Background: Vacuolar ATPase (V-ATPase) proton pumps maintain pH homeostasis.
Results: We discovered new V-ATPase inhibitors that uncouple the proton transport and ATPase activity of the pump.
Conclusion: Residues at the tether connecting V0 subunit a to the membrane give uncoupling potential to V-ATPases.
Significance: The tether may offer new mechanisms to regulate V-ATPase and cellular pH in vivo by uncoupling the pump.
Keywords: ATPases, H+-ATPase, Membrane Proteins, Proton Transport, Vacuolar ATPase, Yeast, V-ATPase, Coupling, Inhibitors, Uncouplers
Abstract
Vacuolar ATPases (V-ATPases) are important for many cellular processes, as they regulate pH by pumping cytosolic protons into intracellular organelles. The cytoplasm is acidified when V-ATPase is inhibited; thus we conducted a high-throughput screen of a chemical library to search for compounds that acidify the yeast cytosol in vivo using pHluorin-based flow cytometry. Two inhibitors, alexidine dihydrochloride (EC50 = 39 μm) and thonzonium bromide (EC50 = 69 μm), prevented ATP-dependent proton transport in purified vacuolar membranes. They acidified the yeast cytosol and caused pH-sensitive growth defects typical of V-ATPase mutants (vma phenotype). At concentrations greater than 10 μm the inhibitors were cytotoxic, even at the permissive pH (pH 5.0). Membrane fractions treated with alexidine dihydrochloride and thonzonium bromide fully retained concanamycin A-sensitive ATPase activity despite the fact that proton translocation was inhibited by 80–90%, indicating that V-ATPases were uncoupled. Mutant V-ATPase membranes lacking residues 362–407 of the tether of Vph1p subunit a of V0 were resistant to thonzonium bromide but not to alexidine dihydrochloride, suggesting that this conserved sequence confers uncoupling potential to V1V0 complexes and that alexidine dihydrochloride uncouples the enzyme by a different mechanism. The inhibitors also uncoupled the Candida albicans enzyme and prevented cell growth, showing further specificity for V-ATPases. Thus, a new class of V-ATPase inhibitors (uncouplers), which are not simply ionophores, provided new insights into the enzyme mechanism and original evidence supporting the hypothesis that V-ATPases may not be optimally coupled in vivo. The consequences of uncoupling V-ATPases in vivo as potential drug targets are discussed.
Introduction
Endogenous V-ATPase proton pumps are present throughout the endomembrane system where they energize the membranes, acidify organelles, and contribute to regulating the cytosolic pH (1–3). V-ATPases carefully control the acidic pH essential for endocytic and exocytic vesicular transport, zymogen activation, and protein sorting and degradation. Cells specialized for active proton secretion also express V-ATPases at the plasma membrane, where the efflux of cytosolic protons sustains the acidic luminal pH necessary for sperm maturation (4), urinary acidification (5), and bone resorption (6).
Maintaining pH homeostasis by V-ATPases6 entails the active transport of protons at the expense of ATP. The two domains forming the V-ATPase complex, V1 and V0, functionally and structurally couple ATP hydrolysis and proton transport via a rotational mechanism of catalysis. V1 is bound peripherally to the cytosolic side of the membrane and catalyzes the hydrolysis of ATP inside a hexameric structure consisting of subunits A and B (A3B3) (1–3). V0 is membrane-bound and forms the path for proton transport, which involves subunit a and a proteolipid ring structure (3, 7). During catalysis, hydrolysis of ATP in A3B3 of V1 drives rotation of a central rotor connected to the proteolipid ring of V0. Protons are transferred from the cytosol to a hemichannel in the V0 subunit a and from subunit a to the proteolipid ring. Hydrolysis of three molecules of ATP in V1 drives a 360° rotation of the ring, allowing the exit of protons. Exiting protons are transferred to a second hemichannel, located in the luminal side of subunit a, and expelled to the other side of the membrane against a concentration gradient.
V-ATPase activity is essential, and blocking of rotational catalysis is lethal. When V-ATPase activity is impaired at physiological (neutral) pH, yeast and most eukaryotic cells are not viable (2). However, yeast have developed a conditionally lethal pH-sensitive growth phenotype in which cells grow under acidic conditions. This phenotypic trait has made yeast an ideal system for the study of the downstream consequences of genetically impairing V-ATPase function in vivo (8). Yeast V-ATPase mutant strains have provided valuable information regarding the molecular mechanism of catalysis and the broad spectrum of physiological processes in which V-ATPases are involved.
It is precisely because V-ATPases are critical for many cellular events that yeast V-ATPase mutants often develop compensatory mechanisms that can mask important V-ATPase functions (9–11). In this context, V-ATPase inhibitors are important research tools for studies requiring sudden inhibition of V-ATPases. The most commonly used V-ATPase inhibitors, bafilomycin A and concanamycin A, have been fundamental to the understanding of V-ATPase catalysis, regulation, and cellular functions (12–15). Bafilomycin A and concanamycin A are now indispensable research tools for the study of important processes in which V-ATPases are involved, including autophagy (16, 17) and membrane fusion (18) under normal physiology and pathophysiology.
It has been proposed that bafilomycin A and concanamycin A act as a stone in a gear, blocking rotation when they bind to the proteolipid ring of V0 (19, 20). A new kind of V-ATPase inhibitors, archazolids (12), also bind to the proteolipid ring to block rotation (21). By ending rotation, bafilomycin A, concanamycin A, and archazolids inhibit ATPase activity in V1 and proton transport in V0 simultaneously. Unfortunately, they cannot offer information regarding the mechanisms by which proton transport and ATP hydrolysis are coupled. Mutagenesis studies of individual V1 and V0 subunits in yeast indicate that coupling is accomplished by the contribution of multiple subunits (22–26). However, exactly how proton transport and ATP hydrolysis are coupled in the V-ATPase complex is not known.
Changing the coupling efficiency of V-ATPase may regulate the pump by offering a mechanism for control of organelle acidification in vivo (27, 28). The hypothesis that V-ATPase may not couple proton transport and ATP hydrolysis optimally is supported by the fact that increased coupling efficiency is observed at lower ATP concentrations (27) and in some genetic mutants (25). An intrinsic uncoupling potential argues against the energetic efficiency of the V-ATPase machine, and intrinsic uncoupling is poorly understood. As such, to gain new insights into this important regulatory mechanism of V-ATPase proton transport, it would be beneficial to find specific V-ATPase inhibitors that modulate coupling of the enzyme.
We took advantage of the fact that V-ATPase inhibition prevents the redistribution of protons and lowers the cytosolic pH (29) to screen the Prestwick Chemical Library of small compounds. We searched for drugs that acidified the yeast cytosol in vivo as a means of identifying new V-ATPase inhibitors. By using Saccharomyces cerevisiae cells expressing a cytosolic pHluorin (29–31) and the HyperCyt® high-throughput flow cytometry platform (32), we identified two V-ATPase inhibitors, alexidine dihydrochloride and thonzonium bromide. They acidified the yeast cytosol, inhibited ATP-dependent proton transport in vacuolar membrane fractions, and caused pH-sensitive growth defects characteristic of yeast cells with impaired V-ATPase function.
We showed that these inhibitors functionally uncoupled V-ATPase pumps and that a mutant V-ATPase lacking the tether (residues 362–407) of V0 subunit a Vph1p was resistant to thonzonium bromide. This finding revealed novel roles for the tether connecting the N- and C-terminal domains of subunit a. The tether confers uncoupling potential to V1V0 complexes and a mechanism for regulating V-ATPase coupling efficiency in vivo. We showed further that alexidine dihydrochloride and thonzonium bromide uncouple V-ATPase pumps of the pathogenic fungi Candida albicans and inhibit cell growth providing evidence for the universal nature of this regulatory mechanism.
EXPERIMENTAL PROCEDURES
Materials and Strains
Zymolase 100T was purchased from Seikagaku (Tokyo), concanamycin A from Wako Biochemicals (Richmond, VA), and Ficoll from United Stated Biologicals (Swampscott, MA). All other reagents were from Sigma. The yeast S. cerevisiae strains referred to throughout the in vivo studies are BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0), vma2Δ (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 VMA2::KanMX), and vma3Δ (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 VMA3::KanMX). For flow cytometry and pH measurements, the cells were transformed by the lithium acetate method (33) with the 2 μ plasmid pHluorin under control of the phosphoglycerate kinase promoter (31); this plasmid was created by Dr. Rajini Rao (Department of Physiology, Johns Hopkins University). After transformation, the strains expressing pHluorin were maintained in fully supplemented synthetic complete (SC) medium lacking uracil (SC-Ura) buffered to pH 5.0 with 50 mm sodium phosphate and 50 mm sodium succinate (SC-Ura pH 5.0). The pHluorin plasmid was a generous gift from Patricia Kane (Upstate Medical University, State University of New York, Syracuse).
The yeast S. cerevisiae strains referred to throughout the in vitro studies involving vacuolar membranes and phenotype analyses are SF838-1Dα (MATα ade6 leu2–3,112 ura3–52 pep4–3 his4–519 gal2), the vph1Δstv1Δ strain, MM112 (MATα his3-Δ200 leu2 lys2 Δstvl::LYS2 ura3–52 Δvphl::LEU2) (34), and the vph1Δstv1Δ strain transformed with the CEN plasmid pRS316 carrying either the wild-type VPH1 gene or the tether-less VPH1 gene (vph1–362-407Δ) (35). MM112 was a gift from Dr. Morris Manolson (University of Toronto, Toronto, Ontario, Canada), and pRS316-HA-VPH1 was a gift from Michael Forgac (Tufts University, Boston). The C. albicans strain referred to throughout the study is DAY185 (ura3::λimm434/ura3::λimm434his1::his6/his1::his6::HIS1arg4::his6/arg4::his6::ARG4::URA3), a generous gift from Aaron Mitchell, Carnegie Mellon University, Pittsburgh. DAY185 cells were maintained in YEPD, pH 5.0 (yeast extract-peptone-2% dextrose medium buffered to pH 5.0 with 50 mm sodium phosphate and 50 mm sodium succinate), supplemented with uridine (80 μg/ml).
