Background: Selenocysteine incorporation requires unique translation factors that interact with the ribosome.
Results: SECIS-binding protein 2 alters ribosome conformation at two discrete sites.
Conclusion: The selenocysteine incorporation reaction requires ribosome modifications in a region known to be required for tRNA accommodation.
Significance: Identifying the ribosomal dynamics required for Sec incorporation will enhance our understanding of the translation elongation reaction.
Keywords: Ribosome Function, Ribosome Structure, Selenium, Selenocysteine, Transfer RNA (tRNA), SBP2, SECIS
Abstract
Sec-tRNASec is site-specifically delivered at defined UGA codons in selenoprotein mRNAs. This recoding event is specified by the selenocysteine insertion sequence (SECIS) element and requires the selenocysteine (Sec)-specific elongation factor, eEFSec, and the SECIS binding protein, SBP2. Sec-tRNASec is delivered to the ribosome by eEFSec-GTP, but this ternary complex is not sufficient for Sec incorporation, indicating that its access to the ribosomal A-site is regulated. SBP2 stably associates with ribosomes, and mutagenic analysis indicates that this interaction is essential for Sec incorporation. However, the ribosomal function of SBP2 has not been elucidated. To shed light on the functional relevance of the SBP2-ribosome interaction, we screened the functional centers of the 28 S rRNA in translationally competent 80 S ribosomes using selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE). We demonstrate that SBP2 specifically alters the reactivity of specific residues in Helix 89 (H89) and expansion segment 31 (ES31). These results are indicative of a conformational change in response to SBP2 binding. Based on the known functions of H89 during translation, we propose that SBP2 allows Sec incorporation by either promoting Sec-tRNASec accommodation into the peptidyltransferase center and/or by stimulating the ribosome-dependent GTPase activity of eEFSec.
Introduction
Protein synthesis involves the sequential incorporation of amino acids by the translational apparatus in response to sense codons in the messenger RNA (mRNA). In eukaryotes, aminoacyl-tRNA molecules (aa-tRNAs)3 are delivered to the ribosomal A-site by the elongation factor, eEF1A (EF-Tu in prokaryotes) in a ternary complex (TC) with GTP. An exception arises in the case of the 21st amino acid, selenocysteine (Sec), which is encoded by a nonsense UGA codon in selenoprotein mRNAs. Sec is biosynthesized on a specialized tRNA (tRNASec) that is recognized and delivered to the ribosome by a dedicated elongation factor, eEFSec (1, 2). However, the eEFSec TC is not sufficient to decode the UGA codon indicating that, unlike the canonical TC, its access to the A-site is restricted.
Interestingly, the serylated precursor from which Sec is biosynthesized (Ser-tRNASec) is capable of suppressing the UGA codon in vitro (3). Ser-tRNASec is presumably bound and delivered by eEF1A because eEFSec exhibits marked specificity for Sec-tRNASec (1, 2, 4). This suggests that the inability of the eEFSec TC to suppress UGA codons randomly is an intrinsic feature of eEFSec, consistent with the presence of an additional fourth domain relative to the canonical elongation factor eEF1A. Canonical aa-tRNAs have been shown to bind the ribosomal A-site with an equal affinity despite the molecular diversity inherent in the various aa-tRNAs. Interestingly, this uniform binding is lost when deacylated tRNAs are used as substrates, suggesting that the amino acid also contributes to the thermodynamic stability of this interaction and highlighting the fact that the ribosome is not blind to the identity of the amino acid (5). Thus, it is also possible that the Sec residue contributes to this regulation by destabilizing the Sec-tRNASec in the absence of the other components of the Sec incorporation complex.
Sec incorporation is specified by the Sec insertion sequence (SECIS) element in the 3′-untranslated region (UTR) of selenoprotein mRNAs and the SECIS-binding protein, SBP2 (6, 7). However, the mechanism through which this information is transmitted to the ribosome is unknown. SBP2 comprises a dispensable N-terminal domain, a central Sec incorporation domain (SID), and a C-terminal RNA binding domain (RBD); together, the SID and the RBD are referred to as CTSBP2. Aside from its essential function in binding the SECIS, SBP2 also interacts with the ribosome, making it a likely candidate for communicating this specificity information. Consistent with this notion, extensive mutagenesis of the RBD in CTSBP2 led to the identification of a discrete stretch of amino acids (647RFQDR651) that when mutated to penta-alanine resulted in a decrease in ribosome binding, and Sec incorporation activity (11). Importantly, this SBP2 mutant retained near wild-type levels of SECIS binding activity allowing us to conclude that ribosome binding is an essential component of the Sec incorporation activity of SBP2.
Despite this finding, we subsequently reported that when expressed as separate proteins and added to a Sec incorporation assay in equimolar amounts, the SID and the RBD retain near wild-type levels of Sec incorporation activity in vitro but lose the ability to associate stably with ribosomes (9). In addition, our attempts to detect an SBP2-SECIS-ribosome complex have been unsuccessful (10, 11). Together with the fact that the SID and RBD as separate proteins could be formaldehyde cross-linked to ribosomes (9), these results indicate that transient SBP2-ribosome interactions are sufficient to promote Sec incorporation and suggest that SBP2 has distinct functions on the SECIS and the ribosome.
Using selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE), we demonstrate that CTSBP2 specifically enhances the reactivity of residues U4071, and C4073 in the apical loop of Helix 89 (H89), and between G3802 and C3812, and G3758 and U3768 on expansion segment 31 (ES31; mouse numbering). Additionally, we demonstrate that the SID and RBD are also able to alter the reactivity of these same residues in ES31, and to a lesser extent H89, indicating that functional interactions persist despite the loss of stable ribosome binding. Based on the known roles of H89, we propose that SBP2-induced conformational changes in this helix promote Sec incorporation by affecting Sec-tRNASec accommodation and/or by stimulating the ribosome-dependent GTPase activation of eEFSec.
EXPERIMENTAL PROCEDURES
Constructs
The CTSBP2 (amino acids 399–846), the SID (amino acids 399–586), and the RBD (amino acids 586–846) were all cloned into pTrcHis (Invitrogen) by TOPO-TA cloning, to generate constructs containing an N-terminal Xpress/His tag. Rat ribosomal protein L30 (rpL30) in pET200 (Invitrogen) was kindly provided by Donna Driscoll. The penta-alanine mutant versions of CTSBP2 and RBD were generated by site-directed mutagenesis of the CTSBP2 and the RBD constructs in pTrcHis. All of the constructs were sequenced completely by automated DNA sequencing.
