separated by native PAGE, transblotted, and then immunoblotted for its component proteins, Sar1, SVIP, Sec13, and FABP1 as shown above the blot. A single membrane was used that was sequentially probed by the indicated antibodies. The membrane was washed between each immunoblot. Identification of the bands was by ECL.
We suspected that FABP1 eluted from the column in a volume suggestive of a protein of ≈75 kDa because it was either part of a multiprotein complex or because FABP1 was present as a pentamer. Performing a nonspecific protein stain on the FABP1-containing fractions from the column would likely have led to a multiplicity of protein bands making interpretation of the results difficult. Therefore, as a first approach to differentiate between a homo- and a multiprotein complex, we performed a pulldown experiment using the column fractions containing the [3H]oleate incubated with anti-FABP1 antibodies attached to beads (Fig. 2B). As shown, we identified three other proteins by immunoblot that interacted with FABP1 by this methodology as follows: Sar1, small VCP/p97 interactive protein (SVIP), and Sec13 (Fig. 2B, IP with FABP1). The identification of these proteins was guided by their kDa and our knowledge of the proteins involved with PCTV budding (14). The same proteins were present in cytosol (Fig. 2B, Cytosol). The total molecular weight of these three proteins plus FABP1 is 77. Each of these proteins was present in the 75-kDa fraction from the column as shown by immunoblot (Fig. 2C).
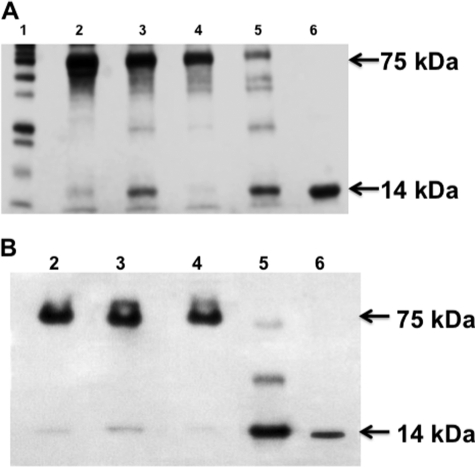
The data from Fig. 2B would suggest that we could concentrate the 75-kDa complex by using an anti-FABP1 antibody adsorption column. In accord with this goal, we collected the [3H]oleate-containing fractions from the Sephacryl S-100 HR column, concentrated them, and passed the concentrate over an anti-FABP1 antibody adsorption column. We used native PAGE to separate the proteins eluted from the column. All the proteins eluted in a single band of ≈75 kDa (Fig. 3A, panel 1). By contrast, on SDS-PAGE, the majority of the proteins from the 75-kDa band now eluted at 50, 33, 21, 14, and 9 kDa by SimplyBlue SafeStain (Fig. 3A, panel 2). That some protein remained at 75 kDa despite treatment with boiling, SDS, and DTT suggests the strength of the protein-protein interactivity. We conclude from these studies that FABP1 is present in cytosol as part of a multiprotein complex and not as a five-membered FABP1 polymer.
Fig. 3.
A, four proteins adhere to FABP1 by anti-FABP1 antibody adsorption chromatography. Anti-FABP1 antibodies were bound to beads (see under “Experimental Procedures”) and a column prepared from the treated beads. The proteins from fractions 3–5 of the Sephacryl S-100 HR column were passed over the column, and the column was washed with buffer and the proteins eluted with glycine buffer (pH 2.5). The eluted samples were rapidly neutralized and the proteins concentrated. A, panel 1, Native Gel. The proteins were separated by native PAGE (30 μg) and identified by SimplyBlue SafeStain. All the proteins migrated at 75 kDa as shown. A, panel 2, SDS-PAGE. The proteins from the adsorption column were separated by SDS-PAGE (30 μg) and identified by SimplyBlue SafeStain. The molecular mass of each band is marked. B, proteins bound to the anti-FABP1 antibody adsorption column were identified as Sec13, Sar1, FABP1, and SVIP. Cytosolic proteins from fractions 3–5 of the Sephacryl S-100 HR column were concentrated and pased over an anti-FABP1 antibody adsorption column. The column was washed, and the adherent proteins were eluted as in A. Sample, proteins (30 μg) eluted from the anti-FABP1 antibody adsorption column (A) were separated by SDS-PAGE, transblotted to a nitrocellulose membrane, and sequentially probed with anti-Sec13, Sar1, FABP1, and SVIP antibodies. The membrane was washed between each antibody useage. The proteins and their molecular mass are shown. Wash, after the proteins were applied to the adsorption column, the column was washed with PBS, and the fractions were collected and concentrated, and the proteins (30 μg) were separated by SDS-PAGE. Sec13, Sar1, FABP1, and SVIP were identified in the wash. The molecular mass of each protein is as shown. Note: the wash contained extraneous proteins in addition to Sec13, FABP1, Sar1, and SVIP. The bands were detected by ECL.
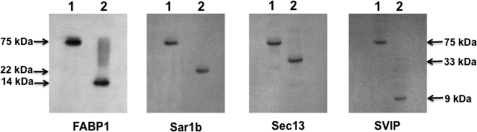
We used immunoblots to confirm the identity of the proteins eluted from the adsorption column. Consistent with the data presented in Fig. 2C, the proteins shown in Fig. 3A, panel 2, were identified as Sec13 (33 kDa), Sar1 (21 kDa), FABP1 (14 kDa), and SVIP (9 kDa) (Fig. 3B, Sample). The 50-kDa protein (Fig. 3A, panel 2) was composed of the heterodimer, Sec13, and FABP1 by immunoblot (data not shown but see FABP1 band at 50 kDa in Fig. 2A, Boiled). Most of the proteins interacted with the adsorption column as relatively few eluted in the column wash (Fig. 3B, Wash) as judged by their respective immunoblots.
