Abstract
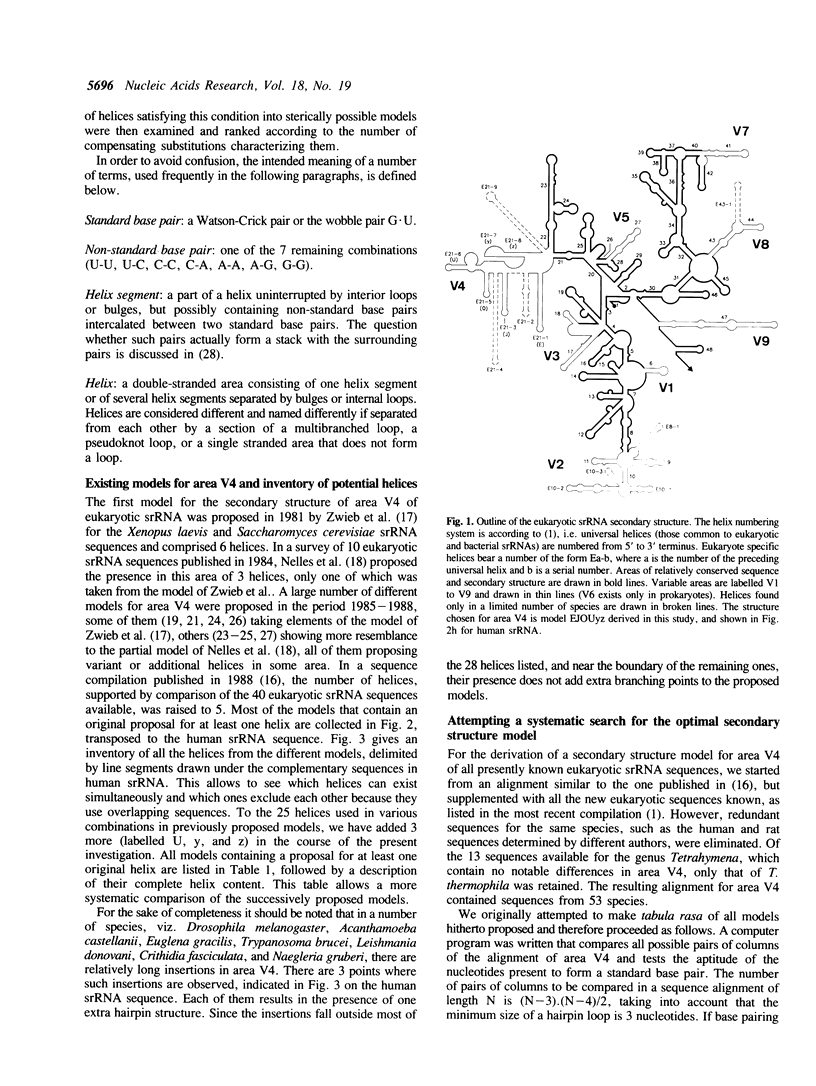
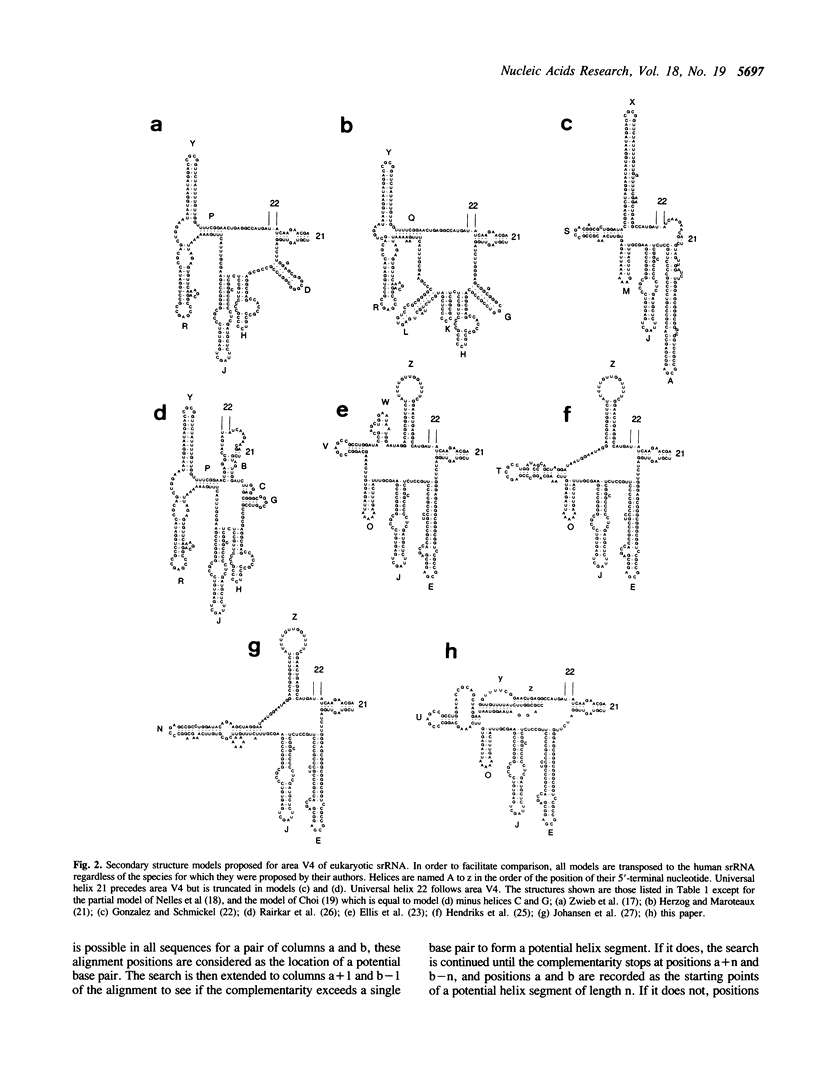
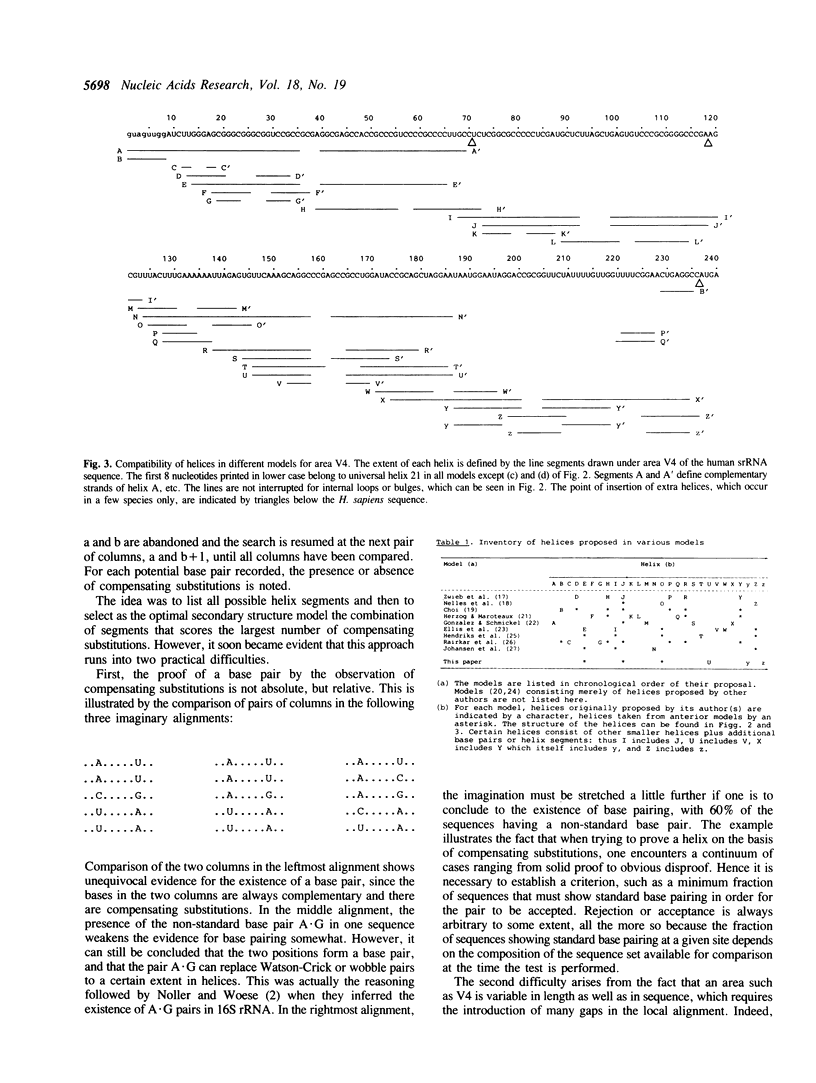
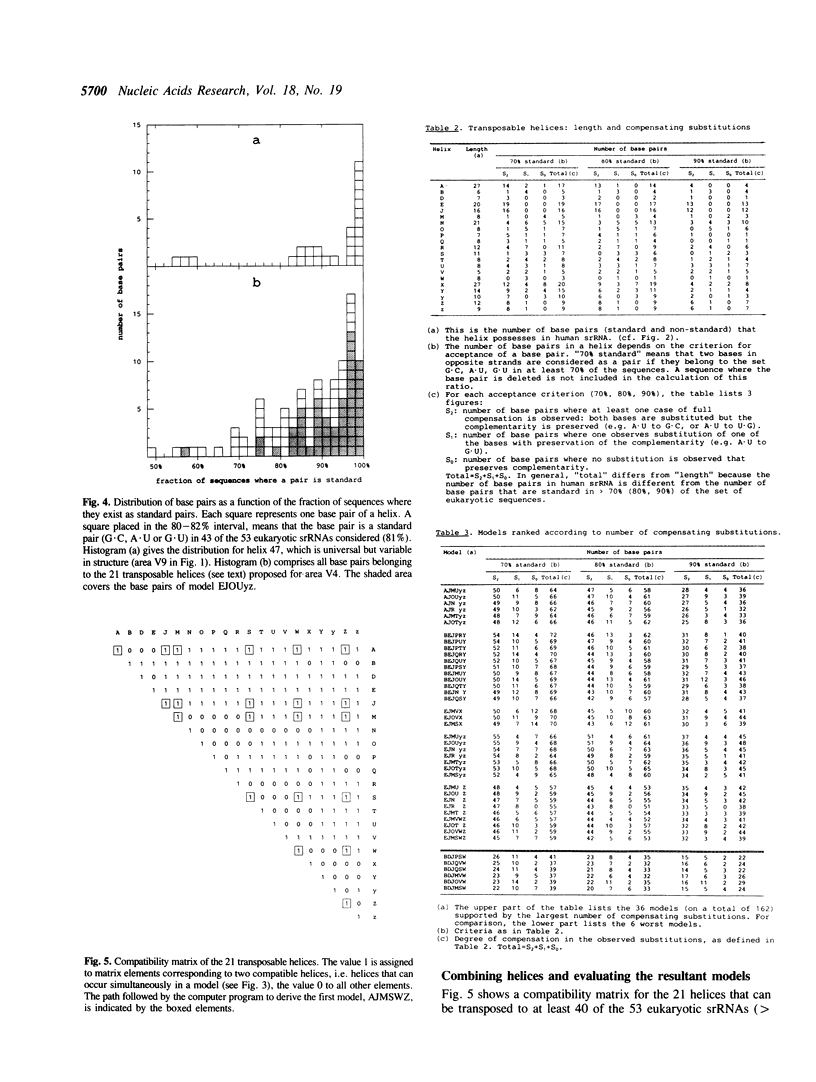
Eukaryotic small ribosomal subunit RNAs contain an area of variable structure, V4, which comprises about 250 nucleotides in most species, whereas the corresponding area in bacterial small ribosomal subunit RNAs consists of about 64 nucleotides folded into a single hairpin. There is no consensus on the secondary structure of area V4 in eukaryotes, about 10 different models having been proposed. The prediction of a model on a comparative basis poses special problems because, due to the variability of the area in length as well as sequence, a dependable alignment is very difficult to achieve. A new model was derived by systematic examination of all combinations of helices that have been hitherto proposed, plus some new ones. The following properties of the helices were examined: transposability to all presently known sequences, presence of compensating substitutions, and thermodynamic stability. A model was selected by ranking all possible combinations of transposable helices according to the number of compensating substitutions scored. The optimal model comprises a pseudoknot and four hairpin structures. Certain species contain additional hairpins inserted between these structural elements, while in others the structure is partially or entirely deleted.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abrahams J. P., van den Berg M., van Batenburg E., Pleij C. Prediction of RNA secondary structure, including pseudoknotting, by computer simulation. Nucleic Acids Res. 1990 May 25;18(10):3035–3044. doi: 10.1093/nar/18.10.3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedergren R., Gray M. W., Abel Y., Sankoff D. The evolutionary relationships among known life forms. J Mol Evol. 1988 Dec;28(1-2):98–112. doi: 10.1007/BF02143501. [DOI] [PubMed] [Google Scholar]
