Background: PRMT1 is highly up-regulated in and plays an important role for developing adult intestinal stem cells during thyroid hormone-dependent Xenopus metamorphosis.
Results: Liganded thyroid hormone receptor activates c-Myc transcription. c-Myc, in turn, activates PRMT1.
Conclusion: Thyroid hormone up-regulates PRMT1 indirectly via c-Myc.
Significance: The finding reveals an important gene regulation pathway by which thyroid hormone controls stem cell development.
Keywords: Intestine, Stem Cells, Thyroid Hormone, Transcription Coactivators, Xenopus, PRMT1, Adult Stem Cells, Metamorphosis, Thyroid Hormone Receptor
Abstract
Adult organ-specific stem cells are essential for organ homeostasis and tissue repair and regeneration. The formation of such stem cells during vertebrate development is poorly understood. Intestinal remodeling during thyroid hormone (T3)-dependent Xenopus metamorphosis resembles postembryonic intestinal maturation in mammals. During metamorphosis, the intestine is remodeled de novo via a yet unknown mechanism. Protein arginine methyltransferase 1 (PRMT1) is up-regulated in and required for adult intestinal stem cells during metamorphosis. PRMT1 up-regulation is the earliest known molecular event for the developing stem cells and is also conserved during zebrafish and mouse intestinal development. To analyze how PRMT1 is specifically up-regulated during the formation of the adult intestinal stem cells, we cloned the Xenopus PRMT1 promoter and characterized it in CaCo-2 cells, a human cell line with intestinal stem cell characteristics. Through a series deletion and mutational analyses, we showed that the stem cell-associated transcription factor c-Myc could bind to a conserved site in the first intron to activate the promoter. Furthermore, we demonstrated that during metamorphosis, both c-Myc and PRMT1 were highly up-regulated, specifically in the remodeling intestine but not the resorbing tail, and that c-Myc was induced by T3 prior to PRMT1 up-regulation. In addition, we showed that T3 directly activated the c-Myc gene during metamorphosis in the intestine via binding of the T3 receptor to the c-Myc promoter. These results suggest that T3 induces c-Myc transcription directly in the intestine, that c-Myc, in turn, activates PRMT1 expression, and that this is an important gene regulation cascade controlling intestinal stem cell development.
Introduction
Adult stem cells have been attracting increasing attention because of their potential applications in tissue-replacement therapies. The intestinal epithelial stem cells have been studied extensively largely because of the constant turnover of the epithelium throughout adult life in vertebrates (1–5, 6). In adult mammals, the epithelial stem cells are localized in the crypts. As they divide, the daughter cells migrate along the crypt-villus axis and gradually differentiate into different types of epithelial cells. At the tip of the villus, the epithelial cells undergo apoptosis and are replaced by the newly arrived differentiated epithelial cells. Despite extensive studies on the epithelial self-renewal from the stem cells, much less is known about how such adult stem cells are formed during development, in part because of the difficulties to study the uterus-enclosed mammalian embryogenesis.
Intestinal remodeling during amphibian metamorphosis offers a unique opportunity to study the development of adult organ-specific stem cells in vertebrates. Amphibian metamorphosis is totally dependent on thyroid hormone (T3)2 and involves drastic changes in essentially every organ of the animal (7, 8). In the South African clawed toad Xenopus laevis, the tadpole intestine consists of largely a monolayer of larval epithelial cells with little connective tissue and muscles (9). During metamorphosis, the larval epithelial cells undergo apoptosis, and, concurrently, adult epithelial stem/progenitor cells appear de novo and proliferate rapidly (9–11). These adult epithelial cells subsequently differentiate to establish a trough-crest axis of epithelial folds by the end of metamorphosis, resembling the crypt-villus axis in the adult mammalian intestine, accompanied by the development of the connective tissue and muscles (9). Just like other processes during metamorphosis, all these changes during intestinal remodeling are controlled by T3 and can be induced even in organ cultures treated with T3 (10, 12, 13), indicating organ-autonomous formation of adult stem/progenitor cells. Interestingly, chronological observations of intestinal metamorphosis (14), and the apparent presence of the proteins of the differentiated epithelial cells in proliferating adult progenitor cells (11, 15) suggests that some differentiated larval epithelial cells may undergo T3-dependent dedifferentiation to become the stem cells during metamorphosis. This was shown conclusively by recent recombinant organ culture studies with intestines from wild-type and transgenic tadpoles expressing GFP (10).
T3 controls metamorphosis by regulating transcription through T3 receptors (TRs) (16–25). TRs form heterodimers with 9-cis retinoic acid receptors, and these dimers bind to T3 response element (TRE) in/around the promoters of T3 target genes (26–29). In the absence of T3, TR/ retinoic acid receptor heterodimers function as repressors by recruiting corepressor complexes, and in the presence of T3, they act as activators by recruiting coactivator complexes (21, 23, 24, 27, 30–45). We have shown recently that protein arginine methyltransferase 1 (PRMT1) plays an important role for gene activation and metamorphosis induced by liganded TR (41, 46). Interestingly, PRMT1 expression is up-regulated in the intestine during both natural and T3-induced metamorphosis (41). Spatiotemporal analyses suggest that at early stages of metamorphosis, PRMT1 is up-regulated in the larval epithelial cells that are destined to become adult epithelial stem cells during metamorphosis (46). More importantly, transgenic studies have shown that overexpression of PRMT1 leads to an increased number of intestinal stem cells during metamorphosis, whereas knocking down endogenous PRMT1 expression reduces the stem cell populations (46), supporting an important role of PRMT1 for the adult intestinal stem cells.
Intestinal metamorphosis resembles intestinal maturation during mammalian postembryonic development, a period around birth in mammals when plasma T3 concentrations are high (5, 6, 47). Furthermore, mammalian intestinal maturation also appears to be dependent on T3 because T3 or TR deficiency leads to abnormal intestinal morphology, a decrease in the number of epithelial cells along the crypt-villus axis and in proliferating crypt cells (48–52). Interestingly, PRMT1 has a similar spatiotemporal expression pattern during postembryonic intestinal development in mouse and zebrafish as that during Xenopus metamorphosis (46). Thus, although neonatal mouse intestine is structurally developed, with the proper crypt-villus organization similar to that in the adult mouse, the embryonic/neonatal mouse intestinal stem cells are molecularly distinct from those in the adult mouse intestine (5, 6, 46). This conclusion is also supported by recent studies on the transcription repressor, B lymphocyte-induced maturation protein 1 (Blimp1) (53, 54). Blimp1 is strongly expressed throughout the intestinal epithelium before birth. During postembryonic development after birth, as the plasma T3 level rises, Blimp1 expression becomes down-regulated only in the intervillus pockets where the embryonic/neonatal intestinal stem cells reside, whereas the other epithelial cells (suckling-type cells) continue to express Blimp1. Eventually, Blimp1 expression is repressed in the entire epithelium as the adult epithelium replaces the neonatal/embryonic intestine because of the proliferation of the Blimp1-negative stem cells. Thus, adult intestinal stem cell formation is conserved during T3-dependent postembryonic development in vertebrates (5, 6).
Given the conserved spatiotemporal expression pattern of PRMT1 during adult intestinal stem cell development and the fact that PRMT1 up-regulation is one of the earliest events in the dedifferentiation of the larval epithelial cells during Xenopus metamorphosis, understanding its regulation by T3 will provide important insight into how T3 induces the formation of the stem cells. Here, we cloned and characterized the Xenopus PRMT1 promoter. Our studies revealed that T3 up-regulates PRMT1 by inducing directly the expression of c-Myc, a transcription factor known to be associated with stem cells and cell proliferation.
EXPERIMENTAL PROCEDURES
Animals
Wild-type tadpoles of Xenopus laevis and Xenopus tropicalis were purchased from Nasco (Fort Atkinson, WI). All animals were maintained and used in accordance with the guidelines established by the National Institute of Child Health and Human Development Animal Use and Care Committee. Developmental stages were assigned according to Ref. 55. Stage 54 X. laevis and X. tropicalis tadpoles were treated with 10 nm T3 for the indicated number of days at 18 °C and 22 °C, respectively.