To supplement the protein identifications by immunoblot, we performed LC-MS/MS on each band except for the band at 9 kDa where MALDI-TOF was used. The LC-MS/MS and MALDI-TOF data confirm the presence of FABP1 with 75% of the molecule covered, Sar1 with 56% covered, and Sec13 with 33% of the molecule covered. The MALDI-TOF spectrum is compatible with SVIP with a Z score of 1.9. The mass spectrometry and MALDI-TOF data and peptide coverage for each heterotetramer component are shown in supplemental Tables 1–3 and supplemental Fig. 1.
We further wished to know the proportionality of the proteins in the complex. As measured from their densities on SimplyBlue SafeStain, 10 μg of complex protein contained 58 pmol of Sec13, 52 pmol of Sar1, 225 pmol of FABP1, and 80 pmol of SVIP. These data suggest a molar ratio of 1:1:4:1.6, respectively. In sum, the data suggest that FABP1 in native intestinal cytosol is present as a component of a 75-kDa protein multimer based on native PAGE, gel permeation chromatography, immunoprecipitation, MS/MS, and antibody adsorption chromatography.
To determine the amount of [3H]oleate bound to FABP1 in the protein complex, we isolated the heterotetramer from the anti-FABP1 column and determined its radioactivity per μg of FABP1 (116,481 dpm/1.45 μg of FABP1). Using the known specific activity of the oleate used in the incubation (“Experimental Procedures”), we calculated that there were 23 pmol of oleate in 104 pmol of FABP1. We do not know the amount of FA bound to FABP1 prior to harvesting the cytosol nor the quantity of [3H]oleate that was exchanged for previously bound FA, so the molar ratio of FA to FABP1 is not known. By using the known dilution of FA radiolabel in enterocyte cytosol of 3:1, however (23), the data suggest that at least one of the two potential FA-binding sites in FABP1 was occupied. We conclude that ligand-bound FABP1 participates in the cytosolic heterotetramer. We do not know the potential participation of apo-FABP1 in the formation of the heterotetramer because recombinant proteins were not used.
75-kDa Complex Can Bud PCTV from Intestinal ER on the Addition of ATP and PKCζ
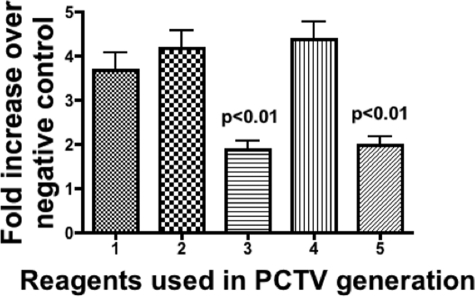
Because there was no free FABP1 in the cytosol, only the 75-kDa multimer, we wished to test if the multimer isolated from the anti-FABP1 antibody adsorption column was active in budding PCTV. As expected, native cytosol plus ATP yielded a robust generation of PCTV on incubation with ER preloaded with [3H]TAG (Fig. 4, bar 1). On addition of 5 μg of PKCζ to the incubation, a modest increase in activity was observed (Fig. 4, bar 2). When only the 75-kDa multimer was incubated with ER, a significantly attenuated PCTV generation was found (Fig. 4, bar 3). By contrast, when the multimer was preincubated with PKCζ and ATP and subsequently with ER, a vigorous PCTV production occurred (Fig. 4, bar 4). However, in the absence of preincubation of the complex with PKCζ, PCTV generation was reduced to a level similar to that found when only the multimer was incubated with the ER (Fig. 4, bar 5). The data suggest that PKCζ and ATP treatment of the 75-kDa complex are required for PCTV budding activity to be fully expressed. They further suggest that the appropriately treated complex retained its PCTV budding activity even after passage over the antibody adsorption column.
Fig. 4.
Proteins eluted from the anti-FABP1 antibody adsorption column are active in generating PCTV. Proteins eluted from the anti-FABP1 antibody adsorption column (150 μg) or native cytosol (1 mg) were incubated with glyceryl [3H]trioleate pre-loaded ER (500 μg), and the PCTV generated was collected by centrifugation (“Experimental Procedures”). ATP and PKCζ were added as indicated. The data are expressed as the fold increase over the PCTV generated from ER incubations with cytosol (1 mg) but without ATP or PKCζ as indicated by [3H]PCTV disintegrations/min. Bar 1, cytosol plus ATP. Bar 2, cytosol plus PKCζ (5 μg) and ATP. Bar 3, eluted proteins without ATP or PKCζ. Bar 4, eluted proteins plus PKCζ (5 μg) and ATP. Bar 5, eluted proteins with ATP but without PKCζ. The p values above the bars test the significance of the differences between the means of bars 3 and 5 against bars 1, 2, and 4. The data are the mean ± S.E. (n = 4).
