Background: ERM protein function is regulated by phosphorylation.
Results: The PP1α isoform specifically dephosphorylates ceramide-induced ERM proteins.
Conclusion: Plasma membrane ceramide activation of PP1α, a novel and a specific mechanism that regulates ERM proteins.
Significance: The mechanisms of regulation of ERM proteins are important to understand and control cancer progression.
Keywords: Ceramide, Cytoskeleton, Ezrin, Phosphatase, PP1, Sphingolipid, Calyculin A, Tautomycin
Abstract
ERM (ezrin, radixin, and moesin) proteins are cytoskeletal interacting proteins that bind cortical actin, the plasma membrane, and membrane proteins, which are found in specialized plasma membrane structures such as microvilli and filopodia. ERM proteins are regulated by phosphatidylinositol 4, 5-biphosphate (PIP2) and by phosphorylation of a C-terminal threonine, and its inactivation involves PIP2 hydrolysis and/or myosin phosphatase (MP). Recently, we demonstrated that ERM proteins are also subject to counter regulation by the bioactive sphingolipids ceramide and sphingosine 1-phosphate. Plasma membrane ceramide induces ERM dephosphorylation whereas sphingosine 1-phosphate induces their phosphorylation. In this work, we pursue the mechanisms by which ceramide regulates dephosphorylation. We found that this dephosphorylation was independent of hydrolysis and localization of PIP2 and MP. However, the results show that ERM dephosphorylation was blocked by treatment with protein phosphatase 1 (PP1) pharmacological inhibitors and specifically by siRNA to PP1α, whereas okadaic acid, a PP2A inhibitor, failed. Moreover, a catalytic inactive mutant of PP1α acted as dominant negative of the endogenous PP1α. Additional results showed that the ceramide mechanism of PP1α activation is largely independent of PIP2 hydrolysis and MP. Taken together, these results demonstrate a novel, acute mechanism of ERM regulation dependent on PP1α and plasma membrane ceramide.
Introduction
ERM (ezrin, radixin, and moesin) proteins serve as regulated linkers between the plasma membrane and the cortical actin cytoskeleton (1). ERM proteins have been shown to provide a platform where different signaling molecules interact to allow signal transduction between the extracellular matrix and the cell (2, 3). Examples of these membrane proteins include CD95 (4), CD44 (5), intercellular adhesion molecule (I-CAM) (6), CD43 (7), cystic fibrosis transmembrane conductance regulator (8), and NHE1 (9). Moreover, ERM family is enriched in specialized cell structures such as filopodia, lamellipodia, membrane ruffles, and other membrane protrusions. It has also been shown that ERM proteins participate in diverse cellular functions such as cell morphology (10), cell polarization (11, 12), plasma membrane vesicular trafficking, adhesion, and migration (3, 13–15). ERM proteins also appear to play important roles in cancer progression involving invasion of tissue surroundings and metastasis (16).
The function of ERM proteins is regulated by a two-step process based on an active (open) and inactive (closed) conformation. In the inactive conformation, the N-terminal domain (FERM) and the C-terminal domain (C-ERMAD) interact with each other in a self-associated conformation, and the protein remains in the cytosol. Phosphorylation on a C-terminal threonine residue (ezrin Thr-567, radixin Thr-564, and moesin Thr-558) provokes a steric impediment for the interaction between FERM and C-ERMAD, leading to an open conformation, and then the FERM domain interacts with the plasma membrane in a process requiring phosphatidylinositol 4,5-biphosphate (PIP2),2 whereas the C-ERMAD domain interacts with the cortical actin cytoskeleton. Several kinases have been shown to phosphorylate the C terminus of ERM, including Rho kinase (17), classical and atypical protein kinase C (18, 19), myotonic dystrophy kinase-related Cdc42-binding kinase (20), and GRK2 (G-protein-coupled receptor kinase 2) (21). However, only myosin phosphatase (MP, also called ERM phosphatase, a term that we will not use in this work to avoid confusion) has been clearly implicated in constitutive ERM dephosphorylation. This process requires the MYPT1-targeting subunit, although other phosphatases have been suggested, including PP2A (22) or PP2C in vitro studies (23). Little is known about mechanisms of regulated dephosphorylation of ERMs.
Interestingly, sphingolipids also have been related to cell migration (24, 25) and regulation of the cytoskeleton (26, 27). Recently, our group showed that ERM proteins were strongly regulated by sphingolipids. Hydrolysis of plasma membrane sphingomyelin producing plasma membrane ceramide induced a rapid and robust ERM dephosphorylation, whereas other SM metabolites failed. Ceramide-induced dephosphorylation could be overcome by hydrolysis of that ceramide using bacterial ceramidase. Moreover, production of sphingosine 1-phosphatre resulted in augmented phosphorylation (28). There is significant literature showing that ceramide functions as a bioactive lipid capable of activating several proteins, including protein phosphatases (29), suggesting that ceramide could directly activate a phosphatase to dephosphorylate ERM proteins in an acute cell response.
In this study, we demonstrate that ceramide generated at the plasma membrane dephosphorylates ERM proteins through PP1α isoform. This establishes a new mechanism of ERM regulation, independent of phospholipase C (PLC), PIP2 hydrolysis, PP1β, and MYPT1.
EXPERIMENTAL PROCEDURES
Materials
High glucose DMEM, fetal bovine serum, and rhodamine-phallodin were obtained from Invitrogen. Sphingomyelinase from Bacillus cereus, BSA (essentially fatty acid free), and anti-β-actin monoclonal antibody were from Sigma. Anti-phospho-ERM protein, anti-ezrin, and PP2AC antibodies were from Cell Signaling Technology (Danvers, MA). Anti-PP2ACα antibody was from BD Biosciences. Anti-PP1α, PP1β, and PP1γ antibodies and protein A/G-agarose beads were from Santa Cruz Biotechnology. Anti-GFP antibody was from Invitrogen. Nuclear stain Draq5 was from Alexis (Carlsbad, CA). Cyclosporin A, calyculin A, okadaic acid, tautomycin, U-73122, and m-3M3FBS were from Calbiochem.
Cell Culture
HeLa cells were purchased originally from American Type Culture Collection (ATCC, Manassas, VA). Cells were grown in DMEM supplemented with 10% FBS and incubated at 37 °C, 5% CO2. When FBS-free medium was used, medium was supplemented with 0.1% of BSA. Testing for the presence of mycoplasma infection was performed routinely on a monthly basis.
RNA Interference
Gene silencing of PP2ACα, PP2ACβ, PP1α, PP1β, PP1γ, and MYPT1 (50 nm) using Oligofectamine was performed according to the manufacturer's instructions (Invitrogen).
Plasmid Constructs
Full-length ezrin cDNA, VSV-G tag pCB6 plasmid has been described previously (Zeidan YH 2008). Single mutation on PP1 isoforms mutants were generated using QuikChange site-directed mutagenesis kit from Stratagene (La Jolla, CA). N-terminal ezrin first 311 amino acids followed by VSV-G tag was cloned in a PCB6 plasmid. All plasmids were sequenced to confirm the sequence in Genewiz (South Plainfield, NJ). The plasmid GFP-C1-PLCδ-PH was obtained from Addgene (Addgene plasmid 21179).