Isolation of Genomic Clones
The transcription start site was determined by a 5′ rapid amplification of cDNA (5′-RACE) (SMART RACE cDNA amplification kit, Clontech). A homozygous diploid X. laevis genomic λ library in the λ GEM-11 vector (56) was used for PCR amplification of the genomic DNA flanking the start site of transcription (+1 position). First, the first intron of X. laevis PRMT1 gene was cloned by using exon1d forward primer KF19 and exon 2 reverse primer KF20. Then the KF17 forward primer for the λ GEM11 vector arm and PRMT1 KF24 reverse primer were used to clone the upstream of the transcription start site. After DNA sequences were determined, the 6 kb (-3092 to +3001) was PCR-amplified from X. laevis wild-type genomic DNA with primers KF73 (bearing KpnI at its 5′ end) and KF71 (bearing BamHI at its 5′ end) (supplemental Table 1), and ligated into KpnI-BamHI-digested pBluescript II KS+ (Agilent Technologies/Stratagene, Santa Clara, CA).
The complete nucleotide sequence of X. laevis PRMT1 genomic DNA obtained in this work has been submitted to GenBankTM (accession no. JQ302819).
Construction of Luciferase Reporter Vectors
The 6-kb (-3092 to +3001) of X. laevis PRMT1 in KpnI-BamHI-digested pBluescript II KS+ was subcloned into a pGL4.10 luciferase vector (Promega, Madison, WI) at the KpnI and BglII sites to construct −3092/+3001-PRMT1-Luc.
The first intron was deleted from the construct −3092/+3001-PRMT1-Luc using KF107 and KF106 to generate −3092/+3001 (Δ+201/+2861)-PRMT1-Luc, KF107 and KF30 for −3092/+3001 (Δ+201/+553)-PRMT1-Luc, and KF26 and KF106 for −3092/+3001 (Δ+554/+2861)-PRMT1-Luc.
A serial deletion construct from the 5′ end of −3092/+3001-PRMT1-Luc and 3092/+3001 (Δ+201/+2861)-PRMT1-Luc were created by PCR using different forward primers and the reverse primer KF92 for the pGL4.10 vector. The forward primers included KF50 for construct −1504, KF105 for −930, KF57 for −660, KF110 for −475, KF111 for −290, KF112 for −145, KF109 for −40, KF108 for −24, and KF101 for +1 (supplemental Table 1).
The internal deletions (Δ-660/-291, Δ-290/-146, and Δ-145/-41) in the promoter region were generated from −3092/+3001-PRMT1-Luc and −3092/+3001 (Δ+201/+2861)-PRMT1-Luc using KF113 and KF111 for the −660/-291 deletion, KF115 and KF112 for the −290/-146 deletion, and KF116 and KF109 for the −145/-41 deletion, respectively.
The c-Myc binding site in the first intron was mutated from the construct −3092/+3001-PRMT1-Luc by using primers KF169 and KF170 to generate −3092/+3001 (Myc-Mut).
Transfection and Luciferase Assays
The human CaCo-2 cell line was purchased from the ATCC and was maintained in Eagle's minimum essential medium (ATCC) supplemented with 20% fetal bovine serum (ATCC). The cells were grown at 37 °C in the presence of 5% CO2. CaCo-2 cells in 24-well plates were transfected with the reporter firefly luciferase plasmid DNA and control Renilla luciferase plasmid phRG-TK (Promega) using FuGENE HD (Roche) according to the instructions of the manufacturer. After transfection, the cells were grown for 24 h, and firefly and Renilla luciferase assays were measured on cell lysates using the dual luciferase assay system (Promega). The ratio of the two was reported as the activity of the reporter construct.
Transcription assay in X. laevis Oocytes and Western Blot Analysis
The transcription assay in the X. laevis oocytes was carried out as described previously (57). To express X. laevis c-Myc protein in the oocyte, X. laevis c-Myc cDNA was cloned into the pSP64 vector (Promega) at the HindIII and BamHI sites using KF198 and KF199 primers, and the expression plasmid was used to synthesize c-Myc mRNA with the mMESSAGE mMACHINE in vitro SP6 transcription system (Applied Biosystems/Ambion). The mRNA (1.15 ng/oocyte) was microinjected into the cytoplasm. Four hours later, the −3092/+3001-PRMT1-Luc (0.33 ng/oocyte) reporter and the control vector phRG-tk (0.03 ng/oocyte) were coinjected into the oocyte nucleus. After incubation at 18 °C overnight, lysates from the injected oocytes were prepared for dual luciferase assay. Eighteen identically injected oocytes for each sample were divided into three groups, and a luciferase assay was done for each group. Data present the average of three groups. A portion of the lysate was used for Western blotting with anti-c-Myc (N-262, Santa Cruz Biotechnology, Inc., Santa Cruz, CA) and anti-β-actin (A1978, Sigma-Aldrich, St. Louis, MO) antibodies.
ChIP Assay
ChIP assays on X. laevis oocytes were done as described previously (41) by using anti-c-Myc antibody (Santa Cruz Biotechnology, Inc.). The immunoprecipitated DNA was analyzed by quantitative PCR performed with gene-specific primer and Taqman probe sets for the PRMT1 c-Myc biding region (5′-GGCGCGTCTGTTCTGAAGAA-3′ (forward) and 5′-CTTTGTGCTACCGGCATGTG-3′ (reverse) and the carboxyfluorescein-labeled Taqman probe 5′-CAGCGGCCACGTGTTC-3′; and the ampicillin resistance gene 5′-GGCCGCAAATGCTAAACCA-3′ (forward) and 5′-CGAAATAGGCAGATCGCTGAGAT-3′ (reverse) and the carboxyfluorescein-labeled Taqman probe 5′-CAAGCACTGGTAACCAC-3′).
ChIP assays on the intestine and tail from X. tropicalis tadpoles were performed as described previously (58). Anti-TR (new PB) antibody was used for TR ChIP (59). Antibody against ID14, an extracellular protein (60), was used as a negative control. The immunoprecipitated DNA was analyzed by quantitative PCR with gene-specific primer sets for the TRE region (5′-CCACCCATCACCCTTTATCCTTTAA-3′ (forward) and 5′-GTTCCCATAGTTTAGGCTTGAGTGA-3′ (reverse) and FAM-labeled Taqman probe 5′-CCTCTACTGACCCAAAGAA-3′, and for exon 3 of X. tropicalis c-Myc gene (5′-CCAGGGTCCTCAAACAGATCA-3′ (forward) and 5′-CTTGTCGTTCTCTTCGGAATCAGA-3′ (reverse) and FAM-labeled Taqman probe 5′-CACTTGCGGTTATTGC-3′).
Quantitative RT-PCR
Total RNA was isolated from the tadpole intestine and tail. First-strand cDNA was prepared from 1 μg of total RNA using the Applied Biosystems high capacity cDNA archive kit, resulting in 20 μl of cDNA solution. Two μl of a 1/10 cDNA dilution were used for each reaction, and EF1α (elongation factor 1α) was analyzed at the same time as the reference control by using SYBR Green quantitative PCR. The KF330 and KF331 primers for PRMT1 were used for SYBR Green PCR. The primers for EF1α and c-Myc were as published (61, 62).
