Background: Nrf2 activation reduces apoptosis that contributes to drug resistance.
Results: Nrf2 binds to Bcl-2 gene antioxidant response element to control antiapoptotic protein Bcl-2 and cellular apoptosis.
Conclusion: Nrf2 up-regulation of Bcl-2 prevents apoptosis that leads to increased drug resistance.
Significance: Nrf2 is a potential target for reducing drug resistance.
Keywords: Antioxidants, Apoptosis, Bcl-2, Gene Expression, Nrf2
Abstract
Nuclear transcription factor Nrf2 regulates the expression and coordinated induction of a battery of genes encoding cytoprotective and drug transporter proteins in response to chemical and radiation stress. This leads to reduced apoptosis, enhanced cell survival, and increased drug resistance. In this study, we investigated the role of Nrf2 in up-regulation of antiapoptotic protein Bcl-2 and its contribution to stress-induced apoptosis and cell survival. Exposure of mouse hepatoma (Hepa-1) and human hepatoblastoma (HepG2) cells to antioxidant tert-butylhydroquinone led to induction of Bcl-2. Mutagenesis and transfection assays identified an antioxidant response element between nucleotides −3148 and −3140 on the reverse strand of the Bcl-2 gene promoter that was essential for activation of Bcl-2 gene expression. Band/supershift and ChIP assays demonstrated binding of Nrf2 to Bcl-2 antioxidant response element. Alterations in Nrf2 led to altered Bcl-2 induction and cellular apoptosis. Moreover, dysfunctional/mutant inhibitor of Nrf2 (INrf2) in human lung cancer cells failed to degrade Nrf2, resulting in an increased Bcl-2 level and decreased etoposide- and UV/γ radiation-mediated DNA fragmentation. In addition, siRNA-mediated down-regulation of Nrf2 also led to decreased apoptosis and increased cell survival. Furthermore, the specific knockdown of Bcl-2 in Nrf2-activated tumor cells led to increased etoposide-induced apoptosis and decreased cell survival and growth/proliferation. These data provide the first evidence of Nrf2 in control of Bcl-2 expression and apoptotic cell death with implications in antioxidant protection, survival of cancer cells, and drug resistance.
Introduction
The Nrf22-inhibitor of Nrf2 (INrf2) complex serves as a sensor of chemical- and radiation-induced oxidative and electrophilic stress (1). Nrf2 resides predominantly in the cytoplasm where it interacts with the actin-associated cytosolic protein INrf2, which is also known as Keap1 (Kelch-like ECH-associated protein 1). INrf2 functions as a substrate adaptor protein for a Cul3/Rbx1-dependent E3 ubiquitin ligase complex to ubiquitinate and degrade Nrf2, thus maintaining a steady-state level of Nrf2 (1). The mechanisms by which Nrf2 is released from INrf2 under stress have been actively investigated. A consensus has emerged that oxidative/electrophilic stress modification of INrf2Cys-151 followed by PKCδ-mediated phosphorylation of Nrf2Ser-40 leads to the release and stabilization of Nrf2 (1–6). Nrf2 is translocated to the nucleus and coordinately activates the transcription of a battery of cytoprotective proteins including NAD(P)H:quinone oxidoreductase 1 (NQO1). This is achieved through Nrf2 binding to antioxidant response element (ARE) present in the promoter regions of cytoprotective genes (1). This is followed by activation of a delayed or postinduction mechanism that controls the switching off of Nrf2 activation of gene expression. GSK3β phosphorylates Fyn and Src at unknown threonine residue(s), leading to nuclear localization of Fyn and Src that phosphorylate Nrf2Tyr-568, resulting in nuclear export and degradation of Nrf2 (7–9). The negative regulation of Nrf2 through the Fyn and Src pathway is important in switching “off” the induction of Nrf2 downstream genes that were switched “on” in response to oxidative/electrophilic stress.
The switching on and off of Nrf2 leads to cell survival and protection against chemical- and radiation-induced oxidative/electrophilic stress, inflammation, and neoplasia (10–13). Indeed, accumulating in vivo evidence has demonstrated the importance of Nrf2 in protecting cells from the toxic and carcinogenic effects of many environmental insults. The Nrf2 knock-out mouse was prone to acute damages induced by acetaminophen, ovalbumin, cigarette smoke, pentachlorophenol, and 4-vinylcyclohexene diepoxide and had increased tumor formation when exposed to carcinogens such as benzo[a]pyrene, diesel exhaust, and N-nitrosobutyl(4-hydroxybutyl)amine (14–21). These observations collectively imply that Nrf2 is a master regulator of ARE-driven transcriptional activation for antioxidant genes in maintaining the homeostasis of redox status within cells and protection against associated diseases.
On the other hand, evidence also suggests that persistent accumulation of Nrf2 in the nucleus is harmful. INrf2-null mice demonstrated persistent accumulation of Nrf2 in the nucleus that led to postnatal death from malnutrition resulting from hyperkeratosis in the esophagus and forestomach (22). The reversal of the phenotype of INrf2-deficient mice by breeding them with Nrf2-null mice suggested that tightly regulated negative feedback might be essential for cell survival (23). The systemic analysis of the INrf2 genomic locus in human lung cancer patients and cell lines showed that deletion, insertion, and missense mutations in functionally important domains of INrf2 result in the reduction of INrf2 affinity for Nrf2 and elevated expression of cytoprotective genes, which resulted in drug resistance and cell survival in lung cancer cells (24, 25). Unrestrained activation of Nrf2 in cells increases the risk of adverse effects including reduced apoptosis, survival of damaged cells, tumorigenesis, and drug resistance (1). A role of Nrf2 in drug resistance is suggested based on its property to induce detoxifying enzymes and antioxidant and drug-transporting proteins (26–30). Although the mechanism of Nrf2 activation of detoxifying, antioxidant, and drug transporter proteins has been investigated as described above, the mechanism of Nrf2-mediated reduced apoptotic cell death remains obscure.
In cancer and drug resistance, apoptosis is a critical process that is deregulated, resulting in tumorigenesis and drug resistance (31). Bcl-2 family proteins, comprising greater than six antiapoptotic members including Bcl-2 and many proapoptotic members, regulate cell death and survival (32–35). Overexpression of Bcl-2 protein is usually associated with poor prognosis in many human cancers. However, in some cancer types, multiple antiapoptotic proteins are overexpressed (36). The mechanism of action of Bcl-2 is complex with many postulated interactions with other proteins, and the role of any single interaction in the final phenotype at the cellular level remains unknown. Recently it has been shown that H2S-mediated stabilization of Nrf2 in the nucleus increased the levels of antiapoptotic protein Bcl-2, which resulted in cardioprotection effects in the mice (37).
In the present study, we investigated the mechanism of Nrf2 up-regulation of antiapoptotic factor Bcl-2 and its role in apoptosis and drug resistance. Promoter analysis, chromatin immunoprecipitation (ChIP) assay, and EMSA identified an antioxidant response element in the reverse strand of the Bcl-2 promoter that was found to be essential for up-regulation of Bcl-2 in response to chemical/radiation. Nrf2 binds to Bcl-2 ARE and regulates expression and induction of the Bcl-2 gene. Nrf2 mediated the up-regulation of Bcl-2, down-regulated the activity of proapoptotic Bax protein and caspases 3/7, and protected cells from etoposide/radiation-mediated apoptosis that leads to drug resistance. Therefore, herein for the first time, we demonstrate that Nrf2-mediated up-regulation of Bcl-2 plays a significant role in prevention of apoptosis, increased cell survival, and drug resistance.
EXPERIMENTAL PROCEDURES
Plasmids
Mouse genomic DNA was isolated from keratinocytes and used to PCR-amplify 3.6 kb of Bcl-2 promoter using the forward primer 5′-AACGCGGGTACCAATTGAAGGCCACCCTGGGCTACATGAGAC-3′, reverse primer 5′-AATGCACTCGAGATAATCCAGCTCTTTTATTGGATGTGC-3′, and Phusion Hot Start high fidelity DNA polymerase (Finnzymes). The PCR-amplified promoter fragment was cloned in pGL2-basic luciferase vector (Promega, Madison, WI) using KpnI and XhoI restriction sites. The resultant plasmid was designated as pGL2b-3.6 kb (−96 to −3607, +1 is A of ATG site). Several 5′ deletions were generated in the 3.6-kb mouse Bcl-2 promoter. The nucleotide sequence of the PCR forward primers for generation of deletion plasmids of Bcl-2 promoter were as follows: 5-AACGCGGGTACCGCCCGATGTGGCAACCTGCTAGCCTGT-3 for 3.3 kb, 5′-AACGCGGGTACCGAGAGCTGATAACATAGTTATCACATA-3′ for 2.9 kb, 5′-AACGCGGGTACCGGTGGCAGGTATCACTCCCTGAGGTCC-3′ for 2.7 kb, 5′-AATTATGGTACCGTGCATTCAAGCAAATTTCATTTCCAG-3′ for 0.44 kb, and 5′-AATTTAGGTACCTTCAGCATTGCGGAGGAAGTAGAC-3′ for 0.3 kb. The same reverse 5′-AATGCACTCGAGATAATCCAGCTCTTTTATTGGATGTGC-3′ primer was used for generation of all deletion plasmids. To generate mutant AREr3 in 3.6-kb constructs, we used the GeneTailor site-directed mutagenesis kit (Invitrogen). The following pair of primers was used: forward primer, 5′-CGGTGTTCTTAACCGCTGAATCATCATTCCAACCACGA-3′; and reverse primer, 5′-TCAGCGGTTAAGAACACCGACTGTTCTTCCGAAGGT-3′. Thirty base pairs of forward and reverse strands of AREr3 and mutant AREr3 were synthesized. The nucleotide sequence of wild type and mutant AREr3 were as follows: AREr3: forward strand, 5′-ATTGCACCCGGGGCTAGCCCGCTGAGCCATCTCACCAACCAC-3′; and reverse strand, 5′-ATTCGGCCCGGGGCTAGCGTGGTTGGTGAGATGGCTCAGCGG-3′; mutant AREr3: forward strand, 5′-ATTGCACCCGGGGCTAGCCCGCTGATTCATCGATCCAACCAC-3′; and reverse strand, 5′-ATTCGGCCCGGGGCTAGCGTGGTTGGATCGATGAATCAGCGG-3′. Both strands of ARE sequences were annealed, digested with SmaI and NheI enzymes, and cloned into pGL2p vector. The sequence accuracy of all constructs was confirmed by DNA sequencing using an ABI3700 capillary sequencer (Applied Biosystems, Foster City, CA). The construction of luciferase plasmid harboring human NQO1 gene ARE and pCMV-FLAG-Nrf2 plasmid was described previously (38).
