Background: In the mammalian ovary, 99% of follicles are removed through follicular atresia caused by granulosa cell apoptosis.
Results: BimEL can induce porcine granulosa cell apoptosis, and its expression is regulated by FSH via the PI3K/Akt/FoxO3a pathway.
Conclusion: The pro-apoptotic protein BimEL is involved in porcine granulosa cell apoptosis.
Significance: This provides novel insights into the molecular mechanisms underlying follicular atresia.
Keywords: Apoptosis, Bcl-2 Family Proteins, Cell Death, Foxo, PI 3-Kinase (PI3K), Bim, FSH, Granulosa Cells, Porcine
Abstract
The pro-apoptotic protein Bim (B-cell lymphoma-2 (Bcl-2)-interacting modulator of cell death) has recently been identified and shown to promote cell death in response to several stimuli. In this report, we investigated the role of Bim in porcine follicular atresia. Initially, Bim cDNA was cloned and characterized from porcine ovarian tissue. Porcine Bim had three alternative splicing variants (Bim-extra long, Bim-long, and Bim-short), all containing the consensus Bcl-2 homology 3 domain. We then found the Bim-extra long (BimEL) protein, the most abundant isoform of Bim, was strongly expressed and co-localized with apoptotic (TUNEL-positive) granulosa cells from porcine atretic follicles. Furthermore, overexpression of BimEL triggered apoptosis in granulosa cells. In primary granulosa cell cultures under basal conditions, we observed that BimEL expression was dampened by treatment with follicle-stimulating hormone (FSH). The role of the PI3K/Akt pathway in the regulation of repression was clarified by the use of the PI3K inhibitor, LY294002, and by transfection with Akt siRNA. Forkhead Box Protein O3a (FoxO3a), a well defined transcriptional activator of Bim, was phosphorylated at Ser-253 and inactivated after FSH stimulation. Also, FSH abolished FoxO3a nuclear accumulation in response to LY294002. Finally, chromatin immunoprecipitation assays demonstrated that FoxO3a directly bound and activated the bim promoter. Taken together, we conclude that BimEL induces porcine granulosa cell apoptosis during follicular atresia, and its expression is regulated by FSH via the PI3K/Akt/FoxO3a pathway.
Introduction
During oogenesis, >99% of developing follicles undergoes atresia (1–4). It is widely accepted that the viability of primordial and primary follicles is determined mainly by survival factors derived from the oocyte, whereas the relative expression level of tumor suppressors, apoptotic proteins, and survival factors in granulosa cells determines whether an ovarian follicle will grow or undergo atresia in the late preantral stage of the follicles (5). FSH functions as a critical survival signal for antral follicles by blocking apoptosis in granulosa cells (6); however, molecules acting as signals to suppress granulosa cell apoptosis and promote growth during this particular stage of follicular development remain largely undetermined.
The B cell lymphoma 2 (Bcl-2) family proteins are considered major regulators of apoptosis (7). These proteins share homology in at least one of four Bcl-2 homology (BH)2 domains and constitute three subfamilies, including Bcl-2 anti-apoptotic proteins (e.g. Bcl-2 and Bcl-xl), Bax pro-apoptotic proteins (e.g. Bax and Bak), and BH3-only proteins (e.g. Bim, Bmf, and Puma) (8). According to data generated from different genetic models, the balance between the pro- and anti-apoptotic Bcl-2 members presumptively determines the fate of a follicle (9–12). Therefore, the “Bcl-2/Bax balance” is proposed to be responsible for the regulation of granulosa cell apoptosis and survival (13). However, there is limited understanding of the involvement of BH3-only proteins, such as Bim, in this process.
The protein Bim (also known as Bcl-2-related ovarian death gene, Bod) was originally identified as a Bcl-2-interacting protein by screening a bacteriophage λ cDNA expression library constructed from a mouse thymic lymphoma (14). Bim promotes apoptosis by binding with high affinity to anti-apoptotic Bcl-2 family members, thereby inducing mitochondrial release of cytochrome c, which subsequently activates caspase-9 and caspase-3 and death effector molecules (15). More than 12 additional Bim mRNA splicing variants have been identified in a wide range of human and rodent tissues (16–19), but most tissues express one predominant isoform of Bim as determined by Western blotting analysis, termed BimEL. Studies on the transcriptional regulation of Bim have mainly focused on FoxO3a, a member of the Forkhead family of transcriptional regulators that directly stimulates Bim transcription (20). Bim proteins were also identified in granulosa cells of primordial, primary, secondary, and mature murine follicles (21); however, their cellular functions and relative contribution to granulosa cell apoptosis remain largely unknown.
It prompted us to investigate the putative role of Bim in porcine granulosa cell apoptosis. Porcine Bim homologues were cloned as the first step toward elucidating differences in BimEL expression between atretic and healthy follicles. An overexpression model was created to determine the pro-apoptotic effect of BimEL in transfected granulosa cells. We then demonstrated that BimEL was down-regulated after FSH treatment in primary cultured granulosa cells. Finally, the mechanism of FSH-regulated BimEL expression was determined.
EXPERIMENTAL PROCEDURES
Unless otherwise specified, all chemicals used in this study were purchased from Sigma.
Classification of Follicles and Recovery of Granulosa Cells
Porcine ovaries were collected at a local abattoir and transported to the laboratory in a vacuum flask (30–35 °C) containing sterile physiological saline within 2–3 h of collection. In the laboratory, ovaries were washed twice with sterile physiological saline (37 °C) containing 100 IU/liter penicillin and 50 mg/liter streptomycin. Follicles were classified as healthy or atretic, according to previously established morphological criteria (22). Briefly, healthy follicles had vascularized theca interna and clear amber follicular fluid with no debris. Follicles lacking any of these criteria were classified as atretic. The slightly atretic and atretic follicles had gray theca interna and flocculent follicular fluid in varying degrees.
Granulosa cells from follicles of different health status were isolated by puncturing follicles with a 25-gauge hypodermic needle and gently expelling the cells into a culture dish containing M199 medium. The cumulus-oocyte complex and ovarian tissue were discarded under a stereo microscope. Granulosa cells were then harvested by centrifuging the media (500 × g for 10 min). Pooled cells were washed three times in PBS for immunoblotting.
Granulosa Cell Culture Conditions and Treatment
For experiments investigating FSH-mediated effects, primary cultures of granulosa cells from healthy follicles (2–5 mm in diameter) were selected, largely due to the fact that the follicles in this phase are highly responsive to FSH in pig (23). Briefly, viable granulosa cells (assessed by trypan blue exclusion) were cultured in 6-well plates (5 × 106/well) with 2 ml of DMEM/Ham's F-12 (Invitrogen) and supplemented with 100 units/ml penicillin and 50 mg/ml streptomycin without (control) or with 0.01, 0.1, and 1 IU/ml porcine FSH (Sioux Biochemical, Inc., Sioux Center, IA). Cells were incubated in a humidified 5% CO2 atmosphere incubator at 37 °C for the indicated time periods. When examining FoxO3a phosphorylation, LY294002 (20 μm) was added 30 min before FSH treatment. Granulosa cells from each treatment were collected from the wells and snap-frozen for subsequent analysis by immunoblotting or caspase-3 activity assay.
For transient transfection, cultured granulosa cells were cultured in 35-mm dishes with 2 ml of DMEM/Ham's F-12 (Invitrogen), supplemented with 100 units/ml penicillin, 50 mg/ml streptomycin, and 10% FBS (Invitrogen). When they reached 70–80% confluence, transient transfection was performed.
Caspase-3 Activity Assay
The caspase-3 activity was used to estimate apoptotic response and determined using caspase-3 activity assay kits (Applygen, Beijing, China). After treatment, granulosa cells were pelleted by centrifugation (1000 × g, 10 min). The cell pellet was lysed in 80 μl of lysis buffer for 20 min on ice. After the lysate was centrifuged at 13,000 × g for 15 min, the supernatant was collected, and the protein concentration was determined with BCA protein assay kit (CW BioTech, Beijing, China). Aliquots (20 μg of protein) of the supernatant in triplicate were transferred to the 96-well plate and incubated in the dark with 100 μl of buffer containing caspase substrates at 37 °C for 4 h. Absorbance at 405 nm was measured using a microplate reader (Bio-Rad).
RNA Isolation
Total cellular RNA (tcRNA) from various tissues (porcine lung, liver, and ovary and murine liver and ovary) was extracted using an RNAprep pure tissue kit (TIANGEN Biotech, Beijing, China) according to the manufacturer's instructions. Total RNA content was determined by spectrophotometry (260 nm).