RESULTS
Genomic Organization of the Xenopus PRMT1 Gene
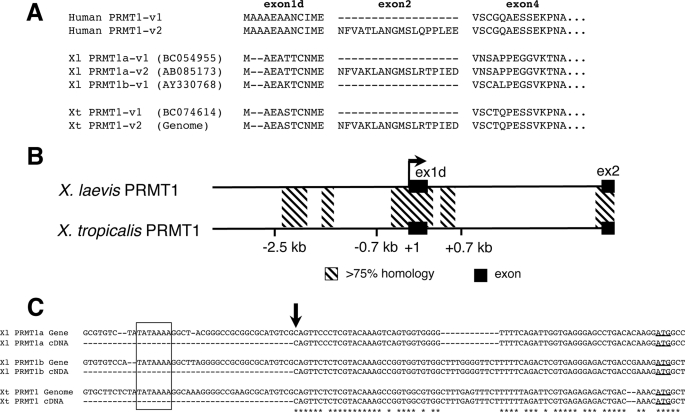
There are two PRMT1 genes in the pseudo-tetraploid X. laevis genome that are highly conserved (63, 64). Two X. laevis PRMT1 genes (originally named xPRMT1 and xPRMT1b, referred here as Xl PRMT1a and 1b) share 94 and 97% identity at the nucleotide and amino acid sequence levels, respectively. To understand the transcriptional regulation of X. laevis PRMT1 genes, we first determined their genomic structures. It has been reported that the human PRMT1 gene has a complex genomic organization at the 5′ end that can produce up to seven splicing variants (v1-v7) from two putative transcription start sites (65). By searching online databases, we found two transcripts of Xl PRMT1a, one of Xl PRMT1b, corresponding to the major splicing variants of human PRMT1 (v1 and v2) produced through alternative splicing of exon 2 and 3 (Fig. 1A). In X. tropicalis, a highly related species, one transcript (v1) was found from cDNA database searches, and the second variant (v2) was obtained on the basis of the genomic sequence (the Joint Genome Institute) and 5′-RACE analysis (Fig. 1A). No additional X. laevis and X. tropicalis PRMT1 isoforms were found in any of the expressed sequence tags databases.
FIGURE 1.
Organization of the Xenopus PRMT1 genes. A, alignment of the deduced N-terminal amino acid sequences of PRMT1 variant 1 (v1) and variant 2 (v2) from human, X. laevis (Xl), and X. tropicalis (Xt). The exons were named on the basis of human PRMT1. There are two duplicated genes in X. laevis that are referred here as Xl PRMT1a and Xl PRMT1b. The GeneBanktm accession numbers of for the Xenopus proteins are shown. Xt PRMT1-v2 was derived from the genomic sequence and 5′-RACE. Note that v-3 transcripts, which include exon 3, were not found in expressed sequence tags and 5′-RACE in X. laevis and X. tropicalis, and exon 3 of the X. tropicalis PRMT1 gene could not be identified in the genome. B, comparison of genomic DNA surrounding the promoter and first intron in the X. laevis PRMT1a gene and the X. tropicalis PRMT1 genes. The transcription start site for X. laevis PRMT1 is set to +1. The black boxes represent exons. The arrow shows the direction of transcription. The conserved regions in the 5′ flanking region and first intron are indicated with hatched bars. C, comparison of X. laevis and X. tropicalis PRMT1 sequences around the transcription start site (arrow). The cDNA sequences obtained by 5′-RACE are aligned with the genomic sequences. The boxed region shows the predicted TATA box. The initiation (ATG) codon is underlined. Note that the 5′-RACE found the transcription start site at the same location for X. laevis and X. tropicalis PRMT1 genes.
By using PCR-based cloning, we obtained two X. laevis PRMT1 genomic clones corresponding to the two PRMT1 genes (Fig. 1, B and C). The two genomic clones, 6 kb for Xl PRMT1a gene and 1 kb for Xl PRMT1b, are highly homologous (data not shown), and for our promoter analysis, we focused on the PRMT1a gene and simply referred to it as the PRMT1 gene. We also obtained X. tropicalis PRMT1 genomic sequences from the Joint Genome Institute database. We did not find the sequence corresponding to the exon 3 of human PRMT1 in the X. tropicalis genome or in the expressed sequence tags database or by 5′-RACE cloning.
The initiation site for the transcription was determined by 5′-RACE. The results revealed a single transcription start site (+1) at the same position in the X. laevis and X. tropicalis PRMT1 gene (Fig. 1C). In addition, a putative TATA box was found at −27 bp upstream of the transcription start site (Fig. 1C). Thus, the Xenopus PRMT1 promoter, or at least the major one, is upstream of this first exon, corresponding to human PRMT1 exon 1d, just like the human gene.
Both X. laevis and X. tropicalis PRMT1 genes have similar expression profiles in the intestine during development (Ref. 41 and data not shown), suggesting conserved transcriptional regulation mechanisms in X. laevis and X. tropicalis, just like all other genes that we and others have studied during metamorphosis (58, 59, 61, 66, 67). Alignment of PRMT1 genomic sequences of X. laevis and X. tropicalis PRMT1 revealed several conserved segments with over 75% identity in the upstream regions and the first intron (Fig. 1B). The splicing donor and acceptor region in the first intron were also highly conserved (Fig. 1B).
The Cis-regulatory Elements Located in the Proximal Promoter and the First Intron of the X. laevis PRMT1 Gene Are Important for Promoter Activity
To characterize the PRMT1 promoter, we placed the full-length promoter encompassing the genomic region from −3092 to +3001, including part of exon 2 (including 27 bp of exon 2), in front of a luciferase reporter gene in the pGL4.10 vector, resulting in the plasmid −3092/+3001 Xl PRMT1-Luc. To investigate the promoter activity, we chose a human intestinal epithelial cell line, CaCo-2, for transient transfection assays because these cells express PRMT1 (68) and have intestinal stem cell-like characteristics. First, CaCo-2 cells were cotransfected with the plasmid −3092/+3001 Xl PRMT1-Luc and the internal control plasmid phRG-TK, which contains the Renilla luciferase under the control of thymidine kinase promoter, and treated cells with 100 nm T3. The result demonstrated that the PRMT1 promoter was functional in CaCo-2 cells and that its activity was not altered by 1- to 3-day T3 treatment (data not shown). This is consistent with fact that PRMT1 is not a direct target gene of T3, as reflected by its slow T3 induction during metamorphosis (41) as well as with our observation that the expression of endogenous human PRMT1 mRNA in Caco-2 was also not altered by T3 (data not shown).
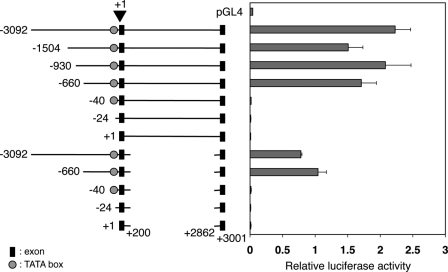
We next generated a series of deletion mutant constructs of −3092/+3001 Xl PRMT1-Luc and analyzed their activities in CaCo-2 cells. As shown in Fig. 2, 5′ deletions up to −660 had little effect on the promoter activity. On the other hand, the activity was completely lost upon deletion from −660 to −41. These results indicate that the distal conserved elements beyond −660 are not important for the promoter activity, whereas the proximal conserved region upstream of the TATA box is essential for the promoter.
FIGURE 2.
Deletion analyses reveal important cis-regulatory elements in the proximal promoter and first intron of the X. laevis PRMT1 gene. CaCo-2 cells were transiently cotransfected with the indicated constructs and the control plasmid phRG-tk. The cells were incubated for 24 h and lysed for dual luciferase assays. The ratio of the firefly luciferase activity to that of the Renilla luciferase was determined as a measure of the PRMT1 promoter activity. Each transfection was performed in duplicate and repeated three times. The error bars indicate mean ± S.E. Note that the activity was completely lost upon deletion to −41 or further from the 5′ end, and the removal of the first intron led to a 2-fold reduction in promoter activity.
To examine the role of the first intron, we made two different types of the first intron deletion constructs. One type retained about 100 bp of 5′-end and 3′-end of the first intron to ensure proper splicing, and the other lacked the first intron or second exon. We confirmed the proper splicing of the first type of constructs after transfection in CaCo-2 cells by RT-PCR (data not shown). When the first intron was deleted, the promoter activities of all constructs were decreased by about 2-fold in the comparison to the corresponding constructs with the first intron (Fig. 2). Similarly, when the entire intron and exon 2 were deleted, the promoter activity was similarly reduced compared with their corresponding promoter constructs (data not shown). This result indicates that the first intron, but not exon 2 or the slicing donor and acceptor regions, is important for the promoter function.
