Background: Nematocysts are the most sophisticated organelles in the animal kingdom and a hallmark of the phylum Cnidaria.
Results: We present the first complete protein map of the Hydra nematocyst.
Conclusion: The nematocyst proteome is highly complex and includes novel structural proteins.
Significance: The proteome data reveal a eukaryotic origin of the nematocyst and point to common molecular features with the extracellular matrix.
Keywords: Animal Models, Evolution, Extracellular Matrix, Invertebrates, Proteomics, Cnidarians, Hydra, Organelle
Abstract
Stinging cells or nematocytes of jellyfish and other cnidarians represent one of the most poisonous and sophisticated cellular inventions in animal evolution. This ancient cell type is unique in containing a giant secretory vesicle derived from the Golgi apparatus. The organelle structure within the vesicle comprises an elastically stretched capsule (nematocyst) to which a long tubule is attached. During exocytosis, the barbed part of the tubule is accelerated with >5 million g in <700 ns, enabling a harpoon-like discharge (Nüchter, T., Benoit, M., Engel, U., Ozbek, S., and Holstein, T. W. (2006) Curr. Biol. 16, R316–R318). Hitherto, the molecular components responsible for the organelle's biomechanical properties were largely unknown. Here, we describe the proteome of nematocysts from the freshwater polyp Hydra magnipapillata. Our analysis revealed an unexpectedly complex secretome of 410 proteins with venomous and lytic but also adhesive or fibrous properties. In particular, the insoluble fraction of the nematocyst represents a functional extracellular matrix structure of collagenous and elastic nature. This finding suggests an evolutionary scenario in which exocytic vesicles harboring a venomous secretome assembled a sophisticated predatory structure from extracellular matrix motif proteins.
Introduction
Venoms are produced in probably all animal phyla (2, 3) and represent a mixture of effector proteins often aimed at tissue integrity by targeting extracellular matrix (ECM)3 and receptor molecules. Intriguingly, many toxins share close structural and functional properties with ECM proteins, indicating a common evolutionary origin or co-evolution (4). It was hitherto unknown whether this process also contributed to ECM formation in providing efficient organ or organelle structures for toxin delivery.
Nematocysts or cnidae are highly complex projectile organelles used for capture of prey and defense in all cnidarians (5, 6). They are produced in specialized stem cell derivatives called nematocytes by a continuous secretory process of proteins into a giant post-Golgi vesicle. Nematocysts are composed of a cylindrical capsule body to which an extended tubule is attached that is often armed with spines. The capsule morphology varies among different cnidarian species, with a general tendency toward higher complexity in medusozoans compared with anthozoans (7). In Hydra, four types of nematocysts are present: the small desmonemes, with a tightly coiled tubule used for prey attachment; the holotrichous and spineless atrichous isorhizas; and the large stenoteles, with a prominent stylet apparatus at the tubule base employed for piercing solid cuticle structures (8–10). The molecular composition of the capsule comprises a highly dense collagenous polymer charged with an osmotic pressure of 150 bars (11, 12). Nematocyst discharge, triggered by the mechanosensory cnidocil apparatus, is one of the fastest processes in biology and creates an impact on the harpoon-like expelled tubule comparable with technical bullets (1). Of the protein components constituting the nematocyst, only a small fraction has hitherto been defined. These include mainly structural proteins like the large family of minicollagens (13–15), the C-type lectin NOWA (16), and spinalin, which is part of the tubule spines (17). Here, we present a proteome analysis of Hydra nematocysts, showing an unusual composition of venomous and structural proteins forming one of the most sophisticated organelles in the animal kingdom.
EXPERIMENTAL PROCEDURES
Animals
Hydra magnipapillata was used for all experiments. Animals were cultured in Hydra medium at 18 °C and fed two to three times a week with freshly hatched Artemia salina nauplii. Animals used for the experiments were starved for 24 h. Intact nematocysts were isolated from whole Hydra tissue as described (18).
One-dimensional Electrophoresis and OrbitrapTM MS
Nematocyst proteins were separated on a one-dimensional NuPAGE 4–12% BisTris gel (pH 6.4) (Invitrogen) and stained with colloidal Coomassie Blue. The resulting gel was cut into 27 pieces. In-gel tryptic digestion and extraction were performed as described previously (19). Peptide separation was achieved using a nano-ACQUITY UPLC® system (Waters). The nano-UPLC system was coupled on-line to an LTQ Orbitrap XL mass spectrometer (Thermo Fisher). Data-dependent acquisition using Xcalibur 2.0.6 (Thermo Fisher) was performed by one Fourier transform MS scan with a resolution of 60,000 and a range from m/z 370 to 2000 in parallel with six MS/MS scans of the most intense precursor ions in the ion trap. The mgf files were used for database searches with the MASCOT search engine (Matrix Science, London, United Kingdom) against Hydra Hma2 protein models (20).
Two-dimensional Electrophoresis and Electrospray Ionization Quadrupole TOF-MS
Protein separation was performed with the PROTEAN system (Bio-Rad). The protein samples were applied to PROTEAN ReadyStrip immobilized pH gradient (IPG) 17-cm pH 3.0–10.0 strips (Bio-Rad). Isolated capsules (6 million) were solubilized in 260 mm DTT and 1× PBS for 3 h at 29 °C. The soluble fraction (supernatant) was precipitated with chloroform/methanol and loaded onto the IPG strip (passive rehydration overnight), and the insoluble fraction (pellet) was further solubilized in lysis buffer (7 m urea, 2 m thiourea, 4% CHAPS, 1% IPG buffer (PharmalyteTM), and 130 mm DTT) at 29 °C for 3 h before precipitation and loading onto the IPG strip. Rehydration was done passively overnight with rehydration buffer (7 m urea, 2 m thiourea, 2% (w/v) CHAPS, 2% (v/v) IPG buffer (IPG pH 3–10 strip), and 18.2 mm DTT). Isoelectric focusing was done with the Bio-Rad IEF system. The second dimension electrophoresis was done on 12% polyacrylamide gels in 30 mm Tris-HCl (pH 8.8) and 0.2% SDS at a constant current mode of 17 mA/gel for 12 h with running buffer (2.5 mm Tris-HCl (pH 8.3), 19.2 mm glycine, and 0.01% SDS) in a Bio-Rad PROTEAN II xi cell system. To visualize the protein spots on the gel, the colloidal Coomassie Blue staining procedure was employed. From the two fractions together, 186 spots were visualized, excised, and digested with trypsin as described (19) Nanoscale LC-electrospray ionization MS/MS analysis of the protein extract was performed using the CapLC capillary LC system (Waters), which was coupled to a Q-Tof Ultima mass spectrometer. Data acquisition was controlled by MassLynxTM 4.0 software (Waters). Data files were processed with Protein Lynx Global Server 2.2 software (Waters). De-isotoping was performed using the MaxEnt3 algorithm. The data obtained were analyzed with the MASCOT algorithm (version 2.1.04, Matrix Science) for protein identification against Hydra Hma2 protein models (20).