Rapid Amplification of cDNA 3′ Ends
The tcRNA (500 ng) from porcine ovarian tissue was used to convert mRNAs into cDNAs, using a 3′-full rapid amplification of cDNA 3′ end core set version 2.0 kit (Takara Biotech, Dalian, China), which included avian myeloblastosis virus reverse transcriptase and a universal oligo(dT) containing-adapter primer. The PCR was conducted in accordance with manufacturer's instructions. The Takara LA Taq polymerase was used in rapid amplification of cDNA 3′ end-PCR. An aliquot of the first PCR (outer PCR) was used for a subsequent nested PCR (inner PCR). Amplified cDNA was gel-purified and subcloned into the pBST-II vector (TIANGEN Biotech, Beijing, China) and sequenced (SUN Biotech, Beijing, China).
Reverse Transcription-PCR (RT-PCR)
In each sample, tcRNA (500 ng) was used for cDNA synthesis. In the first step, secondary structures were removed, and an oligo(dT)18 primer was annealed to the mRNA by heating the sample at 70 °C for 5 min. The reaction was then quenched rapidly on ice, and 5× RT buffer, 10 mm dNTPs, 2 mm dithiothreitol, 20 units of RNase inhibitor (TaKaRa Biotech, Dalian, China), and 200 units of Moloney murine leukemia virus reverse transcriptase (TIANGEN Biotech, Beijing, China) were added (total reaction mixture volume, 25 μl). This mixture was heated at 42 °C for 60 min and then at 70 °C for 15 min to terminate the reaction. Subsequent PCR analysis was carried out with 1 μl of the resulting cDNA, and products were separated by electrophoresis on a 1.5% agarose gel. For analysis of Bim isoform expression profiles, bands were purified and sequenced.
Primers (SUN Biotech, Beijing, China) used were as follows: mouse Bim (accession number NM207680), forward 5′-ACAGAGAAGGTGGACAATTGC-3′ and reverse 5′-AAACACCCTCCTTGTGTAAGTT-3′; porcine Bim (accession number FJ825619), forward 5′-AGCAACCTTCTGATGTAAGTTCT-3′ and reverse 5′-CAGCATTACCCTCCTTGGGTAAT-3′.
Multiple Sequence Alignments
Multiple sequence alignments were done with the ClustalW algorithm in the BCM Search Launcher.
Construction of Fluorescent Protein Expression Vectors and Transfection
BimEL and FoxO3a construct was generated by subcloning PCR-amplified full-length porcine BimEL and FoxO3a cDNA into pEGFP-N1 (Clontech). The reading frame of BimEL and FoxO3a cDNA was connected with enhanced GFP to obtain a recombinant pEGFP-N1-BimEL and pEGFP-N1-FoxO3a plasmid. The plasmid was propagated using DH5α (TIANGEN Biotech, Beijing, China) as the host strain, and purified with an EndoFree Plasmid maxi kit (Qiagen Inc., Hilden, Germany), and sequenced to confirm the open reading frame.
Transient transfection of porcine granulosa cells with plasmids was performed using LipofectamineTM 2000 (Invitrogen), according to the manufacturer's instructions. Expression of the fusion construct was evaluated by fluorescence microscopy after transfection. Meanwhile, at 2 and 8 h after transfection, cells were stained Hoechst 33342 to evaluate nuclear morphology. At the end of the experiments, floating cells were collected from the medium by centrifugation and combined to attached cells harvested by trypsin/EDTA treatment. Cells were then washed in PBS and used for Western immunoblot.
Small Interfering RNAs and Transfection
Control siRNA (siCTR), 5′-UUCUCCGAACGUGUCACGUTT-3′, and two validated siRNA against porcine Akt (siAkt-244, 5′-GGCCCAACACCUUCAUCAUTT-3′; siAkt-1177, 5′-UCUCAGGGCUGCUCAAGAATT-3′) were designed and synthesized by GenePharma (GenePharma, Shanghai, China). Granulosa cells were seeded in a 24-well plate and grown to 30–50% confluence. Cells were then incubated in antibiotic-free medium overnight, followed by transfection with 10 pmol of siRNA duplex using Lipofectamine RNAiMAX according to the manufacturer's instructions (Invitrogen). Total protein was prepared 48 after transfection and was used for Western blotting analysis.
Western Immunoblot
Cells were lysed in Laemmli sample buffer (Bio-Rad). Equal amounts of proteins (15 μg/lane) were separated by SDS-PAGE (7.5% acrylamide running gel) and transferred to a nitrocellulose membrane (BioTraceNT, Pall Corp.). Nonspecific binding to the membrane was blocked with 5% nonfat milk in TBS-T (10 mm Tris (pH 7.5), 150 mm NaCl, and 0.1% Tween 20) for 1 h at room temperature. Membranes were then incubated with primary antibodies (4 °C, overnight), washed in Tris-buffered saline containing 0.1% Tween, and incubated with horseradish peroxidase (HRP)-conjugated secondary antibodies for 1 h at room temperature. Antibodies against Bim (C34C5), FoxO3a (75D8), and phospho-FoxO3a were all purchased from Cell Signaling Technology (Beverly, MA). Anti-GFP, anti-actin, and anti-histone 3.1 antibodies were obtained from Abmart Biotechnology (Shanghai, China). HRP-conjugated anti-rabbit antibody was obtained from Zhongshan Biotechnology (Beijing, China). The protein bands were visualized by enhanced chemiluminescence detection reagents (Applygen Technologies Inc., Beijing, China) and X-Omat BT film (Eastman Kodak Co.), according to the manufacturer's instructions. The films were digitized, and densitometry analysis was performed with ImageJ 1.44p software (National Institutes of Health). The relative intensity of the bands was quantified and normalized to the respective loading control. Nuclear and cytoplasmic protein extracts were prepared using the ProteoJETTM cytoplasmic and nuclear protein extraction kit K0311 (Fermentas, Canada).
Histology
Porcine ovaries were fixed overnight in 4% phosphate-buffered formaldehyde at 4 °C for 7 days and then embedded in paraffin. Randomly selected sections (5 μm each) were mounted on poly-l-lysine-coated slides (Zhongshan Biotechnology, Beijing, China) for subsequent immunohistochemical and TUNEL analyses. Prior to Bim localization, sections were deparaffinized by washing with xylene and then rehydrated in decreasing concentrations of ethanol (100 to 50% ethanol) and washed in PBS. To block endogenous peroxidase activity, sections were incubated for 10 min in 0.5% hydrogen peroxide. After washing with PBS, sections were permeabilized with 0.1% Triton X-100 in PBS for 10 min. Nonspecific binding was blocked by incubation with 10% inactivated goat serum. A monoclonal rabbit Bim (C34C5) antibody (1:200; Cell Signaling Technology, Beverly, MA) was used to detect BimEL protein expression. To eliminate nonspecific binding, nonimmune goat serum was added to the primary antibody. Sections were incubated overnight at 4 °C. The next day, after washing with PBS, the HRP-conjugated anti-rabbit antibody (1:100, Zhongshan Biotechnology, Beijing, China) was applied for 1 h at room temperature. The binding of primary antibody was visualized using diaminobenzidine for 3–5 min. Sections were subsequently washed with twice-distilled water. Finally, sections were rinsed with water, dehydrated in ethanol and toluene, and mounted in Neutral Balsam. Bim labeling was examined using a Leica microscope, and images were recorded (Leica DC 200 digital camera; Leica, Wetzlar, Germany). As a negative control, normal rabbit serum was applied to some sections in lieu of specific antiserum.
Apoptosis was analyzed by TUNEL (in situ cell death detection kit, Roche Applied Science) according to the manufacturer's protocol. Briefly, after deparaffinization and rehydration, sections were incubated with proteinase-K (20 μg/ml) for 15 min at room temperature, quenched in 3% H2O2 in PBS for 10 min (to block endogenous peroxidase), incubated in a humidified chamber with equilibration buffer for 5 min, and finally incubated with terminal deoxynucleotidyltransferase for 1 h at 37 °C. Negative control slides were incubated as described above, without the final addition of terminal deoxynucleotidyltransferase. After TUNEL reactions were complete, slides were washed in PBS, sealed under coverslips with nail varnish, and examined under Leica fluorescence microscopy.