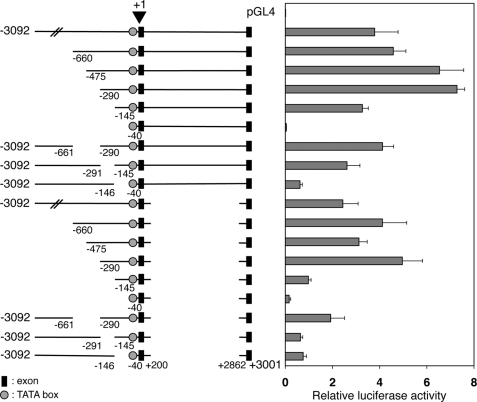
To narrow down the cis-regulatory elements important for the promoter activity, we made additional sequential deletions from the 5′ end to −475, −290, and −145, as well as internal deletions. We generated constructs with these deletions with or without the deletion of the first intron as above. Transfection studies showed that the transcriptional activities were reduced upon deletion from −290 to −146 or from −145 to −41 (Fig. 3). In addition, internal deletion of the region between −290 and −146 or between −145 and −41 yielded a similar outcome, whereas the internal deletion of −660 to −291 had little effect. Thus, two upstream regions, from −290 to −146 and from −145 to −41, are required for promoter function. The same conclusion was reached when these deletions were done in the presence or absence of the first intron, although the absolute promoter activities were lower in the absence of the intron, as found above (Fig. 3). Interestingly, these two regions fall within the proximal conserved region between the X. laevis and X. tropicalis PRMT1 genes (Fig. 1B).
FIGURE 3.
Identification of two regions located at −290/-146 and −145/-41 is important for the promoter activity of the X. laevis PRMT1 gene. The deletion constructs were analyzed by transfection as described in Fig. 2. Note that the promoter activity was dramatically reduced upon deletion between −290 to −146 or −145 to −41. In addition, an internal deletion between −290 to −146 or −145 to −41 but not −661 to −290 also reduced the promoter activity. Similar results were obtained in the presence or absence of the intron sequence.
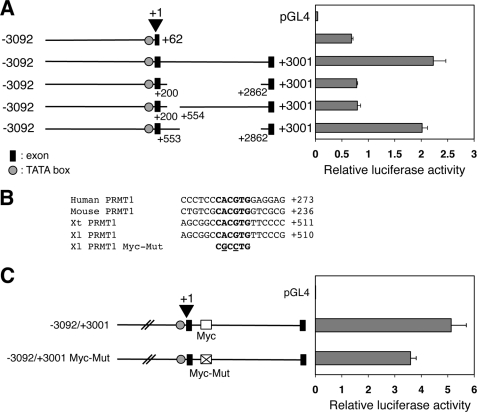
The data above showed that the first intron enhanced the transcriptional activity by about 2-fold in various promoter constructs. To determine whether the activity was due to the conserved region between X. laevis and X. tropicalis PRMT1 within the intron, we next generated internal deletion mutants in the intron by either specifically deleting the conserved region from +201 to +553 or retaining it but deleting most of the rest of the intron (+554 to +2861) (Fig. 4A). Transfection studies showed that the promoter activity was decreased by about 2-fold when the conserved region in the first intron was deleted, similar to the total removal of the intron (Fig. 4A). On the other hand, when the conserved region from +201 to +553 in the first intron was retained but the downstream region from +554 to +2861 was deleted, the activity was comparable with that of the full-length promoter (Fig. 4A). Thus, the conserved region between X. laevis and X. tropicalis in the first intron functions as an enhancer for the PRMT1 promoter.
FIGURE 4.
A c-Myc binding site in the first intron is important for the promoter of X. laevis PRMT1 gene. A, deletion analysis identified the region from +200 to +554 in the first intron as important for the activity of the PRMT1 promoter. The constructs were analyzed as in Fig. 2. B, a conserved c-Myc binding site is present in the first intron of the PRMT1 gene in different vertebrate species. Putative c-Myc binding sites in the first intron of the human, mouse, X. tropicalis, and X. laevis PRMT1 genes were aligned and are shown in boldface. At the bottom were mutations (underlined residues) that inactivate the c-Myc site. Note that the numbers for human and mouse PRMT1 genes were relative to the initiation codon in exon 1d because the exact start site has not been identified. C, mutation of the c-Myc binding site reduces the activity of the PRMT1 promoter. The wild-type and c-Myc binding site mutant (as shown in B) were analyzed as in Fig. 2.
A c-Myc Binding Site within the First Intron Is Important for its Enhanced Activity
To identify potential transcription factors that might contribute to the PRMT1 expression in the adult intestinal stem cells, we searched for putative transcription factor binding sites in the genomic regions shown above to be important for PRMT1 promoter activity by using the rVISTA 2.0 (69). We reasoned that, given the conserved expression pattern of PRMT1 during intestinal development, the important factors should be evolutionally conserved among the vertebrate species. Thus, we also searched for putative transcription factor binding sites in the region from the first intron to up to 3 kb upstream of the start site of the zebrafish PRMT1 gene as well as the regions that are conserved between the mouse and human PRMT1 gene (Fig. 4B). The analysis identified putative binding sites for transcription factors E2F, NF-Y, ccAAT box binding proteins, and SMAD in the proximal conserved promoter region of all PRMT1 genes. In the conserved region of the first intron, there were the putative sites for E2F, cAMP response element-binding protein, SP1, early growth proteins, upstream stimulatory factor, GATA2, activating transcription factor, MYC, Hypoxia-inducible factor 1, clock, and ZF5 (data not shown). Although some of these binding sites may play a role in the promoter function, we were particularly interested in the presence of a conserved binding site for the transcription factor c-Myc. First, c-Myc is a critical activator of cell proliferation and one of the genes most commonly deregulated in cancer (70, 71). Second, it is expressed in different stem cells and is also one of four factors that, when coexpressed, reprogram somatic cells into pluripotent stem cells (72, 73), supporting a likely role in regulating PRMT1 expression during intestinal stem cell development. Finally, our data (see below) indicated that c-Myc is induced during intestinal metamorphosis. Thus, we focused our analysis on the role of c-Myc in PRMT1 regulation.
First, we mutagenized the putative c-Myc binding site and transfected the wild-type and mutant promoter constructs into CaCo-2 cells. The results showed that mutation of the c-Myc binding site in the first intron of X. laevis PRMT1 significantly reduced the promoter activity (Fig. 4C).
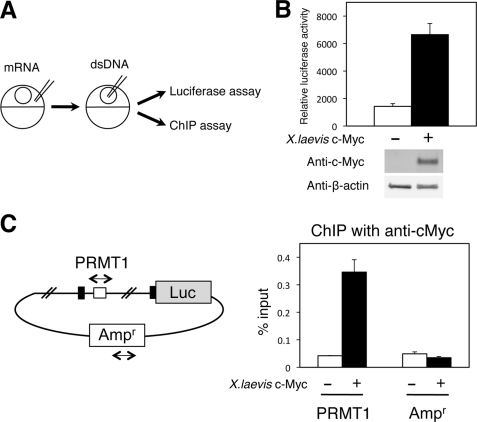
Next, we investigated whether c-Myc can bind to the PRMT1 intron region in vivo. We first used an in vivo ChIP assay to determine the binding of c-Myc to the endogenous gene during metamorphosis. Unfortunately, we could not detect a significant signal with the only commercial antibody available. This was likely due to the fact that only a small number of the cells in the intestine were stem cells that expressed high levels of PRMT1 during metamorphosis, making it difficult to detect c-Myc binding to the PRMT1 gene in the stem cells in the presence of a vast excess of non-stem cells that lacked c-Myc binding to the PRMT1 gene. Thus, we turned to the frog oocyte transcription system, which allows one to study the role of transcription factor function in the context of chromatin (57, 74). To examine the effect of c-Myc on the PRMT1 promoter, we microinjected the mRNA encoding X. laevis c-Myc into the frog oocyte followed by the injection of reporter plasmids (PRMT1-Luc and the control Renilla luciferase vector) (Fig. 5A). Overexpression of c-Myc enhanced luciferase activity (Fig. 5B). When the injected oocytes were subjected to a ChIP assay with the anti-c-Myc antibody, the result clearly showed that c-Myc was bound to the intron region with the c-Myc binding site but not to the ampicillin-resistance gene on the same plasmid (Fig. 5C). Thus, c-Myc can bind to the intronic binding site to enhance the PRMT1 promoter.
