Background: Atg32 is a transmembrane protein essential for mitochondria autophagy in yeast.
Results: Atg32 harbors a module that is crucial for interactions with Atg8 and Atg11, and can even promote other organelle autophagy.
Conclusion: Atg32 acts as a direct initiator at early stages of mitochondria autophagy.
Significance: This might be a common molecular feature in mitochondria autophagy conserved from yeast to humans.
Keywords: Autophagy, Membrane Trafficking, Mitochondria, Mitochondrial Transport, Yeast, Membrane Dynamics, Mitochondrial Biogenesis, Organelle Degradation, Saccharomyces cerevisiae, Selective Autophagy
Abstract
Autophagy-related degradation selective for mitochondria (mitophagy) is an evolutionarily conserved process that is thought to be critical for mitochondrial quality and quantity control. In budding yeast, autophagy-related protein 32 (Atg32) is inserted into the outer membrane of mitochondria with its N- and C-terminal domains exposed to the cytosol and mitochondrial intermembrane space, respectively, and plays an essential role in mitophagy. Atg32 interacts with Atg8, a ubiquitin-like protein localized to the autophagosome, and Atg11, a scaffold protein required for selective autophagy-related pathways, although the significance of these interactions remains elusive. In addition, whether Atg32 is the sole protein necessary and sufficient for initiation of autophagosome formation has not been addressed. Here we show that the Atg32 IMS domain is dispensable for mitophagy. Notably, when anchored to peroxisomes, the Atg32 cytosol domain promoted autophagy-dependent peroxisome degradation, suggesting that Atg32 contains a module compatible for other organelle autophagy. X-ray crystallography reveals that the Atg32 Atg8 family-interacting motif peptide binds Atg8 in a conserved manner. Mutations in this binding interface impair association of Atg32 with the free form of Atg8 and mitophagy. Moreover, Atg32 variants, which do not stably interact with Atg11, are strongly defective in mitochondrial degradation. Finally, we demonstrate that Atg32 forms a complex with Atg8 and Atg11 prior to and independent of isolation membrane generation and subsequent autophagosome formation. Taken together, our data implicate Atg32 as a bipartite platform recruiting Atg8 and Atg11 to the mitochondrial surface and forming an initiator complex crucial for mitophagy.
Introduction
Selective degradation of excess or damaged mitochondria relies on autophagy-related transport pathways (1, 2). This process, called mitophagy, has been conserved during evolution and is thought to be relevant to cellular homeostasis and differentiation (3, 4). One of the critical points in mitophagy is how the autophagy machinery targets to mitochondria to specifically sequester the energy-converting organelles. Recent studies reveal key proteins that localize to the mitochondrial surface and establish the selectivity of mitochondrial degradation in yeast and mammals (5–7). In principle, it has so far been hypothesized that those key molecules directly or indirectly participate in recruiting core Atg proteins required for generation of isolation membranes and subsequent formation of autophagosomes.
Autophagy-dependent transport of mitochondria to lytic compartments takes place in different cell types under various physiological conditions. In the budding yeast Saccharomyces cerevisiae, mitophagy is facilitated during respiratory growth in medium containing glycerol or lactate, a non-fermentable carbon source that is utilized for mitochondrial ATP production (8–10). Under this condition, Atg32 plays a specific and essential role in mitophagy (10, 11). The Atg32 protein levels in glycerol-grown cells are increased 10- to 20-fold compared with log-phase cells grown in medium containing glucose, a fermentable carbon source that is exploited for glycolytic ATP production but does not facilitate degradation of mitochondria (10). Newly synthesized Atg32 is inserted into the outer membrane of mitochondria with its N- and C-terminal domains exposed to the cytosol and mitochondrial intermembrane space (IMS)3, respectively (10, 11). Atg32 interacts with Atg8, a ubiquitin-like protein conjugated to the lipid phosphatidylethanolamine (PE) and localized to the autophagosome (12), and Atg11, a scaffolding protein required for selective autophagy-related pathways that mediates core Atg protein assembly (13). It has not yet been shown whether these interactions are functionally significant and, if so, at which stage during mitophagy Atg32 forms a complex with Atg8 and Atg11. In addition, Uth1, Aup1, and Atg33 are mitochondrial proteins that have been suggested to be involved in mitophagy (8, 14, 15). Whether Atg32-mediated degradation process needs these proteins remains unclear. Alternatively, it is also possible that Atg32 is the sole protein necessary and sufficient for specifying autophagic degradation to mitochondria.
In this study, we demonstrate that the Atg32 cytosol domain is fully capable of targeting the core autophagy machinery to mitochondria. Interactions of Atg32 with Atg8 and Atg11 are important for this process and occur at early stages of mitophagy in a manner independent of isolation membrane generation. On the basis of our findings, we propose that Atg32 acts as an autophagic degron (polypeptide sequence specific for autophagy-dependent degradation) and directly initiates assembly of core Atg proteins on the mitochondrial surface.
EXPERIMENTAL PROCEDURES
Strains and Growth Conditions
Yeast strains and plasmids used in this study are listed in supplemental Tables S1 and S2, respectively. Standard genetic and molecular biology methods were used for S. cerevisiae and Escherichia coli strains. Yeast cells were grown at 30 °C in YPD (1% yeast extract, 2% peptone, 2% dextrose), synthetic medium (0.17% yeast nitrogen base without amino acids and ammonium sulfate, 0.5% ammonium sulfate) containing 2% dextrose or 0.1% dextrose plus 3% glycerol and synthetic medium with 0.5% casamino acids containing 2% dextrose or 0.1% dextrose plus 3% glycerol, supplemented with necessary amino acids and nucleotides. For mitophagy induction, cells grown to mid-log phase in synthetic medium containing 2% dextrose or synthetic medium with 0.5% casamino acids containing 2% dextrose were incubated in synthetic medium containing 0.1% dextrose plus 3% glycerol or synthetic medium with 0.5% casamino acid containing 0.1% dextrose plus 3% glycerol.