Expression of Recombinant Proteins
All of the recombinant proteins in this study were purified as described by Kinzy et al. (10). All of the proteins were buffer-exchanged into 1× SHAPE buffer lacking MgCl2 (80 mm Hepes-KOH, pH 7.5, 100 mm KCl, 2 mm DTT) using a P30 gel filtration column (Bio-Rad) or by overnight dialysis.
Purification of 80 S Ribosomes
80 S ribosomes were purified as described, with minor modifications (12). Briefly, 50 ml of crude rabbit reticulocyte lysate (Green Hectares) was thawed, and one EDTA-free tablet of complete protease inhibitor mixture (Roche Applied Science) was added. Ribosomes were pelleted by ultracentrifugation at 100,000 × g for 2.5 h at 4 °C using a Sorvall S80-AT3 rotor. Ribosome pellets were first washed with Buffer A (50 mm Tris-HCl, pH 7.5, 0.25 m sucrose, 1 mm DTT, and 0.1 mm EDTA) and then gently resuspended in high salt buffer (Buffer A + 0.5 m KCl) using 0.1–0.2 of the original lysate volume. The ribosome solution was allowed to stir for 30–60 min at 4 °C before layering over a 1 m sucrose cushion in high salt buffer (2:1 ratio of ribosome solution to sucrose pad) and centrifuging at 278,000 × g for 2 h at 4 °C. 80 S ribosomes were gently resuspended in 1× SHAPE buffer (80 mm Hepes-KOH, pH 7.5, 100 mm KCl, 2 mm DTT, and 0.5 mm MgCl2). The concentration of 80 S ribosomes was determined using an extinction coefficient at A260 of 50 μm−1 cm−1. 80 S ribosomes were stored in small aliquots at −80 °C. Ribosome function was validated by luciferase mRNA translation in ribosome-depleted rabbit reticulocyte lysate.
SHAPE Reactions
SHAPE reactions were generated by incubating 10 pmol of 80 S ribosomes in the presence or absence of 5 μm CTSBP2 at 37 °C for 5 min. Alternatively, SHAPE reactions contained 10 pmol of ribosomes and 10 μm SID, RBD, or rpL30. All reactions were performed in 1× SHAPE buffer (80 mm Hepes-KOH, pH 7.5, 100 mm KCl, 2 mm DTT, and 0.5 mm MgCl2) in a final volume of 100 μl. Benzoyl cyanide (BzCN; Sigma) or vehicle (DMSO) was added to a final concentration of 80 mm. rRNA was extracted using the RNeasy kit (Qiagen) according to the manufacturer's instructions. Ribosomal RNA was eluted with RNase-free water and either used immediately for primer extension or stored at −80 °C.
Primer Extension and Data Analysis
1 pmol of modified rRNA was incubated with 12.5 pmol of ROX-labeled oligonucleotide (Invitrogen) in a total volume of 6.0 μl at 80 °C and slow cooled to 25 °C using a thermocycler. 3 μl of 4:1:1 buffer (5× Superscript III buffer:100 mm DTT:10 mm dNTP mix) was added and incubated at 52 °C for 1 min before adding 200 units of Superscript III (Invitrogen) and incubating for 20 min at 52 °C. DNA fragments were ethanol-precipitated in the presence of 20 μg of glycogen as a carrier, and DNA pellets were resuspended in 10 μl of deionized formamide (Fermentas). Capillary electrophoresis was performed in house using an ABI 3130XL sequencer. GeneScanTM LIZ®500 was added to each reaction and used as both a size standard to determine the relative position of SHAPE generated peaks and as an internal control to normalize peak areas due to variability in the injection volumes. Capillary electrophoresis-generated traces were analyzed using DAx software (Van Mierlo software consultancy). The positions selected for discussion were altered in an SBP2-dependent fashion with 100% reproducibility for a minimum of five independent experiments in all cases except where the SID and RBD domains were tested as independent domains, in which case three independent experiments were performed.
Fluorescent Primers and End Labeling
The following ROX-labeled primers were used in this study: 4409 (5′-GGATTCTGACTTAGAGGCGTTCAG-3′), 4229 (5′-CACGTTCCCTATTAG-3′), 4168 (5′-CAATGATAGGAAGAGCCGACATCG-3′), 3928 (5′-GACACCTGCGTTACCGTTTGACAG-3′), and 1989 (5′-CCGCTCCCGTCCCGTTCC-3′). The number designation corresponds to the base position on the rat 28 S rRNA (GenBank V01270.1) complementary to the base at the 3′ end of the primer. 4409 covers the sarcin-ricin loop (SRL) on Helix 95 (H95), and the neighboring helices (H94 and H96). 4229 and 4168 cover part of the peptidyltransferase center and the neighboring helices (H88 and H89). 3928 covers Helix 79 and expansion segment 31. 1989 covers the GTPase-associated center (GAC; H43–H44). For sequencing reactions, 5′ 32P-end-labeled primers were generated by mixing 62.5 pmol of DNA primer, 4 μl of [γ-32P]ATP (3000 Ci/mmol; PerkinElmer Life Sciences), 10 units of T4 polynucleotide kinase (New England Biolabs) in a 50-μl reaction containing 1× T4 polynucleotide kinase reaction buffer (70 mm Tris-HCl, pH 7.6, 10 mm MgCl2, 5 mm DTT). Reactions were incubated at 37 °C for 30 min followed by incubation at 65 °C for 20 min. Unincorporated nucleotides were removed using a P30 gel filtration column (Bio-Rad).
Sequencing Reactions and Gel Electrophoresis
For sequencing reactions, rRNA was extracted from 80 S ribosomes using TRIzol (Invitrogen) according to the manufacturer's instructions and used as a template for primer extension. Primer extension was performed by mixing 1 μg of rRNA with 2 μl of end-labeled primer in a total volume of 6 μl. Sequencing reactions also contained 1 μl of ddNTP (10 mm; Roche Applied Science). Reactions were incubated at 80 °C for 2 min and slow cooled to 25 °C in a thermocycler. Next, 3 μl of 4:1:1 buffer (5× Superscript III buffer:100 mm DTT:10 mm dNTP mix) and 200 units of Superscript III was added, and reactions were incubating for 20 min at 52 °C. 10 μl of formamide gel-loading buffer (80% (w/v)) deionized formamide, 10 mm EDTA, pH 8.0, 1 mg/ml xylene cyanol, and 1 mg/ml bromphenol blue) was added directly to the primer extension reactions and incubated at 95 °C for 5 min. 3.5 μl of each reaction was resolved on a 6% denaturing polyacrylamide gel and analyzed by phosphorimaging.