FIGURE 5.
X. laevis c-Myc directly binds to the first intron of the X. laevis PRMT1 gene to activate the promoter in vivo. A, schematic diagram of the Xenopus oocyte transcription system. The mRNA encoding X. laevis c-Myc was injected into the cytoplasm. A few hours later, the double-stranded firefly luciferase reporter and the control Renilla luciferase vector (dsDNA) were injected into the nucleus. After overnight incubation, the oocytes were lysed for luciferase activity measurement, and ChIP assays were carried out. B, c-Myc activates the PRMT1 promoter. The full-length PRMT1 promoter construct was analyzed in the oocyte in the presence or absence of overexpressed c-Myc. A Western blot analysis was also performed on the same lysate with anti-c-Myc and anti-β-actin antibodies, indicating the expected expression of c-Myc in the injected oocytes (bottom panel). C, c-Myc is bound to the first intron of the PRMT1 gene. Oocytes as in B were subjected to a ChIP assay with a c-Myc antibody to determine the association of c-Myc with the first intron (PRMT1) as well as the ampicillin-resistant gene region (Ampr) in the same plasmid as a negative control. The black and white boxes show exons and the putative c-Myc biding site in the X. laevis PRMT1 gene. The immunoprecipitated DNA was analyzed by quantitative PCR.
c-Myc Is Induced by T3 prior to the Up-regulation of the PRMT1 Gene during Metamorphosis
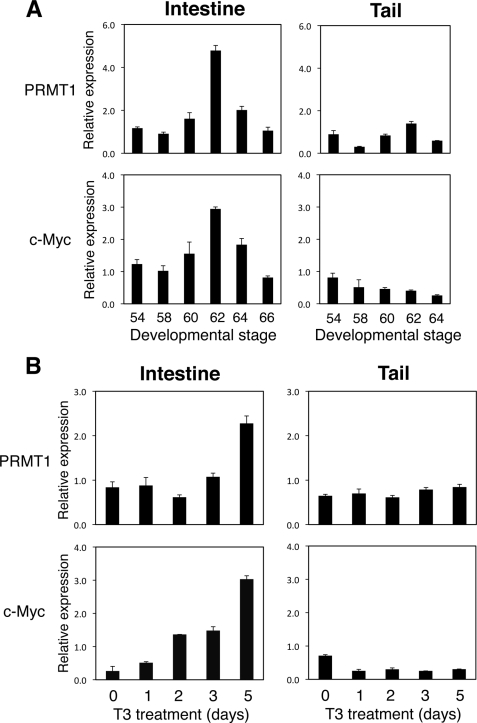
To investigate the role of c-Myc in PRMT1 expression during metamorphosis, we analyzed the expression of c-Myc in the intestine during natural and T3-induced metamorphosis (Fig. 6). For comparison, we also analyzed the tail, which undergoes complete resorption through apoptosis and does not involve stem cell development. As shown in Fig. 6A, both PRMT1 and c-Myc were expressed at low levels in premetamorphic intestine at stage 54. They were up-regulated during natural metamorphosis in the intestine, with their expression peaked at stage 62, the climax of metamorphosis when stem cell number is the highest (9). By the end of metamorphosis at stage 66, their expression returned to the low premetamorphic levels. In contrast, no up-regulation was observed for either gene in the tail during metamorphosis (Fig. 6A).
FIGURE 6.
c-Myc expression is up-regulated in the intestine during metamorphosis. Total RNA was isolated from the intestine and tail of tadpoles from premetamorphosis (stage 54), early metamorphic climax (stage 58), metamorphic climax (stage 60, 62, and 64), and end of metamorphosis (stage 66) (note that the tail is completely resorbed by stage 66 and thus no stage 66 columns for the tail samples) (A) or stage 54 tadpoles treated with 10 nm T3 for indicated numbers of days (B). The expression of PRMT1 and c-Myc was determined by quantitative RT-PCR. The expression was normalized with that of EF1α. Note that in the intestine, but not in the tail, both PRMT1 and c-Myc was up-regulated at the climax of metamorphosis (stage 62). The induction of PRMT1 in the intestine by T3 treatment, however, was delayed compared with that of c-Myc.
When premetamorphic tadpoles were treated with T3 to induce metamorphosis, c-Myc expression was up-regulated within 2 days in the intestine (Fig. 6B). On the other hand, PRMT1 expression was not induced until 5 days. In the tail, the expression of neither c-Myc nor PRMT1 was induced by T3 treatment, similar to that observed during natural metamorphosis (Fig. 6B). These results indicate that c-Myc is up-regulated by T3 before PRMT1 activation, suggesting that T3 induces c-Myc to enhance PRMT1 transcription.
T3 Activates c-Myc Directly via the Binding of TR to the c-Myc Gene during Metamorphosis
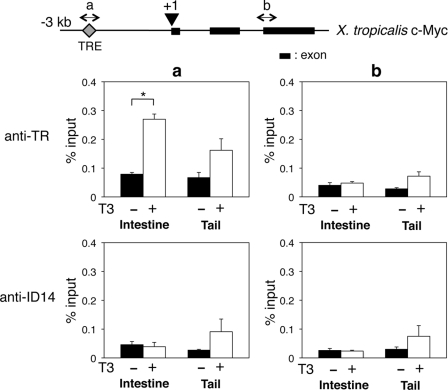
The up-regulation of c-Myc within 2 days of T3 treatment raises the possibility that c-Myc may be directly regulated by TR. To investigate this possibility, we took advantage of the conservation in gene regulation during metamorphosis between X. laevis and X.tropicalis, as the regulation of gene expression during metamorphosis is conserved between the two species for all genes analyzed so far (58, 59, 61, 66, 67). Thus, we searched for the putative TRE in the X. tropicalis c-Myc gene. A putative TRE was found at 2.4 kb upstream of X. tropicalis c-Myc gene transcription start site (Fig. 7). On the other hand, similar analyses failed to identify any TREs in the X. laevis and X. tropicalis PRMT1 genes.
FIGURE 7.
TR binds to the TRE of the X. tropicalis c-Myc gene in vivo. Top panel, schematic diagram of the c-Myc gene showing the presence of the TRE upstream of the transcription start site. The putative transcription start site is shown as +1, and a and b indicate the two regions analyzed by quantitative PCR for the ChIP assay. Bottom panel, tadpoles at stage 54 were treated with or without T3 for 2 days, and the intestine and tail were isolated for a ChIP assay with anti-TR and anti-ID14 (a negative control for specificity) antibodies. The immunoprecipitated DNA was analyzed by quantitative PCR for the presence of the TRE region (a) or a downstream intron region (b) (a negative control for binding specificity). Note that only background signals were found in the absence of T3 at the both the TRE and downstream regions (compared the anti-TR to anti-ID14 ChIP signals) in either the intestine or tail and that T3 treatment led to significant TR binding only to the TRE region in the intestine but not the tail. The error bars indicate mean ± S.E. *, pairs of samples with p < 0.05.
To investigate whether c-Myc is activated by T3 during metamorphosis via direct binding of TR to the putative TRE, we performed a ChIP assay with an anti-TR antibody on intestine and tail isolated from stage-54 premetamorphic X. tropicalis tadpoles treated with 10 nm T3 for 2 days. The results indicated that in the intestine, TR was bound to TRE of the X. tropicalis c-Myc gene after T3 treatment, although only a background signal was found in the absence of T3 (compared with the nonspecific antibody control against ID14, an extracellular protein) (Fig. 7). This binding was specific to the TRE, as no binding was observed in the downstream intron region (Fig. 7). By contrast, in the tail, no significant TR binding to the TRE was detected compared with the control ChIP with the ID14 antibody in the presence or absence of T3 treatment (Fig. 7), in agreement with the lack of T3 induction of the c-Myc gene in the tail during metamorphosis. These results suggest that T3 induces c-Myc directly in the intestine through the direct binding of TR to the TRE.