Immunoblot Analysis
Western blotting, and immunodecoration were performed as described previously (10).
Quantification of Mitophagy
Cells expressing mitochondrial matrix-targeted DHFR-mCherry were grown, and collected at the 3 days point. Whole cell extracts were prepared, and analyzed by SDS-PAGE and Western blotting. After immunodecoration with the anti-RFP antibodies, and treatment with enhanced chemiluminescence reagents, proteins were detected using a luminescent image analyzer (LAS-4000 mini; GE Healthcare). The signal value of free mCherry in wild-type cells was set to 100%. Data represent the averages of three experiments, with bars indicating standard deviations. Quantification was performed using ImageQuant TL (GE Healthcare).
Coimmunoprecipitation
Immunoprecipitation was performed as described previously (10).
X-ray Crystallography
Atg8(1–116) recombinant protein was prepared as described previously (16). Atg32AIM peptide (Ser-Trp-Gln-Ala-Ile-Gln, corresponding to the residues 85–90 of Atg32) was purchased from Sigma. Crystals of the Atg8-Atg32AIM complex were obtained by the sitting drop vapor diffusion method at 20 °C. 0.5 μl of protein solution consisting of 14 mg/ml Atg8 and 1 mg/ml Atg32AIM was mixed with an equal volume of reservoir solution consisting of 2.4 m ammonium sulfate and 0.1 m sodium acetate (pH 4.8) and was equilibrated against the reservoir solution by vapor diffusion. Crystals belong to the cubic space group I23, with unit-cell dimension of a = 104.84 Å. For data collection, crystals were soaked into the reservoir solution supplemented with 25% glycerol, flash-cooled, and kept in a stream of nitrogen gas at −178 °C during data collection. Diffraction data were collected on the Area Detector Systems Corporation Quantum 315 charge-coupled device detector using beamline BL41XU, SPring8, Japan, at a wavelength of 1.00 Å. Diffraction data were processed using the HKL2000 program suite (17). Molecular replacement was performed using the Crystallography and NMR System program (18). The crystal structure of Atg8 complexed with an Atg19-derived peptide (16) (PDB code 2ZPN) was used as a search model. Manual building and modification was performed with the molecular modeling program Crystallographic Object-Oriented Toolkit (19), followed by iterative rounds of refinement using CNS.
Microscopy
Live cell imaging was performed as described previously (10).
RESULTS
The Atg32 IMS Domain Is Dispensable for Mitophagy
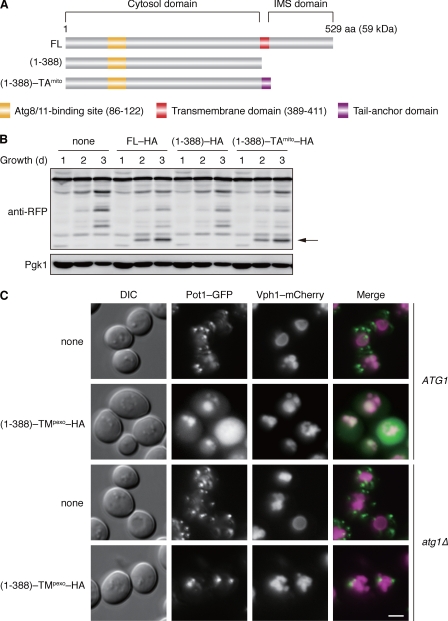
Atg32 is predicted to consist of three major modules: an N-terminal cytosol domain (amino acid residues 1–388), a transmembrane (TM) domain (389–411), and a C-terminal IMS domain (412–529) (Fig. 1A). To investigate the functional relevance of the TM and IMS domains, we constructed GFP- or HA-tagged versions of two variants, Atg32(1–388) lacking the TM and IMS domains, and Atg32(1–388)-TA consisting of the cytosol domain and mitochondrial tail-anchor (TAmito) domain derived from an authentic outer membrane protein (Fig. 1A). When the GFP-tagged variants were expressed under the endogenous promoter, both full-length Atg32 and Atg32(1–388)-TAmito localized to mitochondria (supplemental Fig. S1A). In contrast, Atg32(1–388) dispersed in the cytosol and nucleus. Hence, an exogenous tail anchor can specifically address the Atg32 cytosol domain to the surface of mitochondria.
FIGURE 1.
