Background: Spire is a WH2 domain-containing protein implicated in actin nucleation and critical to oogenesis.
Results: Spire rapidly depolymerizes actin filaments by combining monomer sequestration with weak filament severing, and it nucleates new filaments.
Conclusion: This shows functional and structural variations among actin complexes with Spire.
Significance: Spire-actin structures and actin remodeling by Spir are more complex than originally imagined.
Keywords: Actin, Biophysics, Cytoskeleton, Fluorescence, Microscopy, Spir, WH2, Wasp Homology 2
Abstract
Spire is a WH2 domain-containing actin nucleator essential for establishing an actin mesh during oogenesis. In vitro, in addition to nucleating filaments, Spire can sever them and sequester actin monomers. Understanding how Spire is capable of these disparate functions and which are physiologically relevant is an important goal. To study severing, we examined the effect of Drosophila Spire on preformed filaments in bulk and single filament assays. We observed rapid depolymerization of actin filaments by Spire, which we conclude is largely due to its sequestration activity and enhanced by its weak severing activity. We also studied the solution and crystal structures of Spire-actin complexes. We find structural and functional differences between constructs containing four WH2 domains (Spir-ABCD) and two WH2 domains (Spir-CD) that may provide insight into the mechanisms of nucleation and sequestration. Intriguingly, we observed lateral interactions between actin monomers associated with Spir-ABCD, suggesting that the structures built by these four tandem WH2 domains are more complex than originally imagined. Finally, we propose that Spire-actin mixtures contain both nuclei and sequestration structures.
Introduction
Rapid remodeling of the actin cytoskeleton, involving regulation of its formation and depolymerization, is essential for many cellular processes including motility, cytokinesis, and endocytosis. Actin nucleators are factors that initiate formation of new filaments. To date, three classes of nucleators have been identified: the Arp2/3 complex, the formins, and Wasp homology 2 (WH2)2 nucleators. As the name implies, the third class uses actin-monomer-binding WH2 domains to nucleate. Spire (Spir), JMY, Cordon bleu (Cobl), Leimodin (Lmod), and the bacterial proteins VopF/L/N are grouped together in this class of nucleators, although they are likely to act by distinct mechanisms (1–7). At least two of these proteins, Spir and Cobl, were found to sever filaments and sequester monomers in addition to nucleating new filaments (8, 9). This is not implausible because two other proteins, cofilin and gelsolin, can both sever and nucleate, at least in vitro (10). For Spir and Cobl, it remains to be determined which of these activities are dominant in vivo and/or when the different activities are physiologically relevant. Here we focus on Spir in an effort to better understand how it modulates the actin cytoskeleton.
Spir plays a role in early development of metazoans (11–13). The spir locus was first identified as a Drosophila maternal effect gene essential to establishment of both the anterior/posterior and dorsal/ventral body axes in developing oocytes and embryos (11). Recently a role for the mammalian orthologs, Spir1 and Spir2, in oogenesis was also described (13). An actin mesh that traverses the Drosophila ooctye is absent when the spir gene is mutated; a similar mesh is absent in mouse ooctyes in which both Spir1 and Spir2 are knocked down by RNAi (13–15). These data demonstrate that Spir plays a role in building this mesh, suggesting its in vivo role as a nucleator. In principle, Spir could also enhance polymerization by severing filaments, thereby increasing the concentration of barbed ends available to elongate.