DISCUSSION
PRMT1 is a known coactivator for TR (41, 75). In our studies on the role of TR during X. laevis metamorphosis, we discovered that PRMT1 is up-regulated during intestinal remodeling and, importantly, that its up-regulation in the intestinal larval epithelium is the first known molecular marker induced by T3 in the larval epithelial cells that are destined for dedifferentiation into stem cells. Our findings here have revealed that the induction of PRMT1 by T3 involves a two-step process. T3 activates the transcription factor c-Myc directly through the binding of TR to the TRE upstream of the c-Myc promoter, and c-Myc in turn up-regulates PRMT1 transcription through a binding site in the first intron.
The formation of the adult intestine is highly conserved among vertebrates and takes place through the formation of adult epithelial stem cells during postembryonic development when T3 levels are high (5, 6, 46, 53, 54). In anurans, this corresponds to the metamorphic period. The ability to control and study metamorphosis makes intestinal remodeling in anurans a valuable model to study how organ-specific adult stem cells are formed during development. By studying X. laevis metamorphosis, we have discovered previously that the TR coactivator PRMT1 is important for the development of the adult intestinal stem cells through larval epithelial cell dedifferentiation (10). This, together with the fact that PRMT1 up-regulation is one of the earliest known events induced by T3 in the larval epithelial cells destined to dedifferentiate into adult stem cells, makes it important to understand how PRMT1 is regulated during stem cell development.
Here we have cloned a 6-kb genomic DNA flanking the transcription start site of the X. laevis PRMT1 gene. Making use of the available genomic sequence information for the highly related species X. tropicalis, we have discovered several conserved regions in the PRMT1 genes. Importantly, the conserved regions proximal to the transcription start site and in the first intron are important for the activity of the promoter. The conserved region further upstream of the start site have no detectable role in the promoter function when analyzed in CaCo-2 intestinal cells, although they may play a role in regulation PRMT1 expression under more physiological conditions, such as during development. Bioinformatics analyses of the conserved sequences between the X. laevis and X. tropicalis PRMT1 gene, as well as the mouse, human, and zebrafish PRMT1 gene, have revealed putative bindings sites for a number of transcription factors in all these vertebrate PRMT1 genes. Our interest in PRMT1 expression in stem cell development and other available information prompted us to focus on the binding site for c-Myc in this study. On the other hand, we also identified putative binding sites in the proximal conserved promoter region for transcription factors E2F, NF-Y, ccAAT box binding proteins, and SMAD. Some of these factors may also participate in regulating PRMT1 expression. For example, SMAD, a downstream mediator of bore morphogenetic protein receptors, may be involved in the intestinal stem cell development and PMRT1 expression because the BMP/BMP receptor signaling pathway plays a role in the amphibian metamorphosis (76, 77). Thus, it will be interesting to investigate the roles of these other putative transcription factor binding sites in PRMT1 promoter function in the future.
PRMT1 is up-regulated by T3 during X. laevis metamorphosis (41). Interestingly, although this is an early event observed in the developing adult intestinal stem cells, the induction is kinetically slow, requiring 3–5 days of T3 treatment, suggesting that T3 indirectly regulates the expression of PRMT1. Our identification of a c-Myc binding site within the first intron of the PRMT1 genes provides a likely molecular explanation for this indirect regulation by T3. We have shown that c-Myc binds to the intronic site in the PRMT1 gene in vivo and that mutating this site reduces the promoter activity. On the other hand, c-Myc is quickly induced by T3 in the intestine. More importantly, we have discovered a TRE upstream of the c-Myc promoter and demonstrated that TR is bound to the TRE in the intestine during metamorphosis. These findings suggest that during stem cell formation, T3 first induces the expression of c-Myc directly at the transcription level through TR binding to the TRE in the c-Myc promoter and that c-Myc then activates PRMT1 expression through binding to the intronic enhancer in the PRMT1 gene. PRMT1, in turn, participates in the dedifferentiation of the larval epithelial cells into adult stem cells.
Although PRMT1 is up-regulated in the intestine where stem cell development occurs, it is not altered in the tail during metamorphosis, as the tail undergoes resorption, suggesting that its up-regulation may be specific to the developing adult tissues. Consistently, c-Myc, which is induced by T3 directly in the intestine and in turn activates the PRMT1 promoter, is not regulated by T3 in the tail during metamorphosis. This further supports a role of c-Myc in activating PRMT1 expression to promote adult intestinal stem cell development. The conservation of PRMT1 expression during intestinal stem cell development in zebrafish and mice (46) and the presence of a conserved c-Myc binding site in the first intron of all vertebrate PRMT1 gene argues that c-Myc also plays a role in regulating PRMT1 gene expression during the formation of adult intestinal stem cells in other vertebrates and further supports a conserved molecular pathway governing the development of adult intestinal stem cells during postembryonic development in vertebrates.
Supplementary Material
This work was supported, in whole or in part, by the NICHD, National Institutes of Health intramural research program. This work was also supported by a fellowship by the Japan Society for the Promotion of Sciences (NIH) (to K. F.). This work was also supported by a Fellowship for Japanese Biochemical and Behavioral Researchers at NIH from Japan Society for the Promotion of Science (to K. F.).
The nucleotide sequence(s) reported in this paper has been submitted to the GenBankTM/EBI Data Bank with accession number(s) JQ302819.

This article contains supplemental Table 1.
- T3
- thyroid hormone
- MYC
- v-Myc myelocytomatosis viral oncogene homolog (avian)
- TR
- thyroid hormone receptor
- TRE
- thyroid hormone response element
- Luc
- luciferase
- PRMT1
- protein arginine methyltransferase 1
- SMAD
- vertebrate homolog of Sma from C. elegans and Mad from D. melanogaster.