Proteome Annotation
The annotation of the nematocyst proteome was performed manually for all protein sequences using the EMADT (extensive manual annotation(s) by domain(s) and taxonomy) protocol developed heuristically for this analysis. The protein sequences were analyzed by PSI-BLAST (21) with default algorithm parameters. The E value cutoff was set at 1e−05. The protein domains including the number of repeats identified by the Conserved Domains Database were noted. Domains were also analyzed in parallel using InterProScan, and signal peptide prediction was performed using the SignalP 3.0 server. The redundancy of identified domains was checked, and the annotation was manually curated if necessary. Multidomain proteins were analyzed for every single domain in the sequence. Functional organization of the proteome was done based on the domains identified.
Comparative Genomics
Predicted peptide data sets were retrieved from the DOE Joint Genome Institute (www.jgi.doe.gov/; Nematostella vectensis, Monosiga brevicollis, and Trichoplax adhaerens), from NCBI (ftp.ncbi.nih.gov/genomes/; Strongylocentrus purpuratus), and from the Ensembl Database (Branchiostoma floridae, Caenorhabditis elegans, Drosophila melanogaster, and Homo sapiens) as of June 10, 2010. BLASTp from the NCBI blastall package (21) was used to scan all nematocyst proteins against all reference proteomes mentioned above for the identification of orthologs. BLASTp was run with an E value cutoff of 10−5 and the parameters −F “m S” and −s T for soft filtering of low complexity regions and locally optimal Smith-Waterman alignments, respectively (parameter −F “m S” refers to soft filtering of low complexity regions, and parameter −s T refers to locally optimal Smith-Waterman alignments). The BLAST output was further filtered to include only hits, which correspond to query sequence coverage of at least 30%. Protein domains were annotated based on InterPro (22) domain definitions. Overlapping domain annotations were resolved by removing the annotation with the less significant E value. Putative nematocyst venom proteins were identified based on similarity to known venom proteins stored in the Tox-Prot Database (23). A BLAST search of all nematocyst proteins versus all sequences in the Tox-Prot Database yielded 54 putative nematocyst venom proteins (E < 10−5).
Analysis of Orphan Proteins
The 117 nematocyst orphan proteins (without hits in the reference genomes and without InterPro domains) were clustered based on sequence similarity using BLASTclust (21). Proteins with 25% sequence identity in a region spanning at least 35% of the sequence length were clustered together. This procedure yielded two clusters of sizes 10 and 9, respectively. Both clusters were aligned using T-Coffee (24), and a hidden Markov model based on those alignments was scanned against the NCBI non-redundant protein database using HMMER. This scan yielded significant hits in proteins containing the sweet tooth domain for the cluster of size 3. The three proteins in the nematocyst-specific cluster were predicted to have nucleotide-binding activity based on an analysis with the I-TASSER server (25). Furthermore, the conserved region in the three proteins of the Hydra-specific cluster was predicted to be globular (instead of disordered or fibrous) (26).
Cloning and Recombinant Expression of Cnidoin
Preparation of whole RNA was performed using an RNeasy mini kit (Qiagen) according to the manufacturer's instructions. The isolated RNA was transcribed by reverse transcriptase into cDNA. PCR for the Cnidoin coding sequence was performed using primers ATGTCTCGATTACTACTTC (forward) and TTATCTCTTTTTACCAAAAGCTCC (reverse). The purified PCR product was ligated into the pGEM-T vector (Promega) and confirmed by sequencing. Recombinant expression in Escherichia coli BL21(DE3) cells was performed with a pET-21b vector (Novagen), which introduces a C-terminal polyhistidine tag. Cnidoin protein was exclusively found in inclusion bodies and purified under denaturing conditions (8 m urea) using nickel-nitrilotriacetic acid beads.
Immunostaining
H. magnipapillata animals were relaxed in 2% urethane in Hydra medium and then fixed in freshly prepared 4% paraformaldehyde in Hydra medium for 30 min. The fixative was removed by three 10-min washing steps with PBS and 0.1% Triton X-100. The antibody was diluted 1:250 in PBS and 1% BSA and incubated overnight at 4 °C. In the case of co-staining with two antibodies, both antibodies (anti-minicollagen-1, 1:500 dilution) were incubated simultaneously overnight at 4 °C. To remove unbound antibodies, three 10-min washing steps with PBS and 0.1% Triton X-100 were performed. Incubation with the secondary antibodies was performed for 2 h at room temperature. For detection of Cnidoin, an Alexa 568-conjugated goat anti-guinea pig antibody was used; for detection of minicollagen-1, an Alexa 488-conjugated goat anti-rabbit antibody was used. The secondary antibodies were diluted 1:400 in PBS and 1% BSA. To remove unbound antibodies, the animals were washed three times with PBS and then mounted on object slides with PBS and 90% glycerol.
Microscopy
Fluorescent images were captured with a Nikon A1R confocal laser scanning microscope at the Nikon Imaging Center at the University of Heidelberg. In situ images were captured with a Nikon Eclipse 80i microscope.
