Background: Proteasomal turnover of nucleostemin (NS) in response to low cellular GTP levels was previously shown to be MDM2-dependent without clear mechanisms.
Results: NS is ubiquitinated in cells, but its mycophenolic acid-induced turnover still occurs without ubiquitination and MDM2.
Conclusion: NS is ubiquitinated in cells, and its turnover is ubiquitin- and MDM2-independent.
Significance: NS ubiquitination may be important for regulation of NS function.
Keywords: Nucleolus, Protein Stability, Protein Turnover, Protein-Protein Interactions, Ubiquitination, MDM2, GTP, MPA, Nucleostemin, Proteasomal Turnover, Ubiquitination
Abstract
Nucleostemin (NS) is a nucleolar GTP-binding protein essential for ribosomal biogenesis, proliferation, and animal embryogenesis. It remains largely unclear how this protein is regulated. While working on its role in suppression of MDM2 and activation of p53, we observed that NS protein (but not mRNA) levels decreased drastically in response to GTP depletion. When trying to further elucidate the molecular mechanism(s) underlying this unusual phenomenon, we found that NS was degraded independently of ubiquitin and MDM2 upon GTP depletion. First, depletion of GTP by treating cells with mycophenolic acid decreased the level of NS without apparently affecting the levels of other nucleolar proteins. Second, mutant NS defective in GTP binding and exported to the nucleoplasm was much less stable than wild-type NS. Although NS was ubiquitinated in cells, its polyubiquitination was independent of Lys-48 or Lys-63 in the ubiquitin molecule. Inactivation of E1 in E1 temperature-sensitive mouse embryonic fibroblast (MEF) cells failed to prevent the proteasomal degradation of NS. The proteasomal turnover of NS was also MDM2-independent, as its half-life in p53/MDM2 double knock-out MEF cells was the same as that in wild-type MEF cells. Moreover, NS ubiquitination was MDM2-independent. Mycophenolic acid or doxorubicin induced NS degradation in various human cancerous cells regardless of the status of MDM2. Hence, these results indicate that NS undergoes a ubiquitin- and MDM2-independent proteasomal degradation when intracellular GTP levels are markedly reduced and also suggest that ubiquitination of NS may be involved in regulation of its function rather than stability.
Introduction
Originally known as the factory for ribosomal biogenesis, the nucleolus is increasingly being recognized for its ability to serve as the sensor of cellular stress (1). Many proteins within the nucleolus are able to shuttle dynamically between the nucleoplasm and nucleolus and are thus able to initiate rapid signaling responses to inhibit cell growth and proliferation under unfavorable growth conditions. In particular, nucleostemin (NS),2 a nucleolar GTPase, has been implicated in diverse molecular functions, including ribosomal biogenesis (2), cell cycle progression (3, 4), and stem cell proliferation (3, 5).
The balance of NS protein levels is critical for embryonic development and maintaining cellular homeostasis. Although abundantly expressed in proliferating cancer cells and stem cells, NS protein levels rapidly decrease when the cells exit the mitotic cycle and differentiate (3). Deregulation of the balance of NS protein levels can lead to irregular cell cycle progression, which may contribute to tumorigenesis and other physiological disorders (4, 5).
The role of NS in regulating the cell cycle is partially executed via the ability of this protein to interact with MDM2, the E3 ubiquitin ligase of the p53 tumor suppressor (4). In response to cellular stress signals, p53 tumor suppressor is activated to trigger the transcription of its target genes and promote apoptosis or G1 cell cycle arrest through the functions of these target gene-encoded proteins or microRNAs (6, 7). MDM2 is encoded by one of the p53 target genes and thus acts as a negative feedback regulator (8–10). NS can modulate this p53-MDM2 feedback loop by directly binding to the MDM2 acidic domain and blocking its ubiquitination activity toward p53, thus stabilizing p53 by inhibiting MDM2-mediated proteasomal degradation (4). In addition, the binding of NS to MDM2 prevents MDM2 autoubiquitination and is able to further regulate cell cycle progression (11). Interestingly, siRNA-mediated depletion of NS can also induce p53 levels and activity by causing ribosomal stress that leads to the interaction of ribosomal proteins L5 and L11 with MDM2 (4). These studies suggest that an optimal balance of NS is critical for maintaining cellular homeostasis and normal cell proliferation (12).