Cell Cultures and Generation of Stable Flp-In T-REx HEK293 Cells Expressing Tetracycline-inducible Nrf2
Hepa-1 and HepG2 cells were obtained from the American Type Culture Collection (Manassas, VA). Human embryonic kidney (HEK293) cells were obtained from Invitrogen. Hepa-1 cells and HEK293 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 40 units/ml penicillin, and 40 μg/ml streptomycin. HepG2 cells were grown in α-minimum essential medium containing 10% fetal bovine serum, 40 units/ml penicillin, and 40 μg/ml streptomycin. INrf2 mutant lung cancer A549 cells were grown in F-12/DMEM. We also generated wild type INrf2-expressing stable A549 cells by transfection of pcDNA-INrf2 followed by selection of clones with neomycin (G148). For generation of stable Nrf2-expressing cells, Flp-In T-REx HEK293 cells purchased from Invitrogen were co-transfected with FLAG-Nrf2 cDNA in pcDNA5/FRT/TO and pOG44 plasmids (Invitrogen) using the Effectene (Qiagen, Valencia, CA) method and the manufacturer's instructions. Forty-eight hours after transfection, the cells were grown in medium containing 200 μg/ml hygromycin B (Invitrogen). The 293/FRT/FLAG-Nrf2 cells stably expressing tetracycline-inducible N-terminal FLAG-tagged Nrf2 were selected. The stably selected cells were grown and treated with 2 μg/ml tetracycline (Sigma) for varying periods of time to follow the overexpression of FLAG-tagged Nrf2 protein. The cells were grown in a monolayer in an incubator at 37 °C in 95% air and 5% CO2.
Preparation of Cell Lysates and Western Blotting
Hepa-1 cells were seeded in 100-mm plates and transfected/treated as displayed in the figures. Cells were washed twice with ice-cold phosphate-buffered saline, trypsinized, and centrifuged at 1500 rpm for 5 min. For making whole cell lysates, the cells were lysed in radioimmune precipitation assay buffer (50 mm Tris, pH 8.0, 150 mm NaCl, 0.2 mm EDTA, 1% Nonidet P-40, 0.5% sodium deoxycholate, 1 mm phenylmethylsulfonyl fluoride, and 1 mm sodium orthovanadate supplemented with 1× protease inhibitor mixture (Roche Applied Science)). The protein concentration was determined using Bio-Rad protein assay reagent. Proteins (60–80 μg) were separated by SDS-PAGE and transferred to nitrocellulose membranes. The membranes were blocked with 3% nonfat dry milk in TBS with Tween 20. The antibodies purchased from Santa Cruz Biotechnology and their dilutions for immunoblotting analysis were as follows: anti-INrf2 (E-20) (1:1000), anti-Nrf2 (H-300) (1:500), anti-Bcl-2 (N-19) (1:1000), and anti-Bax (P-20) (1:1000). The antibodies purchased from Cell Signaling Technology and their dilutions in immunoblot analysis were: anti-cytochrome c (1:1000), anti-cytochrome c oxidase IV (1:1000), and anti-caspase 3 (1:1000). Anti-FLAG-HRP and anti-actin antibodies were obtained from Sigma. Immunoreactive bands were visualized using an ECL system (Amersham Biosciences). The intensity of protein bands after immunoblotting was quantitated using Quantity One 4.6.3 imaging software (ChemiDoc XRS, Bio-Rad) and normalized against proper loading controls. Cytoplasmic and nuclear fractions were prepared using the Active Motif nuclear extract kit (Active Motif, Carlsbad, CA) following the manufacturer's protocol. To confirm the purity of nuclear and cytoplasmic fractionations, the membranes were reprobed with cytoplasm-specific anti-lactate dehydrogenase (Chemicon) and nucleus-specific anti-lamin B antibodies (Santa Cruz Biotechnology). In related experiments, the cells were treated with 50 μm t-BHQ or DMSO as a vehicle control for different time intervals.
Transient Transfection and Luciferase Assay
Hepa-1 cells were plated in 100-mm plates at a density of 1 × 106 cells/plate 24 h prior to transfection. In related experiments, the cells were transfected with 1 μg of the indicated plasmids using Effectene transfection reagent (Qiagen) according to the manufacturer's instructions. After 36 h of transfection, cells were harvested, and cell-specific protein regulation was examined by Western blotting. For the luciferase reporter assay, Hepa-1 cells were grown in monolayer cultures in 12-well plates. After 12 h, cells were co-transfected with 0.1 μg of the indicated Bcl-2 promoter ARE-luciferase (Luc) reporter constructs and 10 times less quantities of firefly Renilla luciferase encoded by plasmid pRL-TK. Renilla luciferase was used as the internal control in each transfection. After 12 h of transfection, the cells were treated with DMSO or t-BHQ for 24 h. Cells were washed with 1× phosphate-buffered saline and lysed in 1× Passive lysis buffer from the Dual-Luciferase® reporter assay system kit (Promega). The luciferase activity was measured and plotted.
siRNA Interference Assay
Mouse- and human-specific Nrf2 siRNA and Bcl-2 siRNA were obtained from Applied Biosystems and used to inhibit Nrf2 and Bcl-2 protein. Hepa-1, HepG2, and HEK293 cells were transfected with 50–100 nm Nrf2 siRNA, Bcl-2 siRNA, or GAPDH control siRNA as indicated in the different figures using Lipofectamine RNAiMAX reagent (Invitrogen) according to the manufacturer's instructions. Thirty-two hours after transfection, the cells were harvested, and lysates were immunoblotted for Nrf2, Bcl-2, and NQO1 proteins.
Real Time Quantitative PCR
Hepa-1 cells were treated with t-BHQ for various time intervals or transfected with Nrf2 siRNA as indicated in the figures. Total RNA was isolated using an RNeasy minikit (Qiagen). Total RNA (250 ng) was subjected to reverse transcription using a High Capacity cDNA Reverse Transcription kit (Applied Biosystems). After synthesis of cDNA at 37 °C for 120 min, PCR was performed using a 7500 Real-Time PCR System according to the manufacturer's instructions. Bcl-2 primer and probe amplicon Mm00477531_m1, NQO1 primer and probe amplicon Mm00500821_m1, Nrf2 primer and probe amplicon Mm00477784_m1, and internal control GusB amplicon Mm00446953_m1 (Applied Biosystems) were used. The mixture was run on a 7500 Real-Time PCR System (Applied Biosystems) using relative quantitation according to the manufacturer's protocols.
ChIP Assay
The ChIP assay was performed using a kit from Active Motif as described previously (39). Briefly, 70% confluent Hepa-1 cells were treated with Me2SO or 50 μm t-BHQ for 4 h and then fixed in 1% formaldehyde for 15 min. Cells were lysed, and nuclei were pelleted by centrifugation. Nuclei were resuspended and sheared using a sonicator (Misonix Inc., Farmingdale, NY) with five 20-s pulses at 25% of maximum output. Sheared chromatin was immunoprecipitated with 2 μg of anti-Nrf2 or control IgG antibody. The cross-links were reversed overnight at 65 °C, and the chromatin was deproteinated with 20 μg/ml proteinase K. PCR amplification detected the Bcl-2 promoter region containing AREr3 bound to Nrf2. The following set of primers was used for PCR: forward primer, 5′-GTTCTTAAGCCCGATGTGGCAAC-3′; and reverse primer, 5′-GAGTAGTACCAATATGCTACCCTT-3′. GAPDH PCR was performed as an internal control. Primers used for GAPDH amplification were: forward primer, 5′-ACCACAGTCCATGCCATCAC-3′; and reverse primer, 5′-TCCACCACCCTGTTGCTGTA3′. The relative binding of Nrf2 to the AREr3 region of Bcl-2 promoter was also measured by quantitative real time PCR using custom-made probes and primers obtained from Applied Biosystems (ID 186710084_1). The mixture was run on a 7500 Real-Time PCR System (Applied Biosystems) using relative quantitation according to the manufacturer's protocols. To compare the relative binding of Nrf2 to the Bcl-2 promoter region containing AREr3 in Hepa-1 cells and the binding of Nrf2 to the human NQO1 gene ARE, we also used HepG2 cell chromatin. HepG2 cells were also treated with DMSO and t-BHQ, and sheared chromatin was immunoprecipitated with 2 μg of anti-Nrf2 or control IgG antibody. PCR was performed with a primer pair spanning the human NQO1 gene ARE. The primers were as follows: forward, 5′-CAGTGGCATGCACCCAGGGAA-3′; and reverse, 5′-GCATGCCCCTTTTAGCCTTGGCA-3′. The PCR conditions used for the ChIP assay were 37 cycles of a denaturing step at 94 °C for 30 s, an annealing step at 65 °C for 30 s, and an extension step at 72 °C for 30 s. PCR products were separated on a 2% agarose gel containing ethidium bromide and imaged using Quantity One 4.6.3 imaging software (ChemiDoc XRS, Bio-Rad).
Gel and Supershift Assay
Bcl-2 AREr3 was end-labeled with [γ-32P]ATP and T4 polynucleotide kinase. Labeled AREr3 (100,000 cpm) was incubated with 10 μg of Hepa-1 nuclear extract in the absence and presence of cold AREr3, and a band shift assay was performed according to a previously described procedure (39). In the same experiment, the gel shift mixture was also incubated with 2 μg of control IgG or Nrf2 antibody at 4 °C for 2 h to perform a supershift assay. The mixtures were separated on a 4% polyacrylamide gel and autoradiographed.
Cytochrome c Release and Caspase Activity Measurements
Hepa-1 cells were transfected with FLAG-Nrf2 or FLAG-INrf2 for 24 h followed by treatment with etoposide (20 μm) for 36 h. One set of cells was further treated with t-BHQ for an additional 24 h as indicated in the figures. Cells were harvested, and mitochondria were isolated using a Mitochondria Isolation kit (Thermo Scientific) and lysed. Equal amounts (30 μg) of mitochondrial and cytosolic lysates were immunoblotted with anti-cytochrome c, -cytochrome c oxidase IV, and -actin antibodies. For caspase activity measurements, Hepa-1 cells were transfected with pcDNA or FLAG-Nrf2 for 24 h and treated with 20 μm etoposide for an additional 36 h. One set of cells was further treated with t-BHQ for 24 h. Cells were lysed in the lysis buffer. Cell lysates (20 μg) were mixed with Caspase-Glo 3/7 substrate (Promega), and caspase 3/7 activity was measured according to the manufacturer's instructions and plotted. The cleaved caspase 3 protein was also detected by immunoblotting of the same cell lysates.