RESULTS
Global Annotation of Nematocyst Proteome
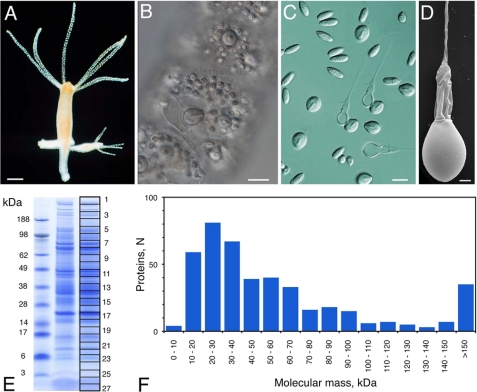
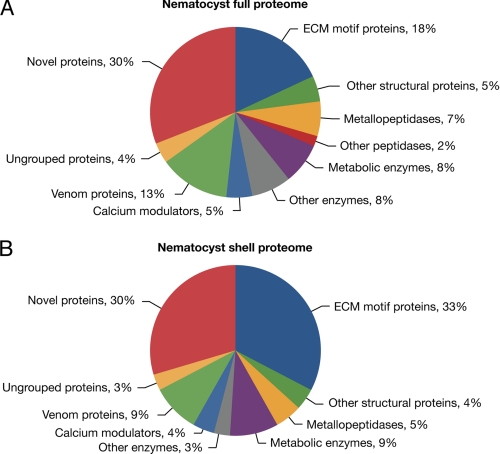
We isolated intact undischarged nematocysts (stenoteles, desmonemes, and atrichous and holotrichous isorhizas) (14) from H. magnipapillata by density gradient centrifugation (18). The purity of the preparation was confirmed by light and electron microscopy (Fig. 1, C and D). The nematocyst preparation was solubilized by DTT/SDS treatment and separated by one-dimensional gel electrophoresis. Gel slices (n = 27) covering the whole molecular mass range (Fig. 1E) were subjected to Orbitrap MS analysis. Fig. 1F shows that the nematocyst proteins exhibited a Gaussian distribution regarding their molecular masses, with the majority ranging between 10 and 40 kDa. One-dimensional gels guaranteed higher protein yields than two-dimensional gels, which were analyzed in parallel (supplemental Fig. 1). Annotation of the obtained nematocyst protein sequences was performed by a manual procedure applying PSI-BLAST and InterProScan analyses in parallel (EMADT; see “Experimental Procedures” for details). By this approach and the application of a MASCOT protein score threshold of 50 to exclude insignificant hits, we identified a total number of 410 unique protein sequences (supplemental Table 1). An additional 25 sequences from the full proteome analysis were categorized as contaminants and included histones, ribosomal proteins, cytoskeletal components, and cytoplasmic enzymes. The fraction of cellular contaminants in the complete proteome was low as estimated from the intensity of the actin spot in the two-dimensional resolution of the nematocyst sample (supplemental Fig. 1). Almost one-third of the protein sequences are lacking a signal peptide, which is supposed to be due to incomplete protein models, unusual secretion as in galectins, or co-purification of cytoplasmic proteins tightly associated with the nematocyst membrane. Functionally, the proteome is characterized by three groups of similar size: enzymes, structural proteins, and novel sequences. Many enzymes are part of the venom proteome, and the group of structural proteins is composed mainly of sequences with ECM motifs. The detailed annotations are shown in Fig. 2A and supplemental Table 1. To obtain the proteome of the components that are firmly integrated into the capsule wall and tubule structures, we prepared nematocyst “shells” by extensive SDS washing of capsules that were discharged in vitro. The shell proteome (supplemental Table 2) contained 98 proteins. As expected, the ratio of structural proteins was substantially increased in this subset of the proteome (Fig. 2B). Interestingly, the ratio of novel proteins did not decrease in the shell proteome, indicating that a significant portion of this group has a structural function. To analyze the percentage of exclusive nematocyst proteins in our data set, we generated nematocyst-free hydras by hydroxyurea treatment (27). This procedure eliminates all stem cells and their progeny, nerve cells, and nematocytes, rendering these “epithelial” animals incapable of active feeding. A lysate from epithelial hydras was subjected to mass spectrometric analysis under identical conditions, yielding 65 proteins, mostly enzymes with broad cellular functions that are shared with the nematocyst proteome (supplemental Table 3). This indicates that the majority of proteins (84%) from the analysis of isolated capsules were nematocyst-specific gene products. Of the nematocyte-specific genes isolated by Hwang et al. (28) by a microarray analysis of cell type-specific cDNAs in Hydra, 36% were covered by our analysis. These constitute only 2.4% of the total nematocyst proteome, which probably reflects a bias of the cDNA screen toward strongly expressed genes with functions in cell differentiation and proliferation.
FIGURE 1.
Nematocyst morphology and protein spectrum. A, H. magnipapillata. B, close-up of undischarged nematocysts assembled in battery cells of the tentacles. C, light microscopic view of isolated nematocyst capsules. D, scanning electron microscopic image of isolated nematocyst. E, one-dimensional SDS-PAGE of a nematocyst sample. The gel slices used for mass spectrometry are indicated. F, molecular mass distribution of the nematocyst proteome. Scale bars = 0.5 mm (A), 15 μm (B and C), and 2 μm (D).
FIGURE 2.
Distribution of functional nematocyst proteome. A, functional protein classes of the total Hydra nematocyst proteome. B, functional protein classes of the reduced proteome from nematocyst shells generated by extensive SDS washing.
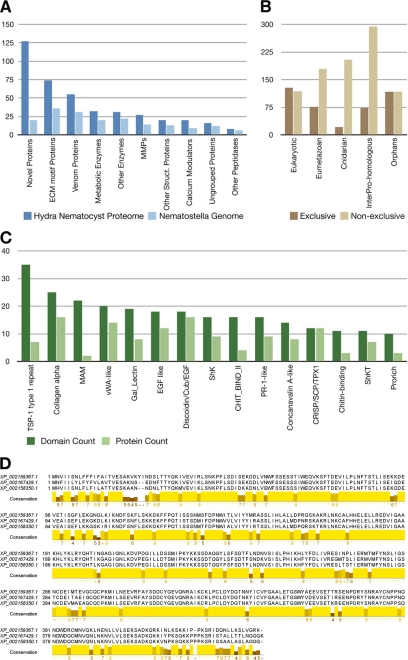
A comparison with homologous sequences in the genome of the starlet sea anemone N. vectensis showed that, in total, 44% of the Hydra nematocyst proteins were represented here (Fig. 3A). Although most of the metabolic proteins are conserved between both species, the number of novel and structural proteins is markedly pronounced in Hydra, which might indicate a large diversity between nematocyst proteins of different taxa or a higher morphological complexity of hydrozoan nematocysts (7). We categorized the novel proteins into four groups: sequences 1) unique to the proteome of Hydra nematocysts, 2) unique to Hydra, 3) unique to Hydra and Nematostella, and 4) not exclusive to cnidarians (supplemental Table 1).