Assay Validation
Overnight mid-log phase cultures of yeast S. cerevisiae (BY4742) expressing pHluorin were resuspended in 2-fold diluted fresh SC medium to an optical density of 0.4 A600/ml (1–2 × 107 cells/ml). Cells (0.004 A600) were transferred to polypropylene 384-well plates (Greiner, Frickenhausen, Germany) containing an equal volume of disulfiram (134 and 26 mm) in 2% DMSO or 2% DMSO alone prepared in the same medium. Wash wells (columns 23 and 24) contained the same volume (10 μl) of 2-fold diluted SC medium plus 0.1% BSA. After incubation (0–120 min) at 30 °C with rotation, the plates were read using a CyAn flow cytometer (Beckman Coulter) and HyperCyt automation (IntelliCyt) (32) on filter sets 1 (ex 488/em 530) and 7 (ex 405/em 530), referred to as FL1 and FL7 hereafter. The ratio of these two measurements (FL1/FL7) was calculated. Cytosolic acidification was monitored as an increase of the FL1/FL7 ratio resulting from a simultaneous increase of the mean intensity at FL1 and a decrease of the mean intensity at FL7.
High-throughput Screening
Polypropylene 384-well plates were configured with 32 control wells and 32 wash wells. Column 1 contained the negative control (1% DMSO), column 2 contained the positive control (67 μm disulfiram), and columns 23 and 24 were the wash wells containing 2-fold diluted SC medium plus 0.1% BSA. This left 320 wells (columns 3–22) to which test compounds were added. The SC medium (2-fold diluted) was distributed into columns 1 and 3–22 (10 μl/well). An equal volume of 134 μm disulfiram or 2% DMSO was distributed into columns 2 and 1, respectively. Compounds of the Prestwick Library were added using the Biomek NX robotic liquid handler with a 200-nl pin tool attachment (V&P Scientific) to a final concentration of 0.1 mg/ml in 1% DMSO. Overnight cultures of the yeast S. cerevisiae expressing pHluorin were pelleted and resuspended in 2-fold diluted fresh SC medium to a cell density of 0.4 A600/ml (1–2 × 107 cells/ml). Cells (0.004 A600) were transferred to columns 1–22 and incubated for 60 min at 30 °C in a rotator, and the FL1/FL7 ratio was calculated as described above. Compounds that lowered the cytosolic pH (increased the FL1/FL7 ratio) were retested in single-point confirmation assays and dose-response assays (between 1 and 100 μm).
Cytosolic pH Measurement
Yeast cells expressing pHluorin were grown overnight to mid-log phase (0.4–0.6 A600) in SC-Ura or SC-Ura, pH 5.0 (vma2Δ and vma3Δ). Cultures were collected, washed three times in SC-Ura, pH 5.0, and resuspended in the same medium to 0.5 A600/μl. Aliquots of cells (10 A600) were treated with 0.2% DMSO or the indicated compounds (100 μm in 0.2% DMSO) for 30 min at 30 °C with shaking. The cells were harvested, washed, resuspended in SC-Ura, pH 5.0 (0.5 A600/ml), and transferred to cuvettes containing 1 mm HEPES/MES, pH 5.0, and 2% glucose (5 A600/ml). The fluorescence intensity was monitored (ex 405, ex 485; em 508) for 10 min at 30 °C with constant stirring, and fluorescence units were converted into pH units by using calibration curves as described previously (29, 30). For calibration curves, the cells were resuspended (5 A600/ml) in calibration buffers (50 mm MES, 50 mm HEPES, 50 mm KCl, 50 mm NaCl, 0.2 m ammonium acetate, 10 mm sodium azide, 10 mm 2-deoxyglucose, 75 μm monensin, 10 μm nigericin) at pH 5.0, 5.5, 6.0, 6.5, 7.0, 7.5, and 8.0 for 30 min at 30 °C after which the fluorescence measured. The background signals (cells without pHluorin) were subtracted, and cytosolic pH values were calculated. A calibration curve was constructed for each strain used.
Proton Transport
Vacuolar membranes were purified by Ficoll density gradient centrifugation (23), and proton transport was measured by monitoring 9-amino-6-chloro-2-methoxyacridine (ACMA) quenching after the addition of MgATP as described previously (36). Briefly, membrane vesicles (10 μg of vacuolar protein) were preincubated with 1% DMSO, 0.1 μm concanamycin A, or the indicated concentrations of the compounds on ice for 10 min. The membranes were transferred to 1.8 ml of reaction buffer (20 mm HEPES, pH 7.0, 50 mm NaCl, 30 mm KCl, 1 μm ACMA), and fluorescence was monitored (ex 410 nm, em 490 nm) for 60 s. MgATP (0.5 mm ATP, 1 mm MgSO4) was added, and fluorescence quenching was monitored for an additional 40 s in a FluoroMax 4 spectrofluorometer (Horiba Jobin Yvon Inc.). The proton transport rates were estimated for the initial 15 s following the addition of MgATP. Proton transport measurements involving C. albicans were conducted as described above, except that 30 μg of vacuolar protein was used. To calculate EC50 values, the vacuolar membranes were preincubated with 0.4, 1.2, 3.6, 11.1, 22.2, 33.3, 50, 100, 200, and 300 μm alexidine dihydrochloride and thonzonium bromide on ice for 10 min, and then proton transport was measured.
ATP Hydrolysis
Vacuolar membranes (4 μg of vacuolar protein) were preincubated with 0.1 μm concanamycin A, 1 μm nigericin, or test compounds (100 μm) on ice for 10 min in the presence of 1% DMSO. ATP hydrolysis was followed spectrophotometrically by using an enzymatic assay coupled to the oxidation of NADH (37). ATPase assays involving C. albicans were conducted as described for yeast, except that each reaction contained 20 μg of vacuolar membrane protein. The protein concentration was measured as described by Bradford (38).
Growth Phenotype
Cell cultures of the yeast S. cerevisiae were grown overnight to 0.5–1.0 A600/ml in YEPD, pH 5.0, at 30 °C. Cells were pelleted, resuspended to 0.5 A600/ml, and incubated in YEPD, pH 5.0 (no treatment), 1% DMSO, or 100, 50, 10, or 1 μm alexidine dihydrochloride, thonzonium bromide, or disulfiram at 30 °C with shaking for 1 h. The cells were harvested, resuspended in 200 μl of sterile ddH2O, and transferred to a 96-well plate. Serial dilutions (1:3) were stamped on YEPD, pH 5.0 and 7.5, plates and incubated at 30 and 37 °C for 48 h. A growth phenotype analysis of C. albicans was conducted as described above, except that the cells were resuspended in 1 ml of sterile H2O before stamping.
Other Methods
Western Blot
Vacuolar membrane protein was separated by SDS-PAGE in 8% polyacrylamide gels and transferred to a nitrocellulose membrane, and V-ATPase subunits a, A, and B were visualized with the monoclonal antibodies 10D7, 8B1, and 13D11 as described previously (35).
Microscopy
S. cerevisiae cells transformed with the pHluorin plasmid were grown to mid-log phase (0.5 A600/ml), harvested, and washed in SC medium three times by centrifugation. Cells (0.5 A600/ml) were transferred to microscope slides and observed with a Zeiss LSM 510 confocal microscope (Zeiss) at ×63 magnification by differential interference contrast microscopy and FITC lens. All images were taken using the same settings on the microscope.
RESULTS
Cytosolic pH Changes Monitored by pHluorin-based High Throughput
V-ATPase inhibition prevents redistribution of cytosolic protons into the vacuolar lumen (29), resulting in acidification of the cytosol. Thus, to search for new V-ATPase inhibitors we used yeast cells expressing a pH-sensitive GFP (pHluorin) in the cytosol (30, 31). We transformed yeast cells with pHluorin, confirming its cytosolic localization by fluorescence microscopy (Fig. 1A). We sorted the cells with the brightest fluorescence signal by flow cytometry (data not shown) and used those cells to develop a pHluorin-based high-throughput flow cytometry assay, which would allow us to identify molecules that acidify the yeast cytosol.
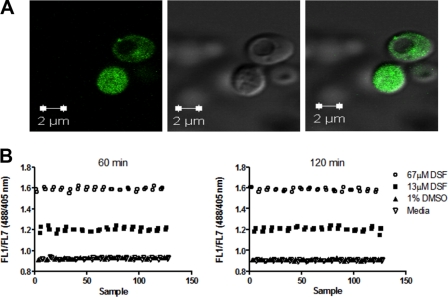
FIGURE 1.
Yeast cytosolic acidification monitored with pHluorin by high throughput. A, stable expression of pHluorin in the cytosol. Wild-type yeast S. cerevisiae cells expressing pHluorin were grown overnight to mid-log phase (0.6 A600/ml) and observed with a Zeiss LSM 520 confocal system. Cells were excited at 488 nm and emission detected with a 505-nm long pass filter (left); the same field was viewed at ×63 differential interference contrast (middle), and with images merged (right). B, pHluorin is responsive to V-ATPase inhibition in vivo. Cells expressing pHluorin were grown overnight to mid-log phase (0.6 A600/ml), harvested, resuspended at 0.4 A600/ml in 2-fold diluted SD medium containing 0.1% BSA, and distributed into 384-well plates to which SD medium alone, 1% DMSO, or disulfiram (DSF) at 13 and 67 μm was added for 60–120 min before sampling started. Plates were rotated at 30 °C and sampled with a HyperCyt® autosampler for flow cytometry pHluorin fluorescence measurements. After excitation with both the 488 and 405 nm lasers, fluorescence emission was collected using a 530/40-nm filter set and data files processed using IDLQuery software programs. The mean fluorescence intensity for each sample was calculated and used for subsequent analysis. Each point represents the ratio of FL1/FL7 from a single well.