Chromatin Immunoprecipitation Assays (ChIP)
Porcine granulosa cells were cultured in 15-cm dishes and transfected using GFP-FoxO3a plasmid as indicated above. To transfer fusion protein into the nucleus, 4-h post-transfection cells were incubated in the presence of LY294002 (20 μm) for 2 h. Then the transiently transfected cells were processed for ChIP assay according to manufacturer's directions in the chromatin immunoprecipitation assay kit (Upstate). Briefly, cells were cross-linked in 1% formaldehyde solution for 10 min at room temperature. Cells were lysed in 500 μl of SDS buffer and sonicated to generate 200–1000 bp of DNA fragments. The sheared chromatin concentration was measured; about 10% of the chromatin was kept as an input. The rest of the chromatin was incubated with a GFP antibody (Abmart Biotechnology, Shanghai, China) for immunoprecipitation. Upon DNA isolation, amplification of bim promoter DNA was assessed by PCR using the following primers: forward (−218/−199), 5′-GATGGGTGCGCACTGAGCCA-3′; reverse (−106/−87), 5′-GCGTGCAGGCTCTGACAGGTA-3′. Analysis of the PCR products was performed on a standard 2% (w/v) agarose gel, by electrophoresis in Tris acetate/EDTA buffer.
Statistical Analyses
Data of the amounts of BimEL, FoxO3a proteins, and caspase-3 activity were analyzed using a general linear model and one-way post hoc test using the MIXED procedure models of SAS (version 9.1, SAS Institute Inc., Cary, NC). Differences were considered statistically significant when p < 0.05.
RESULTS
Complementary DNA Sequence and Structure of Procine Bim
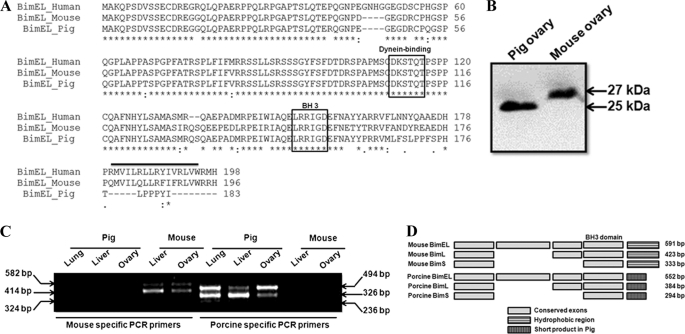
Based on a partial cDNA of porcine BimEL (GenBankTM accession number. AJ606302), a 552-bp open reading frame (ORF) of BimEL cDNA (GenBankTM accession number FJ825619) was obtained from porcine ovarian tissue. The porcine BimEL putative mature protein contains 183 amino acid residues and has amino acid sequence homology of 87.3 and 84.2% with its human and mouse counterparts, respectively (GenBankTM accession numbers AAC39593 and AAC40029) (Fig. 1A). BLAST results indicated that murine and porcine BimEL have similar characteristics. The porcine BimEL has five exons (124, 168, 90, 110, and 60 bp), contains a conserved BH3 domain (LEDIGD; Fig. 1A, boxed), lacks the BH1, BH2, and BH4 domains present in channel-forming BCL-2 family proteins, and possesses a dynein-binding domain (DKSTQT; Fig. 1A, boxed). Unlike the murine BimEL, porcine BimEL did not have a COOH-terminal hydrophobic region (Fig. 1A, overlined). One antibody against both murine and porcine BimEL protein was used for Western blot analysis, and it recognized a 25-kDa protein from porcine ovary extracts and a 27-kDa protein from murine ovary tissue (Fig. 1B). The difference in size may be explained by the fact that porcine BimEL lacked the hydrophobic region.
FIGURE 1.
Complementary DNA sequence and structure of porcine Bim. A, deduced amino acid sequence for porcine BimEL aligned to its human and murine counterparts. Identical amino acids are indicated by asterisks, and amino acids with conserved similarities are indicated by dots or colons. The BH3 and dynein-binding domains are boxed. The COOH-terminal hydrophobic region is overlined. The amino acid numbers are indicated on the right. B, antibody that recognizes both murine and porcine BimEL proteins was used to conduct Western blotting on samples from murine and porcine ovary tissue. C, identification of multiple RT-PCR products of porcine Bim. The coding region of Bim cDNA was amplified by RT-PCR using total RNA from porcine lung, liver, and ovary and murine liver and ovary, using animal-specific primers. The PCR products were subjected to agarose gel electrophoresis and photographed. The segment size is marked on both sides. D, genomic organization of porcine Bim isoforms.
The expression profiles of Bim mRNA in porcine lung, liver, and ovary were determined using RT-PCR with a primer pair that spanned the entire Bim coding sequence. Positive controls were tcRNA isolated from murine liver and ovary. As shown in Fig. 1C, three distinct Bim mRNA products were detected in the porcine lung, liver, and ovary. Nucleotide sequence analysis indicated that the two smaller products were splicing variants from porcine BimEL (Fig. 1D). These isoforms had a similar splicing mode with that of the mouse BimEL and were subsequently named porcine BimL and porcine BimS (GenBankTM accession numbers FJ825620 and FJ825621), respectively.
BimEL Expression Correlates with Atretic Follicles and Apoptotic Granulosa Cells
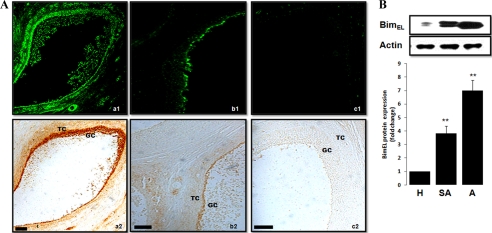
To identify the hypothesis that Bim protein was present and coincident with follicular atresia in vivo, we applied double labeling with TUNEL staining and immunohistochemical staining using antibody against BimEL (our following Western blot experiment showed that the Bim protein recognized by the antibody in our immunohistochemistry assay was BimEL). Atretic follicles were evident by the presence of TUNEL-positive granulosa cells, uneven granulosa cell layers, and exfoliating granulosa cells that could occasionally be detected within the antral cavity (Fig. 2A, panel a1). An adjacent section of the same follicle clearly showed that BimEL immunostaining was concentrated in both granulosa and theca cells, especially in the inner surface of the granulosa layer (Fig. 2A, panel a2). In addition, moderate TUNEL staining and BimEL immunostaining were shown in slightly atretic follicles (Fig. 2A, panels b1 and b2). However, healthy follicles that had a well organized and intact granulosa cell layer showed negative BimEL immunostaining and TUNEL staining (Fig. 2A, panels c1 and c2).
FIGURE 2.
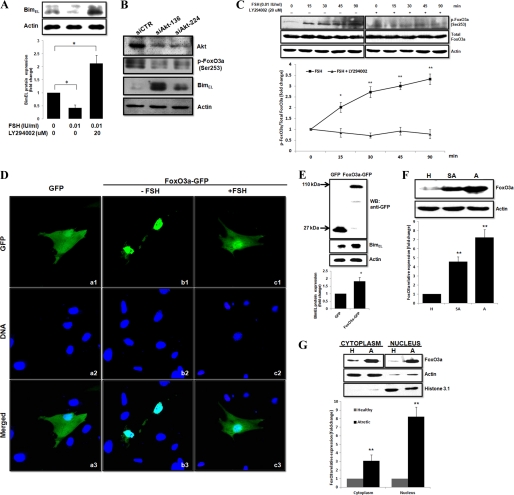
BimEL expression pattern in granulosa cells from healthy and atretic porcine follicles. A, photomicrographs of representative examples of TUNEL and BimEL immunostaining in the porcine atretic (panel a), slightly atretic (panel b), and healthy (panel c) follicles. Cell death was detected by TUNEL (panels a1, b1, and c1), and on a consecutive section, BimEL proteins (panels a2, b2, and c2) were localized with specific antibodies by the diaminobenzidine (DAB) method. GC, granulosa cells; TC, theca cells. Bar, 50 μm. B, isolated granulosa cell lysates, obtained from follicle of different health status (H, healthy follicle; SA, slightly atretic follicle; A, atretic follicle), were subjected to SDS-PAGE and immunoblotting using anti-Bim antibodies. Actin was used as fractionation control. A representative blot is shown. The graph demonstrates the results of the densitometric analysis of BimEL protein levels normalized against loading controls (arbitrary units, healthy follicle = 1). Values represent means ± S.E. of three experiments. **, p < 0.01.
To further verify the results from immunohistochemistry, the relative abundance of the BimEL protein in isolated granulosa cells was quantified by Western blot. Granulosa cells were isolated from freshly procured porcine follicles and classified as healthy, slightly atretic, or atretic follicles according to morphological criteria (see “Experimental Procedures”). As shown in Fig. 2B, granulosa cell BimEL protein expression was indistinguishable in the healthy follicles and increased as atresia progressed. The expression of BimL and BimS, the two other major isoforms of Bim, was not detected in our present Western blot experimental system even with a longer exposure time (data not shown).