FIGURE 3.
A, homologs of Hydra nematocyst proteins identified in the Nematostella genome. MMPs, matrix metalloproteases. B, evolutionary distribution of the 410 nematocyst proteins. Nematocyst proteins have been assigned to categories based on the presence of significant BLAST hits and Pfam domains. The “exclusive” distribution refers to a hierarchical classification in which each protein was assigned to only one category. Proteins with significant BLAST hits in basal eukaryotes (T. adhaerens or M. brevicollis) have been classified as “eukaryotic.” The remaining proteins with significant similarity in H. sapiens, D. melanogaster, Danio rerio, C. elegans, or B. floridae were binned as “eumetazoan.” Proteins that were not classified as either eukaryotic or eumetazoan but with significant hits in the N. vectensis genome were classified as “cnidarian.” The remaining proteins were further classified as “InterPro-homologous” if they had a significant hit against the InterPro Database or as “orphans” otherwise. For the “non-exclusive” distribution, each nematocyst protein was assigned to every category where a significant hit could be identified. C, domain distribution of Hydra nematocyst proteins with a significant Pfam hit but no significant BLAST hit to eukaryotic, eumetazoan, or other cnidarian proteomes. Dark green, total number of occurrences of a domain in the data set; light green, number of proteins in which a domain occurs. vWA, von Willebrand factor A; SCP, sperm coat protein. D, alignment of the proteins corresponding to the second largest cluster of orphan proteins (see “Experimental Procedures”). The conserved region is predicted to be globular and thus likely to represent a previously uncharacterized protein domain. MAM, meprin, A-5 protein, receptor protein tyrosine phosphatase mu; Cub, complement C1r/C1s, Uegf, bone morphogenetic protein; ShkT, Stichodactyla toxin domain; Shk, Stichodactyla-like domain.
Fig. 3B extends this comparison and shows the evolutionary distribution of the proteome among metazoans and eukaryotes. Overall, Hydra nematocysts comprise a gene repertoire of which 5% are cnidarian-specific with homologs exclusively in Nematostella, and 18% have homologs in Eumetazoa and 31% in Eukaryota. An additional 18% have no detectable homologs in other proteomes but contain InterPro domains. 28% of the sequences were classified as orphan genes. Among the proteins with detectable InterPro domains, several structural and enzymatic domains, e.g. α-collagen, AbfB, and von Willebrand factor, are strongly overrepresented (Fig. 3C). Among the remaining orphan proteins, several can be grouped together based on conserved common motifs (see “Experimental Procedures”). Ten proteins in the largest cluster contain the “sweet tooth” domain, which was reported to be among the most abundant extracellular protein domains in the Hydra genome but is not integrated in InterPro yet (30). The three orphan proteins of the second largest cluster share very high sequence identity (>80%) and are predicted to be globular and to have a nucleotide-binding propensity (see “Experimental Procedures”). A hidden Markov model created from their alignment (Fig. 3D) was scanned against the NCBI non-redundant database but returned no significant hit. Consequently, this protein structure seems to be unique for the proteome of the Hydra nematocyst and deserves further functional analysis.
Potential horizontal gene transfer events of bacterial origin comprise three proteins involved in poly-γ-glutamate biosynthesis: CapA, its isoform CapA2, and Hma2.206651, a protein with predicted enzymatic function. Poly-γ-glutamate biosynthesis is a crucial feature of nematocytes, and so far, cnidarians represent the only metazoan organisms with this capacity (29). Recently, Denker et al. (30) characterized CapA as a nematocyte-specific gene of defined bacterial origin, but the low percentage of horizontal gene transfer candidates in our analysis rejects a bacterial origin of the nematocyst.
Venoms and Enzymes Represent Conserved Feature of Nematocysts
The primary function of nematocysts is the release of venoms into the tissue of a prey organism or attacking predator. In Hydra and other cnidarians, polypeptide venoms are expressed mainly in nematocytes and represent a highly complex array of effector molecules aimed at paralyzing a prey and disintegrating its tissue. Our proteome analysis revealed 55 toxin-related sequences, mostly with homologous toxin proteins in diverse phyla (see “Experimental Procedures”). These include neurotoxins, cytolysins, toxic phospholipases, many peptidases, and proteins of the SCP_GAPR-1-like family (supplemental Table 1). Their molecular masses range from 25 to 100 kDa, indicating a lack of small peptide toxins affecting Na+ and K+ channels identified in many sea anemones (31, 32). Four venom sequences are cnidarian-specific (supplemental Table 1), and most venom proteins are part of the soluble nematocyst proteome (Fig. 2).
Enzymes with Lytic Function Constitute Unexpectedly High Proportion of Nematocyst Proteome
The largest group among these is represented by peptidases (supplemental Table 1). This feature is conserved in cnidarians, as 57% of the identified protease genes have a Nematostella homolog compared with 52% of the structural proteins (Fig. 3A). The protease proteome includes several major families, with matrix metalloproteases being the most prominent. Matrix metalloproteases can activate toxins or act as toxins themselves (33). In addition, they are essential factors in ECM remodeling and tissue morphogenesis. We assume that their proteolytic activity is used both for the maturation of the nematocyst and for prey tissue disintegration. Other enzyme classes comprise carbohydrate metabolism and breakdown, lipid hydrolysis, and protein folding and modification.