Immunoprecipitation and Immunoblotting
Cells were treated with vehicle or bacterial sphingomyelinase (bSMase; 50 milliunits/ml, 5 min). Cells were then lysed in a buffer containing 20 mm Tris-HCl, pH 7.5, 100 mm NaCl, 1% Triton X-100, 1 mm EDTA, and protease inhibitor mixture (Sigma). Homogenates were centrifuged at 9,300 × g for 10 min at 4 °C. Nonspecific binding was reduced by adding protein A/G-agarose for 1 h in a circular rotator and centrifuged at 10,000 × g, at 4 °C. The supernatants were used for immunoprecipitation using specific antibodies (1 μg/500 μg lysate) overnight. Immune complexes were isolated by incubation with protein A/G-agarose beads for 3 h. Immunoblotting analysis was carried using similar procedures as described previously (28).
Transfection with VSV.G-tagged Ezrin and GFP-C1-PLCδ-PH Expression Vector
Cells growing on 35-mm dishes were transfected with 1 μg of pCB6-ezrin, pCB6 N-terminal ezrin or GFP-C1-PLCδ-PH plasmid DNA using Lipofectamine transfection reagent (Qiagen) according to the manufacturer's instructions.
Immunofluorescence and Confocal Microscopy
Immunofluorescence analysis was carried using similar procedures as described previously (28). Briefly, HeLa cells, growing on 35-mm confocal dishes, were fixed by addition of warmed (37 °C) 3.7% paraformaldehyde solution for 15 min, and samples were washed with PBS (room temperature). If required, cells were permeabilized with 0.1% Triton X-100 for 10 min and blocked for 20 min at room temperature in PBS containing 2% of human serum (Jackson ImmunoResearch Laboratories). Primary antibodies were diluted in PBS with 2% of human serum and added for 18 h at 4 °C. Samples were examined, and images were collected using a confocal microscope (LSM510 META; Carl Zeiss, Inc.) using a 63× oil objective, and data were imported into Volocity software (Improvision).
RESULTS
Effects of PLC Activator m-3M3FBS on ERM Proteins in HeLa Cells
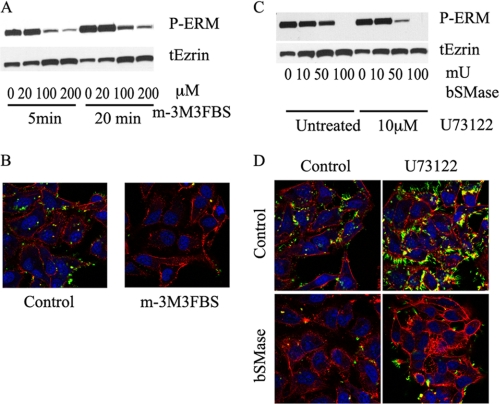
In a recent publication, we showed that ceramide generated at the plasma membrane by hydrolysis of sphingomyelin using bSMase caused a dramatic ERM dephosphorylation (28). However, the only physiologically known mechanism upstream of ERM dephosphorylation reported until now has been activation of PLC, producing hydrolysis of PIP2, followed by detachment of ERMs from the plasma membrane and dephosphorylation. To determine whether this mechanism also operates in HeLa cells, HeLa cells were treated with different doses of a PLC activator (m-3M3FBS) for the indicated durations, and the effects on ERM dephosphorylation were analyzed by Western blotting and confocal microscopy. In corroboration of results published in Jurkat T cells (30), m-3M3FBS was able to induce dephosphorylation of ERM proteins in HeLa cells (Fig. 1, A and B).
FIGURE 1.
Effects of PLC activation and inhibition on ERM dephosphorylation. A, HeLa cells were treated with vehicle, 20, 100, and 200 μm of m-3M3FBS for 5 and 20 min, and dephosphorylation on ERM was measured by Western blotting. Treatment with m-3M3FBS resulted in ERM dephosphorylation starting from 100 μm and 5 min. B, HeLa cells were treated with 100 μm of m-3M3FBS and fixed with paraformaldehyde. The presence and localization of phospho-ERM was visualized using confocal microscope. Nuclei and F-actin were visualized with Draq5 (blue) and rhodamine-phalloidin (red) staining and stained with phospho-ERM antibody (green). C, HeLa cells were pretreated with PLC inhibitor U-73122 (10 μm, 5 min) and treated with bSMase (10, 50, and 100 milliunits/ml (mU), 5 min). Phospho-ERM and total ezrin (tEzrin) were evaluated by Western blotting. D, HeLa cells were treated with U-73122 (10 μm, 5 min) and bSMase (50 milliunits/ml, 5 min) and fixed and stained as described in B.
Effects of PLC Inhibitor U73122 on bSMase- and PLC-induced ERM Dephosphorylation
Hydrolysis of PIP2 is driven by PLC. To study the relation between bSMase- and PIP2-dependent ERM dephosphorylation, we investigated whether inhibition of PLC could block bSMase-induced ERM dephosphorylation. In Jurkat blood T cells, the PLC inhibitor U73122 is able to block the ERM dephosphorylation induced by SDF-1 (30). In HeLa cells, SDF-1 had no effect on ERM proteins,3 but if ceramide generated at the plasma membrane acts via PLC, then inhibiting PLC should block the dephosphorylation induced by bSMase. As shown in Fig. 1, C and D, inhibition of PLC failed to block the ERM dephosphorylation caused by bSMase and actually facilitated ERM dephosphorylation induced by bSMase. These results suggest that bSMase effect on ERM dephosphorylation follows a different path than PLC activation.
bSMase Treatment Does Not Affect PIP2
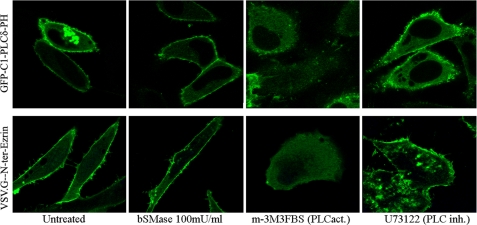
The above results suggested the bSMase effect on ERM phosphorylation was independent of PLC activity. However, we explored whether ceramide generation at the plasma membrane could be affecting ERM phosphorylation by altering the cellular localization of PIP2 or by affecting the interaction of ezrin with PIP2 at the plasma membrane. To investigate these possible effects, we detected cellular PIP2 using the PH domain of PLCδ, which has been shown previously to bind intracellular PIP2 (31) and expressed the N-terminal fragment of ezrin in HeLa cells to confirm plasma membrane localization.
HeLa cells were transiently transfected with GFP-C1-PLCδ-PH plasmid, or with VSV.G-N-ter-Ezrin and treated with bSMase. As a positive control, we used the PLC activator m-3M3FBS (32), and as a negative control, the PLC inhibitor U73122 (33). As shown previously (30), m-3M3FBS caused loss of binding of PH domain of PLCδ to the plasma membrane in Jurkat T cells, and as expected, m-3M3FBS also caused loss of binding to plasma membrane in HeLa cells as well as release of VSV.G-N-ter-Ezrin fragment from the plasma membrane (Fig. 2). Thus, the activator of PLC resulted in loss of both plasma membrane binding and phosphorylation. On the other hand, bSMase treatment did not cause loss of binding of the PH domain of PLCδ, and it also failed to release the N-terminal fragment, providing further evidence that ceramide-driven ERM dephosphorylation does not involve loss of PIP2 and therefore follows an independent mechanism from that of PIP2/PLC.
FIGURE 2.