Overexpression of Porcine BimEL Induces Granulosa Cells Apoptosis in Vitro
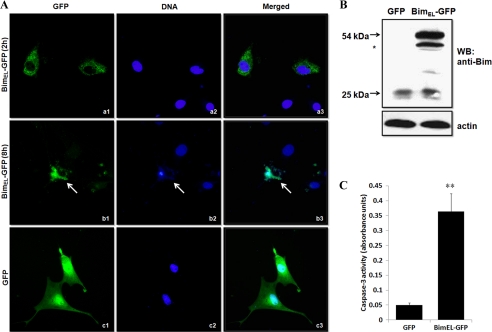
In light of the positive correlation observed between BimEL immunoreactivity and granulosa cell apoptosis, further experiments were done to investigate the proapoptotic potential of BimEL protein in granulosa cells in vitro. Cultured granulosa cells were transiently transfected with an empty EGFP vector or pEGFP-N1-BimEL as a control. Eight hours after transfection, green fluorescence for all constructs was detected in ∼10% of transfected cells (data not shown). At 2 h post-transfection, pEGFP-N1-BimEL-transfected cells exhibited green fluorescence, which was found exclusively in the cytosol (Fig. 3A, panel a). At 8 h post-transfection, fluorescence microscopic examination showed that the majority of granulosa cells overexpressing BimEL exhibited cytomorphological alterations typical for apoptosis, including nuclear fragmentation, cell rounding and shrinkage, plasma membrane blebbing, and apoptotic body formation (Fig. 3A, panel b) (24). In contrast, cells expressing only GFP (control) displayed normal appearing morphology and remained healthy and flat, with the green fluorescence signal located both in the nucleus and cytosol at all times (Fig. 3A, panel c). Western blot analysis of the transfected cells showed that the exogenously expressed BimEL protein levels were much higher than that seen with endogenous BimEL expression (Fig. 3B). Quantitative analysis of apoptotic cells was also assessed by caspase-3 activity assay. Overexpression of EGFP-BimEL caused a 7-fold increase in caspase-3 activity (Fig. 3C), indicating that BimEL can induce porcine granulosa cells apoptosis in vitro.
FIGURE 3.
Overexpression of porcine BimEL induces apoptosis of granulosa cells in vitro. A, cells plated on glass bottom 35-mm dishes were 80% confluent when transfected with pEGFP-N1-BimEL (panels a and b) or pEGFP-N1 (panel c, control) constructs. Note that BimEL-GFP was localized to the cytoplasm at 2 h post-transfection, and apoptotic phenotypes measured using Hoechst 33342 staining could be found in pEGFP-N1-BimEL-transfected cells at 8 h post-transfection (arrow). However, GFP-only expressing cells consistently had intact nuclei, and GFP was distributed throughout the cells. B, Western blot (WB) analysis of protein expression for endogenous and exogenous BimEL in granulosa cells transfected with a full-length BimEL-GFP fusion construct or mock vector. Transfected granulosa cells lysates were subjected to SDS-PAGE and immunoblotting using the anti-Bim antibody. Molecular masses of the protein bands are shown on the left side. Actin was used as fractionation control. * indicates an alternative splicing form of BimEL. C, caspase-3 activity was determined by caspase assay in GFP only or BimEL-GFP transfected granulosa cells (n = 4). Values represent means ± S.E. of three experiments. **, p < 0.01.
BimEL Expression in Granulosa Cell Is Suppressed by FSH in Vitro
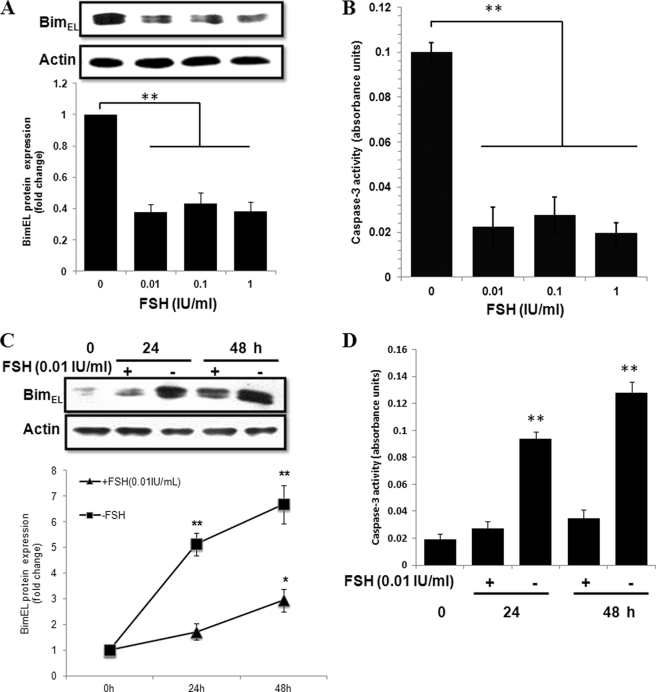
FSH stimulation is the most critical for the survival of antral follicles during follicle selection and development. FSH withdrawal induces granulosa cell apoptosis and follicular atresia (6). To determine whether BimEL expression was associated with FSH, cultured granulosa cells were deprived of FSH and serum for 24 h to induce apoptosis. By immunoblot analysis, high amounts of BimEL proteins were observed within 24 h in cells cultured in FSH and serum-free medium. A decrease in BimEL protein was evident after 24 h of FSH stimulation (Fig. 4A). However, the dose-dependent manner was not observed. The 0.01 IU/ml dose of FSH already yielded maximal protection and thus was the concentration used in subsequent experiments. Next, the time course of repression of BimEL protein by FSH was studied. The primary granulosa cells were cultured in FSH and serum-free medium analyzed as unstimulated (0-h time point) or were analyzed following stimulation with FSH (0.01 IU/ml) or not for 24 or 48 h. In unstimulated cells, the levels of BimEL protein were indistinguishable (Fig. 4C) and consistent with the results from follicles (Fig. 2B). There was a dramatic increase in the amount of BimEL protein at 24 h (5.1-fold) and 48 h (6.7-fold) following serum and FSH-free incubation. When granulosa cells were cultured in medium contained FSH (0.01 IU/ml), the BimEL protein levels increased only 1.7- and 2.9-fold (24 and 48 h, respectively) than in control (Fig. 4C). Thus, FSH could counteract the induction of BimEL after serum deprivation of granulosa cells isolated from FSH-dependent healthy follicles. Also, BimEL protein seemed to migrate with several additional bands, indicating that BimEL could be phosphorylated by FSH-signaling pathways. In accordance with this, as analyzed by caspase-3 activity assay, there was a direct correlation between BimEL expression levels and the extent of FSH-regulated apoptosis (Fig. 4, B and D).
FIGURE 4.
Effect of FSH on apoptosis and BimEL expression in cultured primary granulosa cells. A, granulosa cells were obtained from porcine healthy and 2–5 mm in diameter follicles and cultured for 24 h without or with 0.01, 0.1, and 1 IU/ml porcine FSH. C, granulosa cells cultured without or with 0.01 IU/ml porcine FSH for the indicated times. BimEL protein levels were assessed by immunoblotting. Actin was used as fractionation control. A representative blot is shown. The graph demonstrates the results of the densitometric analysis of BimEL protein levels normalized against loading controls (arbitrary units, without FSH and FSH 0 h = 1, respectively). Values represent means ± S.E. of three experiments. * p < 0.05, **, p < 0.01. B and D, in parallel experiments, granulosa cells from different treatment were harvested to caspase-3 activity assays. **, p < 0.01.
PI3K/Akt/FoxO3a Pathway Is Involved in FSH Regulation of BimEL Expression
The intracellular signaling molecules that mediate FSH-induced granulosa cell survival remain unclear. Accumulating evidence suggests that the PI3K pathway can be activated by FSH signals (25) and subsequently provide a strong survival signal to granulosa cells by partially regulating the activities of the Bcl-2 family of proteins (26). To explore the possibility that FSH activation of PI3K plays a role in repressing Bim expression, in vitro cultured primary granulosa cells were pretreated with LY294002, a specific pharmacological inhibitor for PI3K, followed by FSH stimulation in serum-free medium. The results suggested that the repression effect of FSH on BimEL expression was abolished by PI3K inhibitor at concentrations known to efficiently block PI3K activity (Fig. 5A).
FIGURE 5.