We took advantage of the pH-sensitive emission spectrum of pHluorin in which the dual excitation wavelength (405 and 488 nm) of pHluorin exhibits opposite pH-dependent fluorescence responses despite both emitting at 535 nm (39). As the pH drops, the intensity of the fluorescent signal from 405 nm excitation decreases, whereas the intensity of the signal from excitation at 488 nm increases. Therefore, cytosolic acidification corresponds to a raise in the excitation 488/405 nm ratio (referred to as FL1/FL7). The benefit of a response that changes the signal on two filter sets is an automatic control for false positives in which fluorescent compounds that increase FL1 independent of cytosolic pH changes were excluded.
Concanamycin A and bafilomycin A are not suitable for studies involving high-throughput screenings in vivo (40). Inhibition of yeast V-ATPase pumps with concanamycin A requires 3 orders of magnitude larger concentrations of concanamycin A in vivo (EC50 = 2.10–2.27 μm) (40) than in vitro (EC50 = 1 nm) (20, 21). Likewise, bafilomycin A exhibits poor permeability. We tested the responsiveness of pHluorin to cytosolic acidification by treating yeast cells with disulfiram (tetraethylthiuram disulfide). Disulfiram (EC50 = 26 μm) exhibits both lower specificity and lower potency than concanamycin A and bafilomycin A in vitro (20, 40), but disulfiram is suitable for studies involving high-throughput screening. Disulfiram is stable and readily incorporated into yeast cells (40).
We treated the cells with an inhibitory concentration of disulfiram (67 μm) (40) and monitored the FL1/FL7 ratio. An increased FL1/FL7 ratio relative to control cells treated with 1% DMSO was measured (Fig. 1B). The cells had a FL1/FL7 ratio of 1.2 to 1.6 after 1 h of incubation with disulfiram at 30 °C compared with a FL1/FL7 ratio of 0.9 with 1% DMSO. The FL1/FL7 ratio was dose-dependent, stable for at least 2 h, and exhibited an average Z′ of 0.76 (±0.036) for the 60–120-min time period (supplemental Table 1). The Z′ is a statistical parameter used in high-throughput screening to evaluate the quality of an assay; Z′ establishes whether significant differences exist between the controls. An assay with a Z′ value greater than 0.5 is considered appropriate for screening, that is, where consistent significant differences between the controls, in this case DMSO and disulfiram, are observed. We concluded that cells expressing pHluorin could be used to assay for pharmacological inhibition of V-ATPase pumps in vivo and that disulfiram-treated cells would be an appropriate positive control for identification of molecules (or compounds) that lower the cytosolic pH of yeast cells.
High-throughput Screening of the Prestwick Chemical Library
The yeast cells expressing pHluorin were used to screen a collection of 1120 off-patent drugs (Prestwick Chemical Library). For controls, we treated the cells with either 1% DMSO alone or 67 μm disulfiram. The hit cutoffs were set at a FL1/FL7 ratio of 0.86, which corresponded to the average plus 3-fold the standard deviation of the test compounds. All compounds that resulted in an increased ratio and satisfied the hit cutoff were scrutinized further to confirm that they enhanced fluorescence at 488 nm (FL1) and lowered fluorescence at 405 nm (FL7) concurrently, indicating an actual pH change.
One representative plate is shown in Fig. 2A. We identified nine compounds that met the cutoff criteria. Seven of these hits were validated (hit rate of 0.63%) These drugs differentially altered the magnitudes of the FL1/FL7 ratio, suggesting that some hits acidified the cytosol more significantly than others. Disulfiram, the positive control used, is also a test compound in the Prestwick Chemical Library. The fact that disulfiram itself was identified as a hit (Fig. 2A) validated the cutoff criteria applied and demonstrated the effectiveness of the pHluorin-based assay in high-throughput screening for inhibitors of V-ATPase proton pumps.
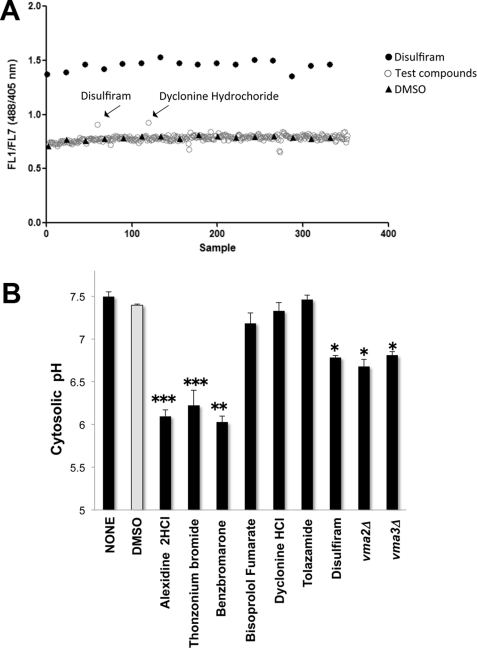
FIGURE 2.
High-throughput screening using a pHluorin-based assay identified drugs that acidify the cytosol. A, high-throughput screening of the Prestwick Chemical Library revealed drugs that increase the FL1/FL7 (488/405 nm) ratio of pHluorin. Mid-log phase cultures of cells expressing pHluorin were distributed in 384-well plates (0.2 A600/ml) to which test compounds, 1% DMSO (negative control), or 67 μm disulfiram (positive control) was added. Test compounds were added by robotic pintool and incubated in a rotator at 30 °C for 60 min, and sampling was started. Sampling, data collection, and processing were as described in the legend for Fig. 1B. One representative plate of compounds that enhanced the FL1/FL7 ratio is shown. The x axis displays each well; time bins were drawn automatically around the clusters by using the IDLQuery software program. B, cytosolic pH measurement. Yeast cells expressing pHluorin were grown overnight to mid-log phase (0.4–0.6 A600/ml) and preincubated with 0.2% DMSO or with 100 μm drug in 0.2% DMSO. Following 30-min incubations at 30 °C, cells were transferred to 1 mm HEPES/MES buffer, pH 5.0, containing 2% glucose at a cell density of 5.0 A600/ml; fluorescence was measured for 6 min at 1-min intervals, and pH values were averaged. The cytosolic pH was estimated using calibration curves made in parallel (pH values ranging from 5.0 to 8.0). Data are presented as average pH values. Error bars = ± S.D., n = 2; *, p < 0.05; **, p < 0.01; and ***, p < 0.001 compared with DMSO as measured by two-tailed paired Student's t test.
Benzbromarone, Alexidine Dihydrochloride, and Thonzonium Bromide Acidify the Yeast Cytosol
To verify that lack of V-ATPase function acidifies the cytosol, we measured fluorometrically the cytosolic pH of yeast V-ATPase mutant strains that do not assemble functional V1V0 complexes and lack all V-ATPase function (2). Yeast vma2Δ (pHcyt 6.68 ± 0.081) and vma3Δ (pHcyt 6.81 ± 0.044) exhibited lower cytosolic pH than wild-type cells (pHcyt 7.5 ± 0.04) (Fig. 2B). These results are consistent with other studies (29) and with the notion that V-ATPases are key players in maintaining yeast cytosolic pH homeostasis.
We treated wild-type cells expressing pHluorin with the hit compounds and measured the cytosolic pH to confirm that the increased FL1/FL7 ratio was the direct outcome of cytosolic acidification. The cells treated with disulfiram (pHcyt 6.78 ± 0.13) resembled vma2Δ and vma3Δ cells (Fig. 2B), whereas DMSO treatment had no effect (pHcyt 7.4 ± 0.01), thus confirming that the positive and negative controls responded properly during the high-throughput screening. Treatment of yeast cells with the hit compounds revealed two groups of drugs. One group, consisting of benzbromarone (pHcyt 6.03 ± 0.07), alexidine dihydrochloride (pHcyt 6.10 ± 0.04), and thonzonium bromide (pHcyt 6.22 ± 0.02), significantly lowered the pH. A second group, consisting of bisoprolol fumarate (pHcyt 7.18 ± 0.08) and dyclonine hydrochloride (pHcyt 7.33 ± 0.05), induced only subtle pH changes (Fig. 2B).
Alexidine Dihydrochloride and Thonzonium Bromide Functionally Uncouple V-ATPase Proton Pumps
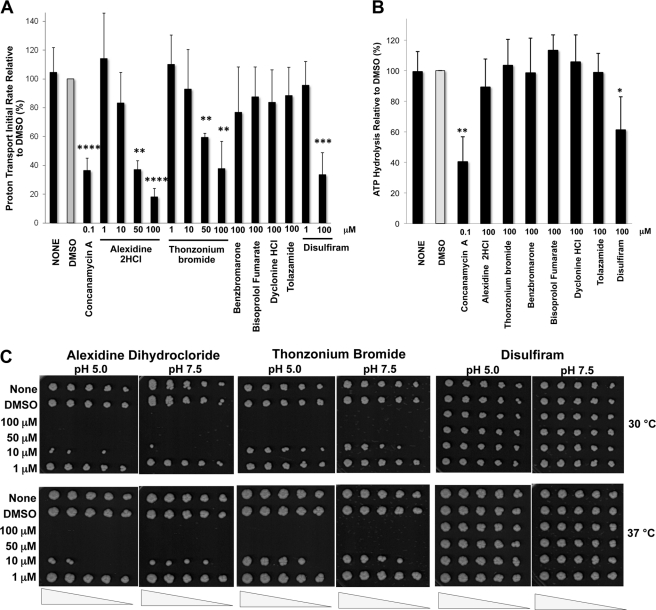
Benzbromarone, alexidine dihydrochloride, and thonzonium bromide acidified the cytosol to different extents, suggesting that they may lower the pH by inhibiting the V-ATPase pump with different efficacy or, alternatively, by affecting different targets. To address these two possibilities, we measured proton transport in vacuolar membrane fractions fluorometrically by monitoring ACMA quenching after the addition of MgATP in the presence of each compound. As anticipated, V-ATPase proton transport was very sensitive to concanamycin A treatment in vitro. At 100 nm concanamycin A, ACMA quenching was blocked, indicating that a pH gradient could not be formed across the vacuolar membrane because the V-ATPase pump was inhibited (Fig. 3A). Alexidine dihydrochloride and thonzonium bromide inhibited proton transport in a dose-dependent manner. We estimated an EC50 at 39 and 69 μm for alexidine dihydrochloride and thonzonium bromide, respectively (Fig. 3B). Benzbromarone, however, did not inhibit proton transport (Fig. 3A), suggesting that it acidified the cytosol by V-ATPase-independent mechanisms. None of the remaining drugs blocked proton transport at the highest concentration tested (100 μm), consistent with their negligible effects on cytosolic pH (Fig. 2B).