Nematocyst Capsule Structure Is Specialized ECM
The largest group of annotated proteins contains molecules with an apparent structural function, including the hitherto identified capsule and spine proteins, with NOWA being one of the most prominent components (supplemental Fig. 1). The structural proteins have well defined domains that are characteristic for multidomain ECM proteins involved in protein-protein and protein-carbohydrate interactions. The identified motifs include von Willebrand factor domains, fibronectin type II and thrombospondin type 1 repeats, LamG domains, and diverse lectins (Fig. 4). As many toxins specifically interact with matrix molecules (4), the similarity of the capsule structure to the ECM might have fostered their storage and delivery. In addition, we cannot exclude evolutionary transitions from venom-related molecules to structural proteins. The SCP domain might serve as an example, as it forms a pathogenesis-related superfamily across the metazoan subkingdom (34) and is part of the major capsule structure protein NOWA.
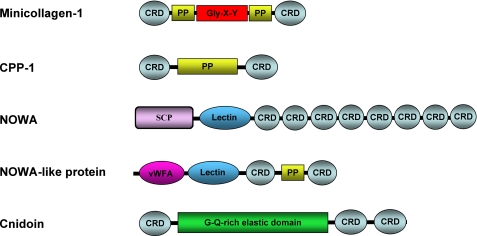
FIGURE 4.
Domain organization of ECM-type nematocyst proteins. PP, polyproline domain; SCP, sperm coat protein-like extracellular domain; vWFA, von Willebrand factor A domain. Gly-X-Y is the collagen triple helix.
Minicollagens constitute a well characterized subgroup among nematocyst ECM proteins (6). Our analysis revealed almost all hitherto characterized minicollagen molecules and four new members of this protein family (NCol18–21). Thus, the heterogeneity of the minicollagen family in nematocysts almost equals the collagen repertoire in the vertebrate ECM. Minicollagen detection by MS is hampered by the limited accessibility to proteolysis and the high similarity of the repetitive protein sequences. However, the majority of the minicollagens could be identified after a collagenase digest of nematocyst shells. Minicollagens are unique constituents of the cnidarian nematocyst and comprise a short central Gly-X-Y repeat sequence (8–16 repeats) flanked by polyproline stretches and terminal cysteine-rich domains (CRDs) (Fig. 4), which have a canonical cysteine pattern (CX3CX3CX3CX3CC) (7, 13). The CRD has been shown to be responsible for intermolecular networks in the capsule structure, which is extremely sensitive to reducing agents (35).
The nematocyst proteome reveals the evolutionary history of the minicollagen family in a broader dimension, as it includes a series of proteins that exhibit minicollagen domains in isolation or combined with unusual motifs (Fig. 4). Non-collagenous proteins containing polyproline stretches and minicollagen CRDs were designated cnidarian proline-rich proteins (CPP) and comprise five members in the capsule proteome (Fig. 4). CRDs are also found autonomously in multiple tandem repeats, often combined with lectin domains as in NOWA (Fig. 4). The CRD thus appears as a widespread linker motif connecting collagenous and non-collagenous structural proteins of the nematocyst.
Capsule Wall Protein Cnidoin Represents Elastic Counterpart of Collagen Network
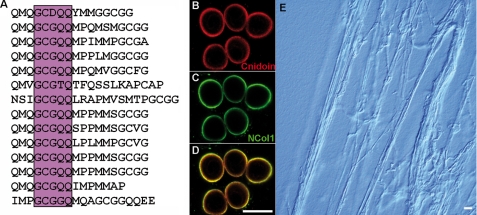
Hydra nematocysts are unique in storing energy for discharge in the capsule wall, which is elastically stretched by an extreme osmotic pressure (1). The molecular basis of the capsule's elastic property has hitherto been elusive, as the collagen polymer alone is unlikely to be responsible for it. An intriguing example of a non-collagenous structural protein containing CRDs is Cnidoin (Fig. 4), which exhibits an extended glycine- and glutamine-rich elastic sequence (Fig. 5A) with sequence similarity to the spider silk protein spidroin-2 (36). A common repetitive element in both proteins is GXGQQ, where X is Cys in Cnidoin and Pro in spidroin-2 (37). Spidroins are composed of amorphous glycine-rich sequences providing elasticity to the silk fiber interspersed by polyalanine crystallites, which provide strength. Poly-Ala blocks are missing in Cnidoin, suggesting that it might serve as a purely elastic counterpart of the collagenous wall matrix, providing the molecular basis for the storage of kinetic energy released in the discharge process. Co-immunostaining for Cnidoin and minicollagen-1 showed overlapping signals in the walls of developing nematocysts (Fig. 5, B–D). Cnidoin appears to be more enriched in the inner wall layer (Fig. 5D), suggesting a late incorporation into the nematocyst vesicle. The resulting wall composition is reminiscent of the vertebrate blood vessel matrix, in which the inner elastin layer is stretched against the collagenous connective tissue. Recombinant expression of Cnidoin in E. coli resulted in a highly viscous protein solution, which precipitated to macroscopic sheet-like structures in aqueous solutions (data not shown). In an 8 m urea solution, purified Cnidoin readily assembled to extended fibers of 2–3-μm thickness (Fig. 5E), indicating a tendency for fibrillogenesis as described for other elastic proteins (38).
FIGURE 5.
Primary sequence and expression pattern of Cnidoin. A, repetitive motif in the glycine-rich elastic domain of Cnidoin. The central consensus sequence showing homology to spidroin-2 is boxed. B–D, whole mount double immunostaining with anti-Cnidoin (red) and anti-minicollagen-1 (green) antibodies. D demonstrates the co-localization of the two signals in the developing capsule walls of nematocysts. Scale bar = 10 μm. E, light microscopic image of fibers formed by purified recombinant Cnidoin in urea. Scale bar = 30 μm.
DISCUSSION
Nematocysts constitute one of the most complex cellular organelles in the animal kingdom. They represent an exclusive feature of cnidarians, and nematocyst proteins like minicollagens serve as a phylogenetic marker for this phylum as exemplified by a recent report on minicollagens in myxosporidians (39). The identification of the nematocyst proteome of Hydra for the first time allows a view into the evolutionary history of this organelle. We can clearly rule out a major transfer event by stable cellular integration of an endosymbiont, as in the case of mitochondria and chloroplasts, although proteins of bacterial origin such as those involved in poly-γ-glutamate synthesis have essentially contributed to nematocyst functions (31).