Although it is clear that NS is essential for ribosomal biogenesis (2), cell proliferation, and embryogenesis (3, 5), it remains largely obscure how this nucleolar protein is regulated. Several studies suggest that the fluctuating intracellular level of guanine nucleotide pools (GTP) may play a role in governing NS levels in cells. For example, it has been shown that the GTP-binding activity of NS is crucial for both its localization in the nucleolus (13) and its protein stability (14). In particular, the N-terminal basic sequence of NS directs the protein to the nucleus, and the GTP-binding motif can retain NS in the nucleolus and prevent it from entering the nucleoplasm (15). Disruption of GTP binding by either depleting intracellular GTP pools or mutating the G1 GTP-binding motifs triggers the relocation of NS from the nucleolus to the nucleoplasm (15), as well as a reduction of NS protein levels independently of mRNA transcription (14). The reduction of NS levels can be rescued by a proteasome inhibitor, such as MG132 (14), suggesting that NS may undergo proteasome-mediated proteolysis. However, it is unknown whether this proteasomal turnover of NS due to lack of GTP binding is ubiquitin-dependent or not. Although the majority of proteins designated for degradation are marked by specific E3 ligases with polyubiquitin, which allows the proteasome to recognize and destroy the proteins, some proteins, such as p21 and MCL-1, have been reported to undergo proteasomal degradation independently of polyubiquitination (16, 17). Because MDM2 can bind to NS (4), it appears probable that MDM2 may also act as the E3 ubiquitin ligase for NS. In support of this assumption, a recent study showed that the overexpression of MDM2 reduces the level of NS, whereas knockdown of MDM2 levels only partially rescues the level of NS (14). Again, it still remains unclear if MDM2 could indeed mediate NS ubiquitination and would be essential for the proteasomal turnover of this nucleolar GTP-binding protein.
In an attempt to address these questions, we examined the possible mechanisms underlying the reduction of NS in response to GTP depletion. We found that although NS was indeed ubiquitinated in cells, the proteasomal degradation of NS triggered by GTP depletion was independent of ubiquitination and MDM2. Degradation of NS induced by GTP depletion still occurred in p53/MDM2 double knock-out mouse embryonic fibroblast (MEF) cells. Furthermore, inactivation of E1 in E1 temperature-sensitive MEF cells failed to rescue NS proteasomal degradation. Our study not only reveals NS as a ubiquitinated protein but also demonstrates that the proteasomal turnover of NS triggered by GTP depletion is independent of MDM2 and ubiquitination. In addition, our results imply that ubiquitination might play a role in regulating the function (but not stability) of NS.
MATERIALS AND METHODS
Cell Lines, Plasmids, and Antibodies
Human non-small cell lung carcinoma H1299 cells (p53-null), human embryonic kidney 293 cells (p53 mutant), MEF cells, and p53/MDM2 double knock-out MEF cells were cultured in DMEM supplemented with 10% FBS and 100 units/ml penicillin/streptomycin at 37 °C and 5% CO2. E1 temperature-sensitive MEFts20 cells were cultured in the abovementioned medium and culture conditions but at 34 or 39 °C (16, 18, 19). The FLAG-NS, FLAG-NS(G261V/G265V), and His-ubiquitin plasmids used in this study were as described previously (4). Monoclonal anti-FLAG, anti-tubulin, and anti-β-actin (Sigma) and polyclonal anti-rabbit NS antibodies were generated in the laboratory using purified His-tagged NS expressed from bacteria as an antigen as described previously (4). Antibodies C23 and B23 and anti-p53 antibody DO-1 were obtained from Santa Cruz Biotechnology.
Transfection and Western Blotting
Cells were transfected with TransFectin reagent (Bio-Rad) along with plasmids as indicated in the figure legends according to the manufacturer's instructions. Cells were harvested 36–48 h after transfection on ice and lysed in cell lysis buffer consisting of 50 mm Tris (pH 8.0), 1 mm EDTA, 150 mm NaCl, 0.5% Nonidet P-40, 2 mm PMSF, and 1 mm DTT with appropriate protease inhibitors. An equal amount of proteins from lysates was subjected to analysis by SDS-PAGE and Western blotting with the antibodies indicated in the figure legends.
In Vivo Ubiquitination Assays
Cells were transfected with the plasmids indicated in the figure legends. Thirty-six hours after transfection, cells were treated with 10 μm MG132 for 8 h and harvested on ice. The collected pellets were aliquoted into two portions. One portion was used for the in vivo ubiquitination assay as described previously (20) with some modifications. The cell pellets were lysed in buffer containing 8 m urea, 0.1 m Tris-Cl, 0.1 m Na2HPO4/NaH2PO4, and 10 mm β-mercaptoethanol (pH 8.0) and incubated with nickel beads for 4 h. The beads were washed twice with the same buffer and then once with the same buffer at pH 6.3. The second aliquot was subjected to Western blotting as described above to determine protein expression.
Immunofluorescence
Twenty-four hours after transfection, cells were washed twice with cold PBS and fixed with cold 20% paraformaldehyde for 20 min. The fixed cells were then blocked with 8% BSA for 1 h and stained with the appropriate antibodies and DAPI, followed by analysis under a fluorescence microscope.