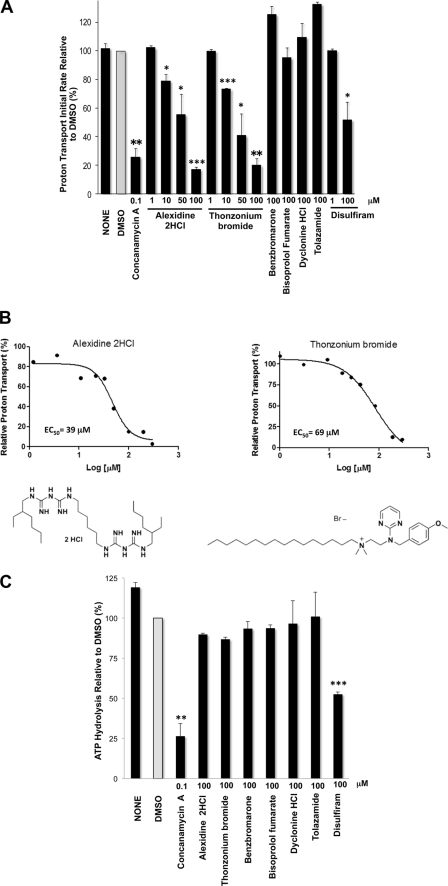
FIGURE 3.
Alexidine dihydrocloride and thonzonium bromide uncouple V-ATPase pumps in vitro. A, ATP-dependent proton transport is inhibited by alexidine dihydrocloride and thonzonium bromide. Purified vacuolar membrane vesicles (10 μg of protein) were preincubated for 10 min on ice with 1% DMSO alone or concanamycin A (100 nm) or with drugs at the indicated concentrations in the presence of 1% DMSO. Fluorescence quenching of ACMA was monitored (ex 410 nm, em 490 nm) upon the addition of 0.5 mm ATP and 1 mm MgSO4. Initial velocities were calculated for 15 s following addition of MgATP (n = 2). The apparently enhanced rate measured with benzbromarone and tolazamide was not dose-dependent and was sustained in the controls, when the drugs were added to the reaction mixture prior to the membrane vesicles. *, p < 0.05; **, p < 0.01; and ***, p < 0.001 for decreased transport compared with DMSO as measured by two-tailed paired Student's t test. B, V-ATPase inhibition by alexidine dihydrocloride and thonzonium bromide is dose-dependent. Purified vacuolar membrane vesicles (10 μg of protein) were incubated with alexidine dihydrochloride or thonzonium bromide at the indicated concentrations for 10 min on ice. ATP-dependent proton transport was measured as described in A. EC50 values were estimated using GraphPad Prism 5.0. The structures of alexidine dihydrochloride (left) and thonzonium bromide (right) are shown. C, ATP hydrolysis is resistant to alexidine dihydrochloride and thonzonium bromide treatments. Purified vacuolar membranes (4 μg of total protein) were preincubated for 10 min on ice with the indicated drug concentrations in 0.2% DMSO or with 0.2% DMSO alone. Following each treatment, ATP hydrolysis was measured spectrophotometrically using an enzymatic assay coupled to oxidation of NADH (340 nm) (59). Data are expressed as the average ± S.D.; n = 3 separate vacuolar purifications. Relative averaged values were used to express percentage activity. The specific activity of the concanamycin A-sensitive ATPase activity of the vacuoles was 1.3–1.5 μmol of ATP/min/mg of protein. **, p < 0.01; and ***, p < 0.001 for decreased ATPase activity (2-fold or more) compared with DMSO as measured by two-tailed paired Student's t test.
Because V-ATPases couple active transport of protons and ATP hydrolysis, we measured ATP hydrolysis as well. We could discriminate between V-ATPase inhibitors, which block both proton transport and ATP hydrolysis, and uncouplers, which block proton transport preferentially. As expected (40), disulfiram inhibited ATP hydrolysis in vacuolar membranes, albeit with less potency than concanamycin A (Fig. 3C). None of the hit compounds inhibited ATP hydrolysis (Fig. 3C), including alexidine dihydrochloride and thonzonium bromide, even at concentrations that completely blocked proton transport (100 μm). Alexidine dihydrochloride and thonzonium bromide therefore uncoupled V-ATPase proton pumps.
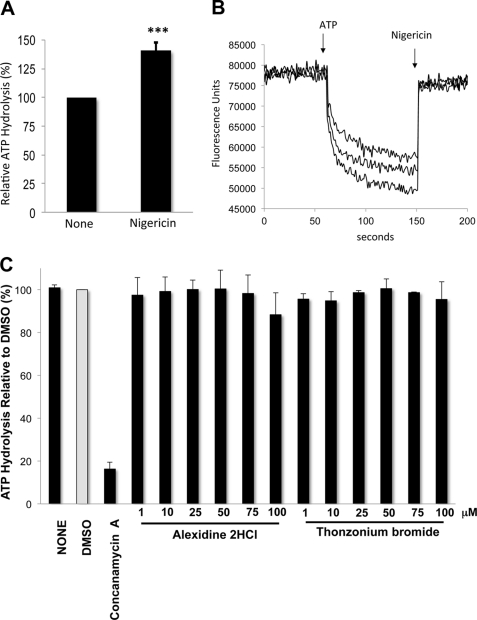
The ATPase activity was not stimulated by alexidine dihydrochloride and thonzonium bromide, suggesting that they do not simply act as ionophores that uncouple V-ATPase pumps by dissipating the proton gradient across the membrane (41). To distinguish between V-ATPase-specific uncoupling and unspecific ionophore effects, we treated vacuolar membranes with 1 μm nigericin. Nigericin also uncoupled the enzyme; it abolished proton transport (Fig. 4B) but stimulated concanamycin A-sensitive ATP hydrolysis (by 1.4-fold) (Fig. 4A). We additionally showed that lower concentrations of alexidine dihydrochloride and thonzonium bromide (1–75 μm) did not stimulate ATPase activity (Fig. 4C). This disproves the possibility that the inhibitors act as proton ionophores at low concentrations while inhibiting activity at high concentrations, such that at 100 μm the two effects (stimulation and inhibition of ATP hydrolysis) cancel each other out. These results indicate that alexidine dihydrochloride and thonzonium bromide did not uncouple V-ATPase pumps by simply dissipating the membrane proton gradient.
FIGURE 4.
Uncoupling by alexidine dihydrocloride and thonzonium bromide differs from ionophore-mediated uncoupling. A, nigericin stimulates ATPase activity. Concanamycin A-sensitive ATPase specific activity was measured as described in the legend for Fig. 3C, except that 1 μm nigericin in 0.1% ethanol was added to the reaction. An equal concentration of ethanol in the reaction had no effect on the activity. Shown is the average activity relative to untreated controls (None) of three independent vacuolar purifications. Error bars = ± S.D. ***, p < 0.001 compared with no treatment as measured by two-tailed paired Student's t test. B, nigericin inhibits proton transport. Proton transport was monitored in three independent vacuolar membrane preparations (10 μg of protein) by measuring ACMA quenching as described in the legend for Fig. 3A, except that nigericin (1 μm) was added 90 s after the addition of MgATP, and florescence was measured for an additional 50 s. C, ATP hydrolysis is not stimulated by lower concentrations of alexidine dihydrochloride and thonzonium bromide. Concanamycin A-sensitive ATPase specific activity was measured as described in the legend to Fig. 3C. Data are expressed as the average ± S.D.; n = 3 separate vacuolar purifications. Relative averaged values were used to express percentage activity.
Yeast Cells Exposed to V-ATPase Uncouplers Develop Growth Defects
To further assess the specificity of alexidine dihydrochloride and thonzonium bromide for V-ATPases in vivo, we asked whether yeast cells treated with the drugs would develop growth defects that could be attributed to impairment of V-ATPase function. Lack of yeast V-ATPase activity leads to pH-sensitive growth (42, 43); cells grow at pH 5.0 but cannot grow at pH 7.5 (Fig. 5A). This growth defect is known as the vma (vacuolar membrane ATPase) mutant growth phenotype (2). We addressed whether exposure to alexidine dihydrochloride and thonzonium bromide leads to a vma growth phenotype by treating the cells with varied concentrations of the drugs for 1 h at 30 °C and plating the cells on medium buffered to pH 5.0 and 7.5. Yeast cell growth was then monitored at 30 and 37 °C for 48 h.
FIGURE 5.