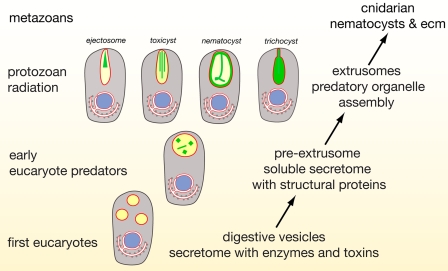
Instead, our data suggest an evolution of nematocysts that was tightly coupled with the invention of ECM-like structural proteins from exocytic vesicles of early eukaryotes specialized in venom secretion (Fig. 6). A conserved portion of the venom-related proteome comprises lytic enzymes and is of a general digestive nature. Extracellular digestion might therefore have been a transitory state in nematocyst evolution leading to phagotrophy and internal digestion (Fig. 6) (40). The close molecular relation of the nematocyst structure to ECM points to a premetazoan role of ECM molecules associated with predatory functions such as entangling prey cells in a fibrous mesh or a more efficient absorption of cellular material by the secretion of adhesive molecules (Fig. 6). We assume that the vesicular environment was optimized for ECM assembly as a basis for an accelerated adaptation (41) under the pressure of a beginning predatory phase among eukaryotes that, in turn, has led to a variety of extrusive organelles exhibiting sophisticated discharge mechanisms (Fig. 6) (42). The existence of nematocyst-like organelles in dinoflagellates (43) and other protozoans argues for an evolutionary origin of nematocysts predating multicellularity (Fig. 6). The nematocyst proteome can therefore shed light into the primordial state of the ECM to which the nematocyst's biomechanical requirements probably contributed.
FIGURE 6.
Schematic representation of hypothetic evolution of cnidarian nematocyst and ECM assembly. The secretome of early heterotrophic eukaryotes exhibited secretory vesicles containing enzymes used for extracellular digestion. On the basis of our findings (Fig. 4), we presume that, in the secretome, structural proteins evolved that finally gave rise to a variety of protozoan extrusive organelles exhibiting a variety of highly sophisticated discharge mechanisms. Some of these extrusomes exhibit a surprising structural similarity to nematocysts, suggesting that nematocysts may have evolved from extrusomes of a non-cnidarian precursor.
Minicollagens are a main molecular feature of nematocysts, but their evolutionary origin has hitherto been elusive. The CPPs constitute a family of non-collagenous proteins with distinct minicollagen features, and our data indicate a scenario of how CPPs might have evolved to minicollagens. CPP-1 shows a perfect minicollagen domain organization with a central polyproline stretch flanked by CRDs instead of the collagen triple helix (Fig. 4). In the C-terminal CRD of minicollagens, mutation of a conserved proline causes a deviated three-dimensional structure (44). In CPP-1, both CRDs have the N-terminal fold shared with all other CRDs outside the minicollagen family. We therefore propose that an early recombination event between polyproline and CRD domains might have produced a founder molecule for minicollagens.
Proline-rich proteins are found in diverse biological contexts and can be extremely polymorphic concerning post-translational modifications. They often fulfill structural functions similar to extensins in plant cell walls and adopt a polyproline II helix structure as the chains in collagen triple helices (45). Interestingly, the sialomes of salivary glands of diverse species, including humans, contain a large fraction of proline-rich proteins supposed to function in tannin binding and interaction with bacteria (46). Proline-rich proteins share sequence similarities and amino acid compositions with collagens but lack repetitive Gly-X-Y sequences (47, 48). A recent analysis of the black fly sialome identified a family of short collagen-like molecules with 7–15 Gly-X-Y repeats supposed to interact with ECM components (49). This opens the attractive perspective that CPPs evolved from secretory proteins with digestive venom-related function to structural proteins, i.e. to minicollagens. Thus, the formation of a complex ECM within the nematocyst vesicle might have been fostered by an extensive domain rearrangement within the secretome of nematocytes. We speculate that this process involved proteins representing a primitive state of the ECM and uncoupled their evolution from those constituting the cellular ECM.
Supplementary Material
Acknowledgments
We thank Jo Adams, Jürgen Engel, Peer Borg, and Jan Lohmann for critical reading of the manuscript and Charles David for fruitful discussions. We thank Tore Kempf for helpful suggestions regarding two-dimensional gels, Agnes Hotz-Wagenblatt for bioinformatics support, and Kerstin Kammerer for excellent technical assistance.
This work was supported by Deutsche Forschungsgemeinschaft Grant HO 1088-4/1-2, the Heidelberg Excellence Cluster CellNetworks, and the Volkswagen Foundation (I184 830).

This article contains supplemental Fig. 1 and Tables 1–3.
- ECM
- extracellular matrix
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- IPG
- immobilized pH gradient
- CRD
- cysteine-rich domain
- CPP
- cnidarian proline-rich protein.