DNA Fragmentation/Cell Death Assay
Hepa-1, HepG2, HEK293, and FRT-FLAG-Nrf2 293 cells were plated at a density of 2000 cells/well in 96-well plates. After 20 h, Hepa-1 and HepG2 cells were treated with DMSO or t-BHQ, and HEK293 and FRT-Nrf2 293 cells were treated with water or 2 μg/ml tetracycline for 24 h. Then cells were exposed to various concentrations of etoposide for 72 h. Cells were harvested, and a photometric enzyme immunoassay was performed for the quantitative in vitro determination of cytoplasmic histone-associated DNA fragments (mono- and oligonucleosomes) using a Cell Death Detection ELISA kit from Roche Applied Science according to the manufacturer's instructions. Each combination of cell line and drug concentration was set up in three replicate wells, and the experiment was repeated thrice. Each data point represents a mean ± S.D. and was normalized to the value of the corresponding control cells.
TUNEL Assay
Hepa-1 cells were grown on coverslips, transfected with FLAG-Nrf2 for 16 h, and treated with 20 μm etoposide for 36 h. This was followed by treatment with DMSO or t-BHQ for an additional 24 h. Cells were fixed and permeabilized, and DNA was labeled with fluorescein-12-dUTP by using a Dead-End Fluorometric TUNEL assay kit (Promega). The fragmented DNA of the apoptotic cells was labeled with fluorescein-12-dUTP at 3′-OH DNA ends by using the terminal deoxynucleotidyltransferase recombinant enzymes according to the manufacturer's protocol. Cells were stained with DAPI and observed under a Nikon fluorescence microscope, photographs were captured, and the percentage of TUNEL-positive cells was quantified and plotted. The experiments were repeated thrice.
MTT Cell Survival Assay
Hepa-1, HepG2, and HEK293 cells were plated at a density of 2000 cells/well in 96-well plates, allowed to recover for 12 h, and then transfected with FLAG-Nrf2 for 16 h or transfected with Bcl-2 siRNA as indicated in the different figures. Control and transfected cells were exposed to 20 μm etoposide for 36 h and further treated with DMSO or t-BHQ for an additional 24 h. Cells were incubated with fresh MTT solution (200 μl/well; stock, 5 mg/ml in PBS) for 2 h, and absorbance at 570 nm was measured. Each combination of cell line and drug concentration was set up in three replicate wells, and the experiment was repeated thrice. Each data point represents a mean ± S.D. and was normalized to the value of the corresponding control cells.
Clonogenic Cell Survival Assay
Hepa-1, HepG2, and HEK293 cells were grown to 70% confluence, transfected with Bcl-2 siRNA or control siRNA, and treated with DMSO or t-BHQ in the presence of etoposide as indicated in the figures (in triplicates). Similarly, A549 and INrf2-A549 cells were grown to 70% confluence and transfected with Bcl-2 siRNA or control siRNA, and cells were treated with DMSO or t-BHQ in the presence of etoposide (20 μm). Cells were trypsinized and reseeded for 9 days. Fresh medium was added at day 5. After 10 days of incubation, 2 ml of freshly prepared clonogenic reagent (0.25% 1,9-dimethylmethylene blue in 50% ethanol) was added into the plates, and plates were kept at room temperature for 45 min. Cells were washed with PBS twice, and blue colonies were counted. Each data point represents a mean ± S.D. and was normalized to the value of the corresponding control cells.
Statistical Analyses
Data from luciferase assays, real time PCR, and immunoblotting band intensities were analyzed using a two-tailed Student's t test. Data are expressed as mean ± S.D. of three independent experiments. Significance values are represented as p < 0.05 (*), p < 0.005 (**), and p < 0.0001 (***) and are shown in the figures.
RESULTS
Antioxidant t-BHQ Induces Bcl-2 Gene Expression
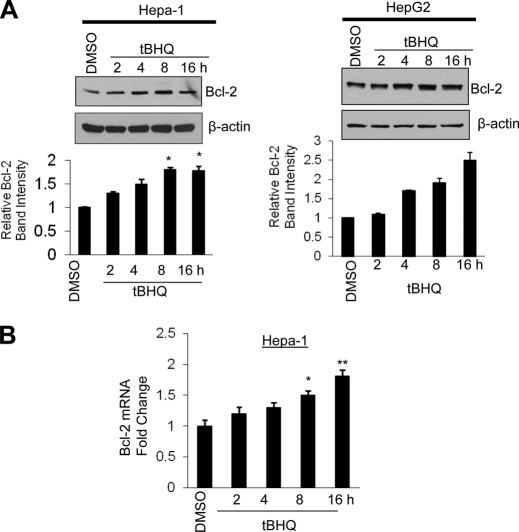
Hepa-1 and HepG2 cells treated with t-BHQ showed a time-dependent increase in Bcl-2 protein (Fig. 1A, left and right upper panels). The Bcl-2 band intensities were measured and plotted (Fig. 1A, left and right lower panels). Real time PCR analysis of t-BHQ-treated cells also demonstrated a time-dependent increase in Bcl-2 mRNA in Hepa-1 cells (Fig. 1B). These results collectively suggested that t-BHQ treatment of Hepa-1 and HepG2 cells leads to increased transcriptional activation of Bcl-2 gene.
FIGURE 1.
Antioxidant t-BHQ induces Bcl-2 gene expression. A, Hepa-1 cells and HepG2 cells were treated with DMSO or the antioxidant t-BHQ (50 μm) for different time periods, and 60 μg of cell extracts were immunoblotted with anti-Bcl-2 and -actin antibodies (left and right upper panels). Densitometry analysis measured Bcl-2 bands (left and right lower panels). B, the effect of t-BHQ on Bcl-2 mRNA expression was analyzed by real time quantitative PCR. All experiments were performed three times, and one set of data is presented (*, p < 0.05; **, p < 0.005). Error bars indicate S.E. of triplicate samples.
ARE on Reverse Strand between Nucleotides −3148 and −3140 of Bcl-2 Gene Promoter Mediates Expression and t-BHQ Induction of Bcl-2 Gene Expression
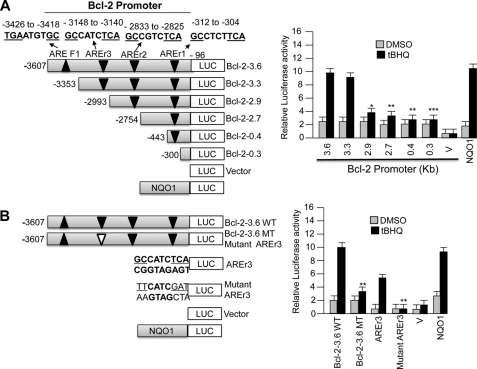
The Invitrogen Vector NTI program was used to analyze mouse Bcl-2 gene promoter for the presence of putative AREs. This analysis revealed the presence of several putative AREs in the Bcl-2 gene promoter (Fig. 2A). Deletion/internal mutagenesis followed by transfection assays investigated the ARE(s) required for the expression and t-BHQ induction of Bcl-2 gene (Fig. 2, A and B). A 3.6-kb Bcl-2 gene promoter attached to the luciferase gene upon transfection in Hepa-1 cells produced luciferase activity that was induced in response to t-BHQ treatment (Fig. 2A, right and left panels). Nucleotide sequence analyses of the 3.6-kb Bcl-2 promoter revealed the presence of four putative AREs (Fig. 2A, left panel). Three of these putative AREs were found located on the reverse strand between nucleotide positions −312 and −304 (AREr1), −2833 and −2825 (AREr2), and −3148 and −3140 (AREr3) from the start site of translation (with A of ATG being +1). The fourth putative ARE (AREF1) was located in the forward strand between nucleotide positions −3426 and −3418. The putative AREs were individually deleted by serial deletions in the 3.6-kb Bcl-2 gene promoter-luciferase plasmid (Fig. 2A). The Bcl-2-ARE-containing plasmids were transfected in Hepa-1 cells and analyzed for luciferase activity in the absence and presence of t-BHQ to determine the role of individual putative AREs in the expression and t-BHQ induction of the Bcl-2 gene (Fig. 2A, right panel). An ARE from the human NQO1 gene cloned into the luciferase reporter plasmid was used as a positive control for t-BHQ-mediated luciferase gene induction (Fig. 2A). Deletion mutagenesis in the 3.6-kb Bcl-2 gene promoter and transfection analysis in Hepa-1 cells revealed that the promoter region between nucleotides −3353 and −2933 containing AREr3 on the reverse strand is required for basal expression and induction in response to t-BHQ (Fig. 2A, left and right panels). The results also revealed that internal deletion of the ARE-r3 element in the 3.6-kb Bcl-2 promoter resulted in a significant reduction in basal expression and abrogation of t-BHQ induction as compared with the 3.6-kb Bcl-2 gene promoter (p > 0.005) (Fig. 2B, left and right panels). The AREr3 and mutated AREr3 were separately cloned in pGL2p vector and transfected in Hepa-1 cells followed by luciferase analysis to further confirm the role of AREr3 in t-BHQ induction of Bcl-2 gene expression (Fig. 2B, left panel). The results demonstrated that AREr3 but not mutated AREr3 efficiently mediated expression and t-BHQ induction of luciferase gene expression. These data suggested that AREr3 between nucleotides −3148 and −3140 of the Bcl-2 promoter was essentially required for expression and t-BHQ induction of the Bcl-2 gene.
FIGURE 2.
ARE between nucleotides −3148 and −3140 on reverse strand of Bcl-2 gene promoter is essential for antioxidant induction of Bcl-2 gene expression. A, systematic representation and cloning strategy of mouse Bcl-2 gene promoter into pGL2B or pGL2P luciferase reporter vectors. Four putative ARE sequences of the Bcl-2 promoter, three on the reverse strand (AREr1, AREr2, and AREr3) and one on the sense strand (ARE-F1), are shown (upper left panel). Mouse Bcl-2 promoter deletions were separately cloned into pGL2B Luc reporter gene, and plasmids were separately transfected in Hepa-1 cells. Cells were treated with DMSO or 50 μm t-BHQ for 24 h, and luciferase activity was measured (right panels). B, Bcl-2-3.6WT (wild type) and Bcl-2–3.6MT (mutated AREr3) promoter plasmids were separately transfected in Hepa-1 cells and analyzed for luciferase gene expression. In the same experiment, AREr3 and mutant AREr3 were attached to SV40 basal promoter hooked to the luciferase reporter gene by cloning in vector pGL2P and transfected in Hepa-1 cells, and cells were treated with DMSO or t-BHQ (50 μm for 24 h) and analyzed for luciferase activity. Human NQO1 ARE-luciferase reporter plasmid was also transfected in Hepa-1 cells as a positive control for t-BHQ-mediated luciferase gene induction. All experiments were performed three times, and one set of data is presented. The data shown are mean ± S.D. of three independent transfection experiments (*, p < 0.05; **, p < 0.005; ***, p < 0.0001). V, vector control. Error bars indicate S.E. of triplicate samples.