RESULTS
Depletion of GTP by Treating Cells with Mycophenolic Acid (MPA) Triggers Reduction of NS Protein Levels
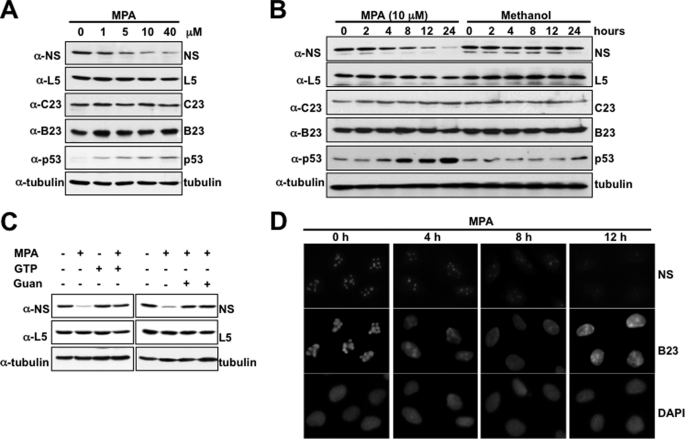
MPA, an inhibitor of the rate-limiting enzyme for de novo guanine nucleotide synthesis known as inosine-monophosphate dehydrogenase, causes a depletion of intracellular GTP pools (21, 22). We reported previously that GTP depletion by MPA treatment causes inhibition of pre-RNA processing and triggers ribosomal stress, leading to activation of p53; furthermore, we also observed that NS protein levels decrease when cells are treated with MPA (23) (data not shown). To further investigate how this might happen upon MPA treatment, we first repeated the experiment with different doses of MPA and for different time points post-MPA treatment. MPA treatment indeed effectively reduced NS protein levels in a dose-dependent (Fig. 1A) or time-dependent (Fig. 1B) manner in U2OS cells. Furthermore, the reduction of NS protein levels appeared to be specific only to NS, as the levels of nucleolar proteins, such as ribosomal protein L5, nucleolin (C23), and nucleophosmin (B23), remained unchanged. The influence of MPA on NS protein levels was based on the drug's ability to inhibit GTP synthesis because supplementing MPA-treated cells with a GTP precursor (guanosine) or GTP itself sufficiently rescued NS depletion (Fig. 1C). These observations further reinforce the fact that NS plays an essential role in ribosomal biogenesis and pre-RNA processing, as the depletion of NS triggered ribosomal stress-induced activation of p53. They also suggest that GTP levels might be critical for maintaining the normal level of NS.
FIGURE 1.
MPA induces NS depletion and ribosomal stress. A, MPA triggers NS depletion and p53 activation at 10 μm. U2OS cells were treated with varying concentrations of MPA for 12 h and then harvested on ice. Equal lysates were used for Western blotting with the indicated antibodies. In each sample, 50 μg of proteins were used (true for the following figures). B, MPA induces NS depletion in a time-dependent manner. U2OS cells were treated with 10 μm MPA and harvested at the indicated time points on ice. Protein expression was analyzed by SDS-PAGE followed by Western blotting with the indicated antibodies. C, U2OS cells were treated with MPA for 12 h in the presence or absence of GTP or guanosine (Guan) and harvested on ice. Equal lysates were used for Western blotting with the indicated antibodies. D, immunofluorescence staining to show NS depletion from the nucleolus. Cells were fixed with methanol and subjected to staining with anti-NS and anti-B23 antibodies. These experiments were repeated in H1299 and HCT116 cells as well (data not shown).
We next explored the possibility that GTP depletion by MPA can influence the localization of nucleolar proteins NS and B23 in the nucleus. U2OS cells were treated with MPA and fixed in methanol at the indicated time points, followed by immunofluorescence staining with DAPI and antibodies specific for NS and B23, respectively. As shown in Fig. 1D, both B23 and NS co-localized to the nucleolus prior to MPA treatment. However, MPA treatment led to a reduction in the nucleolar level of NS. On the other hand, B23 was exported from the nucleolus to the nucleoplasm. Together with the results in Fig. 1 (A and B), which were repeated in other types of human cells as well (data not shown), this result suggests that intracellular GTP levels affect the trafficking of these two nucleolar proteins but differentially regulate their levels, reducing the steady-state level of NS (but not B23), implying that NS is perhaps less stable than B23 under this condition.
GTP Depletion or Mutation of GTP-binding Motif in NS Leads to Destabilization of NS
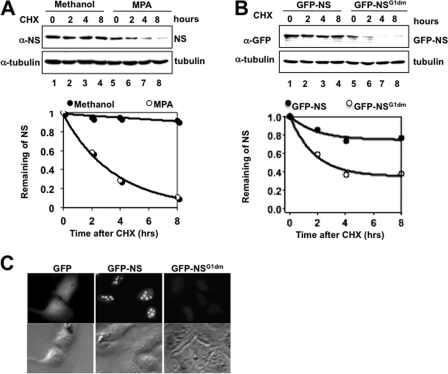
To determine whether the reduction of NS levels upon MPA treatment is due to its destabilization under this conditions, we compared the half-life of NS in H1299 cells treated with MPA or a vehicle control. Indeed, MPA treatment led to a reduction in the half-life of NS from >8 h to <4 h (Fig. 2A). This result was also repeated in different cell lines (data not shown), indicating that depletion of endogenous GTP levels can accelerate the proteasomal turnover of NS. This result also suggests that GTP binding may play a role in stabilizing the NS protein.