REFERENCES
- 1. MacDonald W. C., Trier J. S., Everett N. B. (1964) Cell proliferation and migration in the stomach, duodenum, and rectum of man. Radioautographic studies. Gastroenterology 46, 405–417 [PubMed] [Google Scholar]
- 2. Toner P. G., Carr K. E., Wyburn G. M. (1971) The Digestive System: An Ultrastructural Atlas and Review., Butterworth, London [Google Scholar]
- 3. van der Flier L. G., Clevers H. (2009) Stem cells, self-renewal, and differentiation in the intestinal epithelium. Annu. Rev. Physiol. 71, 241–260 [DOI] [PubMed] [Google Scholar]
- 4. McAvoy J. W., Dixon K. E. (1977) J. Exp. Zool. 202, 129–138 [Google Scholar]
- 5. Ishizuya-Oka A., Shi Y. B. (2011) Evolutionary insights into postembryonic development of adult intestinal stem cells. Cell Biosci. 1, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shi Y. B., Hasebe T., Fu L., Fujimoto K., Ishizuya-Oka A. (2011) The development of the adult intestinal stem cells. Insights from studies on thyroid hormone-dependent amphibian metamorphosis. Cell Biosci. 1, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gilbert L. I., Tata J. R., Atkinson B. G. (1996) Metamorphosis. Post-embryonic Reprogramming of Gene Expression in Amphibian and Insect Cells, Academic Press, New York [Google Scholar]
- 8. Shi Y. B. (1999) Amphibian Metamorphosis. From Morphology to Molecular Biology, John Wiley & Sons, Inc., New York [Google Scholar]
- 9. Shi Y. B., Ishizuya-Oka A. (1996) Biphasic intestinal development in amphibians. Embryogenesis and remodeling during metamorphosis. Curr. Top. Dev. Biol. 32, 205–235 [DOI] [PubMed] [Google Scholar]
- 10. Ishizuya-Oka A., Hasebe T., Buchholz D. R., Kajita M., Fu L., Shi Y. B. (2009) Origin of the adult intestinal stem cells induced by thyroid hormone in Xenopus laevis. FASEB J. 23, 2568–2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schreiber A. M., Cai L., Brown D. D. (2005) Remodeling of the intestine during metamorphosis of Xenopus laevis. Proc. Natl. Acad. Sci. U.S.A. 102, 3720–3725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ishizuya-Oka A., Shimozawa A. (1991) In Vitro Cell. Dev. Biol. 27, 853–857 [DOI] [PubMed] [Google Scholar]
- 13. Ishizuya-Oka A., Ueda S., Damjanovski S., Li Q., Liang V. C., Shi Y. B. (1997) Anteroposterior gradient of epithelial transformation during amphibian intestinal remodeling. Immunohistochemical detection of intestinal fatty acid-binding protein. Dev. Biol. 192, 149–161 [DOI] [PubMed] [Google Scholar]
- 14. Marshall J. A., Dixon K. E. (1978) J. Exp. Zool. 203, 31–40 [Google Scholar]
- 15. Amano T., Noro N., Kawabata H., Kobayashi Y., Yoshizato K. (1998) Metamorphosis-associated and region-specific expression of calbindin gene in the posterior intestinal epithelium of Xenopus laevis larva. Dev. Growth Differ. 40, 177–188 [DOI] [PubMed] [Google Scholar]
- 16. Schreiber A. M., Das B., Huang H., Marsh-Armstrong N., Brown D. D. (2001) Diverse developmental programs of Xenopus laevis metamorphosis are inhibited by a dominant negative thyroid hormone receptor. Proc. Natl. Acad. Sci. U.S.A. 98, 10739–10744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Brown D. D., Cai L. (2007) Amphibian metamorphosis. Dev. Biol. 306, 20–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Buchholz D. R., Hsia S. C., Fu L., Shi Y. B. (2003) A dominant-negative thyroid hormone receptor blocks amphibian metamorphosis by retaining corepressors at target genes. Mol. Cell. Biol. 23, 6750–6758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Buchholz D. R., Tomita A., Fu L., Paul B. D., Shi Y. B. (2004) Transgenic analysis reveals that thyroid hormone receptor is sufficient to mediate the thyroid hormone signal in frog metamorphosis. Mol. Cell. Biol. 24, 9026–9037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Buchholz D. R., Paul B. D., Fu L., Shi Y. B. (2006) Molecular and developmental analyses of thyroid hormone receptor function in Xenopus laevis, the African clawed frog. Gen. Comp. Endocrinol. 145, 1–19 [DOI] [PubMed] [Google Scholar]
- 21. Shi Y. B. (2009) Dual functions of thyroid hormone receptors in vertebrate development. The roles of histone-modifying cofactor complexes. Thyroid 19, 987–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nakajima K., Yaoita Y. (2003) Dual mechanisms governing muscle cell death in tadpole tail during amphibian metamorphosis. Dev. Dyn. 227, 246–255 [DOI] [PubMed] [Google Scholar]
- 23. Denver R. J., Hu F., Scanlan T. S., Furlow J. D. (2009) Thyroid hormone receptor subtype specificity for hormone-dependent neurogenesis in Xenopus laevis. Dev. Biol. 326, 155–168 [DOI] [PubMed] [Google Scholar]
- 24. Bagamasbad P., Howdeshell K. L., Sachs L. M., Demeneix B. A., Denver R. J. (2008) A role for basic transcription element-binding protein 1 (BTEB1) in the autoinduction of thyroid hormone receptor β. J. Biol. Chem. 283, 2275–2285 [DOI] [PubMed] [Google Scholar]
- 25. Schreiber A. M., Mukhi S., Brown D. D. (2009) Cell-cell interactions during remodeling of the intestine at metamorphosis in Xenopus laevis. Dev. Biol. 331, 89–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lazar M. A. (1993) Thyroid hormone receptors. Multiple forms, multiple possibilities. Endocr. Rev. 14, 184–193 [DOI] [PubMed] [Google Scholar]
- 27. Yen P. M. (2001) Physiological and molecular basis of thyroid hormone action. Physiol. Rev. 81, 1097–1142 [DOI] [PubMed] [Google Scholar]
- 28. Mangelsdorf D. J., Thummel C., Beato M., Herrlich P., Schütz G., Umesono K., Blumberg B., Kastner P., Mark M., Chambon P., Evans R. M. (1995) The nuclear receptor superfamily. The second decade. Cell 83, 835–839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tsai M. J., O'Malley B. W. (1994) Molecular mechanisms of action of steroid/thyroid receptor superfamily members. Ann. Rev. Biochem. 63, 451–486 [DOI] [PubMed] [Google Scholar]
- 30. Ito M., Roeder R. G. (2001) The TRAP/SMCC/mediator complex and thyroid hormone receptor function. Trends Endocrinol Metab. 12, 127–134 [DOI] [PubMed] [Google Scholar]
- 31. Rachez C., Freedman L. P. (2000) Mechanisms of gene regulation by vitamin D(3) receptor. A network of coactivator interactions. Gene 246, 9–21 [DOI] [PubMed] [Google Scholar]
- 32. Zhang J., Lazar M. A. (2000) The mechanism of action of thyroid hormones. Annu. Rev. Physiol. 62, 439–466 [DOI] [PubMed] [Google Scholar]
- 33. Burke L. J., Baniahmad A. (2000) Co-repressors 2000. FASEB J. 14, 1876–1888 [DOI] [PubMed] [Google Scholar]
- 34. Jepsen K., Rosenfeld M. G. (2002) Biological roles and mechanistic actions of co-repressor complexes. J. Cell Sci. 115, 689–698 [DOI] [PubMed] [Google Scholar]
- 35. Jones P. L., Shi Y. B. (2003) N-CoR-HDAC in Current Topics in Microbiology and Immunology: Protein Complexes that Modify Chromatin (Workman J. L., ed). Springer-Verlag, Berlin [Google Scholar]
- 36. Rachez C., Freedman L. P. (2001) Mediator complexes and transcription. Curr. Opin. Cell Biol. 13, 274–280 [DOI] [PubMed] [Google Scholar]
- 37. Hu X., Lazar M. A. (2000) Transcriptional repression by nuclear hormone receptors. TEM 11, 6–10 [DOI] [PubMed] [Google Scholar]
- 38. Tomita A., Buchholz D. R., Shi Y. B. (2004) Recruitment of N-CoR/SMRT-TBLR1 corepressor complex by unliganded thyroid hormone receptor for gene repression during frog development. Mol. Cell. Biol. 24, 3337–3346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sachs L. M., Jones P. L., Havis E., Rouse N., Demeneix B. A., Shi Y. B. (2002) Nuclear receptor corepressor recruitment by unliganded thyroid hormone receptor in gene repression during Xenopus laevis development. Mol. Cell. Biol. 22, 8527–8538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sato Y., Buchholz D. R., Paul B. D., Shi Y. B. (2007) A role of unliganded thyroid hormone receptor in postembryonic development in Xenopus laevis. Mech. Dev. 124, 476–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Matsuda H., Paul B. D., Choi C. Y., Hasebe T., Shi Y. B. (2009) Novel functions of protein arginine methyltransferase 1 in thyroid hormone receptor-mediated transcription and in the regulation of metamorphic rate in Xenopus laevis. Mol. Cell. Biol. 29, 745–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Paul B. D., Buchholz D. R., Fu L., Shi Y. B. (2005) Tissue- and gene-specific recruitment of steroid receptor coactivator-3 by thyroid hormone receptor during development. J. Biol. Chem. 280, 27165–27172 [DOI] [PubMed] [Google Scholar]