Spir has four tandem WH2 domains. Constructs containing all four of these domains (the N-terminal half of Spir (Spir-NT) or just the cluster of WH2 domains (Spir-ABCD)) have equivalent, maximal nucleation activity. A construct that contains only the two C-terminal WH2 domains (Spir-CD) is sufficient to nucleate, although at a slower rate (1). Models of the nucleation mechanism vary in detail. A common theme is the idea that the closely spaced WH2 domains bind actin monomers in an elongated structure, as observed by electron microscopy and detected by analytical ultracentrifugation (1, 8). However, the orientation and rigidity of adjacent monomers with respect to each other before elongation begins is an open question. Bosch et al. (8) report that the N-terminal half of human Spir1 (hSpir1) binds actin cooperatively, forming a stable complex with four actin monomers (SA4). They conclude that this structure is not a nucleus of actin filaments; instead it may be a sequesteration complex. Ducka et al. (16) co-crystallized Spir with actin. They observed that the last WH2 domain (Spir-D) binds actin in a conformation that closely resembles other WH2 domains (17), but information about the other WH2 domains and the linkers is absent in their structures. Because of the absence of linkers and the lack of correspondence between WH2 domains and actin monomers in the unit cells, information from these structures about the orientation of actins bound to adjacent WH2 domains is speculative at best. In a crystal of longitudinal actin dimers bound to tethered N-Wasp WH2 domains, the actin monomers are rotated, with respect to each other, ∼30° more than adjacent monomers in a filament (18). Rebowski et al. (18) interpret this structure as an explanation for weak nucleation activity by Spir. Although this may be true, it must be noted that the sequence between WH2 domains, as well as the specific WH2 domains in Spir, contribute significantly to the nucleation activity (1, 6), making it unclear how relevant this crystal structure is to understanding Spir. Thus, more work is required to understand how Spir associates with actin monomers and nucleates filaments.
To study nucleation and severing by Spir, we analyzed the effects of Drosophila melanogaster (Dm) Spir on actin monomers and preformed filaments. We found that Dm Spir induces rapid depolymerization of actin filaments. We confirmed that Spir severs filaments, as reported for hSpir1 (8). However, we found that the severing activity of Spir is weak and conclude that rapid depolymerization is largely due to the sequestering activity of Spir. Polymerization assays show that Spir binds actin in structures that accelerate polymerization but suggest that the mixture is heterogeneous. Notably, velocity sedimentation data show that Dm Spir does not bind readily four actin monomers in a stable complex as described for hSpir1 (8) and confirm that Spir-actin solutions contain a mix of structures. Finally, our data demonstrate differences between Spir-ABCD-actin and Spir-CD-actin complexes, including evidence for Spir-dependent lateral actin interactions in addition to the expected longitudinal structures. Together our data suggest that Spir-actin interactions are more complex than originally believed.
EXPERIMENTAL PROCEDURES
Proteins
All proteins were purified and labeled according to standard published procedures. Details and minor modifications are given in the supplemental methods.
Fluorescence Measurements
Pyrene-actin polymerization and depolymerization assays were performed at 20 °C, using 5 μm actin (2.5% labeled) in 50 mm KCl, 1 mm MgCl2, 1 mm EGTA, 10 mm Hepes, pH 7.0, 0.2 mm ATP unless otherwise noted. Pyrene excimer emission was measured with 5 μm G-actin in the presence of stoichiometric concentrations of Spir. Salts (50 mm KCl and 2 mm MgCl2) were then added to the cuvette and incubated for 15 min before measuring maximum emission under F-actin conditions. Further details are given in the text and the supplemental methods.
Total Internal Reflection Fluorescence Microscopy
Direct visualization of filaments was performed as previously described, with some modifications (19, 20). All proteins were diluted in total internal reflection fluorescence (TIRF) buffer: 50 mm KCl, 1 mm MgCl2, 1 mm EGTA, 10 mm Hepes, pH 7.0, 0.2 mm ATP, 100 mm DTT, 15 mm glucose, 20 μg/ml catalase, and 100 μg/ml glucose oxidase. Either a G-actin mixture or F-actin was labeled with 20% Cy3b-maleimide and 1% biotin-maleimide unless otherwise noted. Filaments were immobilized with streptavidin cross-links to biotin-PEG. See the supplemental methods for further detail.
Crystallization and Structure Determination
Actin was treated with the protease ECP32/grimelysin (ECP) at a 6:1 molar ratio to prevent polymerization. Cleaved actin was mixed with Spir-CD at a 1:1 molar ratio. Crystallography conditions are in the supplemental methods. Data collection and refinement statistics are reported in supplemental Table S1. The coordinates of the final model and the merged structure factors have been deposited in the Protein Data Bank under code 3UE5.