RESULTS
Novel SBP2-dependent Alterations in SHAPE Reactivity in 28 S rRNA
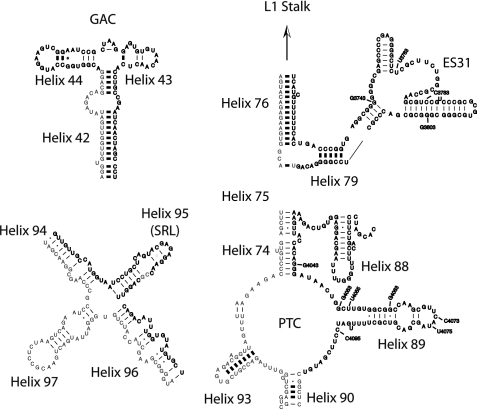
SBP2 binds stably to the large subunit of the ribosome, and preliminary studies indicate that this interaction is mediated by rRNA (13). However, the precise location of SBP2 on the ribosome has not been determined, nor has the mechanistic contribution of this interaction during Sec incorporation been elucidated. The large subunit of the ribosome has two major functional centers: (i) the GAC, and (ii) the peptidyltransferase center (PTC). The GAC is formed by H43 and H44, and along with the universally conserved SRL at the apical loop of H95, is responsible for binding and stimulating the GTPase activity of the translational GTPases, including initiation factor eIF5B (IF2 in prokaryotes), the canonical elongation factors eEF1A and eEF2 (EF-G in prokaryotes), and presumably eEFSec (SelB in prokaryotes). Each of these functional centers is shown in Fig. 1 as a diagram adapted from the Comparative RNA website (14) (Fig. 1). The PTC is the catalytic core of the ribosome; it is responsible for binding and correctly aligning the tRNA substrates in the A- and P-sites and catalyzing peptide bond formation. Due to the large size of 28 S rRNA, we chose to focus our studies on these areas because of their direct role in aa-tRNA selection during canonical translation elongation in all three domains of life. To shed light on the ribosomal function of SBP2, we used SHAPE to probe the 28 S rRNA for SBP2-dependent changes in the reactivity of residues at or near these regions to determine whether SBP2 could induce a unique rRNA conformation that would specify the recruitment of the eEFSec TC.
FIGURE 1.
Mouse 28 S rRNA. The reactivity of bases found at or near the GAC, the SRL, the PTC, and ES31 indicated in bold face type were interrogated with BzCN. The approximate base positions shown in the electrophoretic traces in Figs. 2, 4, 6, and 7 are also indicated. The secondary structures were adapted from the mouse 28 S rRNA structure obtained in the Comparative RNA web site and project.
Probing 28 S Functional Centers
SHAPE makes use of a class of hydroxyl-selective electrophiles to analyze nucleotide dynamics in RNA (15). We used the compound BzCN because of its fast reactivity and hence its potential to capture transient conformational changes (16). Salt-washed 80 S ribosomes were purified from rabbit reticuloctye lysate, and the translational activity of these ribosomes was qualitatively assessed by supplementing rabbit reticuloctye lysate depleted of endogenous ribosomes and testing their ability to translate a luciferase reporter mRNA (data not shown). 80 S ribosomes were treated with 80 mm BzCN in the presence or absence of CTSBP2. Alternatively, ribosomes were treated with vehicle (DMSO) to distinguish primer extension stops arising from RNA cleavages or excessive secondary structure from those resulting from modified RNA. Modified or vehicle-treated rRNA was extracted and used as a template for primer extension using ROX-labeled oligonucleotides downstream of the regions indicated in Fig. 1. DNA fragments were precipitated and resolved by capillary electrophoresis in the presence of an internal size standard (GS500 LIZ). The internal size standard was used to approximate the position of reactive residues and to normalize for variability in the injection volumes of the different samples resolved by capillary electrophoresis. Normalization of the data in this manner significantly reduced any global fluctuation in SHAPE reactivity across samples as evidenced by the high degree of overlap in traces (see Figs. 2, 4, 6, and 7). Visual inspection of the normalized traces revealed that the lowest variability corresponded to a region ∼50 to ∼150 bases from the 5′ end of the primer; for this reason, we limited our analysis to these regions only.
FIGURE 2.
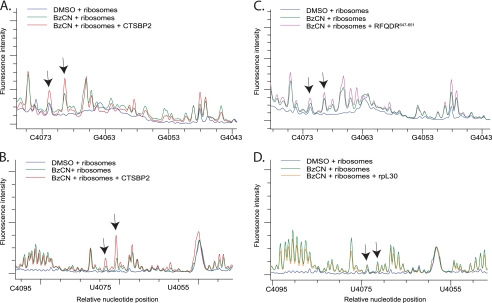
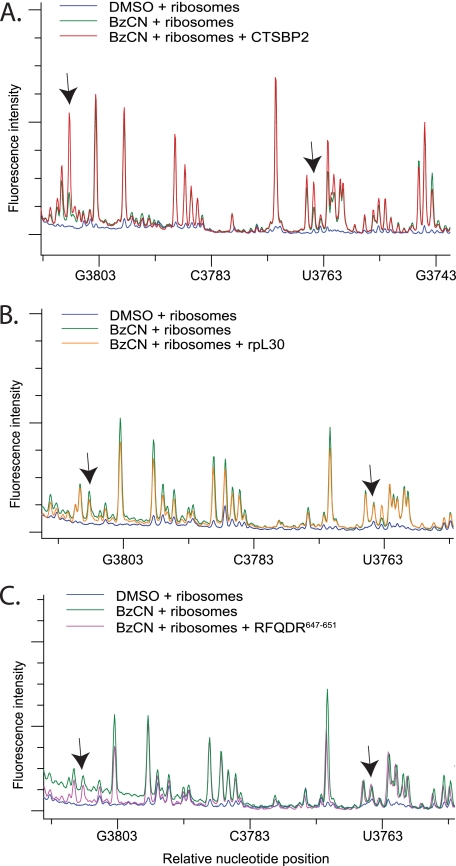
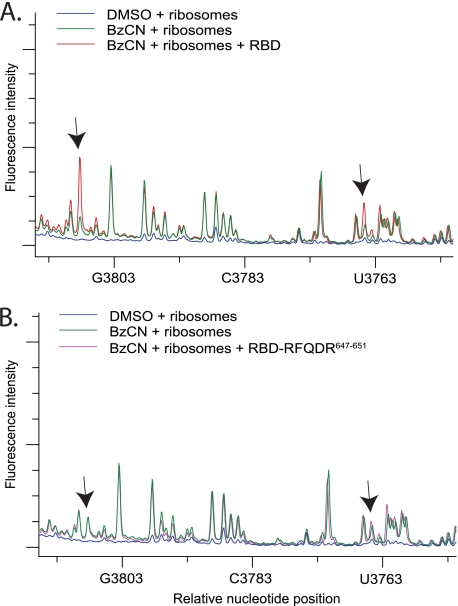
CTSBP2 specifically enhances the reactivity of nucleotides in H89. A, electrophoretic traces obtained from reactions containing 80S ribosomes treated with 80 mm BzCN in the presence (red) or absence (green) of recombinant CT-SBP2, or DMSO (blue). cDNA fragments were generated from primer extension reactions using primer 4168. B, same as in A except primer extension was performed with a second primer (primer 4229) located further downstream (see “Experimental Procedures”). C, Same as A except reactions were performed with the CTSBP2 647RFQDR651 penta-alanine mutant (purple). D, same as B except reactions were performed with ribosomal protein L30 (rpL30, orange). The y axis shows the relative fluorescence intensity, and the x axis shows the approximate nucleotide position on the mouse 28 S rRNA. Arrows indicate the nucleotides displaying enhanced reactivity in the presence of CTSBP2 (A and B) or the corresponding positions in reactions performed with the CTSBP2 647RFQDR651 penta-alanine mutant (C) or rpL30 (D). Each trace is representative of at least three independent SHAPE experiments.