Antioxidant Increases in Vivo Binding of Nrf2 to AREr3 of Bcl-2 Promoter
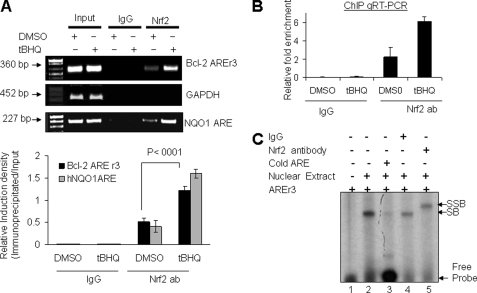
Nuclear transcription factor Nrf2 is known to bind to the ARE in promoter regions of cytoprotective genes including NQO1 gene and activate the downstream gene expression in response to t-BHQ (1). We performed in vivo ChIP and in vitro band/supershift assays to determine whether Nrf2 binds to Bcl-2 gene promoter AREr3, leading to activation of Bcl-2 gene expression. ChIP assay results demonstrated binding of Nrf2 to the AREr3 in Bcl-2 gene promoter (Fig. 3A). The Nrf2 binding to AREr3 was enhanced by 2–3-fold in response to t-BHQ (Fig. 3A). In the same experiment, Nrf2 binding to AREr3 was absent when chromatin was immunoprecipitated with control rabbit IgGs (Fig. 3A, upper and lower panels). Similarly, we also compared the relative binding of Nrf2 to the human NQO1 gene ARE using HepG2 cells promoter. The binding of Nrf2 to the human NQO1 gene ARE also increased by ∼4-fold in HepG2 cells treated with t-BHQ (Fig. 3A, upper and lower panels). Furthermore, using a ChIP assay and quantitative real time PCR, the relative binding of Nrf2 to the Bcl-2 AREr3 was measured and plotted (Fig. 3B). These data from both experiments confirmed a specific interaction of Nrf2 with the AREr3 of the Bcl-2 gene promoter as well as the NQO1 gene ARE that is enhanced upon t-BHQ treatment. It is noteworthy that no other AREs showed binding to Nrf2 in ChIP assays (data not shown). Bcl-2 gene AREr3 was also used in gel/supershift assays to analyze the specificity of Nrf2 binding to AREr3. In vitro band/supershift assays revealed Nrf2 binding to AREr3 that was competed with cold AREr3 and supershifted with Nrf2 antibody (Fig. 3C). Therefore, both ChIP and band/supershift assays indicated that Nrf2 binds to Bcl-2 gene AREr3 to activate ARE-mediated gene expression and induction in response to t-BHQ.
FIGURE 3.
Antioxidant increases binding of Nrf2 to Bcl-2 gene AREr3. A, ChIP assay. Hepa-1 cells were treated with 50 μm t-BHQ for 4 h, fixed with formaldehyde, and cross-linked, and the chromatin was sheared. The chromatin was immunoprecipitated with anti-Nrf2 antibody or control IgG. Nrf2 binding to Bcl-2 promoter was analyzed by PCR with specific primers for the ARE-r3 region of Bcl-2 promoter. GAPDH primers were used as a control. The AREr3 region of Bcl-2 promoter was amplified from 5 μl of purified soluble chromatin before immunoprecipitation to show input DNA (upper two gels). The effect of t-BHQ on the relative binding of Nrf2 to the human NQO1 gene ARE was also determined by the immunoprecipitation of sheared chromatin from HepG2 cells treated with DMSO and t-BHQ followed by a ChIP assay (lower gel). The relative binding of Nrf2 to the Bcl-2 AREr3 and hNQO1 ARE promoter was quantified from the band intensities of three independent experiments and plotted (lower panel). B, ChIP and quantitative (q) RT-PCR. The chromatin was immunoprecipitated as described in A from Hepa-1 cells. The binding of Nrf2 to the AREr3 region of Bcl-2 promoter was measured by quantitative real time PCR using custom-made probes and primers obtained from Applied Biosystems (ID 186710084_1). The mixture was run on a 7500 Real-Time PCR System (Applied Biosystems) using relative quantitation according to the manufacturer's protocols. The amounts of immunoprecipitated DNA were normalized to the inputs and plotted. C, gel and supershift analysis. Bcl-2 AREr3 was end-labeled with [γ-32P]ATP and T4 kinase. Labeled DNA (100,000 cpm) was incubated with 10 μg of Hepa-1 nuclear extract in binding buffer (lanes 2–5). The nuclear proteins binding to AREr3 were either competed with cold AREr3 (lane 3) or supershifted with IgG (lane 4) and Nrf2 antibody (ab) (lane 5). The reaction mixtures were run on a polyacrylamide gel and autoradiographed. SB, shifted band; SSB, supershifted band. All experiments were performed three times, and one set of data is presented. Error bars indicate S.E. of triplicate samples.
Nrf2 Controls AREr3-mediated t-BHQ Induction of Bcl-2 Gene Expression
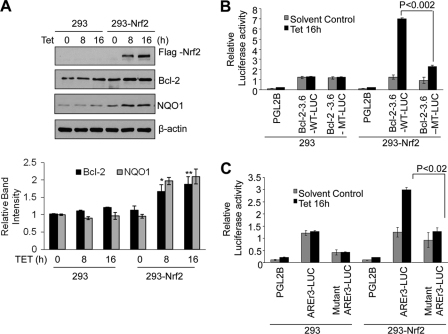
ChIP and band/supershift assays revealed that Nrf2 binds to Bcl-2 AREr3. Next we performed experiments to demonstrate that Nrf2 controls AREr3-mediated Bcl-2 gene expression (Figs. 4 and 5). Nrf2 was either overexpressed or down-regulated by siRNA, and its effect on Bcl-2 gene expression was determined. We successfully generated stable HEK293-FLAG-Nrf2 cell lines that upon exposure to tetracycline overexpressed FLAG-Nrf2 as reported previously (39). HEK293 and HEK293-FLAG-Nrf2 cells were exposed to either solvent control or tetracycline and immunoblotted for FLAG-Nrf2, Bcl-2, NQO1, and loading control β-actin (Fig. 4A). The results demonstrated overexpression of FLAG-Nrf2 in HEK293-FLAG-Nrf2 but not HEK293 cells. Overexpression of FLAG-Nrf2 by tetracycline in HEK293-FLAG-Nrf2 cells led to a significant increase in Bcl-2 and NQO1 proteins (Fig. 4A). These results indicated that, as reported earlier for NQO1, Nrf2 regulated Bcl-2 gene expression. It is noteworthy that minor increases in Bcl-2 and NQO1 gene expression were also observed in HEK293 cells treated with tetracycline. This is presumably through endogenous Nrf2. In the same experiment, the Nrf2 downstream gene NQO1 was also induced (Fig. 4A).
FIGURE 4.
Overexpression of Nrf2 up-regulates endogenous and transfected Bcl-2 gene expression. A, HEK293 and HEK293-Nrf2 cells expressing tetracycline-induced FLAG-tagged Nrf2 were treated with 2.5 μg/ml tetracycline (Tet) for the indicated times. Cell lysates (60 μg) were immunoblotted with anti-FLAG, anti-Bcl-2, anti-NQO1, and anti-actin antibodies (upper panel). The band intensities of Bcl-2 and NQO1 from A were quantified and plotted (lower panel). B and C, HEK293 and HEK293-Nrf2 cells were co-transfected with wild type pGL2B-Bcl-2-3.6WT or AREr3 mutant pGL2B-Bcl-2-3.6MT with the internal control Renilla luciferase plasmid pRL-TK (B) or co-transfected with pGL2P-AREr3 or pGL2P-mutant AREr3 with the internal control Renilla luciferase plasmid pRL-TK (C). pGL2B and pGL2B vectors were also transfected as negative controls. Twenty-four hours after transfection, the cells were treated with 2.5 μg/ml tetracycline for 16 h and analyzed for luciferase activity. The data shown are mean ± S.D. of three independent transfection experiments (*, p < 0.05; **, p < 0.001). Error bars indicate S.E. of triplicate samples.
FIGURE 5.
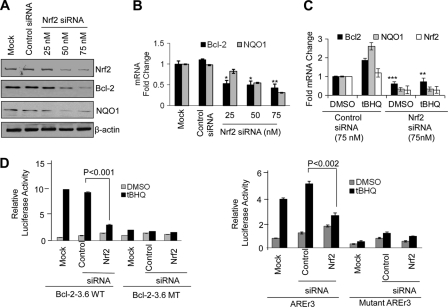
siRNA inhibition of Nrf2 decreases t-BHQ-inducible expression of Bcl-2. A, Western analysis. Hepa-1 cells were transfected with control or 25, 50, or 75 nm Nrf2 siRNA. Forty-eight hours after transfection, cells were harvested, lysed, and immunoblotted with anti-Nrf2, anti-Bcl-2, anti-NQO1, and anti-actin antibodies. B, real time PCR analysis. Hepa-1 cells were transfected with control or Nrf2 siRNA. Twenty-four hours after siRNA transfection, cells were harvested, and total RNA was extracted and converted to cDNA. cDNA (50 ng) was analyzed for mRNA levels using Bcl-2 and NQO1 primers and probes. C, real time PCR analysis. Hepa-1 cells were transfected with control or Nrf2 siRNA (75 nm). Twenty-four hours after siRNA transfection, cells were treated with t-BHQ for an additional 16 h, cells were harvested, and total RNA was extracted. The mRNA levels of Bcl-2, NQO1, and Nrf2 were quantified by real time PCR. D, reporter analysis. Hepa-1 cells were transfected with control or Nrf2 siRNA. Twenty-four hours after transfection, cells were transfected with wild type or mutant Bcl-2-3.6 (left panels) or with AREr3 or mutant AREr3 plasmids (right panels), incubated with DMSO or t-BHQ (50 μm) for 24 h, and analyzed for luciferase activity. All experiments were performed three times. The data shown are mean ± S.D. of three independent transfection experiments (*, p < 0.05; **, p < 0.005; ***, p < 0.0001). Error bars indicate S.E. of triplicate samples.