Atg32 contains a domain compatible for degradation of peroxisomes. A, schematic representation of the domain structures of Atg32 (FL) and its truncated variants. (1–388) lacks the transmembrane and IMS domains, and (1–388) -TAmito consists of the cytosolic domain and a tail-anchor derived from Gem1, a mitochondrial outer membrane protein (39). B, cells expressing the wild-type or truncated variants of HA-tagged Atg32 (FL-HA, (1–388)–HA, or (1–388)-TAmito-HA) were grown in glycerol medium, collected at the indicated time points, and subjected to Western blotting. All strains are atg32-null derivatives expressing a mitochondrial matrix-localized DHFR-mCherry. The arrow depicts free mCherry generated by mitophagy. Pgk1 was monitored as a loading control. C, cells containing or lacking Atg1 (ATG1 or atg1Δ) were transformed with a low-copy, empty plasmid (none) or the one that encodes (1–388)-TMpexo-HA, an Atg32 cytosol domain anchored to the peroxisome via a TM domain derived from Pex15, a peroxisomal membrane protein (40). (1–388)-TMpexo-HA was expressed under the ATG32 promoter. All strains are atg32-null derivatives expressing both Pot1-GFP (peroxisomal marker) and Vph1-mCherry (vacuolar marker). Cells were grown in glycerol medium for 48 h and analyzed by fluorescence microscopy. Scale bar = 2 μm.
Next, we examined the expression of Atg32(1–388)-HA and Atg32(1–388)-TAmito-HA under the endogenous promoter and found their robust induction during respiratory growth (supplemental Fig. S1B). Degradation of mitochondria was then monitored using mito-DHFR-mCherry, a reporter localized in the mitochondrial matrix. Upon mitophagy, this fusion protein is transported and processed to generate free mCherry in the vacuole. We found that cells expressing Atg32(1–388)-TAmito–HA displayed degradation of mitochondria at levels equivalent to the full-length Atg32-HA (Fig. 1B). As expected, mitophagy was almost completely blocked in cells expressing Atg32(1–388)-HA. Together, these data indicate that the Atg32 cytosol domain must be anchored to the surface of mitochondria to promote mitophagy. The TM domain, which can be replaced by other membrane anchors, is unlikely to harbor any specific activities for Atg32 function. Likewise, the IMS domain is fully dispensable for mitophagy.
Pexophagy by the Peroxisome-anchored Atg32 Cytosol Domain
Although previous findings suggest that Atg32 is of central importance for mitophagy in yeast (10, 11), it still remains conceivable that other mitochondrial protein(s) might also be required for the Atg32-mediated degradation process. To test this possibility, we attempted to insert the Atg32 cytosol domain to the surface of peroxisomes via a TM domain derived from an authentic peroxisomal protein (TMpexo) and ask whether this chimeric construct can promote pexophagy (peroxisome autophagy). When expressed under the ATG32 promoter, Atg32(1–388)-TMpexo-HA was strongly induced during respiratory growth (supplemental Fig. S1C). For pexophagy assays, fluorescence microscopy was performed to observe cells expressing both Pot1-GFP, a marker localized in the peroxisomal matrix, and Vph1-mCherry, a marker inserted into the vacuolar membrane (Fig. 1C). Without Atg32(1–388)-TMpexo-HA, Pot1-GFP barely targeted to vacuoles. Remarkably, Pot1-GFP colocalized to Vph1-mCherry in cells expressing Atg32(1–388)-TMpexo-HA, which absolutely depended on Atg1, a protein kinase essential for all autophagy-related processes (20). These results indicate that the peroxisome-anchored version of the Atg32 cytosol domain can recruit core Atg proteins required for autophagosome formation to peroxisomes and facilitate pexophagy. It should be noted that in wild-type cells pexophagy was not stimulated under this growth condition (glycerol medium). Thus, it seems likely that the Atg32 cytosol domain contains a degron-like module capable of promoting other organelle autophagy and that Atg32 is the sole mitochondrial protein necessary and sufficient to directly mediate mitophagy.
The Atg32 AIM Peptide Binds Atg8 in Vitro
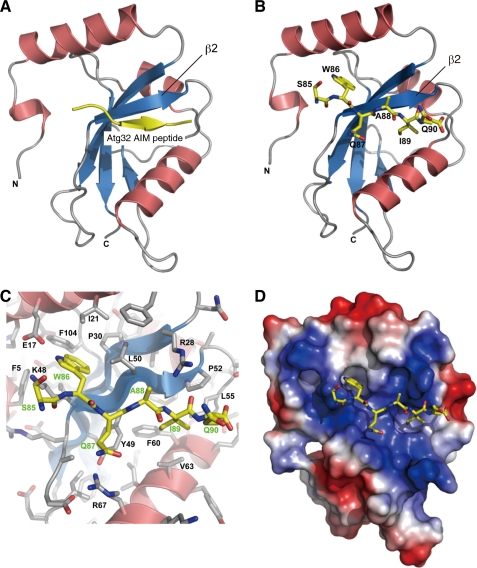
The 43-kDa N-terminal cytosol domain of Atg32 contains an AIM, whose consensus tetrapeptide sequence is W/YXXI/L/V, conserved among proteins that interact with the Atg8 family members (21). The Atg32 AIM consists of WQAI86–89, and Atg32AQAA, a variant containing alanine substitutions for Trp-86 and Ile-89, exhibits a weak defect in mitophagy (10). To verify that this motif directly contributes to Atg8 interaction, we determined the crystal structure of the Atg8-Atg32(SWQAIQ)85–90 peptide complex (Fig. 2, A–D, and supplemental Table S3 and Fig. S2A). Similar to the AIM sequence of Atg19 and Atg3 in yeast and p62, NIX, and Atg4B in mammals (16, 22–25), the Atg32(SWQAIQ)85–90 peptide adopts an extended β conformation in parallel with the Atg8 β2 (Fig. 2, A and B). The side-chains of both W86 and I89 are bound deeply into the hydrophobic pockets of Atg8 (Fig. 2, C and D). We conclude that Atg32 physically associates with Atg8 in a fashion conserved during evolution.
FIGURE 2.