RESULTS
Spir Induces Rapid Depolymerization of Actin Filaments
In this paper we use two constructs of Dm Spir: Spir-ABCD, which has four WH2 domains, and Spir-CD, which has two. Throughout the paper we consider ratios of Spir to actin in terms of the number of WH2 domains in the construct. Thus, we refer to 1 mol of Spir-ABCD:4 mol of actin and 1 mol of Spir-CD:2 mol of actin as stoichiometric concentrations. Other ratios are defined when used.
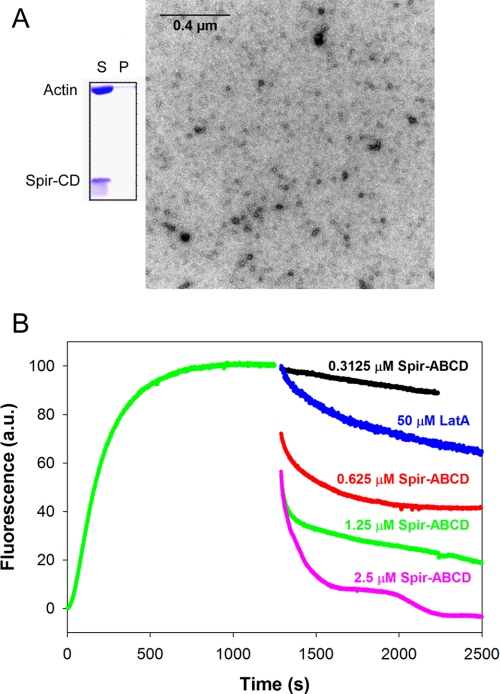
We first observed Spir-induced loss of actin filaments in co-sedimentation assays. We added stoichiometric Spir-CD to actin filaments, centrifuged the solution, and analyzed the supernatant and pellet by SDS-PAGE. All of the Spir and actin was in the supernatant (Fig. 1A). We then measured the time-dependent effect of adding a range of concentrations of Spir-ABCD or Spir-CD to freshly polymerized actin in pyrene-actin assembly assays. At stoichiometric ratios, both Spir-ABCD and Spir-CD depolymerized over 50% of the filaments within 20 s (Fig. 1B and supplemental Fig. S1A). As expected from the loss of pyrene fluorescence, very few filaments could be detected after 15 min when visualized by EM (Fig. 1A). We compared the effect of Spir on F-actin with that of latrunculin A (LatA), a known sequestering agent. At a 10:1 molar ratio to actin, LatA induced depolymerization of F-actin but at a slower rate than Spir (Fig. 1B and supplemental Fig. S1A), suggesting that Spir may be acting by a distinct mechanism. At lower ratios of Spir to actin, depolymerization was still rapid, but the higher plateau of the pyrene fluorescence curves indicated that some filaments remained in solution. At higher ratios (1:1 and higher) of WH2 domains to actin, we observed oscillations in the pyrene signal (see especially for 2.5 μm Spir-CD in supplemental Fig. S1A). We also see oscillations when using light scattering instead of pyrene to detect polymer (supplemental Fig. S1B). This behavior is reminiscent of the “ringing” described for Cobl (9) and may reflect the dual activities of Spir: nucleation and depolymerization, competing under certain conditions.
FIGURE 1.
Rapid actin filament depolymerization by Spir. A, a co-sedimentation assay of 6 μm Spir-CD added to 6 μm F-actin. Spir-CD can be seen in the supernatant (S) (but not in the pellet (P)) with actin that has mostly depolymerized after a 15-min incubation. Representative EM image of actin incubated with a stoichiometric amount of Spir-CD for 15 min. Some short filaments remain, but the majority of the fields imaged have no F-actin. B, fluorescence curves of 5 μm F-actin (2.5% pyrene labeled actin) upon the addition of Spir-ABCD (concentrations as indicated) show rapid depolymerization. The actin polymerization was monitored for 1200 s before Spir was added. Depolymerization is dose-dependent and is complete above stoichiometric Spir concentrations. Filaments exhibit faster depolymerization in the presence of Spir than in the presence of sequestration agent, LatA.