Effects of bSMase on plasma membrane PIP2. HeLa cells were transfected with GFP-C1-PLCδ-PH or with VSV.G-N-ter-Ezrin and treated with vehicle, bSMase (100 milliunits (mU)/ml, 5 min), m-3M3FBS (100 μm, 5 min), or U-73122 (10 μm, 5 min). GFP-C1-PLCδ-PH specifically binds PIP2 and was localized at the plasma membrane in untreated cells. Treatment with bSMase did not affect GFP-C1-PLCδ-PH localization, whereas m-3M3FBS completely depleted GFP-C1-PLCδ-PH from the plasma membrane. U-73122 also kept GFP-C1-PLCδ-PH at the plasma membrane, although enriched GFP-C1-PLCδ-PH spots at the plasma membrane were observed. Expressed VSV.G-N-ter-Ezrin followed the same pattern than GFP-C1-PLCδ-PH. PLCact, PLC activator; PLC inh., PLC inhibitor.
Myosin Phosphatase Is Not Key Mediator in Ceramide-induced ERM Dephosphorylation
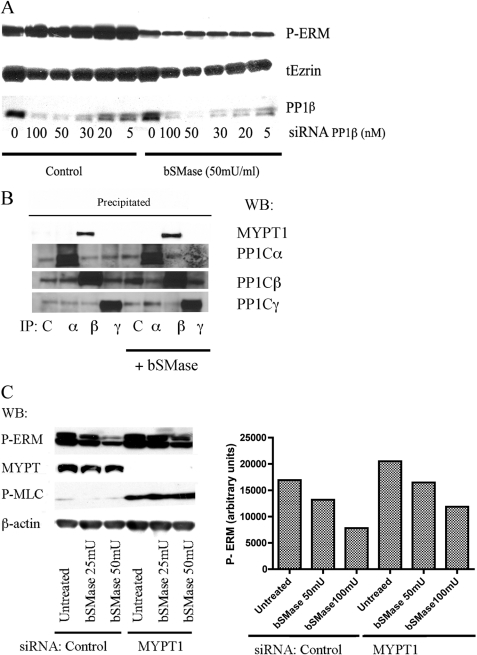
We showed that ceramide-induced ERM dephosphorylation seems to be independent of PIP2 hydrolysis. However, another mechanism, the MYPT1-PP1β-Μ21 complex (also called MP), has been implicated in ERM regulation (34–36) and further explored in additional studies (20, 37). Unlike protein kinases, only a few protein phosphatases are found in the cell; although hundreds of regulatory subunits regulate their specificity, selectivity, localization, and activity (38). As MP has been described as ERM phosphatase, we studied the role of MP on ceramide-induced ERM dephosphorylation. For that purpose, we knocked down the catalytic subunit of MP, the protein phosphatase PP1β, in HeLa cells and then we treated with bSMase (50 milliunits/ml) to induce ERM dephosphorylation. We started observing a decrease in PP1β protein using 5 nm siRNA, but we used increasing concentrations of that siRNA up to 100 nm, with 50 nm as the optimum condition. The effect on ERM dephosphorylation was analyzed by Western blotting (Fig. 3A). As shown previously, bSMase caused a decrease in ERM dephosphorylation; however, knockdown of PP1β could not block this effect.
FIGURE 3.
Study of participation of myosin phosphatase on ceramide-PP1α ERM dephosphorylation. A, HeLa cells were treated with PP1β siRNA for 72 h and then treated with bSMase (50 milliunits (mU)/ml, 5 min) and lysed. PP1β knockdown was confirmed by Western blotting (WB). Phospho-ERM proteins (P-ERM) and total ezrin (tEzrin) were also evaluated by Western blotting. B, HeLa cells were treated with bSMase (50 milliunits/ml, 5 min) and lysed. PP1α, -β, and -γ were immunoprecipitated from the lysate, and co-immunoprecipation of MYPT1 was analyzed. Only PP1β was able to co-immunoprecipitate MYPT1 in control cells and in bSMase-treated cells. C, MYPT1 was knockdown in HeLa cells and treated with bSMase (25 milliunits/ml or 50 milliunits/ml, 5 min) and phospho-ERM (P-ERM) was evaluated by Western blotting. Cells lacking MYPT1 showed a small effect on ERM dephosphorylation after bSMase treatment. Contrarily, MYPT1 knockdown showed a dramatic increase in the phospho-myosin light chain (MLC), although independent of bSMase treatment. β-actin showed equal loading.
MP contains MYPT1 as a regulatory subunit. We could not discard MYPT1 participating in ceramide-induced ERM dephosphorylation. It has already been shown that in basal conditions MYPT1 only binds PP1β (37, 39, 40). However, because PP1 isoforms share a high identity in sequence, and still the key amino acid in PP1 to bind MYPT1 are not clear, MYPT1 might also bind another PP1, at least under some conditions such as in response of bSMase. To address this possibility, HeLa cells were treated with vehicle or bSMase, and the different isoforms of PP1 were immunoprecipitated directly from cell lysates. The immunoprecipitant was analyzed by Western blot to study co-immunoprecipitation of MYPT1 (Fig. 3B). In both, untreated and bSMase-treated cells, there was no evidence of MYPT1 interacting with PP1α or γ but only with PP1β. To completely exclude MYPT1 from ceramide-induced ERM dephosphorylation, MYPT1 was down-regulated in HeLa cells, and the effect of bSMase was studied. As shown in Fig. 3C, knockdown of MYPT1, although increased basal phospho-ERM levels, could not totally block ERM dephosphorylation driven by bSMase, although a small effect was observed. In contradistinction, knock down of MYPT1 resulted in a significant phosphorylation of myosin light chain that was not responsive to bSMase (Fig. 3C). These results support and confirm previous work that has determined specificity of MYPT1 interaction with PP1β and suggesting that ceramide-driven dephosphorylation follows a distinct regulatory mechanism for ERM dephosphorylation.
Effects of Ceramide-activated Phosphatase Inhibitors on bSMase-induced ERM Dephosphorylation
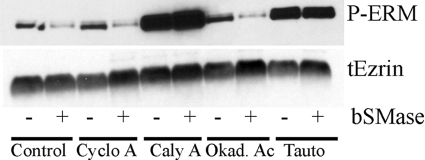
Once we ruled out MP as a ceramide-induced ERM phosphatase, we focused to find which phosphatase dephosphorylated ERM proteins in response of ceramide. Previously, we demonstrated that very short term treatment with bSMase (2–10 min) produced sphingomyelin hydrolysis only at the plasma membrane but did not affect intracellular sphingomyelin. This ceramide generated at the plasma membrane was not further metabolized to other compounds (i.e. complex glycosphingolipids, sphingosine, etc) and was sufficient to induce ERM dephosphorylation in HeLa (28) and other cell lines.3 The major serine/threonine protein phosphatases in eukaryote cells belong to PP1 and PP2A families (41, 42). Moreover, these protein phosphatases have been previously described to be activated in vitro by ceramide by our group (43–46) and others (47, 48). Thus, to determine whether ceramide-driven ERM dephosphorylation was mediated by one of these protein phosphatases, HeLa cells were treated with 50 milliunits/ml of bSMase to induce ERM dephosphorylation, and different protein phosphatase inhibitors were tested for the ability to block ERM dephosphorylation. Of the various inhibitors evaluated, cyclosporine A (PP2B inhibitor), calyculin A (PP1 and PP2A), okadaic acid (PP2A), and tautomycin (PP1 and PP2A at high concentrations), only calyculin A and tautomycin were able to block the dephosphorylation of ERM proteins upon bSMase treatment (Fig. 4). These results pointed to PP1 phosphatases as the candidate mediators, although a PP2A phosphatase could not be completely discarded.