PI3K/Akt/FoxO3a pathway is involved in FSH regulation of BimEL expression. A, granulosa cells preincubated with or without 20 μm LY294002 for 30 min were treated with FSH for 16 h. The LY294002 remained present throughout the FSH stimulation. BimEL expression was assessed by immunoblotting. The graph demonstrates the results of the densitometric analysis of BimEL protein levels normalized against loading controls (arbitrary units, no FSH and LY294002 = 1). B, effect of Akt siRNA on BimEL expression. Granulosa cells were transfected with Akt siRNA (siAkt) or siCTR for 36 h and then were harvested and analyzed by immunoblotting. C, Western blot analysis was completed to determine whether FSH affected FoxO3a phosphorylation status. Granulosa cells were preincubated for 30 min with 20 μm LY294002 or not and then were harvested at specific times after FSH treatment (15–90 min). Phosphorylated (p) Foxo3a and total Foxo3a expression was assessed by immunoblotting. The graph demonstrates the ratio of the densitometric analysis of phosphorylated forms of FoxO3a proteins normalized against its total forms (arbitrary units, 0 h = 1, respectively). D, FSH regulates FoxO3a protein subcellular localization. Granulosa cells plated on glass bottom 35-mm dishes were 80% confluent when transfected with pEGFP-N1 (panel a, control) or pEGFP-N1-FoxO3a (panels b and c) constructs incubated with or without FSH. At 6 h post-transfection, cells were then stained with Hoechst 33342 to visualize nuclei, and the subcellular localization of FoxO3a was monitored by GFP fluorescence. E, Western blot (WB) analysis of fusion protein expression in granulosa cells transfected with a full-length BimEL-GFP construct or mock vector. Transfected granulosa cells lysates were subjected to SDS-PAGE and immunoblotting using the anti-GFP antibody. Molecular masses of the protein bands are shown on the left side. The graph demonstrates the results of the densitometric analysis of BimEL protein levels normalized against loading controls (arbitrary units, GFP = 1). F, FoxO3a is up-regulated in granulosa cells from atretic follicles. Isolated granulosa cell lysates, obtained from follicle of different health status (H, healthy follicle; SA, slightly atretic follicle; A, atretic follicle), were subjected to SDS-PAGE and immunoblotting using anti-FoxO3a antibodies. The graph demonstrates the results of the densitometric analysis of FoxO3a protein levels normalized against loading controls (arbitrary units, H = 1). G, immunoblots showing the expression of FoxO3a in cytosolic and nuclear extracts of granulosa cells from the healthy and atretic follicles. Actin and histone 3.1 were used as fractionation controls, respectively. The graph demonstrates the results of the densitometric analysis of FoxO3a protein levels normalized against loading controls (arbitrary units, H = 1). All values represent means ± S.E. of three experiments. *, p < 0.05; **, p < 0.01.
To further confirm the involvement of PI3K pathway in regulating BimEL expression, siRNA targeting Akt, a downstream substrate of PI3K, was used to transfect granulosa cells. By using two validated Akt siRNA target sequences, we were able to knock down Akt expression in granulosa cells by 75%. Nonsilencing siCTR had no effect on Akt expression. Akt knockdown using siRNA also significantly increased BimEL expression (Fig. 5B). These data clearly show that Akt protein is required for BimEL expression in granulosa cells.
It was recently reported that the Forkhead transcription factor, FoxO3a, the major downstream effector of PI3K/Akt signaling pathway, can bind to the bim promoter region and control Bim expression at the transcriptional level (27, 28). FoxO3a activity and nucleus translocalization is inhibited by PI3K-mediated phosphorylation at three sites (Thr-32, Ser-253, and Ser-315) (29). We hypothesized that in porcine granulosa cells, FSH activates PI3K/Akt, which in turn triggers phosphorylation of FoxO3a and ultimately prevents FoxO3a from inducing Bim expression. Time course experiments demonstrated that FoxO3a (Ser-253) phosphorylation increased dramatically beginning at 15 min after FSH stimulation. The ratio of p-FoxO3a/total FoxO3a (as measured by densitometry) increased more than 2-fold at 15 and 30 min and 3-fold at 45 and 90 min compared with 0 min. However, this induction was consistently blocked by concomitant LY294002 (20 μm) treatment (Fig. 5C).
Having demonstrated FSH stimulation of FoxO3a phosphorylation, we next sought to determine whether FSH regulated the intracellular localization of FoxO3a protein by transfecting cultured granulosa cells with plasmids expressing GFP or FoxO3a-GFP fusion proteins followed by LY294002 (20 μm) treatment. Fluorescence microscope observations showed cells have diffuse fluorescence in both the nucleus and cytoplasm when granulosa cells were transfected with the control GFP-only vector as expected (Fig. 5D, panel a). When granulosa cells were transfected with a FoxO3a-GFP plasmid but left FSH-untreated, the fusion protein was largely in the nucleus (Fig. 5D, panel b). In contrast, treatment of these cells with FSH (0.01 IU/ml) led to relocalization of fusion protein to the cytoplasm, although a portion was still present in the nucleus (Fig. 5D, panel c). Western blot analysis showed that BimEL protein in the FoxO3a-GFP plasmid-transfected cells was about 1.83-fold increased as compared with control cells (Fig. 5E). This result indicated that LY294002 activated FoxO3a protein and sequestered it in the nucleus and thereby induced BimEL expression, which can be reversed by FSH treatment.
To provide in vivo evidence that the FoxO3a protein was involved in follicular atresia, the relative abundance of the FoxO3a protein in isolated granulosa cells from different healthy follicles was quantified. We showed that the amount of the FoxO3a protein was markedly increased in granulosa cells as atresia progressed (4.58- and 7.26-fold higher in lightly atretic and atretic follicles than in healthy follicles, respectively) (Fig. 5F). In addition, the relative cytoplasmic and nuclear distributions of FoxO3a protein were also evaluated in granulosa cells from healthy and atretic follicles. As shown in Fig. 5G, FoxO3a protein in the cytoplasmic fraction from atretic granulosa cells was about 3.08-fold increased as compared with that from healthy granulosa cells, and this increase reached about 8.22-fold in nucleus fraction. The results support our hypothesis that up-regulation and nuclear translocation of the FoxO3a protein is a necessary prerequisite for the activation of pro-apoptotic genes, such as bim, in response to follicular atresia.
FoxO3a Binds to bim Promoter to Activate Transcription
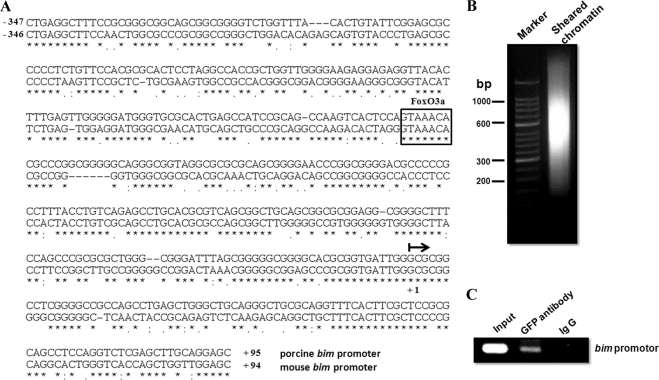
To determine whether FoxO3a activates BimEL expression at the promoter level and to identify the transcriptional element involved, we set out to clone the porcine bim promoter. Essafi et al. (28) showed that the activity of the mouse bim promoter is completely retained in the 500-bp region upstream of the transcriptional start site. Based on the information, we cloned by PCR the porcine bim promoter (442 bp). Sequence analysis of the porcine and mouse promoter regions showed 68% identity and contained a common putative FHRE (Forkhead-responsive element) proximal to the transcription start site (Fig. 6A), which can be involved in the induction of Bim expression.
FIGURE 6.
FoxO3a binds to bim promoter to activate transcription. A, nucleotide sequence of the 442-bp fragment of the porcine bim promoter aligned to the corresponding mouse sequence. Identical bases are indicated by asterisks, and amino acids with conserved similarities are indicated by dots or colons. The arrow indicates the first nucleotide of the transcription start site. The putative Forkhead-responsive element is boxed. B, granulosa cells in Lysis Buffer at a cell concentration of 1 × 107 per ml sheared with six sets of 10-s pulses (40 watts) on wet ice using a high intensity ultrasonic sonicator (Cole Parmer). Sheared chromatin was then electrophoresed through a 2% agarose gel and stained with ethidium bromide. The picture shows that the majority of the DNA has been sheared to a length between 200 and 1000 bp. C, chromatin fragments were subjected to immunoprecipitation with antibodies to IgG (nonspecific) or GFP as indicated. Upon DNA isolation, amplification of the bim promoter DNA was assessed by PCR and gel electrophoresis. Input DNA was used as positive control. A representative graph of three independent experiments is shown.