FIGURE 4.
CTSBP2 specifically enhances the reactivity of bases in ES31. A, electrophoretic traces obtained from reactions containing 80 S ribosomes treated with DMSO (blue), or BzCN in the presence (red) or absence (green) of CTSBP2. cDNA fragments were generated from primer extension reactions using primer 3928. B and C, same as A except reactions were performed with the CTSBP2 647RFQDR651 penta-alanine mutant (purple) or rpL30 (orange). Arrows indicate regions with enhanced reactivity in the presence of CTSBP2 (A), or the corresponding positions in reactions performed with the CTSBP2 647RFQDR651 penta-alanine mutant (B) or rpL30 (C). Each trace is representative of three independent SHAPE experiments.
FIGURE 6.
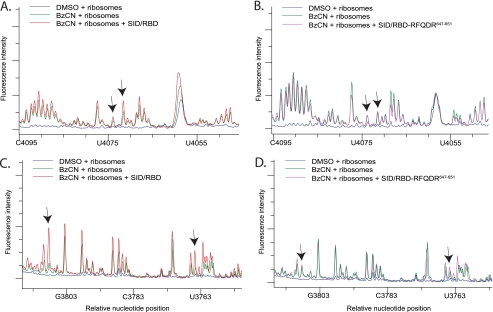
SHAPE analysis of H89 and ES31 with the SBP2 SID/RBD domains. A and C, electrophoretic traces obtained from reactions containing ribosomes treated with DMSO (blue) or BzCN in the presence (red) or absence (green) of the SID and the RBD. cDNA fragments were generated from reactions using primer 4229 to cover H89 (A) and primer 3928 to cover ES31 (C). B and D, same as A and C except reactions were performed with the SID and the RBD 647RFQDR651 penta-alanine mutant (purple). Arrows in A, B and C, D indicate the same nucleotide positions displaying enhanced reactivity in the presence of CTSBP2 seen in Figs. 2B and 3A, respectively. Each trace is representative of three independent SHAPE experiments.
FIGURE 7.
SHAPE analysis of ES31 with the SBP2 RBD domain alone. A, electrophoretic traces obtained from reactions containing ribosomes treated with DMSO (blue), or BzCN in the presence (red) or absence (green) of the RBD. cDNA fragments were generated from reactions using primer 3928 to cover ES31. B, same as A except reactions were performed with the RBD 647RFQDR651 penta-alanine mutant (purple). Arrows indicate the same nucleotide positions displaying enhanced reactivity in the presence of CTSBP2 seen in Fig. 3A. Each trace is representative of three independent SHAPE experiments.
Helix 89 Modifications in Presence of SBP2
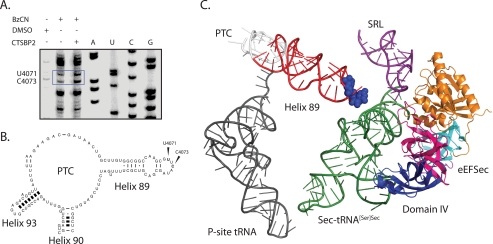
Previous chemical probing studies have correlated a conformational change in the SRL with the selective binding of EF-G over EF-Tu in pretranslocation state ribosomes (17). Additionally, movement of the GAC has also been implicated in EF binding specificity (18). In our analysis, there were no differences in the reactivity of residues in the GAC or the SRL in the presence of SBP2 (data not shown). Instead, we consistently observed an SBP2-dependent enhancement in the reactivity of two nucleotides in H89 (Fig. 2A). Because the reactive residues observed in H89 were in close proximity to the end of the primer, we repeated the primer extension with a second primer located further downstream and confirmed the enhanced reactivity at these positions (Fig. 2B). To confirm the specificity of these results, we performed a control experiment with the ribosome binding-defective SBP2 mutant 647RFQDR651, which lacks Sec incorporation activity as well as the ability to interact stably with ribosomes. In the presence of 647RFQDR651, the reactivity of the aforementioned residues was significantly reduced (compare Fig. 2, A with C). As an additional specificity control, we tested the ability of ribosomal protein L30 (rpL30) to induce similar modifications. Both SBP2 and rpL30 belong to the L7Ae/L30 protein family, and both have been shown to competitively bind to the SECIS element in vitro (8). In the presence of rpL30, no enhanced sensitivity to BzCN was detected (compare Fig. 2, B and D). To identify the exact nucleotide positions in H89 that are affected, cDNA fragments were generated with a radioactively labeled primer and resolved on a 6% sequencing gel with dideoxy sequencing reactions performed in parallel (Fig. 3A). The residues in H89 displaying enhanced reactivity in the presence of CTSBP2 were C4073, and U4071. The location of these residues in the mouse 28 S rRNA secondary structure map and on the high resolution structure of the 70 S ribosome from Thermus thermophilus is indicated in Fig. 3, B and C. We specifically chose this three-dimensional structure because it contains the post-translocation state of the ribosome with the EF-Tu TC in the A-site and thus represents the conformational state of the ribosome that the eEFSec TC is expected to interact with during Sec incorporation. The structure depicted in Fig. 3C has the archaeal eEFSec homolog, aSelB, and the human Sec-tRNA in place of the EF-Tu TC.
FIGURE 3.