Sar1b Is the Substrate for PKCζ in the 75-kDa Complex
Because PKCζ was required when the 75-kDa complex was used to bud PCTV, consistent with our prior findings (16), our next task was to identify the substrate of the kinase. To determine the substrate, we first incubated the multimer with PKCζ and [γ-32P]ATP and separated the proteins by SDS-PAGE (Fig. 5A, blot A). On autoradiography, a single 35-kDa band was evident. Because there were no proteins in the multimer that migrated as a 35-kDa protein, we assumed that the radiolabeled band was a heterodimer. To disassociate the presumed heterodimer, we turned to two-dimensional gel autoradiography because of the more vigorous reducing conditions used with two-dimensional gels. To this end, we incubated the 75-kDa complex with [γ-32P]ATP and PKCζ and separated the resulting proteins on a two-dimensional gel. Autoradiography of the resulting gel shows only one band at 22-kDa (Fig. 5A, blot B). This band reacted with our anti-Sar1 antibody (Fig. 5A, blot C), which does not discriminate between Sar1a and its paralog, Sar1b, suggesting the identity of this band as Sar1. Finally, we wished to know if the amino acid phosphorylated by the kinase was Ser or Thr because PKCζ is a Ser/Thr kinase. Antibodies to Thr(P) were reactive (Fig. 5A, blot D) to the same 22-kDa band as in Fig. 5, A, blots B and C. Antibodies to Ser(P) were nonreactive (data not shown).
As one way of identifying which Sar1 paralog was associated with the 75-kDa multimer, we took advantage of the small difference in pI of the paralogs. Sar1a has a known pI of 6.2 and Sar1b of 5.8. Because our anti-Sar1 antibody cannot discriminate between them, we performed a two-dimensional gel to separate the proteins of native cytosol and the 75-kDa complex. In each case Sar1 was localized by immunoblot. When native cytosol was used, our anti-Sar1 antibodies identified two bands indicating the presence of both paralogs with a pI of 6.2 and 5.8 consistent with Sar1a and Sar1b, respectively (Fig. 5B). By contrast, when the 75-kDa multimer was separated on a two-dimensional gel, the only band identified by the antibody was the more acidic of the two bands (Fig. 5A, blot C, calculated pI = 5.8). These data suggest that only Sar1b is incorporated into the 75-kDa multimer and are consistent with the results of the LC-MS/MS data.
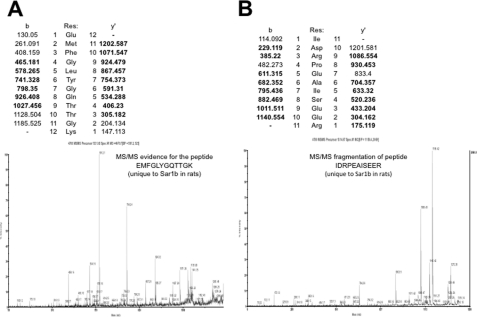
As a more definitive way to establish the identity of the 22-kDa protein on the two-dimensional gel and to determine which Sar1 paralog reacted with our anti-Sar1 antibodies, we performed LC-MS/MS analysis of the band. This identified the Sar1 paralog as Sar1b with 56% of the protein covered (supplemental Table 1). Mass spectroscopy data for two unique peptide sequences (EMFGLYGQTTGK and IDRPEAISEER) that distinguish Sar1a from Sar1b are shown in Fig. 6, A and B.
Fig. 6.
LC-MS/MS of two peptides that are different between Sar1a and Sar1b showing the presence of peptides present in Sar1b. A, spectrum of the peptide EMFGLYGQTTGK, which is unique to Sar1b in rats. B, spectrum of the peptide IDRPEAISEER, which is unique to Sar1b in rats.
Phosphorylation of Sar1b Completely Disrupts the 75-kDa Complex, Releasing FABP1
Next, we turned to the effect of Sar1b phosphorylation on the 75-kDa complex. For these experiments, we incubated the 75-kDa protein complex isolated from the anti-FABP1 antibody adsorption column with or without ATP and PKCζ as indicated. When the untreated complex was incubated at 4 °C and separated by native PAGE, all the proteins migrated at 75 kDa (Fig. 7A, lane 2) consistent with our prior data. On incubation of the untreated complex at 37 °C, a small amount of a protein of 14 kDa split from the complex, which otherwise remained intact (Fig. 7A, lane 3). If the complex were treated with ATP and PKCζ but incubated at 4 °C, the complex again remained intact (Fig. 7A, lane 4). By contrast, if the complex were incubated with ATP and PKCζ at 37 °C, most of the complex was now disrupted with the majority of the proteins migrating at 14 kDa (Fig. 7A, lane 5). In Fig. 7A, because each lane has an equal volume of reaction mixture, the experiments in which apyrase was added (lanes 4 and 5) had effectively less of the proteins of the 75-kDa complex that entered the gel. Apyrase is 200 kDa and consequently did not enter the gel. In Fig. 7A, lane 6, is shown FABP1 migrating at its expected 14 kDa.
Fig. 7.
Treatment of the 75-kDa protein complex with ATP and PKCζ disrupts the complex. A, SimplyBlue SafeStain of the 75-kDa protein complex incubated with ATP and PKCζ as indicated. 15 μg of protein was applied to each lane, and the proteins were separated using native PAGE. Apyrase (2 mg) was added to lanes 4 and 5 to stop additional phosphorylation. B, immunoblot using anti-FABP1 antibodies employing the same reagents as in A. The proteins were separated by native gel. Detection was by ECL. Lane 1, protein markers. Lane 2, untreated 75-kDa complex. Lane 3, 75-kDa complex incubated at 37 °C for 30 min. Lane 4, 75-kDa complex incubated with ATP and PKCζ at 4 °C for 30 min. Lane 5, 75-kDa complex incubated with ATP and PKCζ for 30 min at 37 °C. Lane 6, FABP1 (2 μg) alone. 30 μg of complex protein was applied to each lane. Detection was by ECL.