Severing by Drosophila Spir
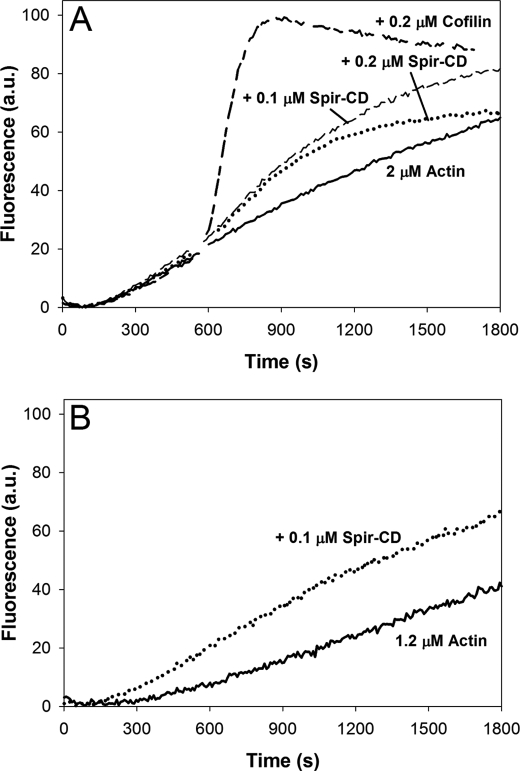
Bosch et al. (8) reported that Spir severs filaments, prompting us to ask whether severing causes the rapid depolymerization observed in this study. To do so, we compared the strength of severing by Spir with that of cofilin. We first tested the effect of adding Spir-CD or cofilin to actin, midpolymerization. Under these conditions cofilin severs filaments, thereby creating new barbed ends and strongly increasing the polymerization rate of actin (21). Indeed, the addition of 0.2 μm cofilin to 2.5 μm actin ∼9 min after polymerization was initiated results in a dramatic increase in the polymerization rate (Fig. 2A). In contrast, the addition of 0.2 μm Spir-CD causes only a modest increase in the polymerization rate (Fig. 2A). Because Spir also nucleates filaments (1, 8), we assessed the contribution of nucleation under these conditions. We calculated that 1.2 μm actin monomer remained at the time that Spir was added and then tested the ability of Spir to nucleate this low concentration of actin. The addition of 0.1 μm Spir causes an increase in polymerization rate and a slight decrease in the lag phase, indicating that Spir can nucleate even at a low actin concentration (Fig. 2B). Thus, nucleation may also contribute to the change seen in our “severing” assay. Taken together, these data suggest that the severing activity of Spir is weak under these conditions.
FIGURE 2.
Spir severs weakly in bulk assays. A, 2.5% pyrene-labeled actin was polymerized for ∼9 min before the addition of either Spir-CD or cofilin. The polymerization rate accelerates strongly after the addition of 0.2 μm cofilin, but only a small rate increase is apparent upon the addition of 0.1 or 0.2 μm Spir-CD. B, weak nucleation is apparent when Spir-CD is added to 1.2 μm G-actin, the amount remaining at the time of addition in A.
The original observation of severing was made with hSpir1 (8), and we were working with Dm Spir. To ascertain whether Dm Spir can in fact sever, we also used EM. Filaments were immobilized on grids and incubated with a 10-fold excess of Spir-CD. After 3 min, the grids were washed and stained with uranyl acetate. We observed fragments of filaments aligned linearly, which suggests that Dm Spir severs actin filaments (supplemental Fig. S2A).