FIGURE 2.
Disruption of NS GTP-binding activity destabilizes NS in nucleoplasm. A, H1299 cells were treated with MPA for 12 h, followed by treatment with cycloheximide (CHX). Cells were harvested at the indicated time points. Equal amounts of proteins from lysates were used for Western blotting. Western blot bands were then quantified with ImageJ software and plotted on a graph. B, H1299 cells were transfected with either GFP-NS or GFP-NSG1dm (NS(G261V/G265V)) expression plasmid. Cells were treated with cycloheximide and harvested at the indicated time points 24 h after transfection. Equal amounts of proteins from cell lysates were used for Western blotting to compare the levels of GFP-NS and GFP-NSG1dm. Western blot bands were quantified and plotted on a graph as described above. C, disruption of GTP binding triggers NS relocalization from the nucleolus to the nucleoplasm. H1299 cells were transfected with GFP-NS or GFP-NSG1dm. Cells were fixed in methanol and subjected to immunofluorescence staining.
Next, we wanted to test if this speculation is true by comparing the half-life of WT NS with that of mutant NS defective in GTP binding but still capable of binding other proteins, such as MDM2 and tumor suppressor p14ARF, as shown previously (15) (data not shown). To do so, we transfected H1299 cells with a plasmid encoding GFP-tagged WT or mutant NS, followed by treatment with cycloheximide. Consistent with GTP depletion, disruption of the GTP-binding motif in NS destabilized NS, reducing the half-life of this relatively stable protein from >8 h to ∼3 h (Fig. 2B), thus confirming the idea that GTP-binding activity is crucial to maintaining NS stability. Immunofluorescence staining (Fig. 2C) revealed that although WT NS was localized in the nucleolus, the GTP-binding mutant NS protein was dispersed in the nucleoplasm, as expected (15). Also, the intensity of the fluorescence signals of WT NS was clearly stronger than that of mutant NS (Fig. 2C). Together, these observations indicate that the GTP-binding activity of NS is critical for both the stability and localization of NS in the nucleus and also suggest that NS might be degraded in the nucleoplasm.
NS Degradation Occurs Independently of Polyubiquitination
Proteasomal degradation can occur through either a ubiquitin-dependent or ubiquitin-independent mechanism. Given that there is no prior evidence of NS ubiquitination, it is unclear whether NS proteasomal degradation is mediated through a ubiquitin-dependent mechanism.
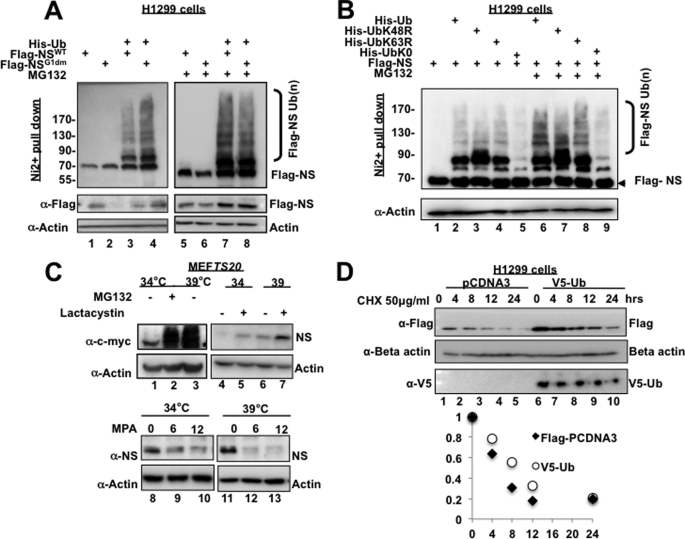
To address this question, we first performed in vivo ubiquitination assays to determine whether both WT NS and its GTP-binding mutant are polyubiquitinated or not. Because the GTP-binding mutant had a much shorter half-life than WT NS, we predicted higher levels of polyubiquitination for mutant NS compared with WT NS. Surprisingly, both WT NS and mutant NS were equally polyubiquitinated in H1299 cells (Fig. 3A), suggesting that polyubiquitination of NS might not contribute to the difference in the stability between WT NS and mutant NS. Consistent with this assumption, polyubiquitination of NS was not dependent on either Lys-48 or Lys-63, as mutation at Lys-48 or Lys-63 to arginine failed to eliminate NS polyubiquitination (Fig. 3B). This result further supports the notion that NS degradation may not be dependent on ubiquitination, as Lys-48 is more important for protein turnover (24, 25). Of note, the expression level of mutant NS was lower than that of wild-type NS (Figs. 2 and 3A). This was likely due to the fact that mutant NS is much less stable than wild-type NS (Fig. 2). Interestingly, ubiquitination appeared to stabilize NS (Fig. 3, A and D; data not shown for endogenous NS). Consistent with these results, when E1 was inactivated at 39 °C, endogenous NS was degraded more rapidly in the presence of MPA (Fig. 3C). These results were reproduced multiple times (data not shown). Although the mechanism underlying the stabilization of NS by ubiquitination remains unclear at the presence, these lines of evidence also support the notion that NS proteasomal turnover is ubiquitination-independent.