Because it would appear from Fig. 7A, lane 5, that FABP1 is split from the complex on phosphorylation of Sar1b, we wished to confirm this by immunoblotting a native PAGE (30 μg used for each lane) for FABP1 using the same protocol for each lane as in Fig. 7A. When the untreated complex was separated by native PAGE, all the FABP1 migrated as a 75-kDa protein (Fig. 7B, lane 2). Most of the FABP1 remained with the multiprotein complex when it was incubated at 37 °C (Fig. 7B, lane 3). When the complex was incubated with ATP and PKCζ at 4 °C, all the FABP1 remained with the complex (Fig. 7B, lane 4). Importantly, when the complex was incubated with ATP and PKCζ at 37 °C, most of the FABP1 migrated at its monomer of 14 kDa (Fig. 7B, lane 5). Fig. 7B, lane 6, is FABP1. These data support the concept that Sar1b phosphorylation splits the multiprotein complex releasing FABP1.
The SimplyBlue SafeStain-stained bands in Fig. 7A suggest that the complex is completely disrupted on phosphorylation of Sar1b. To test this hypothesis, we immunoblotted for the four-component proteins of the complex before and after phosphorylation. As expected, before phosphorylation, all the component proteins migrated at 75 kDa (Fig. 8, lanes 1). By contrast, after phosphorylation by PKCζ and ATP incubated at 37 °C, each of the four-component proteins migrated at their monomer molecular weight as shown by the immunoblots for each protein (Fig. 8, lanes 2). These data confirm the results of Fig. 7A suggesting that on phosphorylation of the 75-kDa complex, the complex is completely disrupted resulting in each component protein migrating at is monomer molecular weight.
Fig. 8.
75-kDa complex is completely disrupted by phosphorylation of the heterotetramer. Untreated 75-kDa protein complex (30 μg) collected from the anti-FABP1 adsorption column was separated by native PAGE (lane 1 of each immunoblot). 75 kDa is indicated by the arrows. The component proteins were identified by immunoblot using antibodies to the proteins as shown below each blot. In lane 2 for each immunoblot, the 75-kDa complex was treated with ATP and PKCζ. 30 μg of the treated protein was separated by native PAGE and transblotted to a nitrocellulose membrane. The proteins identified (FABP1, Sar1b, Sec13, and SVIP) in each gel are shown below the immunoblot, and its molecular mass is indicated by the arrows. The immunoblot for FABP1 was washed and re-immunoblotted for Sar1b. The immunoblot for Sec13 was washed and immunoblotted for SVIP. Detection was by ECL.
FABP1, Released from the 75-kDa Complex, Binds to ER Membranes
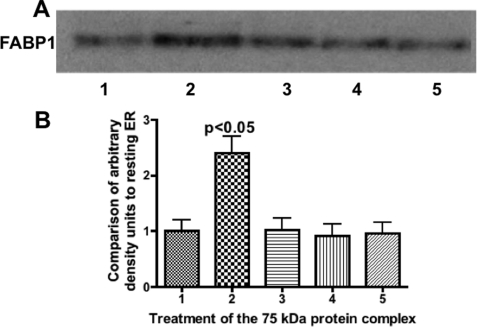
Finally, we wished to test if FABP1, as a component of the 75-kDa complex, is inhibited from binding to the ER, whereas if it were split from the complex, it would be free to bind to the ER in concert with our prior recombinant FABP1 experiments (12). To test this, we performed immunoblots on ER that had been incubated with buffer (Fig. 9A, lane 1), incubated with the 75-kDa complex previously incubated with PKCζ and ATP (Fig. 9A, lane 2), incubated with untreated 75-kDa complex (Fig. 9A, lane 3), or the complex incubated with ATP but without PKCζ (Fig. 9A, lane 4), or the complex incubated with PKCζ but without ATP (Fig. 9A, lane 5). All incubations were performed at 37 °C. Densitometry of the bands is presented in Fig. 9B. The densitometric studies confirm that in the absence of phosphorylation of the complex, FABP1 binding to the ER is not increased over that which is present on resting ER. However, when the complex is phosphorylated and disrupted as a consequence, FABP1 binding to the ER is significantly increased.
Fig. 9.
FABP1, as a member of the 75-kDa complex, has increased binding to intestinal ER only when the complex is incubated with ATP and PKCζ. Intestinal ER (300 μg) was incubated with the 75-kDa protein complex isolated from the anti-FABP1 adsorption column (50 μg). Before incubation with the ER, the complex was treated under various conditions as indicated. After the ER complex incubation, 30 μg of protein was separated by SDS-PAGE and immunoblotted using anti-FABP1 antibodies. Detection was by ECL. Lane 1, native ER. Lane 2, protein complex incubated with ATP and PKCζ at 37 °C. Lane 3, complex incubated with ATP and PKCζ at 4 °C. Lane 4, complex incubated with ATP but without PKCζ at 37 °C. Lane 5, complex incubated with PKCζ but without ATP at 37 °C. A, anti-FABP1 immunoblot. The FABP1 band is indicated. B, band densities from A were analyzed by the Gel Doc XR and recorded as densities compared with the band density of resting ER. The bars are derived from the immunoblot densities in A. The results are the mean ± S.E. of four trials The p value of test differences between the means of lanes 1, 3, 4, and 5 with lane 2 is shown.