- Chan Y. L., Gutell R., Noller H. F., Wool I. G. The nucleotide sequence of a rat 18 S ribosomal ribonucleic acid gene and a proposal for the secondary structure of 18 S ribosomal ribonucleic acid. J Biol Chem. 1984 Jan 10;259(1):224–230. [PubMed] [Google Scholar]
- Choi Y. C. Structural organization of ribosomal RNAs from Novikoff hepatoma. I. Characterization of fragmentation products from 40 S subunit. J Biol Chem. 1985 Oct 15;260(23):12769–12772. [PubMed] [Google Scholar]
- Dams E., Hendriks L., Van de Peer Y., Neefs J. M., Smits G., Vandenbempt I., De Wachter R. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 1988;16 (Suppl):r87–173. doi: 10.1093/nar/16.suppl.r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckenrode V. K., Arnold J., Meagher R. B. Comparison of the nucleotide sequence of soybean 18S rRNA with the sequences of other small-subunit rRNAs. J Mol Evol. 1984;21(3):259–269. doi: 10.1007/BF02102358. [DOI] [PubMed] [Google Scholar]
- Ellis R. E., Sulston J. E., Coulson A. R. The rDNA of C. elegans: sequence and structure. Nucleic Acids Res. 1986 Mar 11;14(5):2345–2364. doi: 10.1093/nar/14.5.2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez I. L., Schmickel R. D. The human 18S ribosomal RNA gene: evolution and stability. Am J Hum Genet. 1986 Apr;38(4):419–427. [PMC free article] [PubMed] [Google Scholar]
- Gray M. W. Organelle origins and ribosomal RNA. Biochem Cell Biol. 1988 May;66(5):325–348. doi: 10.1139/o88-042. [DOI] [PubMed] [Google Scholar]
- Gunderson J. H., Sogin M. L. Length variation in eukaryotic rRNAs: small subunit rRNAs from the protists Acanthamoeba castellanii and Euglena gracilis. Gene. 1986;44(1):63–70. doi: 10.1016/0378-1119(86)90043-0. [DOI] [PubMed] [Google Scholar]