REFERENCES
- 1. Nüchter T., Benoit M., Engel U., Ozbek S., Holstein T. W. (2006) Nanosecond-scale kinetics of nematocyst discharge. Curr. Biol. 16, R316–R318 [DOI] [PubMed] [Google Scholar]
- 2. Fry B. G., Wroe S., Teeuwisse W., van Osch M. J., Moreno K., Ingle J., McHenry C., Ferrara T., Clausen P., Scheib H., Winter K. L., Greisman L., Roelants K., van der Weerd L., Clemente C. J., Giannakis E., Hodgson W. C., Luz S., Martelli P., Krishnasamy K., Kochva E., Kwok H. F., Scanlon D., Karas J., Citron D. M., Goldstein E. J., McNaughtan J. E., Norman J. A. (2009) A central role for venom in predation by Varanus komodoensis (Komodo dragon) and the extinct giant Varanus (Megalania) priscus. Proc. Natl. Acad. Sci. U.S.A. 106, 8969–8974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Fry B. G., Roelants K., Champagne D. E., Scheib H., Tyndall J. D., King G. F., Nevalainen T. J., Norman J. A., Lewis R. J., Norton R. S., Renjifo C., de la Vega R. C. (2009) The toxicogenomic multiverse: convergent recruitment of proteins into animal venoms. Annu. Rev. Genomics Hum. Genet. 10, 483–511 [DOI] [PubMed] [Google Scholar]
- 4. Eble J. A. (2010) Matrix biology meets toxinology. Matrix Biol. 29, 239–247 [DOI] [PubMed] [Google Scholar]
- 5. Mariscal R. N. (1974) Scanning electron microscopy of the sensory surface of the tentacles of sea anemones and corals. Z. Zellforsch. 147, 149–156 [DOI] [PubMed] [Google Scholar]
- 6. Ozbek S., Balasubramanian P. G., Holstein T. W. (2009) Cnidocyst structure and the biomechanics of discharge. Toxicon 54, 1038–1045 [DOI] [PubMed] [Google Scholar]
- 7. David C. N., Ozbek S., Adamczyk P., Meier S., Pauly B., Chapman J., Hwang J. S., Gojobori T., Holstein T. W. (2008) Evolution of complex structures: minicollagens shape the cnidarian nematocyst. Trends Genet. 24, 431–438 [DOI] [PubMed] [Google Scholar]
- 8. Chapman G. B., Tilney L. G. (1959) Cytological studies of the nematocysts of Hydra. I. Desmonemes, isorhizas, cnidocils, and supporting structures. J. Biophys. Biochem. Cytol. 5, 69–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chapman G. B., Tilney L. G. (1959) Cytological studies of the nematocysts of Hydra. II. The stenoteles. J. Biophys. Biochem. Cytol. 5, 79–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Holstein T. W., Campbell R. D., Tardent P. (1990) Identity crisis. Nature 346, 21–22 [Google Scholar]
- 11. Lenhoff H. M., Kline E. S., Hurley R. (1957) A hydroxyproline-rich intracellular, collagen-like protein of Hydra nematocysts. Biochim. Biophys. Acta 26, 204–205 [DOI] [PubMed] [Google Scholar]
- 12. Weber J. (1990) Poly(γ-glutamic acids) are the major constituents of nematocysts in Hydra (Hydrozoa, Cnidaria). J. Biol. Chem. 265, 9664–9669 [PubMed] [Google Scholar]
- 13. Kurz E. M., Holstein T. W., Petri B. M., Engel J., David C. N. (1991) Minicollagens in Hydra nematocytes. J. Cell Biol. 115, 1159–1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Holstein T. W., Benoit M., Herder G. V., David C. N., Wanner G., Gaub H. E. (1994) Fibrous minicollagens in Hydra nematocysts. Science 265, 402–404 [DOI] [PubMed] [Google Scholar]
- 15. Holstein T., Tardent P. (1984) An ultrahigh-speed analysis of exocytosis: nematocyst discharge. Science 223, 830–833 [DOI] [PubMed] [Google Scholar]
- 16. Engel U., Ozbek S., Streitwolf-Engel R., Petri B., Lottspeich F., Holstein T. W. (2002) NOWA, a novel protein with minicollagen Cys-rich domains, is involved in nematocyst formation in Hydra. J. Cell Sci. 115, 3923–3934 [DOI] [PubMed] [Google Scholar]
- 17. Koch A. W., Holstein T. W., Mala C., Kurz E., Engel J., David C. N. (1998) Spinalin, a new glycine- and histidine-rich protein in spines of Hydra nematocysts. J. Cell Sci. 111, 1545–1554 [DOI] [PubMed] [Google Scholar]
- 18. Weber J. (1989) Nematocysts (stinging capsules of Cnidaria) as Donnan potential-dominated osmotic systems. Eur. J. Biochem. 184, 465–476 [DOI] [PubMed] [Google Scholar]
- 19. Schokraie E., Hotz-Wagenblatt A., Warnken U., Mali B., Frohme M., Förster F., Dandekar T., Hengherr S., Schill R. O., Schnölzer M. (2010) Proteomic analysis of tardigrades: towards a better understanding of molecular mechanisms by anhydrobiotic organisms. PLoS ONE 5, e9502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chapman J. A., Kirkness E. F., Simakov O., Hampson S. E., Mitros T., Weinmaier T., Rattei T., Balasubramanian P. G., Borman J., Busam D., Disbennett K., Pfannkoch C., Sumin N., Sutton G. G., Viswanathan L. D., Walenz B., Goodstein D. M., Hellsten U., Kawashima T., Prochnik S. E., Putnam N. H., Shu S., Blumberg B., Dana C. E., Gee L., Kibler D. F., Law L., Lindgens D., Martinez D. E., Peng J., Wigge P. A., Bertulat B., Guder C., Nakamura Y., Ozbek S., Watanabe H., Khalturin K., Hemmrich G., Franke A., Augustin R., Fraune S., Hayakawa E., Hayakawa S., Hirose M., Hwang J. S., Ikeo K., Nishimiya-Fujisawa C., Ogura A., Takahashi T., Steinmetz P. R., Zhang X., Aufschnaiter R., Eder M. K., Gorny A. K., Salvenmoser W., Heimberg A. M., Wheeler B. M., Peterson K. J., Bottger A., Tischler P., Wolf A., Gojobori T., Remington K. A., Strausberg R. L., Venter J. C., Technau U., Hobmayer B., Bosch T. C., Holstein T. W., Fujisawa T., Bode H. R., David C. N., Rokhsar D. S., Steele R. E. (2010) The dynamic genome of Hydra. Nature 464, 592–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Altschul S. F., Madden T. L., Schäffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hunter S., Apweiler R., Attwood T. K., Bairoch A., Bateman A., Binns D., Bork P., Das U., Daugherty L., Duquenne L., Finn R. D., Gough J., Haft D., Hulo N., Kahn D., Kelly E., Laugraud A., Letunic I., Lonsdale D., Lopez R., Madera M., Maslen J., McAnulla C., McDowall J., Mistry J., Mitchell A., Mulder N., Natale D., Orengo C., Quinn A. F., Selengut J. D., Sigrist C. J., Thimma M., Thomas P. D., Valentin F., Wilson D., Wu C. H., Yeats C. (2009) InterPro: the integrative protein signature database. Nucleic Acids Res. 37, D211–D215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jungo F., Bairoch A. (2005) Tox-Prot, the toxin protein annotation program of the Swiss-Prot protein knowledge base. Toxicon 45, 293–301 [DOI] [PubMed] [Google Scholar]