FIGURE 4.
Pharmacological phosphatase inhibitor screening on ceramide-induced ERM dephosphorylation. HeLa cells were pretreated with vehicle or with the protein phosphatase inhibitors cyclosporine A (Cyclo A; 10 μm, 3 h), calyculin A (Caly A; 20 nm, 3 h), okadaic acid (Okad. Ac; 100 nm, 3 h), and tautomycin (Tauto; 200 nm, 3 h) and treated with bSMase (50 milliunits/ml, 5 min). Of the various inhibitors evaluated, calyculin A and tautomycin blocked ERM dephosphorylation induced by bSMase. tEzrin, total ezrin.
Involvement of PP1α Serine/Threonine Protein Phosphatase in bSMase-induced ERM Dephosphorylation
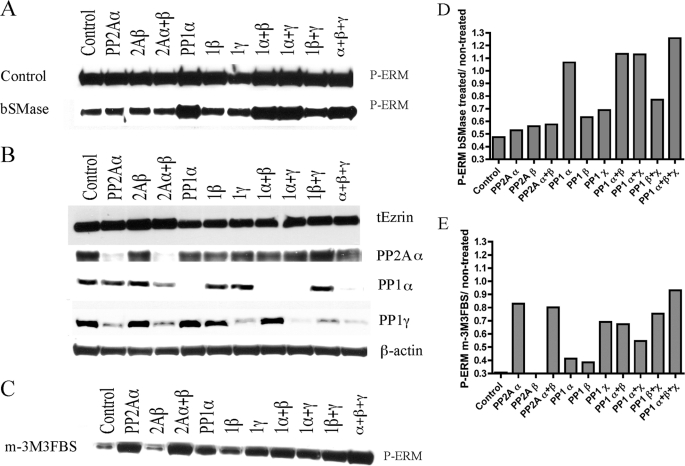
The pharmacological screening pointed to a PP1/PP2A class of phosphatase to block ERM dephosphorylation upon bSMase treatment. To investigate their role in ERM dephosphorylation, individual isoforms of the catalytic subunits of PP2A and PP1 genes were knocked down with specific siRNA previously validated in our group (49). The effects of the siRNA on the ceramide-induced dephosphorylation were evaluated (Fig. 5, A and B, Fig. 5D represents Fig. 5A quantification). In untreated cells, knockdown of all PP1 isoforms resulted in moderate enhancement of basal phosphorylation of ERM proteins and no significant effect by PP2A isoforms. In bSMase-treated cells, only PP1α blocked ERM dephosphorylation, with little or no effect of the other PP1/2A isoforms. These results strongly suggested PP1α as the required phosphatase in bSMase-induced ERM dephosphorylation.
FIGURE 5.
Knockdown of PP1 and PP2A isoforms in ERM dephosphorylation. A, isoforms of PP1 and PP2A phosphatases and combinations within the same family isoforms were knocked down in HeLa cells for 72 h and treated with bSMase (50 milliunits/ml, 5 min) and changes on phospho-ERM (P-ERM) were evaluated by Western blotting. HeLa cells knocked down for PP1α and combination of knockdown containing PP1α blocked bSMase-induced ERM dephosphorylation. B, β-actin showed equal loading, and total ezrin showed that the changes in ERM phosphorylation were due to changes in the phosphorylation state and not because changes in total ezrin (tEzrin) levels. PP2ACα, PP1α, and PP1γ are shown to validate knockdown. Note that there is no PP2ACβ available antibody. C, the PLC activator m-3M3FBS also induced ERM dephosphorylation. Almost all of the knocked down phosphatases tested, with the exception of PP2Aβ, were able to partially block m-3M3FBS-induced ERM dephosphorylation. Western blots from A and C were quantified, and the ratio P-ERM/total ezrin were represented in D and E, respectively.
Effects of PP1 Catalytically Inactive Mutants on bSMase-induced ERM Dephosphorylation
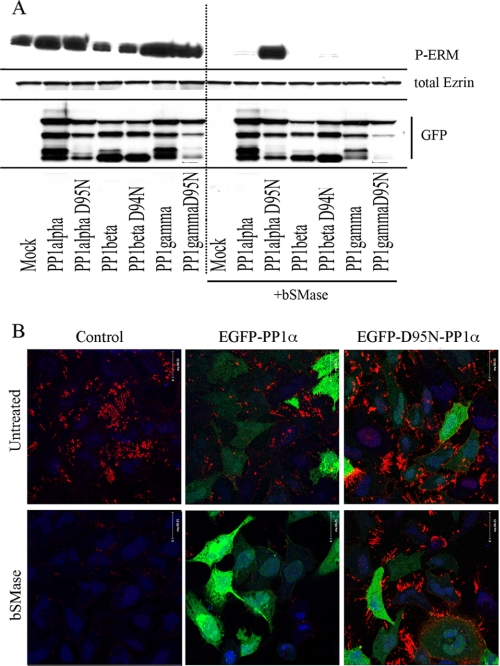
Zhang J et al. (50) published a mutational analysis on PP1α, finding that different single mutations became catalytic inactive. Among those inactive mutants, D95N has been used widely as a dominant negative form (51–53). To further implicate PP1α in ceramide-driven ERM dephosphorylation, D95N PP1α, D94N PP1β, and D95N PP1γ mutants were generated and tested. Because EGFP-PP1 constructs were developed and validated in our group (54, 55), the inactive mutants were generated using EGFP-PP1 as templates. Thus, HeLa cells were transiently transfected with WT EGFP-PP1 isoforms, or with the corresponding catalytically inactive mutants, treated with bSMase and evaluated by Western blot and immunocytochemistry for ERM phosphorylation (Fig. 6, A and B). Before bSMase treatment, all samples showed similar amount of ERM phosphorylation, suggesting that when ceramide was not generated at the plasma membrane none of these phosphatases was especially active toward ERM proteins. However, after bSMase, all the cells transfected with the different PP1 constructs were dephosphorylated excepting the PP1α mutant (Fig. 6A). Thus, D95N EGFP-PP1α blocked bSMase-induced ERM dephosphorylation, whereas the catalytic inactive mutants for PP1β and PP1γ failed. Immunohistochemistry studies also substantiated the ability of the D95N dominant-negative construct to prevent the effects of bSMase on ERM dephosphorylation (Fig. 6B). These results further implicate PP1α and not other PP1 isoforms as mediators of ceramide-induced dephosphorylation of ERM proteins.
FIGURE 6.
Effects of expression of wild type and catalytic inactive mutants PP1 isoforms on ceramide-induced ERM dephosphorylation. A, EGFP-PP1α, -β, and -γ constructs and a respective catalytic inactive mutant were expressed in HeLa cells for 8 h. Next, cells were treated with bSMase (50 milliunits/ml, 5 min), and ERM dephosphorylation was evaluated by Western blotting (phospho-ezrin, P-ERM). Total ezrin was measured to show no changes in total ezrin. GFP was measured to confirm transfection. Cell transfected with PP1α catalytic inactive mutant (PP1α D95N) resulted in insensitivity to bSMase. B, HeLa cells were transfected with WT PP1α or the inactive form, and the effect of bSMase on P-ERM was visualized using a confocal microscope. Phospho-ezrin was visualized in red, EGFP was shown in green, and nuclei were shown in blue.