- Gunderson J. H., Sogin M. L., Wollett G., Hollingdale M., de la Cruz V. F., Waters A. P., McCutchan T. F. Structurally distinct, stage-specific ribosomes occur in Plasmodium. Science. 1987 Nov 13;238(4829):933–937. doi: 10.1126/science.3672135. [DOI] [PubMed] [Google Scholar]
- Gutell R. R., Fox G. E. A compilation of large subunit RNA sequences presented in a structural format. Nucleic Acids Res. 1988;16 (Suppl):r175–r269. doi: 10.1093/nar/16.suppl.r175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell R. R., Noller H. F., Woese C. R. Higher order structure in ribosomal RNA. EMBO J. 1986 May;5(5):1111–1113. doi: 10.1002/j.1460-2075.1986.tb04330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutell R. R., Weiser B., Woese C. R., Noller H. F. Comparative anatomy of 16-S-like ribosomal RNA. Prog Nucleic Acid Res Mol Biol. 1985;32:155–216. doi: 10.1016/s0079-6603(08)60348-7. [DOI] [PubMed] [Google Scholar]
- Hancock J. M., Tautz D., Dover G. A. Evolution of the secondary structures and compensatory mutations of the ribosomal RNAs of Drosophila melanogaster. Mol Biol Evol. 1988 Jul;5(4):393–414. doi: 10.1093/oxfordjournals.molbev.a040501. [DOI] [PubMed] [Google Scholar]
- Hendriks L., De Baere R., Van Broeckhoven C., De Wachter R. Primary and secondary structure of the 18 S ribosomal RNA of the insect species Tenebrio molitor. FEBS Lett. 1988 May 9;232(1):115–120. doi: 10.1016/0014-5793(88)80398-3. [DOI] [PubMed] [Google Scholar]
- Hendriks L., Van Broeckhoven C., Vandenberghe A., Van de Peer Y., De Wachter R. Primary and secondary structure of the 18S ribosomal RNA of the bird spider Eurypelma californica and evolutionary relationships among eukaryotic phyla. Eur J Biochem. 1988 Oct 15;177(1):15–20. doi: 10.1111/j.1432-1033.1988.tb14339.x. [DOI] [PubMed] [Google Scholar]