In sum, the data suggest that the FABP1-containing 75-kDa multimer is completely disrupted by phosphorylation of Sar1b, freeing FABP1 enabling it to bind to the ER membrane.
DISCUSSION
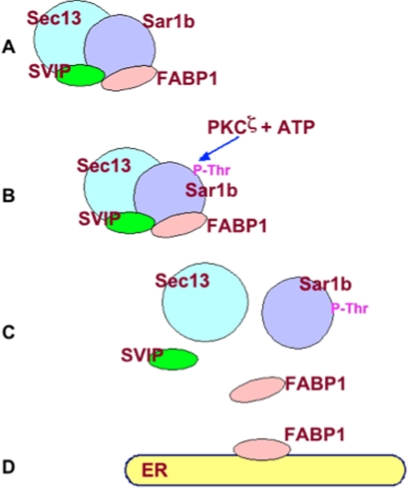
ER to Golgi transport vesicles such as PCTV select specific cargo, are sealed, and transport their cargo in a vectorial manner (8). We found that PCTV only requires FABP1 to organize the PCTV-budding complex (24) and generate the vesicle (12). No ATP is needed. In contrast to these data, when native cytosol is used to bud PCTV, ATP is required (8). In consideration of these data, we postulated that the FABP1 in cytosol (15) must be inhibited from binding to the ER membrane unless the inhibition is relieved by ATP treatment. Here, we provide support for both aspects of this hypothesis. First, we show that FABP1 is present in cytosol as a member of a four-protein complex composed of FABP1, Sar1b, SVIP, and Sec13. The presence of an FABP1-containing protein complex in cytosol is based on protein-protein interactivity as shown by the anti-FABP1 antibody adsorption column and the anti-FABP1 antibody pulldown experiment. A second independent means of support for this thesis is provided by the co-migration of FABP1 on two different gel permeation supports at a suggested mass of 75 kDa. To our knowledge, this is the first time that FABP1 has been proposed to be present as part of a complex in cytosol rather than as a monomer. Importantly, we show that the complex serves to functionally inhibit FABP1 from binding to the ER membrane. Second, we show that on phosphorylation of a member of the complex, Sar1b, the complex is disrupted freeing FABP1 to bind to the ER membrane, the second aspect of our hypothesis. Fig. 10 presents a model incorporating these ideas.
Fig. 10.
Model of the itinerary of FABP1 from cytosol to the ER membrane. A, 75-kDa complex composed of Sar1, FABP1, Sec13, and SVIP. B, PKCζ phosphorylates a Thr on Sar1b. C, complex is completely disrupted after Sar1b phosphorylation releasing FABP1. D, released FABP1 binds to the ER membrane.
FABP1 is thought to be a transporter of FA within the various cell types in which it is expressed, liver, intestine, and kidney (25). This concept is supported by the elegant fluorescent FA studies of Luxon et al. (26, 27) in which FA movement in the cytosol was greatly increased by the presence of FABP1. FABP1 binds two FA and is more promiscuous in ligand binding than intestinal FABP (FABP2), which only binds a single FA (25). FABP1, in contrast to FABP2, has been shown to deliver its FA ligand to membranes by diffusion and not collision (28). This would suggest that there is little interactivity between FABP1 and membranes. However, more recent data have suggested that FABP1 is capable of interacting with membranes (29). This concept is supported by the data reported here and our prior data showing FABP1 binding to the ER (12). By contrast, FABP2 did not bind to ER membranes on incubation (12).
Our current data showing the integration of FABP1 into a multiprotein complex is not necessarily inconsistent with the FA transport thesis of FABP because the FABP1 in the complex could bind FA and transport it to the membrane. We suggest, however, that this role is subsidiary to the ability of FABP1 to organize the PCTV-budding complex and generate PCTV. In further support of the role of FABP1 in association with PCTV budding and/or FA transport, immunohistochemical data show that FABP1 is concentrated near the apical membrane of enterocytes in the fasting state but re-distributes to the cytosol after lipid feeding where it could interact with the ER (30).
There is little information in the literature on the phosphorylation of Sar1b, but phosphorylation of a different COPII protein, Sec24, increases its interactivity with its binding partner, Sec23 (31). Other data suggest that the phosphorylation of Sec24C attenuates its membrane binding, reducing ER to Golgi transport (32). In the intestine, Sec24C appears not to be phosphorylated by PKCζ.
The results of phosphorylation of Sar1b as suggested here might be expected to differ from phosphorylation found for the COPII proteins engaged in budding protein vesicles. Although the COPII proteins, specifically Sec24C (33), are required for Golgi targeting of PCTV, they are not required for PCTV budding (8). GTP is also not needed for PCTV budding (8) thus removing the requirement for the GDP-GTP exchanger, Sec12, to be involved in the budding process. Because the COPII proteins do not play an active role in PCTV budding but do in protein vesicle budding, it suggests that the COPII proteins are attached to the budding PCTV vesicle differently than are the COPII proteins involved with protein vesicle budding. Furthermore, the cargo-selective function of the COPII proteins, especially Sec24 (34, 35), for protein vesicles appears to play a secondary role for PCTV. At least three of the COPII proteins, Sar1b, Sec23, and Sec24C, interact with the quintessential protein of the developing chylomicron, apolipoprotein B48 (36), but this binding is not required for incorporation of the chylomicron cargo into PCTV (12). These data are in contrast to the required engagement of Sec24 with cargo proteins in the protein vesicle system (34).