Distinction between bSMase- and PLC-dependent ERM Dephosphorylation in PP1α Knockdown Cells
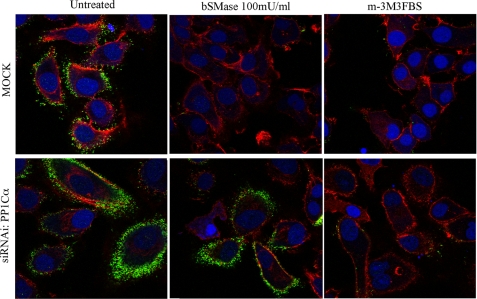
The previous results suggested that ceramide-induced ERM dephosphorylation follows a different mechanism than PIP2-dependent dephosphorylation, and it is independent from MP. We also investigated whether PIP2 hydrolysis-dependent ERM dephosphorylation was regulated by the same phosphatase as ceramide-driven dephosphorylation. For that purpose, several phosphatases were knocked down in HeLa cells and treated with m-3M3FBS, and the effects on ERM dephosphorylation were studied by Western blotting, as shown before for bSMase. Interestingly, as shown in Fig. 5C (quantified in Fig. 5E), there was not a clear phosphatase involved in PIP2 hydrolysis-dependent ERM dephosphorylation. Indeed, knock down of several phosphatases was able to partly overcome dephosphorylation of ERM, unlike the more specific role for PP1α in bSMase-induced dephosphorylation. Moreover, and in specific evaluation of PP1α, we compared ERM dephosphorylation induced by ceramide and by PIP2 hydrolysis after knock down of PP1α (Fig. 7). The results showed that knockdown of PP1α was able to specifically inhibit bSMase but not m-3M3FBS-induced dephosphorylation. These experiments therefore demonstrate that PP1α is required for ceramide-dependent ERM phosphorylation but not for PIP2 hydrolysis-dependent dephosphorylation, whereas other phosphatases were required for PIP2 hydrolysis-dependent ERM dephosphorylation.
FIGURE 7.
PP1α knockdown effects on ceramide- and PIP2-induced ERM dephosphorylation. HeLa cells were knocked down for PP1α and treated with bSMase (100 milliunits (mU)/ml, 5 min) or m-3M3FBS (100 μm, 5 min), and the effect on phospho-ERM was visualized using a confocal microscope. Phospho-ERM was localized at the plasma membrane. Both bSMase and m-3M3FBS induced ERM dephosphorylation. Knocking down PP1α blocked bSMase-induced phosphorylation, but it did not block m-3M3FBS-induced dephosphorylation. Nuclei and F-actin were visualized with Draq5 (blue) and rhodamine-phalloidin (red) staining and stained with phospho-ERM antibody (green).
Ceramide Requirement for PP1α Activation/Recruitment
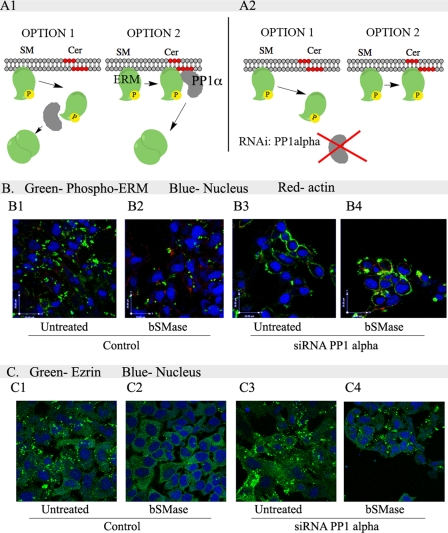
To understand how ceramide generation at the plasma membrane induces ERM dephosphorylation, we evaluated two alternative mechanisms: 1) generation of ceramide at the plasma membrane disrupts the interaction between N-terminal ERM domain and the plasma membrane. Once ERM is detached from the plasma membrane, it is then dephosphorylated by PP1α (Fig. 8, A1, option 1). This mechanism would be similar to what is currently understood for loss of PIP2-induced dephosphorylation. 2) Alternatively, generation of ceramide at the plasma membrane activates PP1α, which then dephosphorylates ERM and subsequently induces ERM detachment from the plasma membrane (Fig. 8, A1, option 2). To determine whether ceramide modulates plasma membrane-ERM binding or activates PP1α, PP1α was down-regulated in HeLa cells and treated with bSMase, and the localization of phospho-ERM and total ezrin was studied. Before bSMase treatment, phospho-ERM was localized at the plasma membrane (Fig. 8, B1), and after bSMase treatment, the majority of ERM proteins were dephosphorylated (Fig. 8, B2). Total ezrin was localized to the cytoplasm and accumulated in plasma membrane protusions (Fig. 8, C1), whereas bSMase treatment led to diminished accumulation at the plasma membrane (Fig. 8, C2). In PP1α knockdown cells, bSMase did not cause ERM dephosphorylation, and phospho-ERM was localized at the plasma membrane and not in the cytoplasm (Fig. 8, B3 and B4). Total ezrin localized in the cytoplasm and at the plasma membrane (Fig. 8, C3 and C4). These results suggest that ceramide is required to activate PP1α toward ERM (Fig. 8, A2, option 2) and not to interfere in ERM-plasma membrane interaction (Fig. 8, A2, option 1).
FIGURE 8.
Role of ceramide in PP1α-dependent ERM dephosphorylation. A1, ceramide generated at the plasma membrane induces ERM dephosphorylation requiring PP1α. We proposed two possible mechanisms of ceramide-PP1α action. In option 1, generation of ceramide (Cer) produces a loss in ERM interaction with the plasma membrane, and then ERM is dephosphorylated at the cytoplasm by PP1α. Option 2 proposes ceramide generation at the plasma membrane activates PP1α that dephosphorylates ERM proteins detaching them form the plasma membrane. A2, each option would respond differently in the localization of phospho-ERM in cells knockdown for PP1α after bSMase treatment. In option 1, after bSMase treatment, phospho-ERM should be localized at the cytosol, whereas in option 2, phospho-ERM would be kept at the plasma membrane. B, HeLa cells were knocked down for PP1α, treated with bSMase and the localization of phospho-ERM was studied by confocal microscopy. Nuclei and F-actin were visualized with Draq5 (blue) and rhodamine-phalloidin (red) staining and stained with phospho-ERM antibody (green). C, also total ezrin, which show clear cytosol localization, forming aggregates at the plasma membrane was visualized in HeLa cells lacking PP1α after bSMase treatment. Total ezrin was stained with specific antibody (green). SM, sphingomyelin. B and C, 1, untreated; 2, bSMase treated; 3, cells knocked down for PP1α; 4, cells knocked down for PP1α and treated with bSMase.
DISCUSSION
In this paper, we present a new mechanism of acute regulation of ERM (ezrin, radixin, and moesin) by ceramide and the PP1α isoform of PP1. Thus, ceramide generated at the plasma membrane induces ERM dephosphorylation through PP1α activation, detaches ERM from the plasma membrane, and leads to inactive ERM. This mechanism is distinct from the previously reported mechanisms of ERM regulation, by either hydrolysis of PIP2 and ERM detachment from the plasma, and/or by dephosphorylation by MYPT1-PP1β (MP). These results have implications for the regulation of ERM proteins, the functions of sphingolipids at the plasma membrane, specifically ceramide, and the regulation of protein phosphatases.