Localization of SBP2-induced modifications in H89. A, sequencing lanes are indicated by A, U, C, or G and were generated from rRNA extracted from ribosomes. Primer extension reactions were generated from rRNA extracted from ribosomes incubated with DMSO, or BzCN in the presence or absence of CTSBP2. Residues U4071 and C4073 (E. coli U2473 and C2475, respectively), which display enhanced reactivity in the presence of CTSBP2, are indicated by the blue box. B, mouse 28 S secondary structure map of H89 with bases U4071 and C4073 indicated. C, PyMOL-generated image of the eEFSec homolog, archaeal SelB (Protein Data Bank code 1WB3), and the human tRNASec (Protein Data Bank code 3A3A), aligned over the crystal structure of the EF-Tu TC bound to the 70 S ribosome (2WRQ and 2WRR) using the align command. Domain I (orange), II (teal), III (magenta), and IV (blue) of eEFSec are also highlighted. H89, which is projected from the PTC (white), is colored in red, and residues U2473 and C2475 are shown as blue spheres. The P-site tRNA (dark gray) and the SRL (purple) are also indicated for reference.
Expansion Segment Modifications
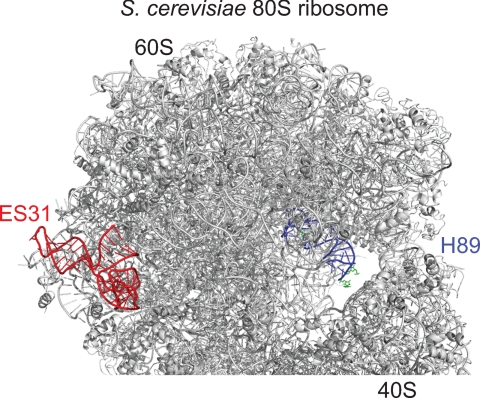
Mammalian 28 S rRNA is ∼1800 nucleotides longer than that in bacteria, and these “extra” highly divergent sequences are termed expansion segments. One of these lies between the PTC and the L1 stalk in eukaryotes and thus was covered by our screen of that region. Interestingly, two striking SBP2-dependent enhancements in BzCN reactivity were detected (Fig. 4A). In control experiments, neither the 647RFQDR651 mutant nor rpL30 was able to induce the same enhancements (Fig. 4, B and C). We were unable to obtain unambiguous sequencing data within ES31, presumably due to the high GC content in this region (>80% in mouse 28 S rRNA). Comparison of the reactive nucleotide positions in H89 identified by gel electrophoresis with those obtained by capillary electrophoresis using the LIZ size standards showed a small margin of error up to 6 bases, which suggests that the latter provides a reliable approximation of the reactive nucleotide position. Based on this observation, we estimate the reactive residues in ES31 to be ∼G3802–C3812 and ∼G3758–U3768. Fig. 5 illustrates the relative position of H89 and ES31 on the crystal structure of the yeast ribosome (29). Although ES31 sequence is not conserved in eukaryotes, the size and overall secondary structure are similar.
FIGURE 5.
Relative positions of ES31(red) and H89 (blue) on the crystal structure of the S. cerevisiae 80 S ribosome. The residues shown in green on H89 displayed enhanced reactivity to BzCN in the presence of SBP2.
SHAPE Reactivity in H89 and ES31 with SID/RBD
We have shown previously that when the SID and the RBD of SBP2 are expressed as separate proteins they retain near wild-type levels of Sec incorporation activity despite losing the ability to stably bind the ribosome in vitro (9). To determine whether the SID and the RBD could interact transiently with the ribosome, we repeated our SHAPE analysis of H89 and ES31 in the presence of wild-type SID and RBD, together or individually. In addition, we tested the 647RFQDR651 to penta-alanine (RBD-647RFQDR651) mutant version of RBD as a control for specificity. When wild-type SID and RBD were incubated with ribosomes, we observed a subtle, but reproducible increase in the reactivity of nucleotides U4071 and C4073 that was absent when the same experiment was repeated with wild-type SID and RBD-647RFQDR651 (compare Fig. 6, A with B). However, wild-type SID and RBD promoted a strong increase in the reactivity of ES31 that was completely abolished when the ribosomes were incubated with wild-type SID and RBD-647RFQDR651 (compare Fig. 6, C with D). When ribosomes were incubated with SID alone, no changes in nucleotide reactivity were observed in H89 or ES31 (data not shown). Interestingly, however, the RBD alone was sufficient to specifically enhance the reactivity of nucleotides in ES31 (compare Fig. 7, A with B). These results confirm that when present in trans, the SID and RBD are still able to interact with the ribosome, despite our inability to detect a stable interaction in vitro.
DISCUSSION
Canonical aa-tRNA Selection and Sec Incorporation
A controversial and unresolved mechanistic question regarding Sec incorporation is how eEFSec delivers the Sec-tRNASec to the ribosome selectively at the UGA codons present in selenoprotein mRNAs. In other words, what prevents the random incorporation of Sec at UGA nonsense codons, and how do the other components of the Sec incorporation complex, namely SBP2 and/or the SECIS element, overcome this to direct its site-specific incorporation? Unraveling this conundrum is essential to our understanding of selenoprotein synthesis and how this process is further regulated by additional cis- and trans-acting factors in vivo.
The canonical aa-tRNA selection pathway has been extensively characterized in prokaryotes and is predicted to follow a similar mechanism in eukaryotes (20). This multistep pathway begins with the docking of a TC to the factor binding site of the ribosome, which allows the aa-tRNA to sample the A-site decoding center in a distorted conformation referred to as the A/T state (21–24). Cognate codon-anticodon interactions induce conformational changes in the ribosome and the TC that lead to GTPase activation (25). Subsequently, ribosome-stimulated GTP hydrolysis induces domain rearrangements in the EF that decreases its affinity for aa-tRNA, resulting in its departure from the ribosome (26). This allows the aa-tRNA to move through a corridor formed by H89–H92 of the large subunit rRNA and into the PTC, where peptide bond formation occurs (27). This newly acquired aa-tRNA conformation is designated the A/A state, and the transition from the A/T to the A/A state is referred to as accommodation. During accommodation, the codon-anticodon interaction is interrogated once again in a separate proofreading step where near cognate aa-tRNAs that are retained during the initial selection step are rejected from the A-site.
When the inability of the eEFSec TC to randomly suppress UGA codons is put into this context, it becomes clear that at least one of the steps in the aa-tRNA delivery pathway is regulated. During canonical translation elongation, the fidelity of aa-tRNA selection is controlled largely by the identity of the anticodon and the induced fit conformations that follow codon recognition (28). Because the anticodon in the Sec-tRNASec is cognate with respect to the UGA codon, it is likely that regulation occurs prior to codon recognition (i.e. TC binding), and/or directly after (i.e. GTPase activation or accommodation).