This study would suggest that the Sar1b synthetic defects due to SAR1B gene mutations would lead to a phenotype in which chylomicrons would be retained in the ER. A phenotype suggestive of this mutation, chylomicron retention disease, was originally described by Anderson et al. in 1961 (37), but the mutated protein was not identified as Sar1b until 42 years later (38). Subsequently, Samson-Bouma and co-workers (17) have systematically collected and identified multiple mutations in the SAR1B gene. Unfortunately, study of the various mutations has not led to a coherent thesis enabling a single functional defect to be pin-pointed. At least some of the defects are in the GTP binding pocket of Sar1b, but its effect on PCTV generation is open to question because PCTV production is independent of GTP/GDP (8).
The data reported here may explain our prior observation that immunodepletion of Sar1 from intestinal cytosol increases PCTV production 6-fold (8). We postulate that on removal of Sar1 from cytosol, FABP1 is freed from the 75-kDa complex and is now able to bind to the ER in an unregulated manner and generate PCTV. If true, this thesis would give further support to the regulatory role of the FABP1 heterotetrameric complex and its phosphorylation in controlling PCTV generation.
The protein heterotetramer proposed here consists of 4 mol of FABP1 compared with 1 or 1.6 mol of the other components. These data, in which one component of a protein complex predominates, are consistent with other protein complexes such as the COPII complex in which Sec13,31 is present as two heterodimers as compared with the other COPII components that are present singly (39) and Hsp90 which is present 4:1 in a complex with the glucocorticoid receptor (40). There are other similar examples (41, 42).
Protein phosphorylation has long been known to both increase or decrease the activity of a variety of proteins, translocate them from cytosol to ER (13, 43) or other membranes (43, 44), bind them to other proteins that recognize the phosphorylated Ser, Tyr, or Thr, induce protein synthesis (45), or target proteins for degradation by the ubiquitin/proteasome pathway. Less well recognized is the promotion of protein aggregation (46). With respect to modulation of ER to Golgi transport vesicle budding, phosphorylation of Sec23p promotes ER to Golgi protein transport as part of the COPII complex (47), and phosphorylation of Sec24 by PKB promotes Sec23–24 binding (31). We describe here a novel result of protein phosphorylation in which the phosphorylation of one protein (Sar1b) in a multiprotein complex disassembles the complex releasing a protein (FABP1) that can initiate the generation of a transport vesicle, PCTV.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant DK074565 from NIDDK (to C. M. M.).

This article contains supplemental Tables 1–3 and Fig. 1.
- FA
- fatty acid
- TAG
- triacylglycerol
- PCTV
- pre-chylomicron transport vesicle
- SVIP
- small VCP/p97-interactive protein
- COPII
- coat protein complex II
- ER
- endoplasmic reticulum.
REFERENCES
- 1. Strålfors P. (1990) Autolysis of isolated adipocytes by endogenously produced fatty acids. FEBS Lett. 263, 153–154 [DOI] [PubMed] [Google Scholar]
- 2. Mansbach C. M., 2nd, Nevin P. (1998) Intracellular movement of triacylglycerols in the intestine. J. Lipid Res. 39, 963–968 [PubMed] [Google Scholar]
- 3. Cartwright I. J., Higgins J. A. (2001) Direct evidence for a two-step assembly of ApoB48-containing lipoproteins in the lumen of the smooth endoplasmic reticulum of rabbit enterocytes. J. Biol. Chem. 276, 48048–48057 [DOI] [PubMed] [Google Scholar]
- 4. Young S. G., Cham C. M., Pitas R. E., Burri B. J., Connolly A., Flynn L., Pappu A. S., Wong J. S., Hamilton R. L., Farese R. V., Jr. (1995) A genetic model for absent chylomicron formation. Mice producing apolipoprotein B in the liver but not in the intestine. J. Clin. Invest. 96, 2932–2946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Jiang Z. G., Liu Y., Hussain M. M., Atkinson D., McKnight C. J. (2008) Reconstituting initial events during the assembly of apolipoprotein B-containing lipoproteins in a cell-free system. J. Mol. Biol. 383, 1181–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zilversmit D. B. (1967) Formation and transport of chylomicrons. Fed. Proc. 26, 1599–1605 [PubMed] [Google Scholar]
- 7. Mahan J. T., Heda G. D., Rao R. H., Mansbach C. M., 2nd (2001) The intestine expresses pancreatic triacylglycerol lipase. Regulation by dietary lipid. Am. J. Physiol. Gastrointest. Liver Physiol. 280, G1187–G1196 [DOI] [PubMed] [Google Scholar]
- 8. Siddiqi S. A., Gorelick F. S., Mahan J. T., Mansbach C. M., 2nd (2003) COPII proteins are required for Golgi fusion but not for endoplasmic reticulum budding of the pre-chylomicron transport vesicle. J. Cell Sci. 116, 415–427 [DOI] [PubMed] [Google Scholar]