FIGURE 3.
NS is degraded independently of polyubiquitination. A, WT NS and GTP-binding mutant NS are polyubiquitinated at similar levels. H1299 cells were transfected with the indicated plasmids. Cells were treated with or without 10 μm MG132 for 6 h and harvested on ice 24 h after transfection. Cell lysates were subjected to His-nickel pulldown assay and Western blotting to detect polyubiquitinated FLAG-NS. B, NS does not require either Lys-48 or Lys-63 for polyubiquitination in cells. The same ubiquitination assay as described above was conducted except that a K48R, K63R, or K0R mutant ubiquitin (Ub) plasmid was used. C, inactivation of E1 ligase rescues proteasomal degradation of c-Myc but not MPA-induced degradation of NS. In lanes 1–3, MEFts20 cells were subjected to incubation at 34 or 39 °C to inactivate E1 ligase in the presence of Me2SO or 10 μm MG132. In lanes 4–7, cells were subjected to incubation at 34 or 39 °C with Me2SO or 10 μm lactacystin. Cells were harvested 12 h later, and equal amounts of lysates were subjected to Western blotting with the indicated antibodies. In lanes 8–13, cells were treated with 10 μm MPA or Me2SO as a control at 34 or 39 °C and harvested at 6 or 12 h later. Western blotting was done with the indicated antibodies. D, H1299 cells were transfected with pcDNA3 control plasmid or plasmid expressing V5-ubiquitin. Twenty-four hours after transfection, cells were treated with cycloheximide (CHX) and harvested at the indicated time points. Equal amounts of lysates were subjected to Western blotting with the indicated antibodies, followed by quantitative and graphic analysis using ImageJ software and Excel.
To validate the aforementioned idea, i.e. the ubiquitin system might not be necessary for MPA-mediated NS degradation, we employed an E1 temperature-sensitive MEF cell line, MEFts20. The inactivation of the E1 ubiquitin ligase by culturing the cells at 39 °C was confirmed by the increase in the level of c-Myc, which is regulated by ubiquitin-mediated proteasomal degradation (19). The inactivation of E1 did not appear to cause a reduction in NS due to cellular stress, as treatment with lactacystin, a specific inhibitor of the 26 S proteasome, was able to rescue NS protein levels in comparison with the Me2SO control regardless of E1 inactivation. The shift in temperature itself appeared to increase the basal level of the NS protein slightly, which may possibly be explained by the rescue of the levels of the c-Myc protein, a transcription factor that has been shown recently to target the promoter of the NS gene (26). However, despite this initial increase in NS levels, the temperature shift from 34 to 39 ºC failed to rescue MPA-induced degradation of NS (Fig. 3C). To further confirm that NS proteasomal degradation is not dependent on NS polyubiquitination, we compared the half-life of FLAG-NS in the presence and absence of V5-ubiquitin. Overexpression of V5-ubiquitin did not reduce the half-life of NS in comparison with the control. Together, these observations further support the notion that NS undergoes proteasomal degradation independently of polyubiquitination.