Mechanism of Rapid F-actin Depolymerization
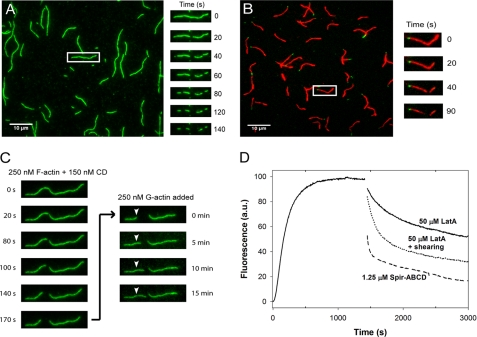
To observe the effect of Spir on actin filaments more directly, we used TIRF microscopy. In each experiment, labeled actin was polymerized and then added to a flow cell. Filaments were held on the surface through biotin-streptavidin bonds. When substoichiometric concentrations of either Spir-ABCD or Spir-CD were added, severing was observed only rarely (data not shown). At approximately stoichiometric concentrations, severing events and depolymerization of actin filaments were both evident (Fig. 3A and supplemental Fig. S2B). When we increased Spir-ABCD or Spir-CD concentrations another 2-fold, filaments disappeared too quickly to distinguish between severing and depolymerization (data not shown). Severing rates at stoichiometric concentrations of Spir-ABCD and Spir-CD were similar (Spir-ABCD: (0.34 ± 0.26) × 10−2 and Spir-CD: (0.14 ± 0.19) × 10−2 cuts/μm/s, n = ∼50 filaments each). The addition of Spir-ABCD and Spir-CD did not affect all filaments. Spir-ABCD did not sever three of the filaments, and Spir-CD did not sever approximately half of the filaments analyzed. Filaments that were not severed were included in the calculations of average severing rates. For comparison, we measured severing rates of yeast cofilin (with yeast actin). When eight times less cofilin was added, the rates were still more than 10 times faster ((4.0 ± 1.4) × 10−2 cuts/μm/s, n = 25), and all of the filaments observed were severed. We also asked whether Spir-ABCD could sever in the presence of actin monomers, to compare with conditions in the pyrene assay. We mixed Spir-ABCD with stoichiometric amounts of G-actin before adding them to filaments in a flow cell (supplemental Fig. S3). Under these conditions, both severing and depolymerization activity were markedly decreased ((0.05 ± 0.11) × 10−2, n = 74; supplemental Fig. S3). Taken together, our results demonstrate that Spir is a weak severer and that this activity alone could not account for the rapid depolymerization we observe.
FIGURE 3.
Spir severs and depolymerizes actin filaments from the barbed end. A, Cy3b-labeled actin filaments were diluted to 200 nm in TIRF buffer before immobilization in flow cells. Time lapse images were recorded for 3 min immediately after 100 nm Spir-ABCD was added. Severing events were observed, and filaments depolymerized faster at one end. Images at 20-s intervals are shown for one filament. B, polarity marked filaments are grown off of AlexaFluor488-phalloidin-stabilized seeds. The addition of 100 nm Spir-ABCD induced both severing and depolymerization of the barbed ends. C, typical Cy3b-labeled actin filament. Time course of filament changes after the addition of 150 nm Spir-CD. Phalloidin was added after 180 s to stabilize existing filaments, and then 250 nm labeled G-actin was added. Growth at one side of the cut site and one end of the filament confirms the polarity of the filament. D, to mimic the effect of severing combined with sequestering, we mechanically sheared 5 μm F-actin by pipetting several times and then adding LatA. The combination of these treatments led to depolymerization that was much faster than the addition of LatA alone. The addition of stoichiometric Spir-ABCD is shown for comparison.
Filament shortening was observed when Spir (ABCD or CD) was added in TIRF experiments. Shortening was also observed when just the buffer was exchanged in the flow chamber as a control. In such control experiments, approximately one-quarter of the filaments shrank, whereas virtually every filament was affected when Spir-ABCD was added. In both cases, shortening occurred predominantly from one end of each filament. We measured depolymerization rates from each end and found that one end shortened ∼15 times faster than the other, consistent with differences in the kinetics of barbed and pointed ends of actin filaments (supplemental Fig. S2C). To confirm that the rapidly shortening end was the barbed end of the filament, we made polarity marked filaments in the flow chamber (see the supplemental methods). Upon addition of Spir-ABCD, we observed rapid depolymerization at the barbed ends as expected (Fig. 3B). Eventually, only the phalloidin-stabilized actin seeds remained, indicating that Spir is not able to depolymerize or sever phalloidin-stabilized filaments (data not shown). Two lines of evidence suggest that stabilizing the pointed ends did not bias our observation. First, polarity marked filaments that were severed also depolymerized predominantly from their barbed ends at the cut point. Second, we confirmed that the shortening end was the barbed end in unmarked filaments. To do so, Spir was added to filaments in a flow chamber for 2 min. Then phalloidin was added to the chamber to halt the reaction. Next, 250 nm labeled G-actin was added, and the growth of filaments was monitored. New growth extended from the depolymerized ends, confirming that barbed end depolymerization was dominant in our assay (Fig. 3C and supplemental Fig. S2D).