Crystal structure of the Atg8-Atg32(SWQAIQ)85–90 peptide complex. A, Atg8 (α helices in red, β sheets in blue) and the Atg32 peptide (yellow) are shown in ribbon models. B, Atg8 and the Atg32(SWQAIQ)85–90 peptide are shown in ribbon and stick models, respectively. C, close-up view of the Atg8-Atg32(SWQAIQ)85–90 interface indicating amino acids of Atg8 (black) and Atg32 (green). D, Atg8 and the Atg32(SWQAIQ)85–90 peptide are shown in surface and stick models, respectively. The surface is colored according to the electrostatic potential (blue, positive; red, negative).
Mutations in the Atg32-Atg8 Interface Affect Mitophagy
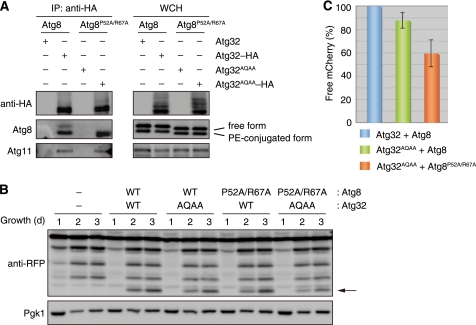
Despite the fact that Atg32AQAA cannot bind Atg8 in yeast two-hybrid systems (10), we found an Atg32AQAA-Atg8 interaction, yet at a reduced level compared with the wild-type protein, in coimmunoprecipitation assays (supplemental Fig. S2B). It is plausible that other protein-protein interfaces could contribute to the interaction between Atg32 and Atg8 in vivo. To clarify the effect of Atg32-Atg8 binding impairment on mitophagy, we sought to introduce mutations into the Atg32 AIM-binding interface of Atg8. Double alanine substitutions for Pro-52 and Arg-67 of Atg8 (Fig. 2C) (16) partially compromised the cytoplasm-to-vacuole targeting (Cvt) pathway (26), an autophagy-related process selective for vacuolar proteins such as Ape1 (supplemental Fig. S2C), but did not alter non-selective autophagy during starvation (supplemental Fig. S2D). We then performed coimmunoprecipitation assays for cells expressing Atg32AQAA-HA and Atg8P52A/R67A. As shown in Fig. 3A, Atg32-HA coprecipitated with both free and PE-conjugated forms of Atg8. Free Atg8 is pooled in the cytosol and serves as a precursor for lipidation. Atg8-PE is present in various intracellular membranes, enriched in autophagosomal structures, and essential for all autophagy-related processes (1, 2). The free form of Atg8P52A/R67A was hardly coprecipitated with Atg32AQAA-HA (Fig. 3A). The coprecipitation of Atg8P52A/R67A-PE with Atg32AQAA-HA was as efficient as that of Atg8-PE with Atg32-HA, raising the possibility that PE-conjugation could affect the affinity of Atg8 for Atg32. As expected, Atg11 was coprecipitated with both Atg32-HA and Atg32AQAA-HA in the presence of Atg8 and Atg8P52A/R67A, respectively.
FIGURE 3.
Atg32-Atg8 interaction contributes to efficient mitophagy. A, coimmunoprecipitation assays for cells expressing the indicated variants of Atg8 and Atg32 grown in glycerol medium for 30 h. All strains are vacuolar protease-deficient, atg8- and atg32-double null derivatives. Mitochondria-enriched fractions were obtained from whole cell homogenates (WCH), solubilized, and subjected to immunoprecipitation using anti-HA antibody-conjugated agarose. The WCH fractions and eluted immunoprecipitates (IP) were analyzed by Western blotting. B, cells expressing the wild-type or mutants of Atg8 (WT or P52A/R67A) and Atg32 (WT or AQAA) were grown in glycerol medium, collected at the indicated time points, and subjected to Western blotting. All strains are atg8- and atg32-double-null derivatives expressing a mitochondrial matrix-localized DHFR-mCherry. The arrow depicts free mCherry generated by mitophagy. Pgk1 was monitored as a loading control. C, the amounts of free mCherry generated in cells expressing the indicated variants of Atg32 and Atg8 at the 3-day time point were analyzed as in B and quantified in three experiments. The signal intensity value of free mCherry in cells expressing wild-type Atg32-HA and Atg8 was set to 100%. Data represent the averages of the all experiments, with bars indicating mean ± S.D.
We next monitored mitophagy in cells expressing both Atg32AQAA and Atg8P52A/R67A using mito-DHFR-mCherry (Fig. 3, B and C). As reported previously (10), cells expressing Atg32AQAA and Atg8 displayed only a weak defect in mitophagy (88% of the wild-type levels). When cells expressing both Atg32AQAA and Atg8P52A/R67A were grown under respiration conditions, mitophagy was modestly decreased (60% of the wild-type levels). Thus, mutations on Atg32 and Atg8 combined in trans cause synthetic defects in mitochondrial degradation. Together, these results are consistent with the notion that the interaction between Atg32 and Atg8 is crucial for mitophagy and that the free form of Atg8 can bind Atg32 on the surface of mitochondria prior to its PE conjugation and membrane anchoring to autophagosomes.