We next performed ChIP assays to determine FoxO3a occupancy of the bim promoter. The chromatins in GFP-FoxO3a plasmid-transfected granulosa cells were sheared into 200–1000-bp fragments (Fig. 6B). As demonstrated in Fig. 6C, the anti-GFP antibody, but not the control antibody (IgG), precipitated the region of the bim promoter containing the FoxO site in GFP-FoxO3a plasmid-transfected granulosa cells. These results strongly suggest that FoxO3a is recruited directly to the FHRE of the bim promoter to activate its transcription. Taken together, these results suggest that the PI3K/Akt/FoxO3a pathway is involved in FSH regulation of BimEL expression.
DISCUSSION
Follicular atresia is a complicated process that requires intrinsic signaling molecules as well as extrinsic regulators such as FSH (31). Some of these factors control follicular atresia by regulating the fate of granulosa cells, specifically granulosa cell apoptosis (5). The Bcl-2 subfamily of proteins represents one of the intrinsic factors that play essential roles in promoting granulosa cell survival, whereas Bax subfamily protein serves as pro-apoptotic signals to induce granulosa cell death (32). It is suggested that the Bcl-2 and Bax protein subfamilies control the balance of “life and death” in granulosa cells. In this study, we examined whether Bim, a BH3-only protein member, had a dynamic expression pattern in atretic follicles and ultimately mediated FSH-regulated granulosa cells apoptosis. To our knowledge, this is the first evidence demonstrating the role of Bim in female reproduction and more specifically in follicular atresia.
It is important to note that, based on the characterization done in this study, porcine Bim does not contain an identifiable COOH-terminal hydrophobic tail. This tail is present in numerous members of the Bcl-2 family (e.g. Bak, Bcl-2, and Bcl-xl) and is used to localize the protein in the membranes of mitochondria, endoplasmic reticulum, or the nuclear envelope (33). In contrast, several pro-apoptotic Bcl-2 family proteins, e.g. Bax, Bad, and Bid, are inactive cytosolic proteins under normal conditions and translocated to the mitochondria via this tail during apoptosis (34). It was recently reported that Bimα1, -α2 (19), and -γ (17), which also lack the COOH-terminal hydrophobic region, were located in the mitochondria, whereas Bimβ1-β4 isoforms (19), which lack both the BH3 domain and the hydrophobic regions, were located only in the cytosol. Perhaps the BH3 domain itself is sufficient for mediating membrane localization, and the COOH-terminal hydrophobic domain is not essential for their apoptotic capability. This model is supported by our overexpression data.
Similar to other members of the Bcl-2 family (35, 36), Bim isoforms show tissue-specific expression patterns (Fig. 1C). There were distinct temporal and spatial roles for specific Bim isoforms during apoptosis (37). However, only the BimEL protein was detected in apoptotic granulosa cells by Western blot in our experiment. Based on our observation that porcine BimEL expression levels in granulosa cells varied with the health status of follicles, we speculated that BimEL might serve to initiate atresia of porcine follicles. Although BimS was considered the most potent compound for promoting apoptosis in human lymphoma cells (38), and its mRNA was detected by RT-PCR in ovary tissue (Fig. 1C), the expression of its protein in porcine granulosa cells was not detected in this study. Therefore, mechanisms governing the tissue specificity of Bim isoform expression require further study.
As a pro-apoptotic protein, Bim was highly expressed in atretic follicles. BimEL immunostaining was barely detectable in stroma cells and concentrated in both granulosa and theca cells, especially in the inner surface of the granulosa layer. Apoptosis in atretic follicles in most mammals was confined to granulosa cells, so we focused on the Bim function in granulosa cells in this study. However, the Bim expression patterns and function in theca cells are also worth further investigation. Our results indicated that there was a clear correlation between follicular atresia and increased BimEL protein levels. To our knowledge, this work is the first to show the Bim protein function in the follicular atresia in any species.
It is well established that FSH is a critical survival factor for granulosa cells during follicular development (39). Sufficient exposure of antral follicles to FSH is essential for the follicles to escape from atresia and to reach preovulatory stage (4). To explore whether BimEL expression is regulated by FSH, an in vitro granulosa cell culture system was utilized. Our results demonstrated that FSH significantly suppressed granulosa cell apoptosis. This result was consistent with other reports (41). Interestingly, granulosa cell apoptosis attenuated by FSH was accompanied by a decrease in BimEL accumulation. This is also the first indication that BimEL accumulation is induced after FSH withdrawal and contributes to FSH withdrawal-induced apoptosis in granulosa cells.
It was recently reported that Forkhead transcription factor FoxO3a has emerged as a major regulator of ovarian function. For instance, gene silencing of FoxO3a in mice causes accelerated follicular initiation, resulting in premature follicular depletion and ovarian failure (42). Matsuda et al. (43) also reported that FoxO3a was expressed in porcine follicles and induced granulosa cell apoptosis. However, the question regarding which protein regulated by FoxO3a could contribute to the above process is unanswered. In this study, we showed that Bim was one candidate that can be up-regulated by FoxO3a overexpression and that is modestly induced in granulosa cells by atresia. However, other FoxO3a-regulated proteins could also be involved in this apoptosis. One method to detect whether other FoxO3a-regulated proteins might participate in this apoptosis would be to assess whether FoxO3a overexpression can induce apoptosis in bim-deficient granulosa cells.
FoxO3a was also expressed and hormonally regulated in the mouse ovary. Both of its transcription and phosphorylation states were determined by FSH in cultured granulosa cells (44). In this study, we found that FSH could rapidly phosphorylate FoxO3a at Ser-253 and ultimately inhibit BimEL expression. In granulosa cells, both phosphorylation of FoxO3a and repression of BimEL expression were diminished after treatment with the PI3K inhibitor, LY294002, suggesting that the PI3K signaling pathway was involved in the regulation of the Bim protein. Indeed, previous studies had shown that LY294002 increases Bim protein levels in Ba/F3 cells (45) or sympathetic neurons (46). It is now clear that the inhibition of PI3K activity by LY294002 allows FoxO3a to translocate into the nucleus, where they are known to be direct activators of bim gene expression (46). Our results indicated that FoxO3a might also regulate Bim expression in granulosa cells using FoxO3a overexpression and ChIP model. Because of this, we speculated that FoxO3a could be translocated to the nucleus to promote Bim expression when follicular atresia originates under either physiological or pathological conditions.
In our studies on the effects of FSH on BimEL expression, we found that FSH not only decreased the levels of BimEL expression but also shifted the molecular weight of the BimEL protein. This finding is in agreement with the results of several previous studies in which changes in the electrophoretic mobility of BimEL were attributed to phosphorylation and dephosphorylation. Möller et al. (47) showed that a stem cell factor-induced band shift of BimEL in mouse mast cells was due to phosphorylation and indicated that the larger band represented phosphorylated BimEL after treatment with alkaline phosphatase. Abayasiriwardana et al. (48) also reported that this shift was abolished by alkaline phosphatase-mediated dephosphorylation but was restored if the phosphatase was inhibited by sodium orthovanadate in malignant mesothelioma cells. Moreover, recent studies have demonstrated that this phosphorylation could regulate the apoptotic function of BimEL. In Ba/F3 cells, Ser-87 of BimEL was found to be an important phosphorylation site targeted by Akt to attenuate the pro-apoptotic function of BimEL (49). Similarly, in a human B lymphoma cell line, phosphorylation of BimEL by Erk1/2 on Ser-69 selectively led to its proteasomal degradation, thereby promoting cell survival (30). However, the JNK-induced phosphorylation of BimEL at Ser-65 promoted the apoptotic effect of BimEL in primary cerebellar granule neurons (40). These results suggested that different kinase pathways might ultimately result in different patterns of BimEL phosphorylation that determine whether phosphorylated BimEL facilitates or antagonizes apoptosis. In this study, our results showed that BimEL phosphorylation status decreased during apoptosis of the porcine granulosa cells, which indicated that phosphorylation of BimEL might protect the porcine granulosa cells from apoptosis. However, the detailed mechanism of BimEL phosphorylation in granulosa cells is as yet unknown and will form the basis of further investigation.
A hypothetical model of Bim protein regulation by FSH in granulosa cells is outlined in Fig. 7. Although there are some questions that still needed to be resolved, in this study, we showed that the level of BimEL expression impacted porcine follicular atresia, and FSH could prevent granulosa cells apoptosis by down-regulating BimEL expression via the PI3K/Akt/FoxO3a pathway. This work provides new insight into how the Bcl-2 protein family, especially Bim, functions in follicular atresia.