In related experiments, Bcl-2-3.6WT-Luc but not mutant Bcl-2-3.6MT-Luc significantly up-regulated luciferase gene expression in HEK293-Nrf2 cells treated with tetracycline (Fig. 4B). The increase in luciferase gene expression was absent in control HEK293 cells (Fig. 4B). In similar experiments, Bcl-2 gene AREr3-Luc but not mutant AREr3-Luc significantly enhanced luciferase gene expression in HEK293-Nrf2 cells exposed to tetracycline (Fig. 4C). In contrast, transfection of wild type or mutant Bcl-2 AREr3 in control HEK293 cells failed to induce tetracycline-mediated luciferase activity (Fig. 4C). These results collectively suggested that Nrf2 controls AREr3-mediated Bcl-2 gene expression.
Next, to support the above data, we used siRNA to inhibit Nrf2 expression in Hepa-1 cells. The transient transfection of Nrf2 siRNA in Hepa-1 cells inhibited (∼80%) the Nrf2 protein level, resulting in decreased Bcl-2 and NQO1 protein levels (Fig. 5A). The real time PCR analysis also clearly showed that knocking down Nrf2 by siRNA significantly deceased Bcl-2 and NQO1 transcripts in Hepa-1 cells (Fig. 5B). Furthermore, knocking down Nrf2 followed by t-BHQ treatments also significantly decreased mRNA levels of Bcl-2, Nrf2, and Nrf2 downstream gene NQO1 (Fig. 5C), suggesting that Nrf2 regulates Bcl-2 expression. In addition, knocking down Nrf2 in Hepa-1 cells resulted in abrogation of antioxidant induction of Bcl-2-3.6WT (wild type) and AREr3-Luc gene expression in transfected Hepa-1 cells (Fig. 5D, left and right panels). In the same experiment, Bcl-2-3.6MT carrying the mutated AREr3 region and mutant AREr3 failed to show an Nrf2 response (Fig. 5D, left and right panels). Together, these results suggested that Nrf2 and AREr3 mediated Bcl-2 gene expression and induction in response to the antioxidant t-BHQ.
Nrf2-induced Antiapoptotic Protein Bcl-2 Contributes to Decrease in Etoposide-mediated Cytochrome c Release from Mitochondria and Caspase 3/7 Activation
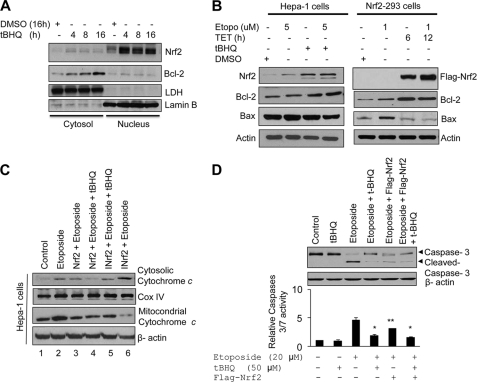
Hepa-1 cells were treated with the antioxidant t-BHQ for different time intervals, and cytosolic and nuclear fractions were prepared and immunoblotted with anti-Nrf2, -Bcl-2, -Lactate dehydrogenase, and -Lamin B antibodies (Fig. 6A). The results demonstrated that t-BHQ-stabilized Nrf2 protein in the nucleus led to increased Bcl-2 in the cytoplasm (Fig. 6A). These along with the above results suggested that t-BHQ-stabilized Nrf2 is translocated into the nucleus, binds to Bcl-2 promoter AREr3, and activates Bcl-2 gene expression. Next we examined the effect of Nrf2-mediated up-regulation of Bcl-2 protein on apoptotic markers as a measure of apoptosis. Hepa-1 cells treated with t-BHQ or HEK293-FLAG-Nrf2 cells overexpressing FLAG-Nrf2 were post-treated with etoposide, an apoptosis-inducing agent, and analyzed for apoptotic markers (Fig. 6, B–D). Antioxidant t-BHQ stabilization of Nrf2 in Hepa-1 cells and overexpression of FLAG-Nrf2 in HEK293-FLAG-Nrf2 cells both increased Bcl-2, decreased Bax (Fig. 6B), and decreased cytochrome c release from mitochondria (Fig. 6C). In addition to this, Hepa-1 cells treated with etoposide showed a 3.5-fold increase in caspase 3/7 activity as compared with the control (Fig. 6D). The increased caspase 3/7 activity and cleaved caspase 3 were significantly reduced in cells treated with t-BHQ or transfected with Nrf2, suggesting that Nrf2 potentially reduced caspase 3 or 3/7 activation (Fig. 6D). These results suggested that Nrf2 activation of Bcl-2 leads to decreased proapoptotic marker activity.
FIGURE 6.
Antioxidant t-BHQ and overexpression of Nrf2 up-regulate Bcl-2, leading to decreased etoposide-mediated cytochrome c release from mitochondria and caspase 3/7 activation. A, Hepa-1 cells were treated with DMSO or the antioxidant t-BHQ for different time periods, and nuclear and cytoplasmic extracts were generated. Nuclear and cytoplasmic extracts (80 μg) were immunoblotted with anti-Nrf2, -Bcl-2, -lactate dehydrogenase (LDH), and -lamin B antibodies. B, Hepa-1 and HEK293-Nrf2 cells were exposed to the indicated concentrations of etoposide (Etopo) followed by treatment with t-BHQ or tetracycline (TET), respectively. The endogenous levels of Nrf2 in Hepa-1 cells, overexpressed FLAG-Nrf2 in HEK293-Nrf2 cells, Bcl-2, Bax, and actin were analyzed by Western blotting. C, Hepa-1 cells were transfected with FLAG-Nrf2 or FLAG-INrf2 constructs, and cells were treated with etoposide (20 μm) for 36 h. Cells were further treated with t-BHQ for an additional 24 h. Cells were harvested, mitochondria were isolated using a Mitochondria Isolation kit (Thermo Scientific), and 30 μg of mitochondrial and cytosolic lysates was immunoblotted with anti-cytochrome c, -cytochrome c oxidase (Cox) IV, and -actin antibodies. D, Hepa-1 cells were transfected with FLAG-Nrf2 or treated with etoposide (20 μm) for 48 h and further treated with 50 μm t-BHQ for an additional 24 h. Cells were harvested and lysed in the lysis buffer. Lysates (60 μg) were immunoblotted for anti-caspase 3 and -actin antibody (upper panel). Cell lysate (20 μg) was mixed with Caspase-Glo 3/7 substrate (Promega), and caspase 3/7 activity was measured (lower panel). The data shown are mean ± S.D. of three independent experiments. All experiments were performed three times, and one set of data is presented (*, p < 0.05; **, p < 0.005). Error bars indicate S.E. of triplicate samples.
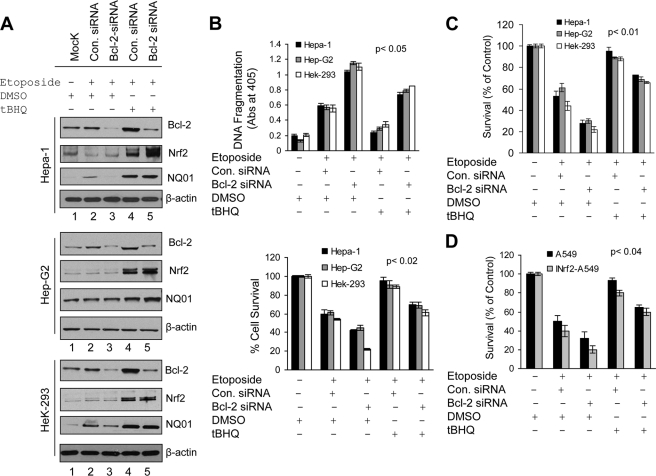
Nrf2-mediated Up-regulation of Bcl-2 Prevents Etoposide-induced DNA Fragmentation/Apoptosis and Promotes Cell Survival
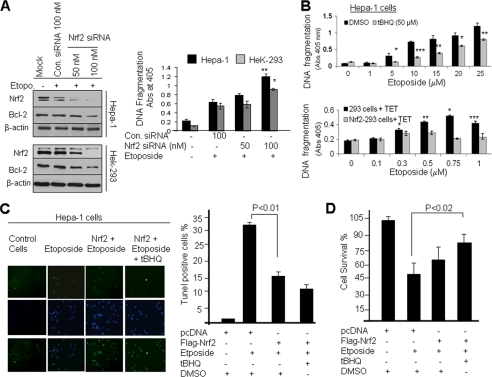
We examined the role of Nrf2 and Bcl-2 in cellular apoptosis and cell survival. First, we knocked down cellular Nrf2 protein in Hepa-1 and HEK293 cells by siRNA, and then cells were treated with etoposide (Fig. 7A). Immunoblotting data clearly demonstrated that Nrf2 knockdown by siRNA decreased Nrf2 and Bcl-2 protein levels in both cell lines (Fig. 7A, left panel). In related experiments, we also measured etoposide-mediated DNA fragmentation. Knocking down Nrf2 levels in Hepa-1 and HEK293 cells increased etoposide-mediated DNA fragmentation significantly (1.5–2-fold) (Fig. 7A, right panel). Next we analyzed the effect of Nrf2 stabilization/overexpression on DNA fragmentation (Fig. 7B). Hepa-1 cells were treated with DMSO or t-BHQ for stabilization of Nrf2. Similarly, control HEK293 and HEK293-FLAG-Nrf2 overexpressing cells were treated with tetracycline for induction of FLAG-Nrf2. The cells were treated with different concentrations of etoposide and analyzed for histone-associated DNA fragmentation (Fig. 7B, upper and lower panels). The results demonstrated that histone-associated DNA fragmentation was significantly decreased by 40% (p > 0.005) in Hepa-1 cells and ∼50% (p > 0.05) in HEK293-FLAG-Nrf2 cells after t-BHQ and tetracycline treatment, respectively (Fig. 7B). These observations suggested that increased Nrf2 down-regulated etoposide-mediated DNA fragmentation. These results were also supported by the TUNEL assay in Hepa-1 cells (Fig. 7C). Etoposide treatment significantly increased TUNEL-positive Hepa-1 cells compared with control DMSO-treated cells, whereas t-BHQ stabilization of Nrf2 significantly deceased the number of TUNEL-positive Hepa-1 cells even after etoposide treatment (Fig. 7C, left and right panels). In addition, we performed an MTT assay to determine cell survival in DMSO- and t-BHQ-treated Hepa-1 cells to determine the role of Nrf2 in cell survival. The results suggested that t-BHQ stabilization of Nrf2 protected cells from etoposide-mediated apoptosis, leading to increased cell survival (∼40%) compared with etoposide treatment alone, and drug resistance (Fig. 7D). The results collectively demonstrate that Nrf2-mediated up-regulation of Bcl-2 protein significantly increased drug resistance and cell survival in cancer cells.