In the past 20 years, understanding the mechanisms of ERM regulation has evolved significantly. ERM are regulated by phosphorylation of one of the most C-terminal threonines, switching them from an inactive conformation to an active conformation. When the N terminus interacts with the plasma membrane, a process requiring PIP2, ERM can be phosphorylated and then interacts with the cortical actin fibers. The only known mechanism of ERM inactivation thus far involves hydrolysis of PIP2 by PLC. This hydrolysis results in detachment of the N terminus of ERM from the plasma membrane and dephosphorylation of the C terminus by an unknown phosphatase. In this mechanism, detachment appears to precede dephosphorylation. Independently, several protein phosphatases have been suggested in ERM dephosphorylation, with MP (MYPT1-PP1β-M20) as the most cited. Recently, we published that ERM proteins are regulated acutely by ceramide and sphingosine 1-phosphate. Ceramide generation at the plasma membrane induced a rapid ERM dephosphorylation, whereas hydrolysis of that ceramide and formation of sphingosine 1-phosphate counteracted this effect. In the present study, we found that ceramide-driven ERM dephosphorylation involves a different and novel mechanism rather than PIP2/MP: a mechanism requiring PP1α.
The first significant finding from this study relates to establishing PP1α as a specific target for ceramide, mediating the effects of plasma membrane ceramide on ezrin. Ceramide has been demonstrated to be a bioactive lipid more than just a structural component of the biological membranes. Indeed, ceramide has been found to activate several proteins due to its capacity to interact with them such as cathepsin D or atypical forms of PKC. The major in vitro defined targets for ceramide have been the protein phosphatases PP2A and PP1. We suspected that ceramide-induced ERM dephosphorylation might be catalyzed by one of these phosphatases. To this end, we employed different techniques to probe a ceramide-activated phosphatase on ceramide-induced ERM dephosphorylation. Thus, pharmacological inhibitors pointed to PP1 and PP2A phosphatases. PP2A, PP1β, and PP1γ were excluded using siRNA. The role of PP1α was determined with siRNA approaches and further confirmed using dominant-negative PP1, defining PP1α as a new player in ERM regulation.
More importantly, the findings from this study addressed the question of how PP1α fits in what is known about ERM regulation. This led to defining a novel mechanism of regulation of ERM that appears to be independent of previously studied mechanisms. As shown by other studies, the cytokine SDF-1 activates PLC, which catalyzes PIP2 hydrolysis, leading to ERM inactivation in Jurkat cells. Other studies also have shown regulation of ERM by PIP2. To determine whether ceramide-PP1α was in the same axis or in a parallel one, we first confirmed that activation of PLC led to ERM dephosphorylation in HeLa cells. However, inhibition of PLC was unable to block ceramide-dependant ERM dephosphorylation. That result suggested that even though ERM proteins can be regulated by PLC activation in HeLa cells, other PLC-independent mechanisms are possible, although not involving or excluding the regulation by PIP2 levels at the plasma membrane. However, we could exclude PIP2 hydrolysis as a mechanism of ceramide-driven dephosphorylation by showing that 1) ceramide production at the plasma membrane did not affect PIP2 localization nor the localization of an N-terminal ezrin fragment at the plasma membrane; 2) knockdown experiments demonstrated that PP1α down-regulation could not block PIP2-dependent dephosphorylation but blocked the ceramide-dependent dephosphorylation; 3) knockdown of PP1α did not result in dissociation of ERM from the plasma membrane; thus, the pathway initiated by ceramide results in dephosphorylation first, whereas PIP2 hydrolysis results in dissociation followed by dephosphorylation. 4) Moreover, PIP2 hydrolysis induced a dephosphorylation that was inhibited partially by knocking down distinct phosphatases, suggesting that once ERM is detached from the PIP2, almost any phosphatase can have access to the C-terminal phosphosite, thus establishing PP1α as a specific phosphatase that can inactivate ERM independently of PLC and PIP2 but is dependent of activation by ceramide.
Continuing that work, we wanted to explore whether ceramide-induced ERM dephosphorylation could be also regulated by MP as MP has been reported widely as an ERM phosphatase. MP is a protein complex containing PP1β and the regulatory subunit MYPT1. As shown previously in this work, PP1β and MYPT1 were excluded by the siRNA experiments. Moreover, MYPT1 has been described recently to bind uniquely the PP1β isoform. We confirmed that specificity of interaction; however, the results showed that PP1β and MYPT1 are not key mediators of the ceramide response, although MYPT1 could have some effect, mainly regulating ERM basal phospholevels, which would modulate the ceramide effect on ERM dephosphorylation.
The results from this study also provide significant support for the emerging concept of compartment-specific functions of ceramide. Ceramide has been considered widely as a bioactive effector without distinguishing among cellular compartments or species. Recent studies, however, have begun to address compartment-specific functions of ceramide (56) .One possible mechanism of the differential response of compartment-specific ceramide may depend on what ceramide-responding proteins are present in that compartment. Concerning plasma membrane ceramide, PP1α could be one of the proteins that translate a specific signal from the plasma membrane, whereas ceramide generated in other compartments may propagate signals through other targets.
The results from this study also raise implications as to the roles of ceramide-mediated pathways and the ERM family in pathobiology and most notably in cancer. Indeed, the ERM proteins have been increasingly implicated in cancer development and metastasis, such that active ERM proteins positively correlate with metastatic events and regulate processes (migration, adhesion) that are intimately related to metastasis. Likewise, ceramide has been shown to reduce cell motility and is involved in cancer regression, leading to the formulation of an anti-tumor function for ceramide. Moreover, PP1 and PP2A inhibitors have been shown to block cell migration and adhesion. In this work, we suggest a mechanism that connects these key players involving ceramide, phosphatases, and ERM proteins.
In conclusion, our results define a novel mechanism of ERM regulation, which is independent of other known ERM regulatory mechanisms, involving ceramide production at the plasma membrane and specifically activation of the PP1α isoform.
Acknowledgments
We thank Dr. Michael Airola, David Perry and María José Hernández-Corbacho for careful review of the manuscript. We thank all members in the Hannun and Obeid laboratories. We also thank the MUSC Cell and Molecular Imaging Core. We thank Tobias Meyer, who is the original producer of the GFP-C1-PLCδ-PH plasmid (31).
This work was supported in part by National Institutes of Health Grants CA87584 and CA97132.
D. Canals, P. Roddy, and Y. A. Hannun, unpublished results.
- PIP2
- phosphatidylinositol 4, 5-biphosphate
- PP1
- protein phosphatase 1
- bSMase
- bacterial sphingomyelinase
- PLC
- phospholipase C.