- 9. Aridor M., Fish K. N., Bannykh S., Weissman J., Roberts T. H., Lippincott-Schwartz J., Balch W. E. (2001) The Sar1 GTPase coordinates biosynthetic cargo selection with endoplasmic reticulum export site assembly. J. Cell Biol. 152, 213–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rothman J. E., Orci L. (1992) Molecular dissection of the secretory pathway. Nature 355, 409–415 [DOI] [PubMed] [Google Scholar]
- 11. Barlowe C., Orci L., Yeung T., Hosobuchi M., Hamamoto S., Salama N., Rexach M. F., Ravazzola M., Amherdt M., Schekman R. (1994) COPII. A membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell 77, 895–907 [DOI] [PubMed] [Google Scholar]
- 12. Neeli I., Siddiqi S. A., Siddiqi S., Mahan J., Lagakos W. S., Binas B., Gheyi T., Storch J., Mansbach C. M., 2nd (2007) Liver fatty acid-binding protein initiates budding of pre-chylomicron transport vesicles from intestinal endoplasmic reticulum. J. Biol. Chem. 282, 17974–17984 [DOI] [PubMed] [Google Scholar]
- 13. Aridor M., Balch W. E. (2000) Kinase signaling initiates coat complex II (COPII) recruitment and export from the mammalian endoplasmic reticulum. J. Biol. Chem. 275, 35673–35676 [DOI] [PubMed] [Google Scholar]
- 14. Siddiqi S., Saleem U., Abumrad N. A., Davidson N. O., Storch J., Siddiqi S. A., Mansbach C. M., 2nd (2010) A novel multiprotein complex is required to generate the prechylomicron transport vesicle from intestinal ER. J. Lipid Res. 51, 1918–1928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bass N. M., Manning J. A., Ockner R. K., Gordon J. I., Seetharam S., Alpers D. H. (1985) Regulation of the biosynthesis of two distinct fatty acid-binding proteins in rat liver and intestine. Influences of sex difference and of clofibrate. J. Biol. Chem. 260, 1432–1436 [PubMed] [Google Scholar]
- 16. Siddiqi S. A., Mansbach C. M., 2nd (2008) PKCζ-mediated phosphorylation controls budding of the pre-chylomicron transport vesicle. J. Cell Sci. 121, 2327–2338 [DOI] [PubMed] [Google Scholar]
- 17. Georges A., Bonneau J., Bonnefont-Rousselot D., Champigneulle J., Rabès J. P., Abifadel M., Aparicio T., Guenedet J. C., Bruckert E., Boileau C., Morali A., Varret M., Aggerbeck L. P., Samson-Bouma M. E. (2011) Molecular analysis and intestinal expression of SAR1 genes and proteins in Anderson disease (chylomicron retention disease). Orphanet J. Rare Dis. 6, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Roy C. C., Levy E., Green P. H., Sniderman A., Letarte J., Buts J. P., Orquin J., Brochu P., Weber A. M., Morin C. L., Marcel Y., Deckelbaum R. J. (1987) Malabsorption, hypocholesterolemia, and fat-filled enterocytes with increased intestinal apoprotein B. Chylomicron retention disease. Gastroenterology 92, 390–399 [DOI] [PubMed] [Google Scholar]
- 19. Shugrue C. A., Kolen E. R., Peters H., Czernik A., Kaiser C., Matovcik L., Hubbard A. L., Gorelick F. (1999) Identification of the putative mammalian orthologue of Sec31P, a component of the COPII coat. J. Cell Sci. 112, 4547–4556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kumar N. S., Mansbach C. M. (1997) Determinants of triacylglycerol transport from the endoplasmic reticulum to the Golgi in intestine. Am. J. Physiol. 273, G18–G30 [DOI] [PubMed] [Google Scholar]
- 21. Siddiqi S. A., Mahan J., Siddiqi S., Gorelick F. S., Mansbach C. M., 2nd (2006) Vesicle-associated membrane protein 7 is expressed in intestinal ER. J. Cell Sci. 119, 943–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hirosawa M., Hoshida M., Ishikawa M., Toya T. (1993) MASCOT. Multiple alignment system for protein sequences based on three-way dynamic programming. Comput. Appl. Biosci. 9, 161–167 [DOI] [PubMed] [Google Scholar]
- 23. Mansbach C. M., 2nd, Parthasarathy S. (1982) A re-examination of the fate of glyceride-glycerol in neutral lipid absorption and transport. J. Lipid Res. 23, 1009–1019 [PubMed] [Google Scholar]
- 24. Siddiqi S. A., Siddiqi S., Mahan J., Peggs K., Gorelick F. S., Mansbach C. M., 2nd (2006) The identification of a novel endoplasmic reticulum to Golgi SNARE complex used by the prechylomicron transport vesicle. J. Biol. Chem. 281, 20974–20982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Storch J., Corsico B. (2008) The emerging functions and mechanisms of mammalian fatty acid-binding proteins. Annu. Rev. Nutr. 28, 73–95 [DOI] [PubMed] [Google Scholar]
- 26. Luxon B. A., Weisiger R. A. (1993) Sex differences in intracellular fatty acid transport. Role of cytoplasmic binding proteins. Am. J. Physiol. 265, G831–G841 [DOI] [PubMed] [Google Scholar]
- 27. Luxon B. A., Milliano M. T. (1997) Cytoplasmic co-diffusion of fatty acids is not specific for fatty acid-binding protein. Am. J. Physiol. 273, C859–C867 [DOI] [PubMed] [Google Scholar]