FIGURE 7.
Antioxidant mediated stabilization and overexpression of Nrf2 and Bcl-2 reduced etoposide-mediated DNA fragmentation, leading to cell survival. A, Nrf2 knockdown increased DNA fragmentation. Hepa-1 and HEK293 cells were transfected with control (Con.) siRNA (100 nm) or different concentrations of Nrf2 siRNA. After 24 h of transfection, Hepa-1 and HEK293 cells were treated with etoposide (Etopo.) (30 and 1 μm concentrations, respectively) for an additional 30 h. Cell lysates (80 μg) were Western blotted with anti-Nrf2, anti-Bcl-2, and anti-actin antibodies (left panels). Similarly, the effect of Nrf2 siRNA on the etoposide-mediated histone-associated DNA fragmentation was analyzed. The cytoplasmic histone-associated DNA fragments (mono- and oligonucleosomes) were quantified using a Cell Death Detection ELISA kit (Roche Applied Science) and plotted (right panel). B, stabilization/overexpression of Nrf2 decreased DNA fragmentation. Hepa-1 cells were treated with increasing concentrations of etoposide for 48 h (in triplicates) followed by treatment with DMSO or t-BHQ for an additional 24 h (upper panel). Control HEK293 and HEK293-Nrf2 cells were treated with increasing concentrations of etoposide for 48 h (in triplicates) followed by treatment with 2.0 μg/ml tetracycline for 24 h (lower panel). The cytoplasmic histone-associated DNA fragments (mono- and oligonucleosomes) were quantified using a Cell Death Detection ELISA kit (Roche Applied Science) and plotted). C, TUNEL assay. Hepa-1 cells were transfected with FLAG-Nrf2 and treated with etoposide (20 μm) for 36 h followed by treatment with t-BHQ for an additional 24 h. The cells were fixed and permeabilized, and a TUNEL assay was performed. TUNEL-positive cells were observed under a fluorescence microscope, and photographs were captured (left panel). The TUNEL-positive cells were quantified and plotted (right panel). The data represented are the mean ± S.D. from three independent experiments. D, cell survival assay. Hepa-1 cells were plated at a density of 5000 cells/well in 24-well plates (in triplicates), transfected with FLAG-Nrf2, and treated with DMSO, etoposide, or etoposide and t-BHQ for 72 h. Cells were incubated with fresh MTT solution for 2 h at 37 °C, and absorbance (Abs) at 570 nm was measured. The experiment was repeated thrice. Each point represents a mean ± S.D. and was normalized to the value of the corresponding control cells (*, p < 0.05; **, p < 0.005; ***, p < 0.0001). Error bars indicate S.E. of triplicate samples.
Dysfunctional INrf2 in Lung Cancer A549 Cells Stabilized Nrf2, Up-regulated Bcl-2, and Decreased Etoposide- and UV and γ Radiation-mediated DNA Fragmentation
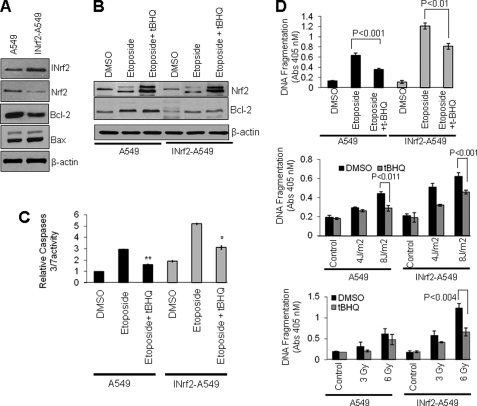
Singh et al. (25) demonstrated that human lung tumor-derived A549 cells possessed a glycine 333 to cysteine (G333C) point mutation in INrf2 protein. This mutant INrf2 was unable to repress Nrf2 activity, which resulted in an increase in drug resistance and cell survival. By stable transfection of wild type pcDNA-INrf2 followed by selection with G148, we generated INrf2-A549 cells that express wild type INrf2 protein along with endogenous mutant INrf2. The role of dysfunctional or mutant INrf2 in A549 cells and wild type INrf2/mutant INrf2 in INrf2-A549 cells in the regulation of Nrf2, Bcl-2, and Bax was examined in the same isogenic cancer cells. Immunoblotting results demonstrated that after stable transfection the level of INrf2 in INrf2-A549 cells was increased by 2.1-fold compared with A549 parent cells (Fig. 8A). The INrf2 expression in INrf2-A549 cells led to a 2–3-fold decrease in Nrf2 and Bcl-2 (Fig. 8A). The level of Bax protein was ∼1.3-fold higher in INrf2-A549 cells compared with A549 cells (Fig. 8A). A549 and INrf2-A549 cells were exposed to different strengths of etoposide or UVB or γ radiation and analyzed for DNA fragmentation (Fig. 8, B–D). The results revealed that all three agents showed concentration/strength-dependent significant increases in apoptosis of INrf2-A549 cells expressing lower levels of Nrf2/Bcl-2 as compared with A549 cells expressing higher levels of Nrf2/Bcl-2. In other words, overexpression of INrf2 led to decreased Nrf2 and Bcl-2 and increased apoptotic cell death.
FIGURE 8.
Dysfunctional INrf2 stabilized Nrf2 and Bcl-2, leading to reduced etoposide and UV/γ radiation-mediated DNA fragmentation in non-small cell lung cancer A549 cells. A, A549 cells expressing mutant INrf2 and cDNA-derived wild type INrf2 (INrf2-A549) were immunoblotted with anti-Nrf2, -INrf2, -Bcl-2, -Bax, and -actin antibodies. B, A549 or INrf2-A549 cells were treated with DMSO, 10 μm etoposide, or etoposide + t-BHQ. The endogenous levels of Nrf2 and Bcl-2 were analyzed by Western blotting. C, the same cell lysates (20 μg) were mixed with Caspase-Glo 3/7 substrate (Promega), and caspase 3/7 activity was measured. D, 5000 A549 or INrf2-A549 cells were plated in 24-well plates (in triplicates) and treated with DMSO or the indicted concentrations of etoposide for 48 h. One set of cells was further treated with t-BHQ (50 μm) for an additional 24 h (upper panel), irradiated with UVB light (middle panel), or irradiated with the indicated grays (Gy) of γ radiation (lower panel). The cells were incubated for 24 h, post-treated with DMSO or t-BHQ for an additional 24 h, and harvested. The cytoplasmic histone-associated DNA fragments (mono- and oligonucleosomes) were quantified using a Cell Death Detection ELISA kit (Roche Applied Science) and plotted. The experiments were repeated three times. The data represented are the mean ± S.D. from three independent experiments (*, p < 0.05; **, p < 0.005). Error bars indicate S.E. of triplicate samples.
Bcl-2 Specifically Contributes to Nrf2-mediated Reduced Apoptosis and Increased Cell Survival/Drug Resistance
Five different cancer cell lines were used in the various experiments to investigate the specific role of Bcl-2 in Nrf2 modulation of apoptosis and cancer cell survival/drug resistance (Fig. 9). Hepa-1, HepG2, and HEK293 cells were either mock-transfected or transfected with control or Bcl-2 siRNA (Fig. 9A, lanes 1–5). The siRNA-transfected cells were either treated with DMSO (control) or t-BHQ and immunoblotted (Fig. 9A, lanes 2–4). The results demonstrated that Bcl-2 knockdown by siRNA reduced Bcl-2 protein more than 80% in Hepa-1 and HEK293 cells and 60% in HepG2 cells (Fig. 9A, lanes 2 and 3). The control and Bcl-2 siRNA-transfected cells upon treatment with t-BHQ showed stabilization of Nrf2 and activation of Nrf2 downstream proteins including NQO1 (Fig. 9A, compare lane 2 with lane 4 and lane 3 with lane 5). t-BHQ-mediated stabilization of Nrf2 also led to increased Bcl-2 (Fig. 9A, compare lane 2 with lane 4). However, t-BHQ induction of Bcl-2 was specifically inhibited in Bcl-2 siRNA-transfected cells (Fig. 9A, compare lane 5 with lane 4). In other words, Bcl-2 was specifically inhibited in t-BHQ/Nrf2-activated cells (Fig. 9A, compare lane 5 with lane 4). These cells were used to determine the specific role of Bcl-2 in etoposide-mediated histone-associated DNA fragmentation and cell survival in t-BHQ/Nrf2-activated cells (Fig. 9B). Bcl-2 siRNA-transfected cells showed a 1.6–2-fold increase in etoposide-mediated DNA fragmentation and a 20–30% decrease in cell survival as compared with control siRNA-transfected cells (Fig. 9B, upper and lower panels). Interestingly, t-BHQ significantly reduced DNA fragmentation (0.6–1.2-fold; p < 0.02) and increased cell survival by 35–40% in control siRNA-transfected cells compared with DMSO-treated control or Bcl-2 siRNA-transfected cells (Fig. 9B, upper and lower panels). Intriguingly, the t-BHQ-mediated reduction in DNA fragmentation and increase in cell survival as observed in control siRNA-transfected/t-BHQ-treated cells were significantly muted in Bcl-2 siRNA-transfected and t-BHQ-treated cells (Fig. 9B, upper and lower panels). In other words, the loss of Bcl-2 in t-BHQ/Nrf2-activated cells specifically increased apoptotic cell death and decreased cell survival in response to etoposide as compared with t-BHQ/Nrf2-activated cells containing increased levels of Bcl-2 (Fig. 9, upper and lower panels). It is noteworthy that all three cell types, Hepa-1, HepG2, and HEK293, showed similar results (Fig. 9).
FIGURE 9.