- 24. Poirot O., O'Toole E., Notredame C. (2003) Tcoffee@igs: a web server for computing, evaluating, and combining multiple sequence alignments. Nucleic Acids Res. 31, 3503–3506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Roy A., Kucukural A., Zhang Y. (2010) I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Linding R., Russell R. B., Neduva V., Gibson T. J. (2003) GlobPlot: Exploring protein sequences for globularity and disorder. Nucleic Acids Res. 31, 3701–3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yaross M. S., Bode H. R. (1978) Regulation of interstitial cell differentiation in Hydra attenuata. III. Effects of I-cell and nerve cell densities. J. Cell Sci. 34, 1–26 [DOI] [PubMed] [Google Scholar]
- 28. Hwang J. S., Ohyanagi H., Hayakawa S., Osato N., Nishimiya-Fujisawa C., Ikeo K., David C. N., Fujisawa T., Gojobori T. (2007) The evolutionary emergence of cell type-specific genes inferred from the gene expression analysis of Hydra. Proc. Natl. Acad. Sci. U.S.A. 104, 14735–14740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Szczepanek S., Cikala M., David C. N. (2002) Poly-γ-glutamate synthesis during formation of nematocyst capsules in Hydra. J. Cell Sci. 115, 745–751 [DOI] [PubMed] [Google Scholar]
- 30. Denker E., Bapteste E., Le Guyader H., Manuel M., Rabet N. (2008) Horizontal gene transfer and the evolution of cnidarian stinging cells. Curr. Biol. 18, R858–R859 [DOI] [PubMed] [Google Scholar]
- 31. Sher D., Zlotkin E. (2009) A Hydra with many heads: protein and polypeptide toxins from Hydra and their biological roles. Toxicon 54, 1148–1161 [DOI] [PubMed] [Google Scholar]
- 32. Bosmans F., Tytgat J. (2007) Sea anemone venom as a source of insecticidal peptides acting on voltage-gated Na+ channels. Toxicon 49, 550–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gutiérrez J. M., Rucavado A., Escalante T., Díaz C. (2005) Hemorrhage induced by snake venom metalloproteinases: biochemical and biophysical mechanisms involved in microvessel damage. Toxicon 45, 997–1011 [DOI] [PubMed] [Google Scholar]
- 34. Henriksen A., King T. P., Mirza O., Monsalve R. I., Meno K., Ipsen H., Larsen J. N., Gajhede M., Spangfort M. D. (2001) Major venom allergen of yellow jackets, Ves v 5: structural characterization of a pathogenesis-related protein superfamily. Proteins 45, 438–448 [DOI] [PubMed] [Google Scholar]
- 35. Ozbek S., Pokidysheva E., Schwager M., Schulthess T., Tariq N., Barth D., Milbradt A. G., Moroder L., Engel J., Holstein T. W. (2004) The glycoprotein NOWA and minicollagens are part of a disulfide-linked polymer that forms the cnidarian nematocyst wall. J. Biol. Chem. 279, 52016–52023 [DOI] [PubMed] [Google Scholar]
- 36. Hinman M. B., Lewis R. V. (1992) Isolation of a clone encoding a second dragline silk fibroin. Nephila clavipes dragline silk is a two-protein fiber. J. Biol. Chem. 267, 19320–19324 [PubMed] [Google Scholar]
- 37. Vollrath F. (2005) Spiders' webs. Curr. Biol. 15, R364–R365 [DOI] [PubMed] [Google Scholar]
- 38. Gosline J., Lillie M., Carrington E., Guerette P., Ortlepp C., Savage K. (2002) Elastic proteins: biological roles and mechanical properties. Philos. Trans. R. Soc. Lond. B Biol. Sci. 357, 121–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Holland J. W., Okamura B., Hartikainen H., Secombes C. J. (2011) A novel minicollagen gene links cnidarians and myxozoans. Proc. R. Soc. B 278, 546–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cavalier-Smith T. (2009) Predation and eukaryote cell origins: a co-evolutionary perspective. Int. J. Biochem. Cell Biol. 41, 307–322 [DOI] [PubMed] [Google Scholar]
- 41. Koonin E. V. (2007) The Biological Big Bang model for the major transitions in evolution. Biol. Direct 2, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hausmann K. (1978) Extrusive organelles in protists. Int. Rev. Cytol. 52, 197–276 [DOI] [PubMed] [Google Scholar]
- 43. Westfall J. A., Bradbury P. C., Townsend J. W. (1983) Ultrastructure of the dinoflagellate Polykrikos. I. Development of the nematocyst-taeniocyst complex and morphology of the site for extrusion. J. Cell Sci. 63, 245–261 [DOI] [PubMed] [Google Scholar]
- 44. Meier S., Jensen P. R., Adamczyk P., Bächinger H. P., Holstein T. W., Engel J., Ozbek S., Grzesiek S. (2007) Sequence-structure and structure-function analysis in cysteine-rich domains forming the ultrastable nematocyst wall. J. Mol. Biol. 368, 718–728 [DOI] [PubMed] [Google Scholar]
- 45. Cassab G. I. (1998) Plant cell wall proteins. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 281–309 [DOI] [PubMed] [Google Scholar]
- 46. Oppenheim F. G., Salih E., Siqueira W. L., Zhang W., Helmerhorst E. J. (2007) Salivary proteome and its genetic polymorphisms. Ann. N.Y. Acad. Sci. 1098, 22–50 [DOI] [PubMed] [Google Scholar]
- 47. Wong R. S., Hofmann T., Bennick A. (1979) The complete primary structure of a proline-rich phosphoprotein from human saliva. J. Biol. Chem. 254, 4800–4808 [PubMed] [Google Scholar]
- 48. Clements S., Mehansho H., Carlson D. M. (1985) Novel multigene families encoding highly repetitive peptide sequences. Sequence analyses of rat and mouse proline-rich protein cDNAs. J. Biol. Chem. 260, 13471–13477 [PubMed] [Google Scholar]
- 49. Andersen J. F., Pham V. M., Meng Z., Champagne D. E., Ribeiro J. M. (2009) Insight into the sialome of the black fly, Simulium vittatum. J. Proteome Res. 8, 1474–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.