Atg32 Recruits Atg11 to the Surface of Mitochondria
Atg11 is a scaffolding protein crucial for selective autophagy-related processes including mitophagy, pexophagy, and the Cvt pathway that acts as an adapter between cargo (or cargo receptor) and core Atg protein assemblies (10, 11, 13). Orthologs of this key protein have been identified in yeast and filamentous fungi but not other higher eukaryotes. Another factor serving similar scaffolding function in yeast is Atg17, a protein specific for starvation-induced autophagy, whose functional counterparts have been suggested in mammals (27–29). Notably, these two proteins share functional redundancy in pexophagy and the Cvt pathway. Cells lacking Atg11 still exhibit degradation of peroxisomes and transport of Ape1 to the vacuole in an Atg17-dependent manner (10, 30, 31). By contrast, Atg11 is absolutely essential for mitophagy under both respiration and starvation conditions (9, 10). Thus, it is conceivable that Atg11 targets to mitochondria via its physical interaction with Atg32, which is indispensable for formation of autophagosomes on mitochondria. Consistent with this hypothesis, we found using fluorescence microscopy that, during respiratory growth, a fraction of GFP-tagged Atg11 expressed under the endogenous promoter localized to mitochondria as discrete foci in a fashion dependent on Atg32 (supplemental Fig. S3, A and B).
Atg32 Mutants Defective in Atg11 Interaction and Mitophagy
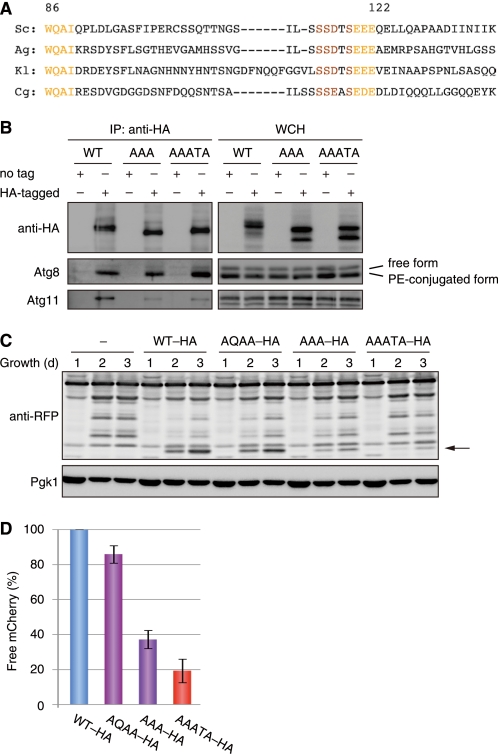
To address whether the Atg32-Atg11 interaction is functionally relevant to mitophagy, we sought to map a region in the Atg32 amino acid sequence that is critical for Atg11 binding in yeast two-hybrid systems. Our truncation analysis indicated the 100–150 amino acid residues of Atg32 as an Atg11 interaction domain (supplemental Fig. S3C). Strikingly, this portion contains an SSD/EXSEE/DE motif conserved among yeast Atg32 homologs, whereas the flanking regions of this sequence vary substantially (Fig. 4A). In addition, Atg19 and Atg34, cargo receptors for the Cvt pathway in yeast (32, 33), have the putative Atg11-interaction motifs DDSSIIST388–395 and DESSIMST380–387, respectively, somewhat resembling the Atg32 SSDTSEEE115–122 sequence. We thus introduced alanine substitutions into the octapeptide motif, generating two variants, Atg32AAATA and Atg32AAA, for the residues SSDTS115–119 and EEE120–122, respectively. When expressed under the endogenous promoter, Atg32AAATA-HA, and Atg32AAA-HA were robustly induced during respiratory growth, as seen in the wild-type protein (supplemental Fig. S3D).
FIGURE 4.
Atg32-Atg11 interaction is crucial for mitochondria autophagy. A, an amino acid sequence alignment of the regions containing putative Atg8- and Atg11-binding domains from yeast Atg32 homologs. Sc, S. cerevisiae; Ag, Ashbya gossypii; Kl, Kluyveromyces lactis; Cg, Candida glabrata. B, coimmunoprecipitation assays for cells expressing untagged and HA-tagged variants of Atg32 (WT), Atg32AAA (AAA) for EEE120–122, or Atg32AAATA (AAATA) for SSDTS115–119 grown in glycerol medium for 30 h. All strains are vacuolar protease-deficient, atg32-null derivatives. Mitochondria-enriched fractions were obtained from whole cell homogenates (WCH), solubilized, and subjected to immunoprecipitation using anti-HA antibody-conjugated agarose. The WCH fractions and eluted immunoprecipitates (IP) were analyzed by Western blotting. C, cells expressing the wild-type or mutants of HA-tagged Atg32 (WT-HA, AQAA-HA, AAA-HA, or AAATA-HA) were grown in glycerol medium, collected at the indicated time points, and subjected to Western blotting. All strains are atg32-null derivatives expressing a mitochondrial matrix-localized DHFR-mCherry. The arrow depicts free mCherry generated by mitophagy. Pgk1 was monitored as a loading control. D, the amounts of free mCherry generated in cells expressing the indicated variants of Atg32 at the 3-day time point were analyzed as in C and quantified in three experiments. The signal intensity value of free mCherry in cells expressing wild-type Atg32-HA was set to 100%. Data represent the averages of the all experiments, with bars indicating mean ± S.D.
We then performed coimmunoprecipitation assays for cells expressing Atg32AAA-HA or Atg32AAATA-HA (Fig. 4B). Remarkably, the coprecipitation efficiency of Atg11-Atg32AAA-HA and Atg11-Atg32AAATA-HA was reduced to 10–20% compared with that of Atg11-Atg32-HA. On the other hand, these alanine substitutions did not affect coprecipitation with Atg8. These results suggest that the SSD/EXSEE/DE motif of Atg32 is specifically crucial for Atg11 interaction.