REFERENCES
- 1. Bretscher A., Edwards K., Fehon R. G. (2002) ERM proteins and merlin: Integrators at the cell cortex. Nat. Rev. Mol. Cell Biol. 3, 586–599 [DOI] [PubMed] [Google Scholar]
- 2. Tsukita S., Yonemura S. (1997) ERM (ezrin/radixin/moesin) family: From cytoskeleton to signal transduction. Curr. Opin. Cell Biol. 9, 70–75 [DOI] [PubMed] [Google Scholar]
- 3. Arpin M., Chirivino D., Naba A., Zwaenepoel I. (2011) Emerging role for ERM proteins in cell adhesion and migration. Cell Adh. Migr. 5, 199–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lozupone F., Lugini L., Matarrese P., Luciani F., Federici C., Iessi E., Margutti P., Stassi G., Malorni W., Fais S. (2004) Identification and relevance of the CD95-binding domain in the N-terminal region of ezrin. J. Biol. Chem. 279, 9199–9207 [DOI] [PubMed] [Google Scholar]
- 5. Tsukita S., Yonemura S. (1997) ERM proteins: Head-to-tail regulation of actin-plasma membrane interaction. Trends Biochem. Sci. 22, 53–58 [DOI] [PubMed] [Google Scholar]
- 6. Helander T. S., Carpén O., Turunen O., Kovanen P. E., Vaheri A., Timonen T. (1996) ICAM-2 redistributed by ezrin as a target for killer cells. Nature 382, 265–268 [DOI] [PubMed] [Google Scholar]
- 7. Yonemura S., Hirao M., Doi Y., Takahashi N., Kondo T., Tsukita S. (1998) Ezrin/radixin/moesin (ERM) proteins bind to a positively charged amino acid cluster in the juxta-membrane cytoplasmic domain of CD44, CD43, and ICAM-2. J. Cell Biol. 140, 885–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Short D. B., Trotter K. W., Reczek D., Kreda S. M., Bretscher A., Boucher R. C., Stutts M. J., Milgram S. L. (1998) An apical PDZ protein anchors the cystic fibrosis transmembrane conductance regulator to the cytoskeleton. J. Biol. Chem. 273, 19797–19801 [DOI] [PubMed] [Google Scholar]
- 9. Denker S. P., Huang D. C., Orlowski J., Furthmayr H., Barber D. L. (2000) Direct binding of the Na–H exchanger NHE1 to ERM proteins regulates the cortical cytoskeleton and cell shape independently of H+ translocation. Mol. Cell 6, 1425–1436 [DOI] [PubMed] [Google Scholar]
- 10. Lamb R. F., Ozanne B. W., Roy C., McGarry L., Stipp C., Mangeat P., Jay D. G. (1997) Essential functions of ezrin in maintenance of cell shape and lamellipodial extension in normal and transformed fibroblasts. Curr. Biol. 7, 682–688 [DOI] [PubMed] [Google Scholar]
- 11. Serrador J. M., Alonso-Lebrero J. L., del Pozo M. A., Furthmayr H., Schwartz-Albiez R., Calvo J., Lozano F., Sánchez-Madrid F. (1997) Moesin interacts with the cytoplasmic region of intercellular adhesion molecule-3 and is redistributed to the uropod of T lymphocytes during cell polarization. J. Cell Biol. 138, 1409–1423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li Q., Nance M. R., Kulikauskas R., Nyberg K., Fehon R., Karplus P. A., Bretscher A., Tesmer J. J. (2007) Self-masking in an intact ERM-merlin protein: An active role for the central α-helical domain. J. Mol. Biol. 365, 1446–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zohar R., Suzuki N., Suzuki K., Arora P., Glogauer M., McCulloch C. A., Sodek J. (2000) Intracellular osteopontin is an integral component of the CD44-ERM complex involved in cell migration. J. Cell. Physiol. 184, 118–130 [DOI] [PubMed] [Google Scholar]
- 14. Endo K., Kondo S., Shackleford J., Horikawa T., Kitagawa N., Yoshizaki T., Furukawa M., Zen Y., Pagano J. S. (2009) Phosphorylated ezrin is associated with EBV latent membrane protein 1 in nasopharyngeal carcinoma and induces cell migration. Oncogene 28, 1725–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Prag S., Parsons M., Keppler M. D., Ameer-Beg S. M., Barber P., Hunt J., Beavil A. J., Calvert R., Arpin M., Vojnovic B., Ng T. (2007) Activated ezrin promotes cell migration through recruitment of the GEF Dbl to lipid rafts and preferential downstream activation of Cdc42. Mol. Biol. Cell 18, 2935–2948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Machesky L. M. (2008) Lamellipodia and filopodia in metastasis and invasion. FEBS Lett. 582, 2102–2111 [DOI] [PubMed] [Google Scholar]
- 17. Matsui T., Maeda M., Doi Y., Yonemura S., Amano M., Kaibuchi K., Tsukita S. (1998) Rho-kinase phosphorylates COOH-terminal threonines of ezrin/radixin/moesin (ERM) proteins and regulates their head-to-tail association. J. Cell Biol. 140, 647–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chuan Y. C., Pang S. T., Cedazo-Minguez A., Norstedt G., Pousette A., Flores-Morales A. (2006) Androgen induction of prostate cancer cell invasion is mediated by ezrin. J. Biol. Chem. 281, 29938–29948 [DOI] [PubMed] [Google Scholar]
- 19. Wald F. A., Oriolo A. S., Mashukova A., Fregien N. L., Langshaw A. H., Salas P. J. (2008) Atypical protein kinase C (ι) activates ezrin in the apical domain of intestinal epithelial cells. J. Cell Sci. 121, 644–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nakamura N., Oshiro N., Fukata Y., Amano M., Fukata M., Kuroda S., Matsuura Y., Leung T., Lim L., Kaibuchi K. (2000) Phosphorylation of ERM proteins at filopodia induced by Cdc42. Genes Cells 5, 571–581 [DOI] [PubMed] [Google Scholar]
- 21. Cant S. H., Pitcher J. A. (2005) G protein-coupled receptor kinase 2-mediated phosphorylation of ezrin is required for G protein-coupled receptor-dependent reorganization of the actin cytoskeleton. Mol. Biol. Cell 16, 3088–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zeidan Y. H., Jenkins R. W., Hannun Y. A. (2008) Remodeling of cellular cytoskeleton by the acid sphingomyelinase/ceramide pathway. J. Cell Biol. 181, 335–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hishiya A., Ohnishi M., Tamura S., Nakamura F. (1999) Protein phosphatase 2C inactivates F-actin binding of human platelet moesin. J. Biol. Chem. 274, 26705–26712 [DOI] [PubMed] [Google Scholar]
- 24. Hla T. (2004) Prostaglandins and other lipid mediators: A new phase, a new team. Prostaglandins Other Lipid Mediat. 73, 1–2 [DOI] [PubMed] [Google Scholar]
- 25. Yu H., Okada T., Kobayashi M., Abo-Elmatty D. M., Jahangeer S., Nakamura S. (2009) Roles of extracellular and intracellular sphingosine 1-phosphate in cell migration. Genes Cells 14, 597–605 [DOI] [PubMed] [Google Scholar]
- 26. Smith E. R., Merrill A. H., Obeid L. M., Hannun Y. A. (2000) Effects of sphingosine and other sphingolipids on protein kinase C. Methods Enzymol. 312, 361–373 [DOI] [PubMed] [Google Scholar]
- 27. Meivar-Levy I., Sabanay H., Bershadsky A. D., Futerman A. H. (1997) The role of sphingolipids in the maintenance of fibroblast morphology. The inhibition of protrusional activity, cell spreading, and cytokinesis induced by fumonisin B1 can be reversed by ganglioside GM3. J. Biol. Chem. 272, 1558–1564 [DOI] [PubMed] [Google Scholar]