Alexidine dihydrochloride and thonzonium bromide treatments lead to growth defects in S. cerevisiae. A, yeast vma mutants cannot grow at neutral pH. Overnight mid-log phase cultures of vma2Δ and vma3Δ cells (lacking all V-ATPase function) and an isogenic wild type were diluted to 0.5 A600/ml in YEPD medium, pH 5.0, and serial dilutions (1:3) were stamped on YEPD plates adjusted to pH 5.0 and 7.5 and incubated at 30 and 37 °C. B, alexidine dihydrochloride and thonzonium bromide prevent growth. Overnight yeast cell cultures were diluted to 0.5 A600/ml in YEPD medium, pH 5.0. Cells were rotated at 30 °C for 1 h with the drugs at the indicated concentrations containing 1% DMSO. Controls included cells resuspended in 1% DMSO alone or an equal volume of YEPD, pH 5.0 (None). Serial dilutions (1:3) were stamped on YEPD, pH 5.0 and pH 7.5, plates and incubated at 30 and 37 °C. Cell growth was monitored for 48 h.
Predictably, cells treated with DMSO and disulfiram did not show a vma phenotype (Fig. 5B), as DMSO did not inhibit the enzyme and disulfiram only partially inhibited V-ATPase activity (by about 50%) at the highest concentration used (Fig. 3, A and C). Cells treated with the lowest concentration (1 μm) of alexidine dihydrochloride and thonzonium bromide exhibited normal cell growth (Fig. 5B). However, exposure to 10 μm alexidine dihydrochloride resulted in a markedly slow growth at pH 5.0 and 7.5, with cells developing the typical vma mutant growth phenotype at 37 °C. Likewise, 10 μm thonzonium bromide reduced cell growth, with the cells developing a mild vma phenotype at 37 °C. Alexidine dihydrochloride and thonzonium bromide completely prevented yeast growth at 50 and 100 μm concentrations. Previous work has shown that yeast cells must lose at least 75% of their wild-type V-ATPase function to develop a vma growth phenotype (23, 37). Thus, we concluded that V-ATPase function was significantly impaired in vivo by treatment with 10 μm uncouplers and that larger concentrations were cytotoxic.
Tether Connecting the Cytosolic Domain of Subunit a (Vph1p) to the Membrane Has Uncoupling Functions
V0 subunit a consists of two domains: a cytosolic N-terminal domain (∼45 kDa) that interacts with several V1 subunits to allow rotation of rotor-forming subunits during catalysis and a membrane-bound C-terminal domain (∼50 kDa) that contributes to the path of proton transport (3). Cleavage of Vph1p at the tether connecting the two domains uncouples yeast V-ATPase complexes (26), suggesting that Vph1-mediated assembly of the two domains is necessary for coupling. We asked whether residues forming the tether were necessary for the observed pharmacological uncoupling.
To gain insight into the mechanisms by which alexidine dyhydrochloride and thonzonium bromide uncouple V-ATPase pumps, we expressed a truncated allele of Vph1p lacking residues 362–407 of the tether (vph1–362-407Δ) in vph1Δstv1Δ cells. Yeast vph1Δstv1Δ does not express endogenous subunit a, because the gene VPH1 and its functional homolog, STV1, are deleted (34). Previously, we showed that biosynthetic assembly of tether-less V1V0 (vph1–362-407Δ) complexes is complete when cells modestly overexpress V0 subunit d (Vma6p) and that the assembled vph1–362-407Δ V1V0 fully retains ATP hydrolytic activity (35). Thus, we simultaneously co-transformed the vph1Δstv1Δ cells with VPH1–362-407Δ and subunit d (VMA6), each of them expressed from a CEN plasmid (pRS316) under control of their natural promoters as described previously (35). We compared the mutant vph1–362-407Δ vacuolar membranes with isogenic wild-type membranes obtained in parallel from vph1Δstv1Δ cells expressing the wild-type allele of VPH1 from pRS316. As expected, the tether-less mutant assembled wild-type levels of V1 (A and B) and V0 (a) subunits at the vacuolar membrane and retained about 90% of the wild-type concanamycin A-sensitive ATP hydrolytic activity (Fig. 6A). The tether-less V1V0 also retained wild-type levels of proton transport (Fig. 6, A and B), suggesting that the length of the tether does not affect coupling efficiency when the physical connection between the N- and C-terminal domains of subunit a is maintained.
FIGURE 6.
The tether of subunit a (Vph1p) is crucial for functional uncoupling by thonzonium bromide. A, tether-less V1V0 complexes are coupled functionally. Left, vacuolar membrane vesicles were purified from vph1Δstv1Δ cells transformed with either wild-type VPH1 or VMA6 plus mutant (Vph1p-362–407Δ). Protein separated by SDS-PAGE (5 μg of total vacuolar protein) was analyzed by Western blot using antibodies against V0 subunit a and V1 subunits A and B. Concanamycin A-sensitive ATPase activity (10 μg of total protein) was measured as described in the legend for Fig. 3C. Right, proton transport was measured as described in the legend for Fig. 3A. B, tether-less V1V0 complexes are resistant to thonzonium bromide-induced uncoupling. Wild-type (Vph1p-WT) and tether-less mutant (vph1p-362–407Δ) vacuolar membranes were treated with 100 μm thonzonium bromide or alexidine dihydrochloride, and ATP hydrolysis and proton transport were measured as described in the legend for Fig. 3, A and C. **, p < 0.01 compared with Vph1p-WT treated with thonzonium bromide, as measured by two-tailed paired Student's t test. C, tether-less V1V0 yeast mutants show growth resistance to thonzonium bromide and alexidine dihydrochloride. Wild-type and tether-less mutant cells were grown overnight to 0.5 A600/ml in SC-Ura medium, pH 5.0, treated with the indicated concentrations of drugs as described in the legend for Fig. 5, and plated on YEPD, pH 7.5 (1:3-fold serial dilutions). Plates were incubated at 37 °C for 72 h.
Tether-less V1V0 complexes were resistant to uncoupling by thonzonium bromide (Fig. 6B). Whereas 80% of the wild-type proton transport was inhibited by 100 μm thonzonium bromide, tether-less V1V0 was inhibited by only 40%, suggesting that the tether is involved in uncoupling by thonzonium bromide. Partial-to-no resistance against alexidine dihydrochloride suggests that uncoupling by alexidine dihydrochloride may entail other uncoupling mechanisms.
Next, we asked whether cells expressing tether-less V1V0 would be resistant to the uncouplers in vivo. We assessed cell growth at pH 7.5, the nonpermissive growth condition for vma mutants, after preincubation with varied concentrations of alexidine dihydrochloride and thonzonium bromide (at 1–100 μm). The vph1–362-407Δ cells required 2–2.5-fold larger concentrations of drugs to decrease growth than the isogenic wild-type cells (Fig. 6C), further suggesting that the tether plays an uncoupling function.
Alexidine Dihydrochloride and Thonzonium Bromide Uncouple C. albicans V-ATPase Proton Pumps and Impair Cell Growth
The tether constitutes the most conserved sequence at the N-terminal end of subunit a. It has 47–51% identity (63–72% conserved) between yeast and human isoforms. Based on the previous results, which indicate that this conserved sequence of Vph1p confers uncoupling potential to V-ATPase pumps, we extended our studies to C. albicans. This pathogenic fungi contains Vph1p with 67% sequence identity (83% conserved) with yeast at the tether region of the protein. We isolated vacuolar membrane fractions from C. albicans and measured proton transport and ATP hydrolysis in the presence of the uncouplers. The results in C. albicans (Fig. 7) mirrored the studies in S. cerevisiae. Alexidine dihydrochloride and thonzonium bromide inhibited proton transport in a dose-dependent manner (Fig. 7A) but had no effect on ATP hydrolysis (Fig. 7B), showing that alexidine dihydrochloride and thonzonium bromide also uncoupled C. albicans V-ATPase pumps.
FIGURE 7.
Alexidine dihydrocloride and thonzonium bromide uncouple C. albicans V-ATPase pumps and prevent growth. A, C. albicans ATP-dependent proton transport is inhibited by the uncouplers. Fluorescence quenching of ACMA was measured as described in Fig. 3A, except that 30 μg of vacuolar membrane protein was used for each treatment. B, ATPase activity is resistant to the uncouplers. ATP hydrolysis was measured as described in the legend for Fig. 3C, except that 20 μg of purified vacuolar membrane protein was used for each treatment. In both A and B, the indicated concentrations of drugs were present in 1% DMSO. Data are expressed as the average ± S.D. for n = 3–5 independent vacuolar purifications. The specific activity of the concanamycin A-sensitive ATPase activity of the vacuoles was 0.2–0.4 μmol of ATP/min/mg of protein. *, p < 0.05; **, p < 0.01; ***, p < 0.001; and ****, p < 0.0001 by two-tailed paired Student's t test. C, uncouplers reduced the growth of C. albicans. Cultures of C. albicans were grown overnight and treated as described in the legend for Fig. 5B.
For in vivo analyses, C. albicans was exposed to various concentrations of the inhibitors for 1 h at 30 °C, and growth was assessed at pH 5.0 and 7.5 at both 30 and 37 °C (Fig. 7C). Like the yeast S. cerevisiae, C. albicans cells exposed to 1 μm alexidine dihydrochloride, 1 μm thonzonium bromide, and up to 100 μm disulfiram mimicked the untreated and DMSO control cells. Although the cells did not develop a vma growth phenotype, they exhibited significant general growth defects after treatment with 10 μm alexidine dihydrochloride or thonzonium bromide at both pH 5.0 and 7.5. Alexidine dihydrochloride was more toxic than thonzonium bromide, but both drugs completely blocked growth at the higher concentrations tested (50 and 100 μm).
DISCUSSION
We screened a chemical library using a pHluorin-based high-throughput flow cytometry assay to identify compounds that acidify the yeast cytosol as a means of identifying new in vivo inhibitors of S. cerevisiae V-ATPase proton pumps. We identified two drugs, alexidine dihydrochloride and thonzonium bromide, which selectively target the V-ATPase complex in vitro and in vivo by blocking proton transport. Our studies indicate that they are bona fide uncouplers of V-ATPase pumps.