NS Degradation Occurs Independently of MDM2 E3 Ligase
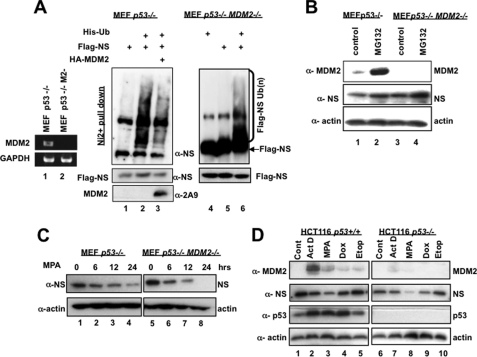
Because MDM2 can directly bind to the coiled-coiled domain of NS through its acidic domain (4) and because we also previously observed the inverse correlation of the MDM2 level and the NS level upon MPA treatment (data not shown) (23), it was possible that MDM2 might mediate NS degradation after GTP depletion. Indeed, overexpression of MDM2 was later shown to lead to a decrease in NS levels (14). However, it remained unclear if this decrease is due to ubiquitination-dependent degradation of NS by MDM2. To address this issue, we first tested if NS polyubiquitination is MDM2-dependent or not in cells. As shown in Fig. 4A, NS was still polyubiquitinated in p53 single knock-out or p53/MDM2 double knock-out MEF cells regardless of the presence or absence of MDM2. Furthermore, overexpression of MDM2 in p53 single knock-out cells appeared to inhibit NS polyubiquitination, possibly by competing with other E3 ligases for interaction with NS or for the limited pool of His-ubiquitin molecules in the cells. This suggests that although MDM2 can interact with NS, there appear to be alternative E3 ligases that may mediate NS polyubiquitination independently of MDM2. Because MDM2 has been demonstrated to facilitate the proteasomal degradation of p21 independently of polyubiquitination (17), we wanted to determine whether MPA-induced NS degradation is MDM2-dependent or not. First, we compared the levels of NS in p53 single knock-out MEF cells with those in p53/MDM2 double knock-out MEF cells in response to treatment with the proteasome inhibitor MG132. As shown in Fig. 4B, MG132 treatment was able to induce the endogenous level of NS in these MEF cells regardless of the presence or absence of MDM2, suggesting that the proteasome-mediated degradation of NS is independent of MDM2. To verify this conclusion, we examined the time-dependent decay of NS after MPA treatment in these two cell lines. Again, endogenous NS decreased in a similar time course in both p53−/− and p53−/−/MDM2−/− MEF cells upon MPA treatment (Fig. 4C), indicating that MPA-induced NS degradation is MDM2-independent. Consistent with these results, NS levels were still drastically reduced by MPA, doxorubicin, or actinomycin D, all of which were reported recently to decrease NS levels as well (27), even when endogenous MDM2 levels were considerably low in p53-null HCT116 cells (Fig. 4D, compare left and right panels). Together, these results strongly demonstrate that the polyubiquitination and MPA-induced proteasomal degradation of NS are independent of MDM2.
FIGURE 4.
MPA induces NS degradation independently of MDM2. A, NS polyubiquitination occurs independently of MDM2. p53−/−/MDM2−/− MEF cells were transfected with or without a His-ubiquitin (Ub) expression plasmid. Twenty-fours hours after transfection, cells were treated with 10 μm MG132 for 6 h. Cell lysates were subjected to His-nickel bead pulldown assay, followed by SDS-PAGE and Western blotting to detect polyubiquitinated NS species and input. B, NS degradation can be rescued by MG132 independently of MDM2. p53−/− or p53−/−/MDM2−/− MEF cells were treated with 10 μm MG132 for 8 h and harvested, followed by SDS-PAGE and Western blotting. C, MPA induces NS degradation independently of MDM2. p53−/− or p53−/−/MDM2−/− MEF cells were treated with 40 μm MPA and harvested at the indicated time points. Cell lysates were analyzed by SDS-PAGE and Western blotting with the indicated antibodies. D, MDM2-independent decrease in NS levels upon treatment with various reagents. p53+/+ and p53−/− HCT116 cells were treated with Me2SO as a control (Cont), 5 nm actinomycin D (ActD), 40 μm MPA, 1 mm doxorubicin (Dox), and 10 μm etoposide (Etop) for 12 h and harvested for Western blot analyses using the indicated antibodies.
DISCUSSION
The GTP-binding activity of NS is crucial for regulating the stability and nuclear localization of NS (14, 28). However, it has been unclear how the fluctuations of intracellular GTP levels may potentially regulate NS protein stability or function. In this study, we found that the shuttling of NS from the nucleolus to the nucleoplasm in response to GTP depletion or disruption of the GTP-binding motif results in NS degradation in a ubiquitin-independent manner.
How would the shuttling of the nucleolar protein from one nuclear compartment to the other facilitate the degradation of NS? One explanation would be that NS is maintained in a stable conformation in the nucleolus. In a previous study (2), biochemical purification of NS from HeLa cells revealed that the 70-kDa nucleolar protein exists in a 700-kDa large complex consisting of a multitude of ribosomal proteins, pre-RNA-processing factors, and other cofactors. NS itself is a rather stable protein with an extended half-life of 12 h in cancer cells (Fig. 2) and normal MEF cells (data not shown). It is possible that the association of NS with a multitude of NS-interacting partners or the GTP binding of NS itself may stabilize this GTP-binding protein within the nucleolus by blocking its interaction with the proteasome. Alternatively, the dispersion of NS into the nucleoplasm through the depletion or hydrolysis of GTP itself may promote NS interaction with the proteasome by changing the nuclear compartmentalization of the protein. At least two pieces of evidence support this hypothesis. First, depletion of GTP by MPA treatment led to the reduction of only the NS (but not B23) level, as shown by both Western blotting (Fig. 1, A and B) and fluorescence images (Fig. 1C). Consistent with this observation, NS was hardly detected in the nucleoplasm, whereas B23 was dispersed in this cellular compartment upon MPA treatment (Fig. 1C), suggesting that NS might undergo a quick proteasomal turnover once localized from the nucleolus to the nucleoplasm, whereas B23 does not. In line with these results, the NS mutant that is defective in GTP binding and excluded from the nucleolus (15) is clearly a short-lived protein, and its staining in the nucleoplasm was also drastically reduced (Fig. 2). This is probably why either depletion of endogenous GTP by MPA or mutation of the GTP-binding sites of NS leads to drastic degradation of NS.