Our SHAPE analysis showed a specific set of SBP2-dependent changes in the reactivity of nucleotides ∼G3758–U3768 and ∼G3802–C3812 in ES31, and U4071 and C4073 in H89, consistent with a conformational change in these regions. Interestingly, our data indicate that the RBD alone was sufficient for the enhanced reactivity in ES31, but we see a large discrepancy in the reactivity in H89 when comparing CTSBP2 and the SID/RBD. Our interpretation for this result is that the putative conformational change in H89 is dependent on an allosteric signal between the SID and the RBD, which occurs more readily when the two domains are physically linked, thus increasing the likelihood of capturing this conformational state. Below, we speculate on the functional relevance of these SBP2-induced conformational rearrangements.
Expansion Segment 31 and E-site/A-site Allostery
ES31 is a eukaryotic-specific expansion segment located near the E-site of the ribosome. The functional role of ES31 during translation is unknown, but the crystal structure of the Saccharomyces cerevisiae 80 S ribosome indicates that it forms part of a eukaryotic-specific intersubunit bridge (eukaryotic bridge 8) with a previously unidentified small subunit ribosomal protein near the L1 stalk (H76–H78) (29). This intersubunit bridge was also observed in the structure of the mammalian 80 S ribosome obtained by cryoelectron microscopy (30). The L1 stalk has been implicated in the movement and release of deacylated tRNAs from the E-site (31, 32). In addition, E-site clearance is allosterically modulated by the tRNA status of the A-site (33). This reciprocal linkage between the A- and E-sites initially described in bacteria has also been observed in the ribosomes of higher eukaryotes (34). Given its location near the L1 stalk, the authors suggest that eukaryotic bridge 8 may be important in restoring the ribosome conformation competent for aa-tRNA binding in eukaryotic ribosomes (30). Consistent with the allosteric communication between the A- and E-sites, a deletion that shortened the size of H89 led to an increase in the reactivity of nucleotide A2225 (Escherichia coli numbering/mouse A3822) in H75 (35). Interestingly, H75 is located at the base of ES31 in eukaryotes (Fig. 1), further implicating this region in A site/E site allostery.
Helix 89 and aa-tRNA Accommodation
H89 is located near the A-site pocket where it forms part of the accommodation corridor that directs the movement of aa-tRNAs to the PTC following GTP hydrolysis. A computer simulation of the accommodation pathway revealed that nucleotide U2473 (E. coli numbering/mouse U4071) in H89 makes direct contacts with the elbow region of the aa-tRNA in the early stages of the A/T to A/A transition and continuously associates with the backbone of the aa-tRNA throughout this process (27). Yet another simulation showed that H89 is displaced during accommodation, and it was suggested that residues in this helix can acquire alternative conformations for different aa-tRNAs (36, 37). Comparison of x-ray structures of the 70 S ribosome with cognate or near cognate tRNA reveal a network of rRNA helices (including H89) and ribosomal protein contacts that stabilize the elbow of cognate tRNA in the A/A state (38). The absence of these stabilizing contacts when a near cognate tRNA is bound to the A-site may explain why these tRNAs are rejected during proofreading. Unlike canonical aa-tRNAs, the Sec-tRNASec is unusually large, has a reduced number of post-transcriptional modifications, a long variable arm, as well as a highly reactive selenium atom. Given its unique nature, a possible function for SBP2 may be to modulate the conformation of H89 to allow the accommodation and stable binding of the Sec-tRNASec to the ribosomal A-site.
Helix 89 and GTPase Activation
Aside from its function in tRNA accommodation, H89, which is projected from the PTC toward the elongation factor binding site, also interacts with IF2 and its eukaryotic homolog eIF5B (39, 40). Chemical probing studies show that IF2 protects nucleotides A2476 and A2478 (E. coli numbering/mouse A4074 and A4076) on the apical loop of H89, and cryo-EM studies indicate that these contacts are mediated through domain IV of IF2 (39, 41). Interestingly, a C2475G (mouse C4075G) mutation in H89 caused a decrease in the affinity of IF2 for the ribosome but increased its GTP hydrolysis activity; this same mutation had no impact on the function of the elongation factors EF-Tu and EF-G (35). This indicates that interactions between IF2 domain IV and H89 can influence the GTPase activation of IF2, possibly by modulating the interaction between the G-domain and the SRL. This interdomain communication is not unprecedented because the nucleotide state of the G-domain has been shown to alter the position of IF2 domain IV nearly 90 Å away (42).
The crystal structure of aSelB revealed that, despite its sequence homology with EF-Tu in domains I–III, it has an overall shape that strikingly resembles that of IF2/eIF5B (43). Interestingly, both eEFSec and IF2 recognize a specialized tRNA (Sec-tRNASec and fMet-tRNAMet, respectively), and both factors have an additional, flexible, C-terminal extension (domain IV) relative to EF-Tu. Despite the low sequence homology between domain IV in IF2 and aSelB, these domains are structurally, and thus possibly functionally similar.
This information is suggestive of a role for SBP2 in modulating the conformation of H89 and promoting the ribosome-dependent GTPase activation of eEFSec through its fourth domain. This possibility is appealing because it may allow SBP2 to enhance eEFSec function without interfering with the activity of the canonical elongation factors during translation. Further evidence, albeit indirect, for GTPase activation being a limiting step, comes from the Sec incorporation mechanism in prokaryotes where interactions between the prokaryotic SECIS and SelB domain IV are required for its robust ribosome-dependent GTPase activation (44). It is important to note, however, that the fourth domains of eEFSec and SelB share no sequence or structural homology, and unlike the prokaryotic SECIS, the eukaryotic SECIS is predicted to be a kink-turn (19). In addition, despite multiple attempts, we have been unable to observe ribosome- or SBP2-dependent stimulation of eEFSec GTPase activity, likely because eEFSec lacks an intrinsic ribosome binding activity. Thus, the fundamental and so far intractable barrier to testing the mechanism of SBP2 in Sec incorporation is the lack of a Sec incorporation system composed of purified components.