- Herzog M., Maroteaux L. Dinoflagellate 17S rRNA sequence inferred from the gene sequence: Evolutionary implications. Proc Natl Acad Sci U S A. 1986 Nov;83(22):8644–8648. doi: 10.1073/pnas.83.22.8644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen T., Johansen S., Haugli F. B. Nucleotide sequence of the Physarum polycephalum small subunit ribosomal RNA as inferred from the gene sequence: secondary structure and evolutionary implications. Curr Genet. 1988 Sep;14(3):265–273. doi: 10.1007/BF00376747. [DOI] [PubMed] [Google Scholar]
- Michot B., Qu L. H., Bachellerie J. P. Evolution of large-subunit rRNA structure. The diversification of divergent D3 domain among major phylogenetic groups. Eur J Biochem. 1990 Mar 10;188(2):219–229. doi: 10.1111/j.1432-1033.1990.tb15393.x. [DOI] [PubMed] [Google Scholar]
- Neefs J. M., Van de Peer Y., Hendriks L., De Wachter R. Compilation of small ribosomal subunit RNA sequences. Nucleic Acids Res. 1990 Apr 25;18 (Suppl):2237–2317. doi: 10.1093/nar/18.suppl.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelles L., Fang B. L., Volckaert G., Vandenberghe A., De Wachter R. Nucleotide sequence of a crustacean 18S ribosomal RNA gene and secondary structure of eukaryotic small subunit ribosomal RNAs. Nucleic Acids Res. 1984 Dec 11;12(23):8749–8768. doi: 10.1093/nar/12.23.8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninio J. Prediction of pairing schemes in RNA molecules-loop contributions and energy of wobble and non-wobble pairs. Biochimie. 1979;61(10):1133–1150. doi: 10.1016/s0300-9084(80)80227-6. [DOI] [PubMed] [Google Scholar]
- Noller H. F., Woese C. R. Secondary structure of 16S ribosomal RNA. Science. 1981 Apr 24;212(4493):403–411. doi: 10.1126/science.6163215. [DOI] [PubMed] [Google Scholar]