- 28. Canals D., Jenkins R. W., Roddy P., Hernández-Corbacho M. J., Obeid L. M., Hannun Y. A. (2010) Differential effects of ceramide and sphingosine 1-phosphate on ERM phosphorylation: Probing sphingolipid signaling at the outer plasma membrane. J. Biol. Chem. 285, 32476–32485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hannun Y. A., Obeid L. M. (2008) Principles of bioactive lipid signaling: Lessons from sphingolipids. Nat. Rev. Mol. Cell Biol. 9, 139–150 [DOI] [PubMed] [Google Scholar]
- 30. Hao J. J., Liu Y., Kruhlak M., Debell K. E., Rellahan B. L., Shaw S. (2009) Phospholipase C-mediated hydrolysis of PIP2 releases ERM proteins from lymphocyte membrane. J. Cell Biol. 184, 451–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stauffer T. P., Ahn S., Meyer T. (1998) Receptor-induced transient reduction in plasma membrane PtdIns(4,5)P2 concentration monitored in living cells. Curr. Biol. 8, 343–346 [DOI] [PubMed] [Google Scholar]
- 32. Bae Y. S., Lee T. G., Park J. C., Hur J. H., Kim Y., Heo K., Kwak J. Y., Suh P. G., Ryu S. H. (2003) Identification of a compound that directly stimulates phospholipase C activity. Mol. Pharmacol. 63, 1043–1050 [DOI] [PubMed] [Google Scholar]
- 33. Bleasdale J. E., Bundy G. L., Bunting S., Fitzpatrick F. A., Huff R. M., Sun F. F., Pike J. E. (1989) Inhibition of phospholipase C-dependent processes by U-73, 122. Adv. Prostaglandin Thromboxane Leukot. Res. 19, 590–593 [PubMed] [Google Scholar]
- 34. Fukata Y., Kimura K., Oshiro N., Saya H., Matsuura Y., Kaibuchi K. (1998) Association of the myosin-binding subunit of myosin phosphatase and moesin: Dual regulation of moesin phosphorylation by Rho-associated kinase and myosin phosphatase. J. Cell Biol. 141, 409–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kimura K., Fukata Y., Matsuoka Y., Bennett V., Matsuura Y., Okawa K., Iwamatsu A., Kaibuchi K. (1998) Regulation of the association of adducin with actin filaments by Rho-associated kinase (Rho-kinase) and myosin phosphatase. J. Biol. Chem. 273, 5542–5548 [DOI] [PubMed] [Google Scholar]
- 36. Kawano Y., Fukata Y., Oshiro N., Amano M., Nakamura T., Ito M., Matsumura F., Inagaki M., Kaibuchi K. (1999) Phosphorylation of myosin-binding subunit (MBS) of myosin phosphatase by Rho-kinase in vivo. J. Cell Biol. 147, 1023–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Eto M., Kirkbride J. A., Brautigan D. L. (2005) Assembly of MYPT1 with protein phosphatase-1 in fibroblasts redirects localization and reorganizes the actin cytoskeleton. Cell Motil. Cytoskeleton 62, 100–109 [DOI] [PubMed] [Google Scholar]
- 38. Virshup D. M., Shenolikar S. (2009) From promiscuity to precision: Protein phosphatases get a makeover. Mol. Cell 33, 537–545 [DOI] [PubMed] [Google Scholar]
- 39. Alessi D., MacDougall L. K., Sola M. M., Ikebe M., Cohen P. (1992) The control of protein phosphatase-1 by targetting subunits. The major myosin phosphatase in avian smooth muscle is a novel form of protein phosphatase-1. Eur. J. Biochem. 210, 1023–1035 [DOI] [PubMed] [Google Scholar]
- 40. Scotto-Lavino E., Garcia-Diaz M., Du G., Frohman M. A. (2010) Basis for the isoform-specific interaction of myosin phosphatase subunits protein phosphatase 1c β and myosin phosphatase targeting subunit 1. J. Biol. Chem. 285, 6419–6424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Oliver C. J., Shenolikar S. (1998) Physiologic importance of protein phosphatase inhibitors. Front. Biosci. 3, D961–972 [DOI] [PubMed] [Google Scholar]
- 42. Shi Y. (2009) Assembly and structure of protein phosphatase 2A. Sci. China C Life Sci. 52, 135–146 [DOI] [PubMed] [Google Scholar]
- 43. Dobrowsky R. T., Hannun Y. A. (1992) Ceramide stimulates a cytosolic protein phosphatase. J. Biol. Chem. 267, 5048–5051 [PubMed] [Google Scholar]
- 44. Dobrowsky R. T., Kamibayashi C., Mumby M. C., Hannun Y. A. (1993) Ceramide activates heterotrimeric protein phosphatase 2A. J. Biol. Chem. 268, 15523–15530 [PubMed] [Google Scholar]
- 45. Galadari S., Kishikawa K., Kamibayashi C., Mumby M. C., Hannun Y. A. (1998) Purification and characterization of ceramide-activated protein phosphatases. Biochemistry 37, 11232–11238 [DOI] [PubMed] [Google Scholar]
- 46. Chalfant C. E., Kishikawa K., Mumby M. C., Kamibayashi C., Bielawska A., Hannun Y. A. (1999) Long chain ceramides activate protein phosphatase-1 and protein phosphatase-2A. Activation is stereospecific and regulated by phosphatidic acid. J. Biol. Chem. 274, 20313–20317 [DOI] [PubMed] [Google Scholar]
- 47. Kowluru A., Metz S. A. (1997) Ceramide-activated protein phosphatase-2A activity in insulin-secreting cells. FEBS Lett. 418, 179–182 [DOI] [PubMed] [Google Scholar]
- 48. Leoni L. M., Shih H. C., Deng L., Tuey C., Walter G., Carson D. A., Cottam H. B. (1998) Modulation of ceramide-activated protein phosphatase 2A activity by low molecular weight aromatic compounds. Biochem. Pharmacol. 55, 1105–1111 [DOI] [PubMed] [Google Scholar]
- 49. Kitatani K., Idkowiak-Baldys J., Bielawski J., Taha T. A., Jenkins R. W., Senkal C. E., Ogretmen B., Obeid L. M., Hannun Y. A. (2006) Protein kinase C-induced activation of a ceramide/protein phosphatase 1 pathway leading to dephosphorylation of p38 MAPK. J. Biol. Chem. 281, 36793–36802 [DOI] [PubMed] [Google Scholar]
- 50. Zhang J., Zhang Z., Brew K., Lee E. Y. (1996) Mutational analysis of the catalytic subunit of muscle protein phosphatase-1. Biochemistry 35, 6276–6282 [DOI] [PubMed] [Google Scholar]
- 51. Gallego M., Kang H., Virshup D. M. (2006) Protein phosphatase 1 regulates the stability of the circadian protein PER2. Biochem. J. 399, 169–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Luo W., Peterson A., Garcia B. A., Coombs G., Kofahl B., Heinrich R., Shabanowitz J., Hunt D. F., Yost H. J., Virshup D. M. (2007) Protein phosphatase 1 regulates assembly and function of the β-catenin degradation complex. EMBO J. 26, 1511–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hutchinson J. A., Shanware N. P., Chang H., Tibbetts R. S. (2011) Regulation of ribosomal protein S6 phosphorylation by casein kinase 1 and protein phosphatase 1. J. Biol. Chem. 286, 8688–8696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Jones J. A., Rawles R., Hannun Y. A. (2005) Identification of a novel phosphatidic acid binding domain in protein phosphatase-1. Biochemistry 44, 13235–13245 [DOI] [PubMed] [Google Scholar]
- 55. Marchesini N., Jones J. A., Hannun Y. A. (2007) Confluence-induced threonine 41/serine 45 phospho-β-catenin dephosphorylation via ceramide-mediated activation of PP1cγ. Biochim. Biophys. Acta 1771, 1418–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hannun Y. A., Obeid L. M. (2011) Many ceramides. J. Biol. Chem. 286, 27855–27862 [DOI] [PMC free article] [PubMed] [Google Scholar]