What protein may mediate the proteasomal turnover of NS triggered by loss of its GTP-binding activity due to either GTP depletion or mutations at its GTP-binding site? MDM2 was previously suggested to potentially do the job (14), as ectopic expression of MDM2 can reduce the levels of exogenous and endogenous NS. However, siRNA knockdown of MDM2 or C464A mutation in the Ring domain of MDM2 (critical for its E3 ligase activity) only partially rescues NS levels in cells (14). Thus, we were motivated to determine whether MDM2 is required for NS degradation induced by MPA by using p53/MDM2 double knock-out MEF cells, which do not express endogenous MDM2. To our surprise, we found that MDM2 was not necessary for either NS polyubiquitination (Fig. 4A) or GTP depletion-induced NS degradation (Fig. 4, B--D). Our study demonstrates that MPA-induced NS proteasomal turnover is not only ubiquitin-independent but also MDM2-independent, although it remains possible that MDM2 might regulate NS turnover under a specific condition or in a specific cell type. Our data also suggest that there are other yet unidentified factors or proteins that may mediate the proteasomal degradation and ubiquitination of NS.
Our study presents the first piece of evidence demonstrating that NS can be polyubiquitinated in cells. Surprisingly, this ubiquitination does not appear to contribute to the proteasomal turnover of NS, as NS polyubiquitination does not require ubiquitin Lys-48, and NS proteasomal degradation is E1-independent (Fig. 3). These data also suggest that this type of post-translational modification may regulate the function, instead of stability, of NS (24, 25). How would this modification contribute to the regulation of NS function? It is possible the polyubiquitination forms a scaffold in which both ribosomal proteins and pre-RNA-processing factors can bind to form a stable functional complex. Alternatively, polyubiquitination may influence NS activity by recruiting other proteins, which may post-translationally modify NS. Also, polyubiquitination of NS may modulate its role in mediating pre-RNA processing and ribosomal biogenesis, such as by influencing its interaction with known pre-RNA-processing factors, including DDX21, EBP2, and Pes1 (2), or affect downstream ribosomal assembly and protein synthesis. The identification of a NS-specific E3 ligase would certainly facilitate the characterization of these possible regulations. Although mapping the ubiquitination target lysine residues within NS would be informative for this study, it would be quite challenging to mutate single lysine residues, as there are >60 lysines in NS. Deletion mutation was not fruitful either because we could detect NS ubiquitination only when its intact form was expressed in cells, suggesting that direct binding of an unknown cellular E3 ubiquitin ligase to NS is essential for executing its activity toward NS. More intensive endeavors will be necessary to decipher the role of ubiquitination in regulating NS function.
Acknowledgments
We thank Qi Zhang for technique advice and help and members of the Lu laboratory for active discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants CA095441, CA079721, and CA129828 from NCI (to H. L.).
- NS
- nucleostemin
- MEF
- mouse embryonic fibroblast
- MPA
- mycophenolic acid.