CONCLUSION
Our SHAPE analysis of the SBP2-ribosome interaction has led to the identification of a specific set of rRNA residues in H89 and ES31 that are modified by SBP2 as reflected by their increased reactivity to BzCN. As discussed above, the literature on H89 provides interesting insights on the putative ribosomal function(s) of SBP2 during Sec incorporation. Although the scarcity of information on ES31 makes it difficult to predict the relevance of modifications in this region, the reciprocal linkage between the A- and E-sites provide a rational explanation for the observed modifications in this region despite their large distance from the A-site. These results invoke a more dynamic role for SBP2 during Sec incorporation than that of a static anchor that simply recruits eEFSec to the SECIS element. The mechanistic framework that is beginning to emerge is consistent with a ribosome-based function for SBP2, but how this fits in with its role in SECIS binding and eEFSec recruitment is not clear. A deeper understanding of the basic mechanism of eukaryotic Sec incorporation awaits elucidation of the precise functional role of both of these interactions and the interplay between them.
Acknowledgments
We thank Jesse Donovan and Jonathan Gonzales-Flores for critical reading of this manuscript.
This work was supported, in whole or in part, by National Research Service Award F31GM081920 from the National Institutes of Health (to K. C.). This work was also supported by Public Health Service Grants GM077073 and GM094833 (to P. R. C.).
- aa-tRNA
- aminoacyl-tRNA
- BzCN
- benzoyl cyanide
- CTSBP2
- C-terminal SBP2
- eEFSec
- eukaryotic elongation factor for selenocysteine
- DMSO
- dimethyl sulfoxide
- EF
- elongation factor
- ES31
- expansion segment 31
- GAC
- GTPase-associated center
- H
- Helix
- PTC
- peptidyltransferase center
- RBD
- RNA binding domain
- SBP2
- SECIS-binding protein 2
- Sec
- selenocysteine
- SECIS
- selenocysteine insertion sequence
- Ser-tRNASec
- serylated Sec tRNA
- SHAPE
- selective 2′-hydroxyl acylation analyzed by primer extension
- SID
- Sec incorporation domain
- SRL
- sarcin-ricin loop
- TC
- ternary complex.
REFERENCES
- 1. Fagegaltier D., Hubert N., Yamada K., Mizutani T., Carbon P., Krol A. (2000) Characterization of mSelB, a novel mammalian elongation factor for selenoprotein translation. EMBO J. 19, 4796–4805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Tujebajeva R. M., Copeland P. R., Xu X. M., Carlson B. A., Harney J. W., Driscoll D. M., Hatfield D. L., Berry M. J. (2000) Decoding apparatus for eukaryotic selenocysteine insertion. EMBO Rep. 1, 158–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Diamond A., Dudock B., Hatfield D. (1981) Structure and properties of a bovine liver UGA suppressor serine tRNA with a tryptophan anticodon. Cell 25, 497–506 [DOI] [PubMed] [Google Scholar]
- 4. Jung J. E., Karoor V., Sandbaken M. G., Lee B. J., Ohama T., Gesteland R. F., Atkins J. F., Mullenbach G. T., Hill K. E., Wahba A. J. (1994) Utilization of selenocysteyl-tRNA[Ser]Sec and seryl-tRNA[Ser]Sec in protein synthesis. J. Biol. Chem. 269, 29739–29745 [PubMed] [Google Scholar]
- 5. Fahlman R. P., Dale T., Uhlenbeck O. C. (2004) Uniform binding of aminoacylated transfer RNAs to the ribosomal A and P sites. Mol. Cell 16, 799–805 [DOI] [PubMed] [Google Scholar]
- 6. Berry M. J., Banu L., Chen Y. Y., Mandel S. J., Kieffer J. D., Harney J. W., Larsen P. R. (1991) Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3′-untranslated region. Nature 353, 273–276 [DOI] [PubMed] [Google Scholar]
- 7. Copeland P. R., Fletcher J. E., Carlson B. A., Hatfield D. L., Driscoll D. M. (2000) A novel RNA-binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs. EMBO J. 19, 306–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chavatte L., Brown B. A., Driscoll D. M. (2005) Ribosomal protein L30 is a component of the UGA-selenocysteine recoding machinery in eukaryotes. Nat. Struct. Mol. Biol. 12, 408–416 [DOI] [PubMed] [Google Scholar]
- 9. Donovan J., Caban K., Ranaweera R., Gonzalez-Flores J. N., Copeland P. R. (2008) A novel protein domain induces high affinity selenocysteine insertion sequence binding and elongation factor recruitment. J. Biol. Chem. 283, 35129–35139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kinzy S. A., Caban K., Copeland P. R. (2005) Characterization of the SECIS- binding protein 2 complex required for the co-translational insertion of selenocysteine in mammals. Nucleic Acids Res. 33, 5172–5180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Caban K., Kinzy S. A., Copeland P. R. (2007) The L7Ae RNA binding motif is a multifunctional domain required for the ribosome-dependent Sec incorporation activity of Sec insertion sequence binding protein 2. Mol. Cell. Biol. 27, 6350–6360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Merrick W. C. (1979) Purification of protein synthesis initiation factors from rabbit reticulocytes. Methods Enzymol. 60, 101–108 [DOI] [PubMed] [Google Scholar]
- 13. Copeland P. R., Stepanik V. A., Driscoll D. M. (2001) Insight into mammalian selenocysteine insertion: domain structure and ribosome binding properties of Sec insertion sequence binding protein 2. Mol. Cell. Biol. 21, 1491–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cannone J. J., Subramanian S., Schnare M. N., Collett J. R., D'Souza L. M., Du Y., Feng B., Lin N., Madabusi L. V., Müller K. M., Pande N., Shang Z., Yu N., Gutell R. R. (2002) The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wilkinson K. A., Merino E. J., Weeks K. M. (2006) Selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 1, 1610–1616 [DOI] [PubMed] [Google Scholar]
- 16. Mortimer S. A., Weeks K. M. (2009) Time-resolved RNA SHAPE chemistry: quantitative RNA structure analysis in one-second snapshots and at single-nucleotide resolution. Nat. Protoc. 4, 1413–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yu H., Chan Y. L., Wool I. G. (2009) The identification of the determinants of the cyclic, sequential binding of elongation factors Tu and G to the ribosome. J. Mol. Biol. 386, 802–813 [DOI] [PubMed] [Google Scholar]
- 18. Sergiev P. V., Lesnyak D. V., Burakovsky D. E., Kiparisov S. V., Leonov A. A., Bogdanov A. A., Brimacombe R., Dontsova O. A. (2005) Alteration in location of a conserved GTPase-associated center of the ribosome induced by mutagenesis influences the structure of peptidyltransferase center and activity of elongation factor G. J. Biol. Chem. 280, 31882–31889 [DOI] [PubMed] [Google Scholar]