Nrf2-regulated induction of Bcl-2 specifically contributed to Nrf2-mediated decreased apoptosis and increased cell survival. A, Western analysis. Hepa-1, HepG2, and HEK293 cells were transfected with control (Con.) siRNA (50 nm) or Bcl-2 siRNA (50 nm) for 24 h and treated with DMSO or t-BHQ in the presence of etoposide (Hepa-1, 30 μm; HepG2, 25 μm; HEK293, 1 μm) for an additional 30 h as indicated in the figure. Cell extracts (80 μg) were immunoblotted with anti-Nrf2, anti-Bcl-2, anti-NQO1, and anti-β-actin antibodies. B, cell death/DNA fragmentation assay. Five thousand Hepa-1, HepG2, and HEK293 cells were plated in 24-well plates and transfected/treated as indicated in A (in triplicates). The cytoplasmic histone-associated DNA fragments (mono- and oligonucleosomes) were quantified using a Cell Death Detection ELISA kit (Roche Applied Science) and plotted (upper panel). After transfections/treatments, Hepa-1, HepG2, and HEK293 cells were incubated with fresh MTT solution for 2 h at 37 °C, and absorbance (Abs) at 570 nm was measured. The experiment was repeated thrice. Each point represents a mean ± S.D. and was normalized to the value of the corresponding control cells. C and D, clonogenic cell survival assay. Hepa-1, HepG2, and HEK293 cells were grown to 70% confluence, transfected with control or Bcl-2 siRNA, and treated with DMSO or t-BHQ in the presence of etoposide for 30 h in triplicates (C). Similarly, A549 and INrf2-A549 cells were also transfected with Bcl-2 or control siRNA and treated with etoposide (20 μm) in the presence of DMSO or t-BHQ for 30 h in triplicates (D). Cells were trypsinized, 1000 cells were reseeded in 100-mm tissue culture dishes (in triplicate) and incubated for 9 days, and a clonogenic cell survival assay was performed as described under “Experimental Procedures.” Each data point represents a mean ± S.D. and was normalized to the value of the corresponding control cells. All experiments were repeated three times, and one set of data is presented. Error bars indicate S.E. of triplicate samples.
In addition, we performed clonogenic cell survival assays to obtain further support of the above conclusions and analyze the effects of Bcl-2 and Nrf2 in tumor cell growth and survival (Fig. 9C). The results demonstrated that siRNA-mediated knockdown of Bcl-2 increased the sensitivity of tumor cells to the etoposide-induced cell death and decreased cell growth by 15–20% compared with control siRNA-transfected cells. The t-BHQ treatment reversed the effect and promoted cell survival in Hepa-1, HepG2, and HEK293 cells (Fig. 9C). However, Bcl-2 knockdown cells upon treatment with t-BHQ showed significantly lower recovery than control siRNA-transfected and t-BHQ-treated cells. In other words, the loss of Bcl-2 specifically reduced cell survival in Nrf2-activated/t-BHQ-treated cells. The same results were observed in Hepa-1, HepG2, and HEK293 cells (Fig. 9C). The results also suggested that not only Bcl-2 but several other antiapoptotic and cytoprotective proteins may be involved in cell survival. Finally, we also analyzed the role of Nrf2, Bcl-2, INrf2, and etoposide in cell survival/growth in A549 and A459 cells stably transfected with INrf2 (INrf2-A459). The data from the clonogenic cell survival assay demonstrated that siRNA-mediated knockdown of Bcl-2 and INrf2-mediated Nrf2 degradation both led to a significant reduction (75%; p < 0.04) in cell survival compared with control cells (∼15–20% in A549 cells) (Fig. 9D). The exposure of cells to t-BHQ promoted cell growth but to a lesser extent in the Bcl-2-inhibited cells as compared with cells expressing Nrf2-induced Bcl-2. This again suggested that Nrf2 up-regulates several cytoprotective proteins including NQO1 as well as an antiapoptotic factor such as Bcl-2 that promotes cell survival/drug resistance in tumor cells.
DISCUSSION
Nrf2 is considered a double edge sword (1). On one hand, Nrf2 is known to regulate expression and coordinated induction of a battery of cytoprotective genes that contribute to cellular protection and prevention of inflammation, neoplasia, and neurodegeneration (1). On the other hand, persistent accumulation/activation of Nrf2 in the nucleus is known to enhance oncogenesis and induce drug resistance (1). Factors like mutations in INrf2 in cancer cells lead to inactivation of INrf2, resulting in persistent nuclear accumulation of Nrf2 and promotion of cell survival. Recent studies have reported increased stabilization/accumulation of Nrf2 due to mutations in INrf2, resulting in loss of function in lung tumors (24, 25). Lung cancer cell line A549 contains INrf2G333C mutant protein that has lost its capacity to bind/degrade Nrf2, leading to accumulation of Nrf2 (25). It has been suggested that higher levels of Nrf2 in A549 cells might have contributed to the survival of these cells in lung cancer (25). In addition, a role of Nrf2 in drug resistance is suggested based on its property to induce detoxifying enzymes and antioxidant and drug-transporting proteins (25–30). However, information on the mechanism of the role of Nrf2 in oncogenesis and drug resistance is limited. We demonstrate here that Nrf2-mediated reduced apoptotic cell death plays a significant role in the survival of oncogenic cells and drug resistance.
The current studies demonstrated that Nrf2-mediated up-regulation of antiapoptotic protein Bcl-2 led to a decrease in Bax, cytochrome c release from mitochondria, activation of caspases, decreased DNA fragmentation, a significant reduction in etoposide-induced apoptotic cell death, and increased cell survival. In addition, expression of wild type INrf2 in lung carcinoma A549 cells resulted in a significant decrease in Nrf2 and Bcl-2 that decreased apoptotic cell death in response to exposure to etoposide and UVB and γ radiation. These results collectively demonstrated that Nrf2-mediated up-regulation of Bcl-2 and reduced apoptosis contribute to the enhanced cell survival and drug resistance. Five different cell lines and two different assays including apoptotic cell death/cell survival and clonogenic analysis demonstrated that inhibition of Bcl-2 in t-BHQ/Nrf2-activated cells increased cell death and reduced cell survival compared with t-BHQ/Nrf2/Bcl-2-activated cells. These experiments clearly suggested that Nrf2 activation of Bcl-2 contributes to Nrf2 regulation of cellular apoptotic cell death and survival. The studies also determined the mechanism of Nrf2 up-regulation of Bcl-2. Nrf2 up-regulated transcription of Bcl-2 gene. Deletion mutagenesis and transfection studies identified AREr3 between nucleotides −3148 and −3140 of the Bcl-2 gene promoter as the DNA element that bound to Nrf2 and activated Bcl-2 gene expression.
It is noteworthy that Nrf2 coordinately up-regulates Bcl-2 gene expression along with a battery of cytoprotective genes encoding detoxifying enzymes, antioxidant proteins, and drug transporters (1). Therefore, it is a reasonable interpretation that the coordinated activation of cytoprotective proteins and antiapoptotic protein Bcl-2 contributed to the reduced apoptosis and enhanced cell survival.
Previous studies have shown that INrf2 in association with Cul3/Rbx1 ubiquitinates and degrades Bcl-2 (40). Antioxidants antagonize this interaction. Bcl-2 is released from INrf2 and stabilized, leading to decreased etoposide-mediated apoptotic cell death and increased cell survival. These observations together with the present study suggest that release of Bcl-2 from INrf2 and Nrf2-mediated transcriptional activation of Bcl-2 both lead to a significant accumulation of Bcl-2 that reduces apoptosis and increases cell survival. The studies also raise interesting questions regarding the role of other antiapoptotic proteins such as Bcl-xL and Mcl-1 in the Nrf2-mediated decrease in apoptosis and increase in cell survival. Preliminary studies indicated that t-BHQ coordinately induces the expression of Bcl-2 along with the other antiapoptotic proteins Bcl-xL and Mcl-1, suggesting a role of these proteins in Nrf2 regulation of apoptosis/cell survival (supplemental Fig. S1). However, this clearly remains to be determined by further experiments.
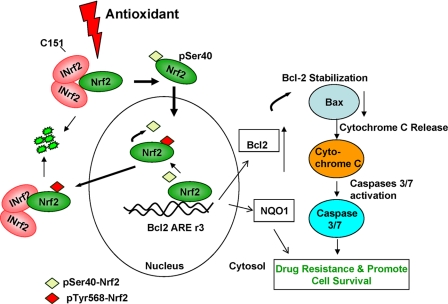
A model showing the role of Nrf2 and Bcl-2 in reduced apoptosis and increased cell survival is shown in Fig. 10. Exposure to antioxidant leads to dissociation of Nrf2 from INrf2. Nrf2 is stabilized and translocated to the nucleus where it binds to AREr3 in the promoter region of Bcl-2 gene, resulting in increased transcription of Bcl-2 gene. Increased transcripts of Bcl-2 gene are translated to increase Bcl-2 protein. Increased Bcl-2 leads to decreased Bax, cytochrome c release, activation of caspase 3/7, reduced apoptosis, and increased cell survival.
FIGURE 10.
Model showing antioxidant/Nrf2-mediated regulation of Bcl-2 and cellular apoptosis.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant RO1 GM047466.

This article contains supplemental Fig. S1.
- Nrf2
- NF-E2-related factor
- INrf2
- inhibitor of Nrf2, also known as Keap1
- ARE
- antioxidant response element
- NQO1
- NAD(P)H:quinone oxidoreductase
- t-BHQ
- tert-butyl hydroquinone
- Luc
- luciferase
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide.