REFERENCES
- 1. Boulon S., Westman B. J., Hutten S., Boisvert F. M., Lamond A. I. (2010) The nucleolus under stress. Mol. Cell 40, 216–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Romanova L., Grand A., Zhang L., Rayner S., Katoku-Kikyo N., Kellner S., Kikyo N. (2009) Critical role of nucleostemin in pre-rRNA processing. J. Biol. Chem. 284, 4968–4977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tsai R. Y., McKay R. D. (2002) A nucleolar mechanism controlling cell proliferation in stem cells and cancer cells. Genes Dev. 16, 2991–3003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dai M. S., Sun X. X., Lu H. (2008) Aberrant expression of nucleostemin activates p53 and induces cell cycle arrest via inhibition of MDM2. Mol. Cell. Biol. 28, 4365–4376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Beekman C., Nichane M., De Clercq S., Maetens M., Floss T., Wurst W., Bellefroid E., Marine J. C. (2006) Evolutionarily conserved role of nucleostemin: controlling proliferation of stem/progenitor cells during early vertebrate development. Mol. Cell. Biol. 26, 9291–9301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Raver-Shapira N., Marciano E., Meiri E., Spector Y., Rosenfeld N., Moskovits N., Bentwich Z., Oren M. (2007) Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol. Cell 26, 731–743 [DOI] [PubMed] [Google Scholar]
- 7. He L., He X., Lim L. P., de Stanchina E., Xuan Z., Liang Y., Xue W., Zender L., Magnus J., Ridzon D., Jackson A. L., Linsley P. S., Chen C., Lowe S. W., Cleary M. A., Hannon G. J. (2007) A microRNA component of the p53 tumor suppressor network. Nature 447, 1130–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Haupt Y., Maya R., Kazaz A., Oren M. (1997) Mdm2 promotes the rapid degradation of p53. Nature 387, 296–299 [DOI] [PubMed] [Google Scholar]
- 9. Kubbutat M. H., Jones S. N., Vousden K. H. (1997) Regulation of p53 stability by Mdm2. Nature 387, 299–303 [DOI] [PubMed] [Google Scholar]
- 10. Honda R., Tanaka H., Yasuda H. (1997) Oncoprotein MDM2 is a ubiquitin ligase E3 for tumor suppressor p53. FEBS Lett. 420, 25–27 [DOI] [PubMed] [Google Scholar]
- 11. Meng L., Lin T., Tsai R. Y. (2008) Nucleoplasmic mobilization of nucleostemin stabilizes MDM2 and promotes G2-M progression and cell survival. J. Cell Sci. 121, 4037–4046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lo D., Lu H. (2010) Nucleostemin: another nucleolar “Twister” of the p53-MDM2 loop. Cell Cycle 9, 3227–3232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Meng L., Zhu Q., Tsai R. Y. (2007) Nucleolar trafficking of nucleostemin family proteins: common versus protein-specific mechanisms. Mol. Cell. Biol. 27, 8670–8682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huang M., Itahana K., Zhang Y., Mitchell B. S. (2009) Depletion of guanine nucleotides leads to the Mdm2-dependent proteasomal degradation of nucleostemin. Cancer Res. 69, 3004–3012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tsai R. Y., McKay R. D. (2005) A multistep, GTP-driven mechanism controlling the dynamic cycling of nucleostemin. J. Cell Biol. 168, 179–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stewart D. P., Koss B., Bathina M., Perciavalle R. M., Bisanz K., Opferman J. T. (2010) Ubiquitin-independent degradation of anti-apoptotic MCL-1. Mol. Cell. Biol. 30, 3099–3110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jin Y., Lee H., Zeng S. X., Dai M. S., Lu H. (2003) MDM2 promotes p21waf1/cip1 proteasomal turnover independently of ubiquitylation. EMBO J. 22, 6365–6377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chowdary D. R., Dermody J. J., Jha K. K., Ozer H. L. (1994) Accumulation of p53 in a mutant cell line defective in the ubiquitin pathway. Mol. Cell. Biol. 14, 1997–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Salghetti S. E., Kim S. Y., Tansey W. P. (1999) Destruction of Myc by ubiquitin-mediated proteolysis: cancer-associated and transforming mutations stabilize Myc. EMBO J. 18, 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dai M. S., Zeng S. X., Jin Y., Sun X. X., David L., Lu H. (2004) Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol. Cell. Biol. 24, 7654–7668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Huang M., Ji Y., Itahana K., Zhang Y., Mitchell B. (2008) Guanine nucleotide depletion inhibits preribosomal RNA synthesis and causes nucleolar disruption. Leuk. Res. 32, 131–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lowe J. K., Brox L., Henderson J. F. (1977) Consequences of inhibition of guanine nucleotide synthesis by mycophenolic acid and Virazole. Cancer Res. 37, 736–743 [PubMed] [Google Scholar]
- 23. Sun X. X., Dai M. S., Lu H. (2008) Mycophenolic acid activation of p53 requires ribosomal proteins L5 and L11. J. Biol. Chem. 283, 12387–12392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Haglund K., Dikic I. (2005) Ubiquitylation and cell signaling. EMBO J. 24, 3353–3359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Yang W. L., Zhang X., Lin H.K. (2010) Emerging role of Lys-63 ubiquitination in protein kinase and phosphatase activation and cancer development. Oncogene 29, 4493–4503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zwolinska A. K., Heagle Whiting A., Beekman C., Sedivy J. M., Marine J. C. (2011) Oncogene 10.1038/onc.2011.507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Siddiqi S., Gude N., Hosoda T., Muraski J., Rubio M., Emmanuel G., Fransioli J., Vitale S., Parolin C., D'Amario D., Schaefer E., Kajstura J., Leri A., Anversa P., Sussman M. A. (2008) Myocardial induction of nucleostemin in response to postnatal growth and pathological challenge. Circ. Res. 103, 89–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Meng L., Yasumoto H., Tsai R. Y. (2006) Multiple controls regulate nucleostemin partitioning between nucleolus and nucleoplasm. J. Cell Sci. 119, 5124–5136 [DOI] [PMC free article] [PubMed] [Google Scholar]