New Inhibitors of V-ATPase Pumps Acidify the Cytosol and Uncouple the Enzyme
This unbiased screen identified six drugs that lower the yeast cytosolic pH, albeit to different extents (Fig. 2B). Benzbromarone, alexidine dihydrochloride, thonzonium bromide, and disulfiram showed the strongest effects, whereas dyclonine hydrochloride and bisoprolol fumarate induced smaller pH changes. Only disulfiram, alexidine dihydrochloride, and thonzonium bromide acidified the cytosol through inhibition of V-ATPase pumps. The remaining drugs did not inhibit ATP hydrolysis and/or proton transport in purified yeast vacuolar membranes, indicating that they act on different targets and acidify the cytosol through V-ATPase-independent mechanisms.
Yeast cytosolic pH is maintained through a concerted movement of protons out of the cytosol by the V-ATPase and Pma1p pumps, the major electrogenic pumps at the vacuolar and plasma membranes, respectively (13, 29). Whereas V-ATPase transfers protons into the vacuolar lumen, Pma1p moves cytosolic protons out of the cell, helping yeast to sustain a favorable acidic growth environment. Therefore, by using the pHluorin system to monitor cytosolic acidification, we anticipated the identification of inhibitors of Pma1p in addition to V-ATPase inhibitors. We also most likely identified general inhibitors of protonophore activity, because they would be expected to dissipate cellular proton gradients, accelerate the leakage of protons from organelles, and result in net cytosolic acidification.
Disulfiram was identified previously as a V-ATPase inhibitor in a high-throughput screen of the Prestwick Chemical Library, and we showed that disulfiram treatment alkalinizes the yeast vacuoles (40). In that study, we characterized the inhibitory effect of disulfiram on ATP hydrolysis (40). Here, we have further shown that disulfiram treatment acidified the yeast cytosol in vivo (Fig. 2B) and inhibited V-ATPase proton transport (Figs. 3A and 7A) in addition to ATP hydrolysis (Figs. 3C and 7B) in vitro.
Alexidine dihydrochloride and thonzonium bromide inhibited proton transport in vacuolar membrane fractions without inhibiting ATP hydrolysis. At high concentrations (100 μm), proton transport was significantly impaired (by 80–90%), whereas ATP hydrolysis remained intact. Our results indicate that alexidine dihydrochloride and thonzonium bromide are not simply ionophores, because they do not stimulate ATP hydrolysis, in contrast with nigericin, which both inhibits proton transport and stimulates hydrolysis of ATP (Fig. 4). Alexidine dihydrochloride and thonzonium bromide selectively uncouple V-ATPase pumps.
Although less potent than concanamycin A (EC50 = 1 nm), uncouplers such as alexidine dihydrochloride (EC50 = 39 μm) and thonzonium bromide (EC50 = 69 μm) are anticipated to be highly toxic. In addition to disturbing cytosolic and vacuolar pH homeostasis, alexidine dihydrochloride and thonzonium bromide should deplete the energy reserves of the cell, because uncoupled V-ATPase pumps will hydrolyze cytosolic ATP continuously. In a global sense, V-ATPase uncouplers hinder multiple cellular processes and constitute a quick acting way to block vital functions.
As anticipated, yeast was more sensitive to treatment with the uncouplers in vivo than in vitro. Treatment with alexidine dihydrochloride and thonzonium bromide was cytotoxic at concentrations (50 μm) that inhibited proton transport by only 40% in vitro (Figs. 3A and 5B). Yeast cells treated with a lower concentration of the drugs (10 μm) developed a vma growth phenotype, despite the fact that proton transport was barely inhibited (by 20–25%) in vitro. Therefore, a fine threshold (20–40% uncoupling) delineates conditional growth (vma phenotype) and cell death following uncoupling of the proton transport and ATPase activity by alexidine dihydrochloride and thonzonium bromide.
Yeast V-ATPase mutants develop the vma phenotype when V-ATPase function (proton transport and ATP hydrolysis) is impaired by about 75% (23, 37). Yet, a more modest loss of proton transport (by about 60%) leads to vma growth phenotype if the pump is uncoupled (44). The fact that yeast died after 40% proton transport inhibition by the uncouplers (Figs. 3 and 5) indicates that the cells are more susceptible to pharmacological (acute) than genetic (chronic) V-ATPase uncoupling. Perhaps, V-ATPase mutants can develop compensatory ATP conservation mechanisms that will allow yeast to survive in the face of uncoupling. In addition, alexidine dihydrochloride and thonzonium bromide at concentrations greater than 50 μm could have additional targets and effects unrelated to V-ATPase that could also contribute to cytotoxicity.
Nonetheless, the vma growth phenotype as defined by significantly slow growth (thonzonium bromide) or no growth (alexidine dihydrochloride) at pH 7.5 showed selectivity of the drugs for V-ATPase (Fig. 5B). The vma phenotype is the hallmark trait of yeast cells with impaired V-ATPase function (8, 43) and is a legitimate indicator of V-ATPase defects in yeast. In line with this notion, we found that 1 μm alexidine dihydrochloride or thonzonium bromide does not inhibit the pumps or affect cell growth at pH 7.5. Additionally, benzbromarone (100 μm), which does not inhibit V-ATPase pumps (Fig. 3, A and Fig. C), does not inhibit cell growth at pH 7.5 (data not shown), despite acidifying the cytosol (Fig. 2B). Thus, cytosolic acidification alone is not enough to develop vma mutant growth phenotype; V-ATPase function also must be impaired.
Uncoupling Roles of the Cytosolic Tether Connecting Vph1p to the Membrane
V0 subunit a structurally bridges the V0 and V1 domains of the V-ATPase complex. Subunit a interacts with V1 subunits E, G, C, H, and A at its N terminus (cytosolic domain) and the proteolipid ring at its C terminus (integral membrane domain) (45–48). Its unique topography makes subunit a an attractive candidate for the coupling of proton transport in V0 and ATP hydrolysis in V1. Subunit a is the only stator subunit anchored in the membrane. By holding the three peripheral stators (EG dimers) at its cytosolic domain, subunit a secures the steady catalytic sites in the A3B3 hexamer while transferring protons through its membrane domain. Providing evidence that the V1V0 association facilitated by subunit a is necessary for coupling of the enzyme, separation of the N terminus from the C terminus of Vph1 by cleavage at residue 376 of the tether partially uncouples V-ATPases (26). Cleavage reduces proton transport by 2-fold without affecting ATPase activity. We showed in an earlier study that the tether of the subunit a Vph1p is a flexible element and that Vph1p tether-less mutant V1V0 complexes are functional, as they retain concanamycin A-sensitive ATP hydrolytic activity and disassemble and reassemble normally (35).
Here we show that tether-less V1V0 completely retained wild-type levels of proton transport activity. Wild-type levels of both proton transport and ATPase activity in tether-less V1V0 indicate that the length of the tether is not essential for functional coupling of the enzyme. Our results indicate instead that the intact tether facilitates uncoupling. Amino acid residues 362–407 of the tether conferred uncoupling potential to V-ATPases, as their truncation made the enzyme resistant to treatment with thonzonium bromide. In the presence of thonzonium bromide, the coupling efficiency (the ratio of relative proton transport to relative ATPase activity) of tether-less V1V0 was greater than that of the wild-type enzyme (Fig. 6B).
In line with the idea that the tether of Vph1p confers sensitivity to thonzonium bromide, wild-type strains SF838-1Dα (Fig. 5B) and Vph1p-WT (Fig. 6C), which express Vph1p, showed similar sensitivity to thonzonium bromide. Both strains had reduced growth at pH 7.5 and 37 °C (vma phenotype) after exposure to 10 μm thonzonium bromide. The additional finding that only SF838-1Dα developed the vma growth phenotype when the cells were challenged with alexidine dihydrochloride further supports this notion. The major difference between these two strains is that SF838-1Dα expresses both isoforms of subunit a (the vacuolar isoform, Vph1p, and the Golgi isoform, Stv1p), whereas Vph1p-WT expresses only Vph1p. Thus, if alexidine dihydrochloride additively uncoupled Stv1p- and Vph1p-containing V-ATPases, the absence of Stv1p could explain Vph1p-WT growth resistance.
The presence of a structural element that gives uncoupling potential to V-ATPase complexes is consistent with the hypothesis that wild-type V-ATPase pumps may not be coupled optimally (27, 28) and that the tightness of coupling may be adjusted in vivo to modulate organelle acidification (2, 28). The tightness of coupling of proton transport and ATPase activity has been shown to be modulated by ATP concentrations (27) and by the nonhomologous region of V1 subunit A (25). The mutation P217V in the nonhomologous region of subunit A generates more tightly coupled complexes because it stimulates proton transport and reduces ATPase activity. As the nonhomologous region of the V1 subunit A interacts with V0 subunit a (25), such tightness of coupling could be explained if structural changes are transmitted from subunit A to the tether of subunit a.
Other uncoupling elements may exist in addition to the tether of the V0 subunit a. Our results indicate that alexidine dihydrochloride uncouples the enzyme by a mechanism unique from thonzonium bromide, despite their similar EC50. Alexidine dihydrochloride is an amphipathic bisbiguanide that binds to lipopolysaccharides (49) and is used as an antibacterial agent (50), whereas thonzonium bromide is a monocationic detergent surface-active agent. Although both may have adverse effects on lipid domains (51), and we cannot eliminate the possibility that they could affect V-ATPase function by altering V0 interactions with membrane lipids, our results indicate that thonzonium bromide and alexidine dihydrochloride selectively inhibited V-ATPase proton transport in vitro. Arguing against unspecific effects are the fact that ATP hydrolysis was kept intact even at high concentrations of the drugs (100 μm); that uncoupling by thonzonium bromide and alexidine dihydrochloride was demonstrated in two different sources of vacuolar membranes (S. cerevisiae and C. albicans); that uncoupling by thonzonium bromide and alexidine dihydrochloride differed from uncoupling by ionophores such as nigericin; and that vph1–362-407Δ mutants showed selective resistance to uncoupling by thonzonium bromide only.