Given the fact that Atg11 bridges between cargo (or cargo receptor) and core Atg proteins, we assumed that mutations in the SSD/EXSEE/DE motif of Atg32 severely impair degradation of mitochondria. Indeed, cells expressing Atg32AAA or Atg32AAATA displayed a strong defect in mitophagy (38 and 20%, respectively, of the wild-type levels) (Fig. 4, C and D). Hence, Atg32-Atg11 interaction is a primarily crucial step to accelerate degradation of mitochondria.
Atg32 Is a Phosphoprotein
During the course of this study, we noticed that multiple protein bands of Atg32 appeared during respiratory growth (supplemental Fig. S4A). Those patterns were markedly detected in the absence of vacuolar protease activities and diminished with protein phosphatase treatment in vitro (supplemental Fig. S4B), indicating that Atg32 is phosphorylated and transported to the vacuole. A recent study has also described similar results, suggesting that phosphorylation is a regulatory step crucial for Atg32 function and mitophagy (34). On the basis of our finding that Atg32 variants defective in Atg11 interaction are barely phosphorylated during respiratory growth (supplemental Fig. S3D), it is possible that the interaction with Atg11 may be a prerequisite step for Atg32 phosphorylation. Alternatively, this posttranslational modification may be critical for Atg32 to interact with Atg11. Notably, the two upper bands of Atg32-HA are hardly seen in the absence of Atg1 or Atg11 (supplemental Fig. S4C). In addition, these putative phosphorylated bands of Atg32-HA were not detected in cells expressing Atg1D211A, a kinase-dead mutant (35) (supplemental Fig. S4D). These findings, together with the previous report that Atg11 interacts with Atg1 (13), raise the possibility that Atg1 directly or indirectly phosphorylates Atg32 via interaction with Atg11.
Atg32 Interacts with Atg8 and Atg11 at Early Stages of Mitophagy
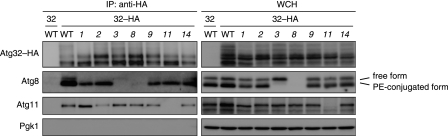
To test whether Atg32 interacts with Atg8 and Atg11 in a manner independent of the downstream events of mitophagy, we performed coimmunoprecipitation assays for cells lacking each of core Atg proteins (Fig. 5). As reported previously (10), Atg32-HA expressed under the endogenous promoter coprecipitated with the chromosomally encoded Atg8 and Atg11. Although the coprecipitation efficiencies varied among wild-type and mutant cells, the interactions of Atg8 and Atg11 with Atg32-HA were seen in the autophagy-deficient cells. We also found that, in addition to the PE-conjugated form, the free form of Atg8 was coprecipitated with Atg32-HA (Fig. 5, lane 2), but the coprecipitation efficiency was lower than that of Atg8-PE (5–10%). Moreover, an interaction between Atg32-HA and free Atg8 was slightly detected in cells lacking Atg3, an enzyme that catalyzes conjugation of Atg8 to PE (Fig. 5, lane 5), indicating that membrane anchoring via PE phospholipids is not essential for Atg8 to interact with Atg32. Strikingly, the Atg11-Atg32-HA coprecipitation in the absence of Atg1 was 4- to 8-fold more efficient than that in the presence of this protein kinase (Fig. 5, lane 3). The reason for this alteration is not entirely clear. However, it is possible that loss of Atg1 halts mitochondrial sequestration, leading to accumulation of Atg11. In agreement with this idea, a previous study has reported that Atg11 oligomerization is enhanced in the absence of Atg1 kinase activity (36).
FIGURE 5.
Atg32-Atg8 and -Atg11 interactions occur before isolation membrane generation. Coimmunoprecipitation assays for autophagy-competent (WT) and atg1∼14 null mutant (1Δ∼14Δ) cells expressing Atg32 (32) or Atg32-HA (32-HA) grown in glycerol medium for 30 h. All strains are vacuolar protease-deficient, atg32-null derivatives. Mitochondria-enriched fractions were obtained from whole cell homogenates (WCH), solubilized, and subjected to immunoprecipitation using anti-HA antibody-conjugated agarose. The WCH fractions and eluted immunoprecipitates (IP) were analyzed by Western blotting.
Together, our results suggest that Atg32-Atg8 and -Atg11 interactions are early steps linked to, but separable from, isolation membrane generation and subsequent autophagosome formation.
DISCUSSION
Our data implicate the mitophagy protein Atg32 as a membrane-anchored degradation landmark that directly recruits Atg8 and Atg11 to the surface of mitochondria. These initial interactions act in parallel but overlapping and are most likely to be prerequisite for the activation of subsequent steps in mitophagy. We do not exclude the possibility that some or all of the other core Atg proteins might loosely and/or dynamically associate with the Atg32-Atg8-Atg11 ternary complex, forming an multimeric protein assembly similar to the pre-autophagosomal structure, which is comprised of core Atg proteins, a putative center for autophagosome formation (37).
The Atg32 cytosol domain contains conserved binding motifs for Atg8 and Atg11 and harbors activities necessary and sufficient for degradation of mitochondria. Our microscopic observations that the peroxisome-anchored Atg32 cytosol domain can act as an autophagic degron and mediate pexophagy raises the possibility that faithful mitochondrial targeting of this degradation landmark is crucial for protection of other organelles against deregulated turnover and that the TM and IMS domains could serve as a membrane anchor specific to mitochondria.