- 43. Paul B. D., Fu L., Buchholz D. R., Shi Y. B. (2005) Coactivator recruitment is essential for liganded thyroid hormone receptor to initiate amphibian metamorphosis. Mol. Cell. Biol. 25, 5712–5724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Paul B. D., Buchholz D. R., Fu L., Shi Y. B. (2007) SRC-p300 coactivator complex is required for thyroid hormone-induced amphibian metamorphosis. J. Biol. Chem. 282, 7472–7481 [DOI] [PubMed] [Google Scholar]
- 45. Havis E., Sachs L. M., Demeneix B. A. (2003) Metamorphic T3-response genes have specific co-regulator requirements. EMBO Rep. 4, 883–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Matsuda H., Shi Y. B. (2010) An essential and evolutionarily conserved role of protein arginine methyltransferase 1 for adult intestinal stem cells during postembryonic development. Stem Cells 28, 2073–2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Tata J. R. (1993) Gene expression during metamorphosis. An ideal model for post-embryonic development. BioEssays 15, 239–248 [DOI] [PubMed] [Google Scholar]
- 48. Plateroti M., Gauthier K., Domon-Dell C., Freund J. N., Samarut J., Chassande O. (2001) Functional interference between thyroid hormone receptor α (TRα) and natural truncated TRΔα isoforms in the control of intestine development. Mol. Cell. Biol. 21, 4761–4772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Flamant F., Poguet A. L., Plateroti M., Chassande O., Gauthier K., Streichenberger N., Mansouri A., Samarut J. (2002) Congenital hypothyroid Pax8(−/−) mutant mice can be rescued by inactivating the TRα gene. Mol. Endocrinol. 16, 24–32 [DOI] [PubMed] [Google Scholar]
- 50. Kress E., Rezza A., Nadjar J., Samarut J., Plateroti M. (2009) The frizzled-related sFRP2 gene is a target of thyroid hormone receptor α1 and activates β-catenin signaling in mouse intestine. J. Biol. Chem. 284, 1234–1241 [DOI] [PubMed] [Google Scholar]
- 51. Plateroti M., Chassande O., Fraichard A., Gauthier K., Freund J. N., Samarut J., Kedinger M. (1999) Involvement of T3Rα- and β-receptor subtypes in mediation of T3 functions during postnatal murine intestinal development. Gastroenterology 116, 1367–1378 [DOI] [PubMed] [Google Scholar]
- 52. Plateroti M., Kress E., Mori J. I., Samarut J. (2006) Thyroid hormone receptor α1 directly controls transcription of the β-catenin gene in intestinal epithelial cells. Mol. Cell. Biol. 26, 3204–3214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Harper J., Mould A., Andrews R. M., Bikoff E. K., Robertson E. J. (2011) The transcriptional repressor Blimp1/Prdm1 regulates postnatal reprogramming of intestinal enterocytes. Proc. Natl. Acad. Sci. U.S.A. 108, 10585–10590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Muncan V., Heijmans J., Krasinski S. D., Büller N. V., Wildenberg M. E., Meisner S., Radonjic M., Stapleton K. A., Lamers W. H., Biemond I., van den Bergh Weerman M. A., O'Carroll D., Hardwick J. C., Hommes D. W., van den Brink G. R. (2011) Blimp1 regulates the transition of neonatal to adult intestinal epithelium. Nat. Commun. 2, 452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Nieuwkoop P. D., Faber J. (1956) Normal Table of Xenopus laevis., 1st. Ed., North Holland Publishing, Amsterdam [Google Scholar]
- 56. Shi Y. B., Yaoita Y., Brown D. D. (1992) Genomic organization and alternative promoter usage of the two thyroid hormone receptor β genes in Xenopus laevis. J. Biol. Chem. 267, 733–788 [PubMed] [Google Scholar]
- 57. Wong J., Shi Y. B. (1995) Coordinated regulation of and transcriptional activation by Xenopus thyroid hormone and retinoid X receptors. J. Biol. Chem. 270, 18479–18483 [DOI] [PubMed] [Google Scholar]
- 58. Matsuura K., Fujimoto K., Fu L., Shi Y. B. (2011) Endocrinology 153, 961–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang X., Matsuda H., Shi Y. B. (2008) Developmental regulation and function of thyroid hormone receptors and 9-cis retinoic acid receptors during Xenopus tropicalis metamorphosis. Endocrinology 149, 5610–5618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Buchholz D. R., Ishizuya-Oka A., Shi Y. B. (2004) Spatial and temporal expression pattern of a novel gene in the frog Xenopus laevis. Correlations with adult intestinal epithelial differentiation during metamorphosis. Gene. Expr. Patterns 4, 321–328 [DOI] [PubMed] [Google Scholar]
- 61. Das B., Heimeier R. A., Buchholz D. R., Shi Y. B. (2009) Identification of direct thyroid hormone response genes reveals the earliest gene regulation programs during frog metamorphosis. J. Biol. Chem. 284, 34167–34178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Christen B., Robles V., Raya M., Paramonov I., Izpisúa Belmonte J. C. (2010) Regeneration and reprogramming compared. BMC Biology 8, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Aoki K., Ishii Y., Matsumoto K., Tsujimoto M. (2002) Methylation of Xenopus CIRP2 regulates its arginine- and glycine-rich region-mediated nucleocytoplasmic distribution. Nucleic Acids Res. 30, 5182–5192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Batut J., Vandel L., Leclerc C., Daguzan C., Moreau M., Néant I. (2005) The Ca2+-induced methyltransferase xPRMT1b controls neural fate in amphibian embryo. Proc. Natl. Acad. Sci. U.S.A. 102, 15128–15133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Goulet I., Gauvin G., Boisvenue S., Côté J. (2007) Alternative splicing yields protein arginine methyltransferase 1 isoforms with distinct activity, substrate specificity, and subcellular localization. J. Biol. Chem. 282, 33009–33021 [DOI] [PubMed] [Google Scholar]
- 66. Bilesimo P., Jolivet P., Alfama G., Buisine N., Le Mevel S., Havis E., Demeneix B. A., Sachs L. M. (2011) Specific histone lysine 4 methylation patterns define TR-binding capacity and differentiate direct T3 responses. Mol. Endocrinol. 25, 225–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Fujimoto K., Nakajima K., Yaoita Y. (2006) One of the duplicated matrix metalloproteinase-9 genes is expressed in regressing tail during anuran metamorphosis. Dev. Growth Differ. 48, 223–241 [DOI] [PubMed] [Google Scholar]
- 68. Barrero M. J., Malik S. (2006) Two functional modes of a nuclear receptor-recruited arginine methyltransferase in transcriptional activation. Mol. Cell 24, 233–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Loots G., Ovcharenko I. (2007) ECRbase. Database of evolutionary conserved regions, promoters, and transcription factor binding sites in vertebrate genomes. Bioinformatics 23, 122–124 [DOI] [PubMed] [Google Scholar]
- 70. Eilers M., Eisenman R. N. (2008) Myc's broad reach. Genes Dev. 22, 2755–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Meyer N., Penn L. Z. (2008) Reflecting on 25 years with MYC. Nat. Rev. Cancer 8, 976–990 [DOI] [PubMed] [Google Scholar]
- 72. Pei D. (2009) Regulation of pluripotency and reprogramming by transcription factors. J. Biol. Chem. 284, 3365–3369 [DOI] [PubMed] [Google Scholar]
- 73. Takahashi K., Yamanaka S. (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 74. Wong J., Shi Y. B., Wolffe A. P. (1995) A role for nucleosome assembly in both silencing and activation of the Xenopus TR β A gene by the thyroid hormone receptor. Genes Dev. 9, 2696–2711 [DOI] [PubMed] [Google Scholar]
- 75. Chen D., Ma H., Hong H., Koh S. S., Huang S. M., Schurter B. T., Aswad D. W., Stallcup M. R. (1999) Regulation of transcription by a protein methyltransferase. Science 284, 2174–2177 [DOI] [PubMed] [Google Scholar]
- 76. Ishizuya-Oka A., Ueda S., Amano T., Shimizu K., Suzuki K., Ueno N., Yoshizato K. (2001) Thyroid-hormone-dependent and fibroblast-specific expression of BMP-4 correlates with adult epithelial development during amphibian intestinal remodeling. Cell Tissue Res. 303, 187–195 [DOI] [PubMed] [Google Scholar]
- 77. Ishizuya-Oka A., Hasebe T., Shimizu K., Suzuki K., Ueda S. (2006) Shh/BMP-4 signaling pathway is essential for intestinal epithelial development during Xenopus larval-to-adult remodeling. Dev. Dyn. 235, 3240–3249 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.