V-ATPase Uncouplers as Potential Anti-V-ATPase Drugs
Alexidine dihydrochloride and thonzonium bromide effectively inhibit V-ATPase proton transport in vacuolar membranes from C. albicans (Fig. 7). Although uncoupling in vitro is very similar between C. albicans and S. cerevisiae, C. albicans does not develop the vma growth phenotype. In vivo, the uncouplers significantly reduce cell growth at 10 μm and are cytotoxic at greater concentrations. This may be because complete inactivation of V-ATPase activity appears to be necessary for development of the vma phenotype in C. albicans (52), as cells expressing genetic mutations that partially inactivate V-ATPase function do not develop pH-sensitive growth.7 Our results indicate that V-ATPase uncoupling must be lethal, because deletions of V-ATPase subunits that prevent enzyme function leading to the vma growth phenotype do not cause general cytotoxicity in C. albicans (52).
Fungi need vacuolar and trafficking functions to infect their hosts, and genetic inactivation of V-ATPase prevents virulence of C. albicans and other pathogenic fungi (52–54). We have provided evidence that V-ATPase uncoupling would be a highly effective mechanism to control virulence, because in addition to preventing proton transport, uncouplers are expected to deplete the energy reserve of the cell. Although V-ATPase inhibitors that have therapeutic applications have yet to be identified, the Prestwick Chemical Library is a useful resource for drug repurposing, as most of the compounds have known safety and toxicity profiles. In fact, cytotoxicity by alexidine dihydrochloride has been shown previously in Cryptococcus neoformans, C. albicans, and Paracoccidioides brasiliensis (55, 56). Together, those studies and our current findings make alexidine dihydrochloride and thonzonium bromide attractive candidates to evaluate for repurposing as potential antifungal therapeutic drugs. Further studies will specifically assess the anti-virulence mechanisms behind these drugs and the efficacy of combining these uncouplers with other antifungal therapeutics such as azoles, which also alter V-ATPase activity (57).
In conclusion, alexidine dihydrochloride and thonzonium bromide disengaged proton transport and ATPase activity, and to our knowledge, they are the only uncouplers of V-ATPase proton pumps described. This study revealed new information regarding the mechanisms of V-ATPase coupling, a region within V0 subunit a (the tether) that confers “uncoupling potential” to V-ATPase pumps, supporting the hypothesis that wild-type V-ATPase complexes may not be optimally coupled in vivo (27, 28). Uncouplers of V-ATPase pumps may offer new knowledge to the understanding of the mechanisms governing rotational catalysis and V-ATPase regulation, and they may be useful as potential therapeutic drugs. Because alexidine dihydrochloride and thonzonium bromide uncouple C. albicans V-ATPase, additionally they can help to elucidate the cellular processes pertaining to fungal infection and virulence in pathogens like C. albicans, the single most important cause of opportunistic mycoses worldwide for which available treatment methods remain suboptimal (58).
Acknowledgments
We thank Juan Strouse, Anna Waller, Mark Carter, and other researchers and staff at the University of New Mexico Center for Molecular Discovery for helpful discussions. Images in this article were generated at the University of New Mexico Cancer Center Fluorescence Microscopy Shared Resource.
This work was supported, in whole or in part, by National Institutes of Health Grants 5R01GM086495 (to K. J. P.), 1U54MH084690 (to L. A. S., C. P., and C. A.), and CA118100 (to the University of New Mexico Cancer Center).

This article contains supplemental Table 1.
S. M. Raines, S. A. Lee, and K. J. Parra, unpublished results.
- V-ATPase
- vacuolar proton-translocating ATPase
- DMSO
- dimethyl sulfoxide
- SC
- synthetic complete
- ACMA
- 9-amino-6-chloro-2-methoxyacridine.
REFERENCES
- 1. Nishi T., Forgac M. (2002) The vacuolar (H+)-ATPases: nature's most versatile proton pumps. Nat. Rev. Mol. Cell Biol. 3, 94–103 [DOI] [PubMed] [Google Scholar]
- 2. Kane P. M. (2006) The where, when, and how of organelle acidification by the yeast vacuolar H+-ATPase. Microbiol. Mol. Biol. Rev. 70, 177–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Forgac M. (2007) Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell Biol. 8, 917–929 [DOI] [PubMed] [Google Scholar]
- 4. Shum W. W., Ruan Y. C., Da Silva N., Breton S. (2011) Establishment of cell-cell cross-talk in the epididymis: control of luminal acidification. J. Androl. 32, 576–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brown D., Breton S., Ausiello D. A., Marshansky V. (2009) Sensing, signaling and sorting events in kidney epithelial cell physiology. Traffic 10, 275–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ochotny N., Flenniken A. M., Owen C., Voronov I., Zirngibl R. A., Osborne L. R., Henderson J. E., Adamson S. L., Rossant J., Manolson M. F., Aubin J. E. (2011) The V-ATPase a3 subunit mutation R740S is dominant negative and results in osteopetrosis in mice. J. Bone Miner. Res. 26, 1484–1493 [DOI] [PubMed] [Google Scholar]
- 7. Nakanishi-Matsui M., Sekiya M., Nakamoto R. K., Futai M. (2010) The mechanism of rotating proton pumping ATPases. Biochim. Biophys. Acta 1797, 1343–1352 [DOI] [PubMed] [Google Scholar]
- 8. Kane P. M. (2007) The long physiological reach of the yeast vacuolar H+-ATPase. J. Bioenerg. Biomembr. 39, 415–421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Forster C., Kane P. M. (2000) Cytosolic Ca2+ homeostasis is a constitutive function of the V-ATPase in Saccharomyces cerevisiae. J. Biol. Chem. 275, 38245–38253 [DOI] [PubMed] [Google Scholar]
- 10. Zhang J. W., Parra K. J., Liu J., Kane P. M. (1998) Characterization of a temperature-sensitive yeast vacuolar ATPase mutant with defects in actin distribution and bud morphology. J. Biol. Chem. 273, 18470–18480 [DOI] [PubMed] [Google Scholar]
- 11. Bishop A. L., Rab F. A., Sumner E. R., Avery S. V. (2007) Phenotypic heterogeneity can enhance rare cell survival in “stress-sensitive” yeast populations. Mol. Microbiol. 63, 507–520 [DOI] [PubMed] [Google Scholar]
- 12. Huss M., Wieczorek H. (2009) Inhibitors of V-ATPases: old and new players. J. Exp. Biol. 212, 341–346 [DOI] [PubMed] [Google Scholar]
- 13. Bowman E. J., O'Neill F. J., Bowman B. J. (1997) Mutations of pma-1, the gene encoding the plasma membrane H+-ATPase of Neurospora crassa, suppress inhibition of growth by concanamycin A, a specific inhibitor of vacuolar ATPases. J. Biol. Chem. 272, 14776–14786 [DOI] [PubMed] [Google Scholar]
- 14. Bowman E. J., Bowman B. J. (2000) Cellular role of the V-ATPase in Neurospora crassa: analysis of mutants resistant to concanamycin or lacking the catalytic subunit A. J. Exp. Biol. 203, 97–106 [DOI] [PubMed] [Google Scholar]
- 15. Huss M., Vitavska O., Albertmelcher A., Bockelmann S., Nardmann C., Tabke K., Tiburcy F., Wieczorek H. (2011) Vacuolar H(+)-ATPases: intra- and intermolecular interactions. Eur. J. Cell Biol. 90, 688–695 [DOI] [PubMed] [Google Scholar]
- 16. Mijaljica D., Prescott M., Devenish R. J. (2011) V-ATPase engagement in autophagic processes. Autophagy 7, 666–668 [DOI] [PubMed] [Google Scholar]
- 17. Klionsky D. J., Elazar Z., Seglen P. O., Rubinsztein D. C. (2008) Does bafilomycin A1 block the fusion of autophagosomes with lysosomes? Autophagy 4, 849–950 [DOI] [PubMed] [Google Scholar]
- 18. El Far O., Seagar M. (2011) A role for V-ATPase subunits in synaptic vesicle fusion? J. Neurochem. 117, 603–612 [DOI] [PubMed] [Google Scholar]
- 19. Bowman B. J., McCall M. E., Baertsch R., Bowman E. J. (2006) A model for the proteolipid ring and bafilomycin/concanamycin-binding site in the vacuolar ATPase of Neurospora crassa. J. Biol. Chem. 281, 31885–31893 [DOI] [PubMed] [Google Scholar]
- 20. Bowman E. J., Bowman B. J. (2005) V-ATPases as drug targets. J. Bioenerg. Biomembr. 37, 431–435 [DOI] [PubMed] [Google Scholar]
- 21. Bockelmann S., Menche D., Rudolph S., Bender T., Grond S., von Zezschwitz P., Muench S. P., Wieczorek H., Huss M. (2010) Archazolid A binds to the equatorial region of the c-ring of the vacuolar H+-ATPase. J. Biol. Chem. 285, 38304–38314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xu T., Forgac M. (2000) Subunit D (Vma8p) of the yeast vacuolar H+-ATPase plays a role in coupling of proton transport and ATP hydrolysis. J. Biol. Chem. 275, 22075–22081 [DOI] [PubMed] [Google Scholar]