Several lines of evidence support the idea that Atg32 forms an initiator complex with Atg8 and Atg11 and localizes as discrete foci on the mitochondrial surface. First, when tagged with GFP, Atg8 and Atg11 exhibit dot-like patterns along with mitochondria (10) (supplemental Fig. S3A). Second, Atg32-GFP expressed under the endogenous promoter distributed throughout mitochondria with one to three brighter spots (supplemental Fig. S5). Third, formation of these mitochondria-specific puncta does not occur in cells grown in glucose media, even with Atg32 overexpression (10). Fourth, Atg32-Atg8 and -Atg11 interactions do not require other core Atg proteins, indicating that it takes place independently of later steps of mitophagy.
It has recently been reported that two MAPK signaling pathways are involved in mitophagy and that the Atg32 serine 114 residue is critical for Atg11 interaction (34, 38). The phosphorylation of Atg30, a pexophagy receptor, is also critical for recruiting Atg11 to peroxisomes (30), supporting the idea that this posttranslational modification is a common regulatory mechanism for selective organelle autophagy. Identification of Atg32 phosphorylation site(s) relevant to its function, and protein kinase(s) and phosphatase(s) directly responsible for its posttranslational modification is a primary issue to be explored. In addition, future studies will be needed to address the issue of how the selectivity and efficiency of Atg32-mediated mitophagy are regulated in response to cellular needs.
Supplementary Material
Acknowledgments
We thank Yoshiaki Kamada and Hayashi Yamamoto for kind gifts of plasmids and experimental supports and Ralf Erdmann for helpful suggestions on protein targeting to peroxisomes. We also thank Kuninori Suzuki and all members of the Tamotsu Yoshimori and Yoshinori Ohsumi laboratories for valuable discussions.
This work was supported in part by Grant-in-Aid 22770124 from the Ministry of Education, Culture, Science, and Technology of Japan for Young Scientists (B) (to N. K. O.), the Targeted Proteins Research Program (to F. I.), Specially Promoted Research Grant 19002015 (to Y. O.), Scientific Research (C) Grant 20570144 (to K. O.), Scientific Research on Priority Areas Grants 20059034 and 22020012 (to K. O.), Scientific Research on Innovative Areas Grant 23113717 (to K. O.), Challenging Exploratory Research Grant 23657090 (to K. O.), special coordination funds from the Osaka University Life Science Young Independent Researcher Support Program to Disseminate Tenure Tracking System (to K. O.), and by grants from the Mochida Memorial Foundation for Medical and Pharmaceutical Research (to K. O.) and the Sumitomo Foundation (to K. O.).

This article contains supplemental Figs. S1–S5, Tables S1–S3, data, methods, and references.
- IMS
- intermembrane space
- PE
- phosphatidylethanolamine
- AIM
- Atg8 family-interacting motif
- TM
- transmembrane
- Cvt
- cytoplasm-to-vacuole targeting.
REFERENCES
- 1. Weidberg H., Shvets E., Elazar Z. (2011) Biogenesis and cargo selectivity of autophagosomes. Annu. Rev. Biochem. 80, 125–156 [DOI] [PubMed] [Google Scholar]
- 2. Mizushima N., Yoshimori T., Ohsumi Y. (2011) Annu. Rev. Cell Dev. Biol. 27, 107–132 [DOI] [PubMed] [Google Scholar]
- 3. Youle R. J., Narendra D. P. (2011) Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 12, 9–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Okamoto K., Kondo-Okamoto N. (2012) Biochim. Biophys. Acta in press [DOI] [PubMed] [Google Scholar]
- 5. Zhang J., Ney P. A. (2011) Mechanisms and biology of B-cell leukemia/lymphoma 2/adenovirus E1B-interacting protein 3 and Nip-like protein X. Antioxid. Redox Signal. 14, 1959–1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kanki T., Klionsky D. J., Okamoto K. (2011) Mitochondria autophagy in yeast. Antioxid. Redox Signal. 14, 1989–2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Narendra D. P., Youle R. J. (2011) Targeting mitochondrial dysfunction. Role for PINK1 and Parkin in mitochondrial quality control. Antioxid. Redox Signal. 14, 1929–1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tal R., Winter G., Ecker N., Klionsky D. J., Abeliovich H. (2007) Aup1p, a yeast mitochondrial protein phosphatase homolog, is required for efficient stationary phase mitophagy and cell survival. J. Biol. Chem. 282, 5617–5624 [DOI] [PubMed] [Google Scholar]
- 9. Kanki T., Klionsky D. J. (2008) Mitophagy in yeast occurs through a selective mechanism. J. Biol. Chem. 283, 32386–32393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Okamoto K., Kondo-Okamoto N., Ohsumi Y. (2009) Mitochondria-anchored receptor Atg32 mediates degradation of mitochondria via selective autophagy. Dev. Cell 17, 87–97 [DOI] [PubMed] [Google Scholar]
- 11. Kanki T., Wang K., Cao Y., Baba M., Klionsky D. J. (2009) Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev. Cell 17, 98–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kirisako T., Baba M., Ishihara N., Miyazawa K., Ohsumi M., Yoshimori T., Noda T., Ohsumi Y. (1999) Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 147, 435–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kim J., Kamada Y., Stromhaug P. E., Guan J., Hefner-Gravink A., Baba M., Scott S. V., Ohsumi Y., Dunn W. A., Jr., Klionsky D. J. (2001) Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J. Cell Biol. 153, 381–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kissová I., Deffieu M., Manon S., Camougrand N. (2004) Uth1p is involved in the autophagic degradation of mitochondria. J. Biol. Chem. 279, 39068–39074 [DOI] [PubMed] [Google Scholar]