REFERENCES
- 1. Kaspar J. W., Niture S. K., Jaiswal A. K. (2009) Nrf2:INrf2 (Keap1) signaling in oxidative stress. Free Radic. Biol. Med. 47, 1304–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wakabayashi N., Dinkova-Kostova A. T., Holtzclaw W. D., Kang M. I., Kobayashi A., Yamamoto M., Kensler T. W., Talalay P. (2004) Protection against electrophile and oxidant stress by induction of the phase 2 response: fate of cysteines of the Keap1 sensor modified by inducers. Proc. Natl. Acad. Sci. U.S.A. 101, 2040–2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Eggler A. L., Liu G., Pezzuto J. M., van Breemen R. B., Mesecar A. D. (2005) Modifying specific cysteines of the electrophile-sensing human Keap1 protein is insufficient to disrupt binding to the Nrf2 domain Neh2. Proc. Natl. Acad. Sci. U.S.A. 102, 10070–10075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bloom D. A., Jaiswal A. K. (2003) Phosphorylation of Nrf2 at Ser40 by protein kinase C in response to antioxidants leads to the release of Nrf2 from INrf2, but is not required for Nrf2 stabilization/accumulation in the nucleus and transcriptional activation of antioxidant response element-mediated NAD(P)H:quinone oxidoreductase-1 gene expression. J. Biol. Chem. 278, 44675–44682 [DOI] [PubMed] [Google Scholar]
- 5. Huang H. C., Nguyen T., Pickett C. B. (2002) Phosphorylation of Nrf2 at Ser-40 by protein kinase C regulates antioxidant response element-mediated transcription. J. Biol. Chem. 277, 42769–42774 [DOI] [PubMed] [Google Scholar]
- 6. Niture S. K., Jain A. K., Jaiswal A. K. (2009) Antioxidant-induced modification of INrf2 cysteine 151 and PKC-δ-mediated phosphorylation of Nrf2 serine 40 are both required for stabilization and nuclear translocation of Nrf2 and increased drug resistance. J. Cell Sci. 122, 4452–4464 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7. Jain A. K., Jaiswal A. K. (2006) Phosphorylation of tyrosine 568 controls nuclear export of Nrf2. J. Biol. Chem. 281, 12132–12142 [DOI] [PubMed] [Google Scholar]
- 8. Jain A. K., Jaiswal A. K. (2007) GSK-3β acts upstream of Fyn kinase in regulation of nuclear export and degradation of NF-E2 related factor 2. J. Biol. Chem. 282, 16502–16510 [DOI] [PubMed] [Google Scholar]
- 9. Niture S. K., Jain A. K., Shelton P. M., Jaiswal A. K. (2011) Src subfamily kinases regulate nuclear export and degradation of transcription factor Nrf2 to switch off Nrf2-mediated antioxidant activation of cytoprotective gene expression. J. Biol. Chem. 286, 28821–28832 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10. Jaiswal A. K. (2004) Nrf2 signaling in coordinated activation of antioxidant gene expression. Free Radic. Biol. Med. 36, 1199–1207 [DOI] [PubMed] [Google Scholar]
- 11. Zhang D. D. (2006) Mechanistic studies of the Nrf2-Keap1 signaling pathway. Drug Metab. Rev. 38, 769–789 [DOI] [PubMed] [Google Scholar]
- 12. Kobayashi M., Yamamoto M. (2006) Nrf2-Keap1 regulation of cellular defense mechanisms against electrophiles and reactive oxygen species. Adv. Enzyme Regul. 46, 113–140 [DOI] [PubMed] [Google Scholar]
- 13. Copple I. M., Goldring C. E., Kitteringham N. R., Park B. K. (2008) The Nrf2-Keap1 defence pathway: role in protection against drug-induced toxicity. Toxicology 246, 24–33 [DOI] [PubMed] [Google Scholar]
- 14. Enomoto A., Itoh K., Nagayoshi E., Haruta J., Kimura T., O'Connor T., Harada T., Yamamoto M. (2001) High sensitivity of Nrf2 knockout mice to acetaminophen hepatotoxicity associated with decreased expression of ARE-regulated drug metabolizing enzymes and antioxidant genes. Toxicol. Sci. 59, 169–177 [DOI] [PubMed] [Google Scholar]
- 15. Chan K., Han X. D., Kan Y. W. (2001) An important function of Nrf2 in combating oxidative stress: detoxification of acetaminophen. Proc. Natl. Acad. Sci. U.S.A. 98, 4611–4616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rangasamy T., Guo J., Mitzner W. A., Roman J., Singh A., Fryer A. D., Yamamoto M., Kensler T. W., Tuder R. M., Georas S. N., Biswal S. (2005) Disruption of Nrf2 enhances susceptibility to severe airway inflammation and asthma in mice. J. Exp. Med. 202, 47–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Iizuka T., Ishii Y., Itoh K., Kiwamoto T., Kimura T., Matsuno Y., Morishima Y., Hegab A. E., Homma S., Nomura A., Sakamoto T., Shimura M., Yoshida A., Yamamoto M., Sekizawa K. (2005) Nrf2-deficient mice are highly susceptible to cigarette smoke-induced emphysema. Genes Cells 10, 1113–1125 [DOI] [PubMed] [Google Scholar]
- 18. Hu X., Roberts J. R., Apopa P. L., Kan Y. W., Ma Q. (2006) Accelerated ovarian failure induced by 4-vinyl cyclohexene diepoxide in Nrf2 null mice. Mol. Cell. Biol. 26, 940–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Aoki Y., Sato H., Nishimura N., Takahashi S., Itoh K., Yamamoto M. (2001) Accelerated DNA adduct formation in the lung of the Nrf2 knockout mouse exposed to diesel exhaust. Toxicol. Appl. Pharmacol. 173, 154–160 [DOI] [PubMed] [Google Scholar]
- 20. Ramos-Gomez M., Kwak M. K., Dolan P. M., Itoh K., Yamamoto M., Talalay P., Kensler T. W. (2001) Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor-deficient mice. Proc. Natl. Acad. Sci. U.S.A. 98, 3410–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Iida K., Itoh K., Kumagai Y., Oyasu R., Hattori K., Kawai K., Shimazui T., Akaza H., Yamamoto M. (2004) Nrf2 is essential for the chemopreventive efficacy of oltipraz against urinary bladder carcinogenesis. Cancer Res. 64, 6424–6431 [DOI] [PubMed] [Google Scholar]
- 22. Wakabayashi N., Itoh K., Wakabayashi J., Motohashi H., Noda S., Takahashi S., Imakado S., Kotsuji T., Otsuka F., Roop D. R., Harada T., Engel J. D., Yamamoto M. (2003) Keap1-null mutation leads to postnatal lethality due to constitutive Nrf2 activation. Nat. Genet. 35, 238–245 [DOI] [PubMed] [Google Scholar]
- 23. Kwak M. K., Wakabayashi N., Itoh K., Motohashi H., Yamamoto M., Kensler T. W. (2003) Modulation of gene expression by cancer chemopreventive dithiolethiones through the Keap1-Nrf2 pathway. Identification of novel gene clusters for cell survival. J. Biol. Chem. 278, 8135–8145 [DOI] [PubMed] [Google Scholar]
- 24. Padmanabhan B., Tong K. I., Ohta T., Nakamura Y., Scharlock M., Ohtsuji M., Kang M. I., Kobayashi A., Yokoyama S., Yamamoto M. (2006) Structural basis for defects of Keap1 activity provoked by its point mutations in lung cancer. Mol. Cell 21, 689–700 [DOI] [PubMed] [Google Scholar]
- 25. Singh A., Misra V., Thimmulappa R. K., Lee H., Ames S., Hoque M. O., Herman J. G., Baylin S. B., Sidransky D., Gabrielson E., Brock M. V., Biswal S. (2006) Dysfunctional KEAP1-NRF2 interaction in non-small-cell lung cancer. PLoS Med. 3, e420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vollrath V., Wielandt A. M., Iruretagoyena M., Chianale J. (2006) Role of Nrf2 in the regulation of the Mrp2 (ABCC2) gene. Biochem. J. 395, 599–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kim Y. J., Ahn J. Y., Liang P., Ip C., Zhang Y., Park Y. M. (2007) Human prx1 gene is a target of Nrf2 and is up-regulated by hypoxia/reoxygenation: implication to tumor biology. Cancer Res. 67, 546–554 [DOI] [PubMed] [Google Scholar]
- 28. Okawa H., Motohashi H., Kobayashi A., Aburatani H., Kensler T. W., Yamamoto M. (2006) Hepatocyte-specific deletion of the keap1 gene activates Nrf2 and confers potent resistance against acute drug toxicity. Biochem. Biophys. Res. Commun. 339, 79–88 [DOI] [PubMed] [Google Scholar]
- 29. Wang X. J., Sun Z., Villeneuve N. F., Zhang S., Zhao F., Li Y., Chen W., Yi X., Zheng W., Wondrak G. T., Wong P. K., Zhang D. D. (2008) Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of Nrf2. Carcinogenesis 29, 1235–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Goel A., Aggarwal B. B. (2010) Curcumin, the golden spice from Indian saffron, is a chemosensitizer and radiosensitizer for tumors and chemoprotector and radioprotector for normal organs. Nutr. Cancer 62, 919–930 [DOI] [PubMed] [Google Scholar]
- 31. Cory S., Adams J. M. (2002) The Bcl2 family: regulators of the cellular life-or-death switch. Nat. Rev. Cancer 2, 647–656 [DOI] [PubMed] [Google Scholar]
- 32. Danial N. N., Korsmeyer S. J. (2004) Cell death: critical control points. Cell 116, 205–219 [DOI] [PubMed] [Google Scholar]
- 33. Boise L. H., González-García M., Postema C. E., Ding L., Lindsten T., Turka L. A., Mao X., Nuñez G., Thompson C. B. (1993) bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell 74, 597–608 [DOI] [PubMed] [Google Scholar]
- 34. Aravind L., Dixit V. M., Koonin E. V. (2001) Apoptotic molecular machinery: vastly increased complexity in vertebrates revealed by genome comparisons. Science 291, 1279–1284 [DOI] [PubMed] [Google Scholar]
- 35. Muchmore S. W., Sattler M., Liang H., Meadows R. P., Harlan J. E., Yoon H. S., Nettesheim D., Chang B. S., Thompson C. B., Wong S. L., Ng S. L., Fesik S. W. (1996) X-ray and NMR structure of human Bcl-xL, an inhibitor of programmed cell death. Nature 381, 335–341 [DOI] [PubMed] [Google Scholar]
- 36. Chao D. T., Linette G. P., Boise L. H., White L. S., Thompson C. B., Korsmeyer S. J. (1995) Bcl-XL and Bcl-2 repress a common pathway of cell death. J. Exp. Med. 182, 821–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Calvert J. W., Jha S., Gundewar S., Elrod J. W., Ramachandran A., Pattillo C. B., Kevil C. G., Lefer D. J., (2009) Hydrogen sulfide mediates cardioprotection through Nrf2 signaling. Circ. Res. 105, 365–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Niture S. K., Jaiswal A. K. (2009) Prothymosin-α mediates nuclear import of the INrf2/Cul3 Rbx1 complex to degrade nuclear Nrf2. J. Biol. Chem. 284, 13856–13868 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39. Lee O. H., Jain A. K., Papusha V., Jaiswal A. K. (2007) An auto-regulatory loop between stress sensors INrf2 and Nrf2 controls their cellular abundance. J. Biol. Chem. 282, 36412–36420 [DOI] [PubMed] [Google Scholar]
- 40. Niture S. K., Jaiswal A. K. (2011) INrf2 (Keap1) targets Bcl-2 degradation and controls cellular apoptosis. Cell Death Differ. 18, 439–451 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.