FIGURE 7.
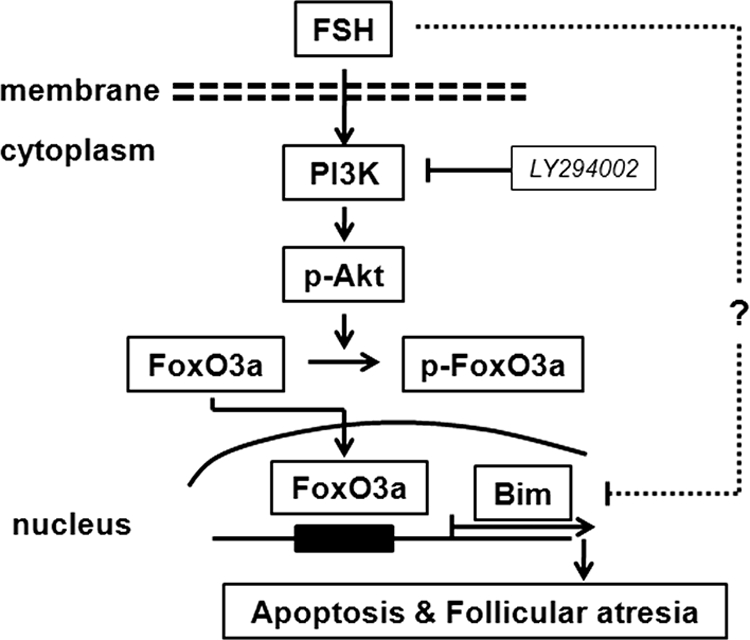
Diagram of Bim expression regulated by FSH in granulosa cells. FoxO3a transcriptional activity is inhibited by FSH via PI3K/Akt signaling-mediated phosphorylation. FSH withdrawal or LY294002 inhibit PI3K/Akt signaling, resulting in FoxO3a activation and subsequent activation of Bim expression, possibly leading to follicular atresia. Furthermore, our results do not rule out the possibility that other signaling pathways might also be involved in regulation of Bim expression in granulosa cells.
Acknowledgments
We thank Dr. Dongbao Chen (Dept. of Ob/Gyn, University of California, Irvine) and Dr. Qien Yang (Center for Reproductive Biology, School of Molecular Biosciences, Washington State University, Pullman) for critical reading and comments.
This work was supported in part by National Natural Science Foundation of China Grant 31072031, Key Project of the Ministry of Education of China Grant 109018, and the Graduate Scientific Research Innovation Special Fund of the China Agricultural University.
- BH3
- Bcl-2 homology 3
- tcRNA
- total cellular RNA.
REFERENCES
- 1. Hughes F. M., Jr., Gorospe W. C. (1991) Biochemical identification of apoptosis (programmed cell death) in granulosa cells. Evidence for a potential mechanism underlying follicular atresia. Endocrinology 129, 2415–2422 [DOI] [PubMed] [Google Scholar]
- 2. Tilly J. L., Kowalski K. I., Johnson A. L., Hsueh A. J. (1991) Involvement of apoptosis in ovarian follicular atresia and postovulatory regression. Endocrinology 129, 2799–2801 [DOI] [PubMed] [Google Scholar]
- 3. Billig H., Furuta I., Hsueh A. J. (1994) Gonadotropin-releasing hormone directly induces apoptotic cell death in the rat ovary. Biochemical and in situ detection of deoxyribonucleic acid fragmentation in granulosa cells. Endocrinology 134, 245–252 [DOI] [PubMed] [Google Scholar]
- 4. Chun S. Y., Eisenhauer K. M., Minami S., Billig H., Perlas E., Hsueh A. J. (1996) Hormonal regulation of apoptosis in early antral follicles. Follicle-stimulating hormone as a major survival factor. Endocrinology 137, 1447–1456 [DOI] [PubMed] [Google Scholar]
- 5. Asselin E., Xiao C. W., Wang Y. F., Tsang B. K. (2000) Mammalian follicular development and atresia: role of apoptosis. Biol. Signal. Recept. 9, 87–95 [DOI] [PubMed] [Google Scholar]
- 6. Peluso J. J., Steger R. W. (1978) Role of FSH in regulating granulosa cell division and follicular atresia in rats. J. Reprod. Fertil. 4, 275–278 [DOI] [PubMed] [Google Scholar]
- 7. Kaipia A., Hsueh A. J. (1997) Regulation of ovarian follicle atresia. Annu. Rev. Physiol. 59, 349–363 [DOI] [PubMed] [Google Scholar]
- 8. Cory S., Adams J. M. (2002) The Bcl2 family. Regulators of the cellular life-or-death switch. Nat. Rev. Cancer 2, 647–656 [DOI] [PubMed] [Google Scholar]
- 9. Johnson A. L., Bridgham J. T., Jensen T. (1999) Bcl-X(LONG) protein expression and phosphorylation in granulosa cells. Endocrinology 140, 4521–4529 [DOI] [PubMed] [Google Scholar]
- 10. Hsu S. Y., Lai R. J., Finegold M., Hsueh A. J. (1996) Targeted overexpression of Bcl-2 in ovaries of transgenic mice leads to decreased follicle apoptosis, enhanced folliculogenesis, and increased germ cell tumorigenesis. Endocrinology 137, 4837–4843 [DOI] [PubMed] [Google Scholar]
- 11. Ratts V. S., Flaws J. A., Kolp R., Sorenson C. M., Tilly J. L. (1995) Ablation of bcl-2 gene expression decreases the numbers of oocytes and primordial follicles established in the pos-natal female mouse gonad. Endocrinology 136, 3665–3668 [DOI] [PubMed] [Google Scholar]
- 12. Leo C. P., Hsu S. Y., Chun S. Y., Bae H. W., Hsueh A. J. (1999) Characterization of the antiapoptotic Bcl-2 family member myeloid cell leukemia-1 (Mcl-1) and the stimulation of its message by gonadotropins in the rat ovary. Endocrinology 140, 5469–5477 [DOI] [PubMed] [Google Scholar]
- 13. Hsu S. Y., Hsueh A. J. (2000) Tissue-specific Bcl-2 protein partners in apoptosis. An ovarian paradigm. Physiol. Rev. 80, 593–614 [DOI] [PubMed] [Google Scholar]
- 14. O'Connor L., Strasser A., O'Reilly L. A., Hausmann G., Adams J. M., Cory S., Huang D. C. (1998) Bim. A novel member of the Bcl-2 family that promotes apoptosis. EMBO J. 17, 384–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hsu S. Y., Lin P., Hsueh A. J. (1998) BOD (Bcl-2-related ovarian death gene) is an ovarian BH3 domain-containing proapoptotic Bcl-2 protein capable of dimerization with diverse antiapoptotic Bcl-2 members. Mol. Endocrinol. 12, 1432–1440 [DOI] [PubMed] [Google Scholar]
- 16. Chen J. Z., Ji C. N., Gu S. H., Li J. X., Zhao E. P., Huang Y., Huang L., Ying K., Xie Y., Mao Y. M. (2004) Overexpression of Bim α3, a novel isoform of human Bim, results in cell apoptosis. Int. J. Biochem. Cell Biol. 36, 1554–1561 [DOI] [PubMed] [Google Scholar]
- 17. Liu J. W., Chandra D., Tang S. H., Chopra D., Tang D. G. (2002) Identification and characterization of Bimγ, a novel proapoptotic BH3-only splice variant of Bim. Cancer Res. 62, 2976–2981 [PubMed] [Google Scholar]
- 18. Marani M., Tenev T., Hancock D., Downward J., Lemoine N. R. (2002) Identification of novel isoforms of the BH3 domain protein Bim which directly activate Bax to trigger apoptosis. Mol. Cell. Biol. 22, 3577–3589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Miyashita U. M., Shikama Y., Tadokoro K., Yamada M. (2001) Molecular cloning and characterization of six novel isoforms of human Bim, a member of the proapoptotic Bcl-2 family. FEBS Lett. 509, 135–141 [DOI] [PubMed] [Google Scholar]
- 20. Urbich C., Knau A., Fichtlscherer S., Walter D. H., Brühl T., Potente M., Hofmann W. K., de Vos S., Zeiher A. M., Dimmeler S. (2005) FOXO-dependent expression of the proapoptotic protein Bim. Pivotal role for apoptosis signaling in endothelial progenitor cells. FASEB J. 19, 974–976 [DOI] [PubMed] [Google Scholar]
- 21. O'Reilly L. A., Cullen L., Visvader J., Lindeman G. J., Print C., Bath M. L., Huang D. C., Strasser A. (2000) The proapoptotic BH3-only protein bim is expressed in hematopoietic, epithelial, neuronal, and germ cells. Am. J. Pathol. 157, 449–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jolly P. D., Smith P. R., Heath D. A., Hudson N. L., Lun S., Still L. A., Watts C. H., McNatty K. P. (1997) Morphological evidence of apoptosis and the prevalence of apoptotic versus mitotic cells in the membrana granulosa of ovarian follicles during spontaneous and induced atresia in ewes. Biol. Reprod. 56, 837–846 [DOI] [PubMed] [Google Scholar]