- 19. Driscoll D. M., Copeland P. R. (2003) Mechanism and regulation of selenoprotein synthesis. Annu. Rev. Nutr. 23, 17–40 [DOI] [PubMed] [Google Scholar]
- 20. Agirrezabala X., Frank J. (2009) Elongation in translation as a dynamic interaction among the ribosome, tRNA, and elongation factors EF-G and EF-Tu. Q. Rev. Biophys. 42, 159–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Valle M., Zavialov A., Li W., Stagg S. M., Sengupta J., Nielsen R. C., Nissen P., Harvey S. C., Ehrenberg M., Frank J. (2003) Incorporation of aminoacyl-tRNA into the ribosome as seen by cryo-electron microscopy. Nat. Struct. Biol. 10, 899–906 [DOI] [PubMed] [Google Scholar]
- 22. Geggier P., Dave R., Feldman M. B., Terry D. S., Altman R. B., Munro J. B., Blanchard S. C. (2010) Conformational sampling of aminoacyl-tRNA during selection on the bacterial ribosome. J. Mol. Biol. 399, 576–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Schuette J. C., Murphy F. V., 4th, Kelley A. C., Weir J. R., Giesebrecht J., Connell S. R., Loerke J., Mielke T., Zhang W., Penczek P. A., Ramakrishnan V., Spahn C. M. (2009) GTPase activation of elongation factor EF-Tu by the ribosome during decoding. EMBO J. 28, 755–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schmeing T. M., Voorhees R. M., Kelley A. C., Gao Y. G., Murphy F. V., 4th, Weir J. R., Ramakrishnan V. (2009) The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science 326, 688–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Voorhees R. M., Schmeing T. M., Kelley A. C., Ramakrishnan V. (2010) The mechanism for activation of GTP hydrolysis on the ribosome. Science 330, 835–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dell V. A., Miller D. L., Johnson A. E. (1990) Effects of nucleotide- and aurodox-induced changes in elongation factor Tu conformation upon its interactions with aminoacyl transfer RNA: a fluorescence study. Biochemistry 29, 1757–1763 [DOI] [PubMed] [Google Scholar]
- 27. Sanbonmatsu K. Y., Joseph S., Tung C. S. (2005) Simulating movement of tRNA into the ribosome during decoding. Proc. Natl. Acad. Sci. U.S.A. 102, 15854–15859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zaher H. S., Green R. (2009) Fidelity at the molecular level: lessons from protein synthesis. Cell 136, 746–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ben-Shem A., Jenner L., Yusupova G., Yusupov M. (2010) Crystal structure of the eukaryotic ribosome. Science 330, 1203–1209 [DOI] [PubMed] [Google Scholar]
- 30. Chandramouli P., Topf M., Ménétret J. F., Eswar N., Cannone J. J., Gutell R. R., Sali A., Akey C. W. (2008) Structure of the mammalian 80 S ribosome at 8.7 Å resolution. Structure 16, 535–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cornish P. V., Ermolenko D. N., Staple D. W., Hoang L., Hickerson R. P., Noller H. F., Ha T. (2009) Following movement of the L1 stalk between three functional states in single ribosomes. Proc. Natl. Acad. Sci. U.S.A. 106, 2571–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fei J., Bronson J. E., Hofman J. M., Srinivas R. L., Wiggins C. H., Gonzalez R. L. (2009) Allosteric collaboration between elongation factor G and the ribosomal L1 stalk directs tRNA movements during translation. Proc. Natl. Acad. Sci. U.S.A. 106, 15702–15707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wilson D. N., Nierhaus K. H. (2006) The E-site story: the importance of maintaining two tRNAs on the ribosome during protein synthesis. Cell. Mol. Life Sci. 63, 2725–2737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Budkevich T. V., El'skaya A. V., Nierhaus K. H. (2008) Features of 80 S mammalian ribosome and its subunits. Nucleic Acids Res. 36, 4736–4744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Burakovskiı̆ D. E., Smirnova A. S., Lesniak D. V., Kiparisov S. V., Leonov A. A., Sergiev P. V., Bogdanov A. A., Dontsova O. A. (2007) Interaction of 23S ribosomal RNA helices 89 and 91 of Escherichia coli contributes to the activity of IF2 but is insignificant for elongation factors functioning. Mol. Biol. 41, 1031–1041 [PubMed] [Google Scholar]
- 36. Whitford P. C., Geggier P., Altman R. B., Blanchard S. C., Onuchic J. N., Sanbonmatsu K. Y. (2010) Accommodation of aminoacyl-tRNA into the ribosome involves reversible excursions along multiple pathways. RNA 16, 1196–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Rakauskaite R., Dinman J. D. (2011) Mutations of highly conserved bases in the peptidyltransferase center induce compensatory rearrangements in yeast ribosomes. RNA 17, 855–864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jenner L., Demeshkina N., Yusupova G., Yusupov M. (2010) Structural rearrangements of the ribosome at the tRNA proofreading step. Nat. Struct. Mol. Biol. 17, 1072–1078 [DOI] [PubMed] [Google Scholar]
- 39. La Teana A., Gualerzi C. O., Dahlberg A. E. (2001) Initiation factor IF 2 binds to the α-sarcin loop and helix 89 of Escherichia coli 23 S ribosomal RNA. RNA 7, 1173–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Unbehaun A., Marintchev A., Lomakin I. B., Didenko T., Wagner G., Hellen C. U., Pestova T. V. (2007) Position of eukaryotic initiation factor eIF5B on the 80 S ribosome mapped by directed hydroxyl radical probing. EMBO J. 26, 3109–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Myasnikov A. G., Marzi S., Simonetti A., Giuliodori A. M., Gualerzi C. O., Yusupova G., Yusupov M., Klaholz B. P. (2005) Conformational transition of initiation factor 2 from the GTP- to GDP-bound state visualized on the ribosome. Nat. Struct. Mol. Biol. 12, 1145–1149 [DOI] [PubMed] [Google Scholar]
- 42. Roll-Mecak A., Cao C., Dever T. E., Burley S. K. (2000) X-ray structures of the universal translation initiation factor IF2/eIF5B: conformational changes on GDP and GTP binding. Cell 103, 781–792 [DOI] [PubMed] [Google Scholar]
- 43. Leibundgut M., Frick C., Thanbichler M., Böck A., Ban N. (2005) Selenocysteine tRNA-specific elongation factor SelB is a structural chimaera of elongation and initiation factors. EMBO J. 24, 11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hüttenhofer A., Böck A. (1998) Selenocysteine inserting RNA elements modulate GTP hydrolysis of elongation factor SelB. Biochemistry 37, 885–890 [DOI] [PubMed] [Google Scholar]