- 28. Thumser A. E., Storch J. (2000) Liver and intestinal fatty acid-binding proteins obtain fatty acids from phospholipid membranes by different mechanisms. J. Lipid Res. 41, 647–656 [PubMed] [Google Scholar]
- 29. Falomir-Lockhart L. J., Franchini G. R., Guerbi M. X., Storch J., Córsico B. (2011) Interaction of enterocyte FABPs with phospholipid membranes. Clues for specific physiological roles. Biochim. Biophys. Acta 1811, 452–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Alpers D. H., Bass N. M., Engle M. J., DeSchryver-Kecskemeti K. (2000) Intestinal fatty acid binding protein may favor differential apical fatty acid binding in the intestine. Biochim. Biophys. Acta 1483, 352–362 [DOI] [PubMed] [Google Scholar]
- 31. Sharpe L. J., Luu W., Brown A. J. (2011) Akt phosphorylates Sec24. New clues into the regulation of ER-to-Golgi trafficking. Traffic 12, 19–27 [DOI] [PubMed] [Google Scholar]
- 32. Dudognon P., Maeder-Garavaglia C., Carpentier J. L., Paccaud J. P. (2004) Regulation of a COPII component by cytosolic O-glycosylation during mitosis. FEBS Lett. 561, 44–50 [DOI] [PubMed] [Google Scholar]
- 33. Siddiqi S., Siddiqi S. A., Mansbach C. M., 2nd (2010) Sec24C is required for docking the prechylomicron transport vesicle with the Golgi. J. Lipid Res. 51, 1093–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Miller E. A., Beilharz T. H., Malkus P. N., Lee M. C., Hamamoto S., Orci L., Schekman R. (2003) Multiple cargo-binding sites on the COPII subunit Sec24p ensure capture of diverse membrane proteins into transport vesicles. Cell 114, 497–509 [DOI] [PubMed] [Google Scholar]
- 35. Miller E., Antonny B., Hamamoto S., Schekman R. (2002) Cargo selection into COPII vesicles is driven by the Sec24p subunit. EMBO J. 21, 6105–6113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Siddiqi S., Mani A. M., Siddiqi S. A. (2010) The identification of the SNARE complex required for the fusion of VLDL-transport vesicle with hepatic cis-Golgi. Biochem. J. 429, 391–401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Anderson C. M., Townley R. R., Freemanm, Johansen P. (1961) Unusual causes of steatorrhoea in infancy and childhood. Med. J. Aust. 48, 617–622 [DOI] [PubMed] [Google Scholar]
- 38. Jones B., Jones E. L., Bonney S. A., Patel H. N., Mensenkamp A. R., Eichenbaum-Voline S., Rudling M., Myrdal U., Annesi G., Naik S., Meadows N., Quattrone A., Islam S. A., Naoumova R. P., Angelin B., Infante R., Levy E., Roy C. C., Freemont P. S., Scott J., Shoulders C. C. (2003) Mutations in a Sar1 GTPase of COPII vesicles are associated with lipid absorption disorders. Nat. Genet. 34, 29–31 [DOI] [PubMed] [Google Scholar]
- 39. Miller E. A., Barlowe C. (2010) Regulation of coat assembly-sorting things out at the ER. Curr. Opin. Cell Biol. 22, 447–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bresnick E. H., Dalman F. C., Pratt W. B. (1990) Direct stoichiometric evidence that the untransformed Mr 300,000, 9 S, glucocorticoid receptor is a core unit derived from a larger heteromeric complex. Biochemistry 29, 520–527 [DOI] [PubMed] [Google Scholar]
- 41. Sellin M. E., Sandblad L., Stenmark S., Gullberg M. (2011) Deciphering the rules governing assembly order of mammalian septin complexes. Mol. Biol. Cell 22, 3152–3164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Allen J. R., Ensign S. A. (1997) Purification to homogeneity and reconstitution of the individual components of the epoxide carboxylase multiprotein enzyme complex from Xanthobacter strain Py2. J. Biol. Chem. 272, 32121–32128 [DOI] [PubMed] [Google Scholar]
- 43. Anandatheerthavarada H. K., Biswas G., Mullick J., Sepuri N. B., Otvos L., Pain D., Avadhani N. G. (1999) Dual targeting of cytochrome P4502B1 to endoplasmic reticulum and mitochondria involves a novel signal activation by cyclic AMP-dependent phosphorylation at Ser-128. EMBO J. 18, 5494–5504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fujiwara H., Hasegawa M., Dohmae N., Kawashima A., Masliah E., Goldberg M. S., Shen J., Takio K., Iwatsubo T. (2002) α-Synuclein is phosphorylated in synucleinopathy lesions. Nat. Cell Biol. 4, 160–164 [DOI] [PubMed] [Google Scholar]
- 45. Ding L., Ma W., Littmann T., Camp R., Shen J. (2011) The P2Y(2) nucleotide receptor mediates tissue factor expression in human coronary artery endothelial cells. J. Biol. Chem. 286, 27027–27038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wallace B. G. (1994) Staurosporine inhibits agrin-induced acetylcholine receptor phosphorylation and aggregation. J. Cell Biol. 125, 661–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Palmer K. J., Konkel J. E., Stephens D. J. (2005) PCTAIRE protein kinases interact directly with the COPII complex and modulate secretory cargo transport. J. Cell Sci. 118, 3839–3847 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.