- Pleij C. W., Rietveld K., Bosch L. A new principle of RNA folding based on pseudoknotting. Nucleic Acids Res. 1985 Mar 11;13(5):1717–1731. doi: 10.1093/nar/13.5.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rairkar A., Rubino H. M., Lockard R. E. Chemical probing of adenine residues within the secondary structure of rabbit 18S ribosomal RNA. Biochemistry. 1988 Jan 26;27(2):582–592. doi: 10.1021/bi00402a013. [DOI] [PubMed] [Google Scholar]
- Schnare M. N., Collings J. C., Gray M. W. Structure and evolution of the small subunit ribosomal RNA gene of Crithidia fasciculata. Curr Genet. 1986;10(5):405–410. doi: 10.1007/BF00418414. [DOI] [PubMed] [Google Scholar]
- Spangler E. A., Blackburn E. H. The nucleotide sequence of the 17S ribosomal RNA gene of Tetrahymena thermophila and the identification of point mutations resulting in resistance to the antibiotics paromomycin and hygromycin. J Biol Chem. 1985 May 25;260(10):6334–6340. [PubMed] [Google Scholar]
- Studnicka G. M., Eiserling F. A., Lake J. A. A unique secondary folding pattern for 5S RNA corresponds to the lowest energy homologous secondary structure in 17 different prokaryotes. Nucleic Acids Res. 1981 Apr 24;9(8):1885–1904. doi: 10.1093/nar/9.8.1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y., Neefs J. M., De Wachter R. Small ribosomal subunit RNA sequences, evolutionary relationships among different life forms, and mitochondrial origins. J Mol Evol. 1990 May;30(5):463–476. doi: 10.1007/BF02101118. [DOI] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R., Gutell R. R. Evidence for several higher order structural elements in ribosomal RNA. Proc Natl Acad Sci U S A. 1989 May;86(9):3119–3122. doi: 10.1073/pnas.86.9.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwieb C., Glotz C., Brimacombe R. Secondary structure comparisons between small subunit ribosomal RNA molecules from six different species. Nucleic Acids Res. 1981 Aug 11;9(15):3621–3640. doi: 10.1093/nar/9.15.3621. [DOI] [PMC free article] [PubMed] [Google Scholar]