- 23. Owegi M. A., Pappas D. L., Finch M. W., Jr., Bilbo S. A., Resendiz C. A., Jacquemin L. J., Warrier A., Trombley J. D., McCulloch K. M., Margalef K. L., Mertz M. J., Storms J. M., Damin C. A., Parra K. J. (2006) Identification of a domain in the V0 subunit d that is critical for coupling of the yeast vacuolar proton-translocating ATPase. J. Biol. Chem. 281, 30001–30014 [DOI] [PubMed] [Google Scholar]
- 24. Liu M., Tarsio M., Charsky C. M., Kane P. M. (2005) Structural and functional separation of the N- and C-terminal domains of the yeast V-ATPase subunit H. J. Biol. Chem. 280, 36978–36985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shao E., Forgac M. (2004) Involvement of the nonhomologous region of subunit A of the yeast V-ATPase in coupling and in vivo dissociation. J. Biol. Chem. 279, 48663–48670 [DOI] [PubMed] [Google Scholar]
- 26. Qi J., Forgac M. (2008) Function and subunit interactions of the N-terminal domain of subunit a (Vph1p) of the yeast V-ATPase. J. Biol. Chem. 283, 19274–19282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Arai H., Pink S., Forgac M. (1989) Interaction of anions and ATP with the coated vesicle proton pump. Biochemistry 28, 3075–3082 [DOI] [PubMed] [Google Scholar]
- 28. Moriyama Y., Nelson N. (1988) The vacuolar H+-ATPase, a proton pump controlled by a slip. Prog. Clin. Biol. Res. 273, 387–394 [PubMed] [Google Scholar]
- 29. Martínez-Muñoz G. A., Kane P. (2008) Vacuolar and plasma membrane proton pumps collaborate to achieve cytosolic pH homeostasis in yeast. J. Biol. Chem. 283, 20309–20319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Brett C. L., Tukaye D. N., Mukherjee S., Rao R. (2005) The yeast endosomal Na+K+/H+ exchanger Nhx1 regulates cellular pH to control vesicle trafficking. Mol. Biol. Cell 16, 1396–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tarsio M., Zheng H., Smardon A. M., Martínez-Muñoz G. A., Kane P. M. (2011) Consequences of loss of Vph1 protein-containing vacuolar ATPases (V-ATPases) for overall cellular pH homeostasis. J. Biol. Chem. 286, 28089–28096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Edwards B. S., Oprea T., Prossnitz E. R., Sklar L. A. (2004) Flow cytometry for high-throughput, high-content screening. Curr. Opin. Chem. Biol. 8, 392–398 [DOI] [PubMed] [Google Scholar]
- 33. Elble R. (1992) A simple and efficient procedure for transformation of yeasts. BioTechniques 13, 18–20 [PubMed] [Google Scholar]
- 34. Manolson M. F., Wu B., Proteau D., Taillon B. E., Roberts B. T., Hoyt M. A., Jones E. W. (1994) STV1 gene encodes functional homologue of 95-kDa yeast vacuolar H(+)-ATPase subunit Vph1p. J. Biol. Chem. 269, 14064–14074 [PubMed] [Google Scholar]
- 35. Ediger B., Melman S. D., Pappas D. L., Jr., Finch M., Applen J., Parra K. J. (2009) The tether connecting cytosolic (N terminus) and membrane (C terminus) domains of yeast V-ATPase subunit a (Vph1) is required for assembly of V0 subunit d. J. Biol. Chem. 284, 19522–19532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Forgac M., Cantley L., Wiedenmann B., Altstiel L., Branton D. (1983) Clathrin-coated vesicles contain an ATP-dependent proton pump. Proc. Natl. Acad. Sci. U.S.A. 80, 1300–1303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Owegi M. A., Carenbauer A. L., Wick N. M., Brown J. F., Terhune K. L., Bilbo S. A., Weaver R. S., Shircliff R., Newcomb N., Parra-Belky K. J. (2005) Mutational analysis of the stator subunit E of the yeast V-ATPase. J. Biol. Chem. 280, 18393–18402 [DOI] [PubMed] [Google Scholar]
- 38. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochim. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 39. Mahon M. J. (2011) chlorine: an enhanced, radiometric, pH-sensitive green florescent protein. Adv. Biosci. Biotechnol. 2, 132–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Johnson R. M., Allen C., Melman S. D., Waller A., Young S. M., Sklar L. A., Parra K. J. (2010) Identification of inhibitors of vacuolar proton-translocating ATPase pumps in yeast by high-throughput screening flow cytometry. Anal. Biochem. 398, 203–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vasilyeva E., Forgac M. (1998) Interaction of the clathrin-coated vesicle V-ATPase with ADP and sodium azide. J. Biol. Chem. 273, 23823–23829 [DOI] [PubMed] [Google Scholar]
- 42. Ohya Y., Umemoto N., Tanida I., Ohta A., Iida H., Anraku Y. (1991) Calcium-sensitive cls mutants of Saccharomyces cerevisiae showing a Pet− phenotype are ascribable to defects of vacuolar membrane H(+)-ATPase activity. J. Biol. Chem. 266, 13971–13977 [PubMed] [Google Scholar]
- 43. Nelson H., Nelson N. (1990) Disruption of genes encoding subunits of yeast vacuolar H(+)-ATPase causes conditional lethality. Proc. Natl. Acad. Sci. U.S.A. 87, 3503–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Shao E., Nishi T., Kawasaki-Nishi S., Forgac M. (2003) Mutational analysis of the non-homologous region of subunit A of the yeast V-ATPase. J. Biol. Chem. 278, 12985–12991 [DOI] [PubMed] [Google Scholar]
- 45. Zhang Z., Zheng Y., Mazon H., Milgrom E., Kitagawa N., Kish-Trier E., Heck A. J., Kane P. M., Wilkens S. (2008) Structure of the yeast vacuolar ATPase. J. Biol. Chem. 283, 35983–35995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Landolt-Marticorena C., Williams K. M., Correa J., Chen W., Manolson M. F. (2000) Evidence that the NH2 terminus of vph1p, an integral subunit of the V0 sector of the yeast V-ATPase, interacts directly with the Vma1p and Vma13p subunits of the V1 sector. J. Biol. Chem. 275, 15449–15457 [DOI] [PubMed] [Google Scholar]
- 47. Inoue T., Forgac M. (2005) Cysteine-mediated cross-linking indicates that subunit C of the V-ATPase is in close proximity to subunits E and G of the V1 domain and subunit a of the V0 domain. J. Biol. Chem. 280, 27896–27903 [DOI] [PubMed] [Google Scholar]
- 48. Kawasaki-Nishi S., Nishi T., Forgac M. (2003) Interacting helical surfaces of the transmembrane segments of subunits a and c′ of the yeast V-ATPase defined by disulfide-mediated cross-linking. J. Biol. Chem. 278, 41908–41913 [DOI] [PubMed] [Google Scholar]
- 49. Zorko M., Jerala R. (2008) Alexidine and chlorhexidine bind to lipopolysaccharide and lipoteichoic acid and prevent cell activation by antibiotics. J. Antimicrob. Chemother. 62, 730–737 [DOI] [PubMed] [Google Scholar]
- 50. Roberts W. R., Addy M. (1981) Comparison of the bisbiguanide antiseptics alexidine and chlorhexidine. I. Effect on plaque accumulation and salivary bacteria. J. Clin. Periodontol. 8, 213–219 [DOI] [PubMed] [Google Scholar]
- 51. Chawner J. A., Gilbert P. (1989) Interaction of the bisbiguanides chlorhexidine and alexidine with phospholipid vesicles: evidence for separate modes of action. J. Appl. Bacteriol. 66, 253–258 [DOI] [PubMed] [Google Scholar]
- 52. Poltermann S., Nguyen M., Günther J., Wendland J., Härtl A., Künkel W., Zipfel P. F., Eck R. (2005) The putative vacuolar ATPase subunit Vma7p of Candida albicans is involved in vacuole acidification, hyphal development, and virulence. Microbiology 151, 1645–1655 [DOI] [PubMed] [Google Scholar]
- 53. Hilty J., Smulian A. G., Newman S. L. (2008) The Histoplasma capsulatum vacuolar ATPase is required for iron homeostasis, intracellular replication in macrophages, and virulence in a murine model of histoplasmosis. Mol. Microbiol. 70, 127–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Erickson T., Liu L., Gueyikian A., Zhu X., Gibbons J., Williamson P. R. (2001) Multiple virulence factors of Cryptococcus neoformans are dependent on VPH1. Mol. Microbiol. 42, 1121–1131 [DOI] [PubMed] [Google Scholar]
- 55. Ganendren R., Widmer F., Singhal V., Wilson C., Sorrell T., Wright L. (2004) In vitro antifungal activities of inhibitors of phospholipases from the fungal pathogen Cryptococcus neoformans. Antimicrob. Agents Chemother. 48, 1561–1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Soares D. A., de Andrade R. V., Silva S. S., Bocca A. L., Soares Felipe S. M., Petrofeza S. (2010) Extracellular Paracoccidioides brasiliensis phospholipase B involvement in alveolar macrophage interaction. BMC Microbiol. 10, 241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Zhang Y. Q., Gamarra S., Garcia-Effron G., Park S., Perlin D. S., Rao R. (2010) Requirement for ergosterol in V-ATPase function underlies antifungal activity of azole drugs. PLoS Pathog. 6, e1000939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ostrosky-Zeichner L., Casadevall A., Galgiani J. N., Odds F. C., Rex J. H. (2010) An insight into the antifungal pipeline: selected new molecules and beyond. Nat. Rev. Drug Discov. 9, 719–727 [DOI] [PubMed] [Google Scholar]
- 59. Lötscher H. R., deJong C., Capaldi R. A. (1984) Interconversion of high and low adenosinetriphosphatase activity forms of Escherichia coli F1 by the detergent lauryldimethylamine oxide. Biochemistry 23, 4140–4143 [DOI] [PubMed] [Google Scholar]