- 23. Guthrie H. D. (2005) The follicular phase in pigs: Follicle populations, circulating hormones, follicle factors and oocytes. J. Anim. Sci. 83, E79–E89 [Google Scholar]
- 24. Sasaki H., Sheng Y., Kotsuji F., Tsang B. K. (2000) Down-regulation of X-linked inhibitor of apoptosis protein induces apoptosis in chemoresistant human ovarian cancer cells. Cancer Res. 60, 5659–5666 [PubMed] [Google Scholar]
- 25. Alam H., Maizels E. T., Park Y., Ghaey S., Feiger Z. J., Chandel N. S., Hunzicker-Dunn M. (2004) Follicle-stimulating hormone activation of hypoxia-inducible factor-1 by the phosphatidylinositol 3-kinase/AKT/Ras homolog enriched in brain (Rheb)/mammalian target of rapamycin (mTOR) pathway is necessary for induction of select protein markers of follicular differentiation. J. Biol. Chem. 279, 19431–19440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jin X., Xiao L. J., Zhang X. S., Liu Y. X. (2011) Apotosis in ovary. Front. Biosci. 3, 680–697 [DOI] [PubMed] [Google Scholar]
- 27. Stahl M., Dijkers P. F., Kops G. J., Lens S. M., Coffer P. J., Burgering B. M., Medema R. H. (2002) The forkhead transcription factor FoxO regulates transcription of p27Kip1 and Bim in response to IL-2. J. Immunol. 168, 5024–5031 [DOI] [PubMed] [Google Scholar]
- 28. Essafi A., Fernández de Mattos S., Hassen Y. A., Soeiro I., Mufti G. J., Thomas N. S., Medema R. H., Lam E. W. (2005) Direct transcriptional regulation of Bim by FoxO3a mediates STI571-induced apoptosis in Bcr-Abl-expressing cells. Oncogene 24, 2317–2329 [DOI] [PubMed] [Google Scholar]
- 29. Brunet A., Bonni A., Zigmond M. J., Lin M. Z., Juo P., Hu L. S., Anderson M. J., Arden K. C., Blenis J., Greenberg M. E. (1999) Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 96, 857–868 [DOI] [PubMed] [Google Scholar]
- 30. Fukazawa H., Noguchi K., Masumi A., Murakami Y., Uehara Y. (2004) BimEL is an important determinant for induction of anoikis sensitivity by mitogen-activated protein/extracellular signal-regulated kinase kinase inhibitors. Mol. Cancer Ther. 3, 1281–1288 [PubMed] [Google Scholar]
- 31. Hsueh A. J., Billig H., Tsafriri A. (1994) Ovarian follicle atresia. A hormonally controlled apoptotic process. Endocr. Rev. 15, 707–724 [DOI] [PubMed] [Google Scholar]
- 32. Kim M. R., Tilly J. L. (2004) Current concepts in Bcl-2 family member regulation of female germ cell development and survival. Biochim. Biophys. Acta 1644, 205–210 [DOI] [PubMed] [Google Scholar]
- 33. Reed J. C. (1997) Double identity for proteins of the Bcl-2 family. Nature 387, 773–776 [DOI] [PubMed] [Google Scholar]
- 34. Adams J. M., Cory S. (1998) The Bcl-2 protein family. Arbiters of cell survival. Science 281, 1322–1326 [DOI] [PubMed] [Google Scholar]
- 35. Sun Y. F., Yu L. Y., Saarma M., Timmusk T., Arumae U. (2001) Neuron-specific Bcl-2 homology 3 domain-only splice variant of Bak is anti-apoptotic in neurons but pro-apoptotic in non-neuronal cells. J. Biol. Chem. 276, 16240–16247 [DOI] [PubMed] [Google Scholar]
- 36. Song Q., Kuang Y., Dixit V. M., Vincenz C. (1999) Boo, a novel negative regulator of cell death, interacts with Apaf-1. EMBO J. 18, 167–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Miao J., Chen G. G., Yun J. P., Chun S. Y., Zheng Z. Z., Ho R. L., Chak E. C., Xia N. S., Lai P. B. (2007) Identification and characterization of BH3 domain protein Bim and its isoforms in human hepatocellular carcinomas. Apoptosis 12, 1691–1701 [DOI] [PubMed] [Google Scholar]
- 38. Harris C. A., Johnson E. M., Jr. (2001) BH3-only Bcl-2 family members are coordinately regulated by the JNK pathway and require Bax to induce apoptosis in neurons. J. Cell Biol. 276, 37754–37760 [DOI] [PubMed] [Google Scholar]
- 39. Markström E., Svensson E. Ch., Shao R., Svanberg B., Billig H. (2002) Survival factors regulating ovarian apoptosis. Dependence on follicle differentiation. Reproduction 123, 23–30 [DOI] [PubMed] [Google Scholar]
- 40. Becker E. B., Howell J., Kodama Y., Barker P. A., Bonni A. (2004) Characterization of the c-Jun N-terminal kinase-BimEL signaling pathway in neuronal apoptosis. J. Neurosci. 24, 8762–8770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Guthrie H. D., Garrett W. M., Cooper B. S. (1998) Follicle-stimulating hormone and insulin-like growth factor-I attenuate apoptosis in cultured porcine granulosa cells. Biol. Reprod. 58, 390–396 [DOI] [PubMed] [Google Scholar]
- 42. Castrillon D. H., Miao L., Kollipara R., Horner J. W., DePinho R. A. (2003) Suppression of ovarian follicle activation in mice by the transcription factor Foxo3a. Science 301, 215–218 [DOI] [PubMed] [Google Scholar]
- 43. Matsuda F., Inoue N., Maeda A., Cheng Y., Sai T., Gonda H., Goto Y., Sakamaki K., Manabe N. (2011) Expression and function of apoptosis initiator FOXO3 in granulosa cells during follicular atresia in pig ovaries. J. Reprod. Dev. 57, 151–158 [DOI] [PubMed] [Google Scholar]
- 44. Richards J. S., Sharma S. C., Falender A. E., Lo Y. H. (2002) Expression of FKHR, FKHRL1, and AFX genes in the rodent ovary. Evidence for regulation by IGF-I, estrogen, and the gonadotropins. Mol. Endocrinol. 16, 580–599 [DOI] [PubMed] [Google Scholar]
- 45. Dijkers P. F., Medema R. H., Lammers J. W., Koenderman L., Coffer P. J. (2000) Expression of the pro-apoptotic Bcl-2 family member Bim is regulated by the forkhead transcription factor FKHR-L1. Curr. Biol. 10, 1201–1204 [DOI] [PubMed] [Google Scholar]
- 46. Gilley J., Coffer P. J., Ham J. (2003) FOXO transcription factors directly activate bim gene expression and promote apoptosis in sympathetic neurons. J. Cell Biol. 162, 613–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Möller C., Alfredsson J., Engström M., Wootz H., Xiang Z., Lennartsson J., Jönsson J. I., Nilsson G. (2005) Stem cell factor promotes mast cell survival via inactivation of FOXO3a-mediated transcriptional induction and MEK-regulated phosphorylation of the proapoptotic protein Bim. Blood 106, 1330–1336 [DOI] [PubMed] [Google Scholar]
- 48. Abayasiriwardana K. S., Barbone D., Kim K. U., Vivo C., Lee K. K., Dansen T. B., Hunt A. E., Evan G. I., Broaddus V. C. (2007) Malignant mesothelioma cells are rapidly sensitized to TRAIL-induced apoptosis by low-dose anisomycin via Bim. Mol. Cancer Ther. 6, 2766–2776 [DOI] [PubMed] [Google Scholar]
- 49. Qi X. J., Wildey G. M., Howe P. H. (2006) Evidence that Ser-87 of BimEL is phosphorylated by Akt and regulates BimEL apoptotic function. J. Biol. Chem. 281, 813–823 [DOI] [PubMed] [Google Scholar]