- 15. Kanki T., Wang K., Baba M., Bartholomew C. R., Lynch-Day M. A., Du Z., Geng J., Mao K., Yang Z., Yen W. L., Klionsky D. J. (2009) A genomic screen for yeast mutants defective in selective mitochondria autophagy. Mol. Biol. Cell 20, 4730–4738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Noda N. N., Kumeta H., Nakatogawa H., Satoo K., Adachi W., Ishii J., Fujioka Y., Ohsumi Y., Inagaki F. (2008) Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells 13, 1211–1218 [DOI] [PubMed] [Google Scholar]
- 17. Otwinowski Z., Minor W. (1997) Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 18. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Crystallography and NMR system. A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 19. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 20. Kamada Y., Funakoshi T., Shintani T., Nagano K., Ohsumi M., Ohsumi Y. (2000) Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 150, 1507–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Noda N. N., Ohsumi Y., Inagaki F. (2010) Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. 584, 1379–1385 [DOI] [PubMed] [Google Scholar]
- 22. Yamaguchi M., Noda N. N., Nakatogawa H., Kumeta H., Ohsumi Y., Inagaki F. (2010) Autophagy-related protein 8 (Atg8) family interacting motif in Atg3 mediates the Atg3-Atg8 interaction and is crucial for the cytoplasm-to-vacuole targeting pathway. J. Biol. Chem. 285, 29599–29607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ichimura Y., Kumanomidou T., Sou Y. S., Mizushima T., Ezaki J., Ueno T., Kominami E., Yamane T., Tanaka K., Komatsu M. (2008) Structural basis for sorting mechanism of p62 in selective autophagy. J. Biol. Chem. 283, 22847–22857 [DOI] [PubMed] [Google Scholar]
- 24. Novak I., Kirkin V., McEwan D. G., Zhang J., Wild P., Rozenknop A., Rogov V., Löhr F., Popovic D., Occhipinti A., Reichert A. S., Terzic J., Dötsch V., Ney P. A., Dikic I. (2010) Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 11, 45–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Satoo K., Noda N. N., Kumeta H., Fujioka Y., Mizushima N., Ohsumi Y., Inagaki F. (2009) The structure of Atg4B-LC3 complex reveals the mechanism of LC3 processing and delipidation during autophagy. EMBO J. 28, 1341–1350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lynch-Day M. A., Klionsky D. J. (2010) The Cvt pathway as a model for selective autophagy. FEBS Lett. 584, 1359–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kabeya Y., Kamada Y., Baba M., Takikawa H., Sasaki M., Ohsumi Y. (2005) Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol. Biol. Cell 16, 2544–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cheong H., Yorimitsu T., Reggiori F., Legakis J. E., Wang C. W., Klionsky D. J. (2005) Atg17 regulates the magnitude of the autophagic response. Mol. Biol. Cell 16, 3438–3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hara T., Takamura A., Kishi C., Iemura S., Natsume T., Guan J. L., Mizushima N. (2008) FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 181, 497–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Farré J. C., Manjithaya R., Mathewson R. D., Subramani S. (2008) PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev. Cell 14, 365–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nazarko T. Y., Farré J. C., Subramani S. (2009) Peroxisome size provides insights into the function of autophagy-related proteins. Mol. Biol. Cell 20, 3828–3839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Shintani T., Huang W. P., Stromhaug P. E., Klionsky D. J. (2002) Mechanism of cargo selection in the cytoplasm to vacuole-targeting pathway. Dev. Cell 3, 825–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Suzuki K., Kondo C., Morimoto M., Ohsumi Y. (2010) Selective transport of α-mannosidase by autophagic pathways: identification of a novel receptor, Atg34p. J. Biol. Chem. 285, 30019–30025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Aoki Y., Kanki T., Hirota Y., Kurihara Y., Saigusa T., Uchiumi T., Kang D. (2011) Phosphorylation of Serine 114 on Atg32 mediates mitophagy. Mol. Biol. Cell 22, 3206–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Matsuura A., Tsukada M., Wada Y., Ohsumi Y. (1997) Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene 192, 245–250 [DOI] [PubMed] [Google Scholar]
- 36. Yorimitsu T., Klionsky D. J. (2005) Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol. Biol. Cell 16, 1593–1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Suzuki K., Kirisako T., Kamada Y., Mizushima N., Noda T., Ohsumi Y. (2001) The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 20, 5971–5981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mao K., Wang K., Zhao M., Xu T., Klionsky D. J. (2011) Two MAPK-signaling pathways are required for mitophagy in Saccharomyces cerevisiae. J. Cell Biol. 193, 755–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Frederick R. L., McCaffery J. M., Cunningham K. W., Okamoto K., Shaw J. M. (2004) Yeast Miro GTPase, Gem1p, regulates mitochondrial morphology via a novel pathway. J. Cell Biol. 167, 87–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Halbach A., Landgraf C., Lorenzen S., Rosenkranz K., Volkmer-Engert R., Erdmann R., Rottensteiner H. (2006) Targeting of the tail-anchored peroxisomal membrane proteins PEX26 and PEX15 occurs through C-terminal PEX19-binding sites. J. Cell Sci. 119, 2508–2517 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.