The combination of TIRF experiments and bulk assays suggests that depolymerization, whether by sequestration or active depolymerization, is the major activity causing rapid loss of filaments. This activity is enhanced by weak severing producing additional barbed ends for depolymerization under these conditions. To test this hypothesis, we returned to the pyrene assay. This time, when adding LatA, we mixed the solution by pipetting several times with the expectation that we would shear actin filaments, mimicking the effect of a severer. Indeed, the depolymerization rate was much greater with pipetting compared with adding LatA with gentle mixing (Fig. 3D). This observation demonstrates that synergy between sequestering and severing results in rapid filament disassembly and supports our hypothesis that Spir is acting by combining these mechanisms.
Spir-Actin Complexes Include Nuclei
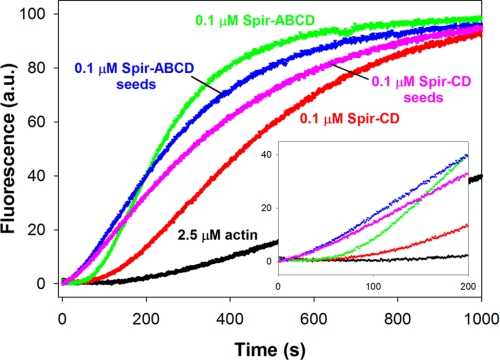
To better understand how Spir can nucleate, sever, depolymerize, and/or sequester actin, we examined the arrangement of actin monomers in Spir-actin complexes. We first mixed stoichiometric concentrations of Spir (ABCD or CD) with actin under polymerizing conditions (50 mm KCl and 1 mm MgCl2) and then added this solution to monomeric actin (2.5% pyrene-actin), such that the final concentrations were 0.1 μm Spir and 2.5 μm actin. We observed polymerization with virtually no lag time (Fig. 4), suggesting that small seeds or nuclei were present in this Spir-actin mixture. This observation was surprising based on findings with hSpir1 (8). That study reported cooperative binding of four actin monomers to Spir and a stable structure described as a sequestration complex, as opposed to a nucleus. For comparison, we added 0.1 μm Spir (ABCD or CD) to 2.5 μm actin and monitored these standard polymerization reactions. As expected, in these cases there was a detectable acceleration of actin nucleation, with the lag time reduced to ∼50 s (versus ∼100 s for actin alone) and polymerization rates faster than for actin alone. To further compare these five conditions, we calculated the concentration of barbed ends at half-maximal polymerization. In all cases where Spir was present, there were at least four times more barbed ends compared with spontaneous polymerization, consistent with improved nucleation of actin leading to a greater number of filaments. However, because polymerization rates are not explosive when we add either Spir-actin complexes or just Spir to monomeric actin, we assume that there is a mixture of species present in the solution. Again, based on the barbed end concentration at half-maximal polymerization, we estimate that ∼1/100th of the Spir molecules form nuclei and speculate that many of the remaining molecules bind actin in sequestration structures based on our earlier EM studies (1) and findings with hSpir1 (8).
FIGURE 4.
Spir-ABCD and Spir-CD seeds enhance actin polymerization. Spir-ABCD and Spir-CD seeds, created by incubating stoichiometric ratios of Spir and actin in 2 mm MgCl2 and 50 mm KCl (blue and magenta traces), exhibit faster nucleation of filaments as indicated by the shorter lag times compared with actin alone (black) and actin mixed with either Spir-ABCD or Spir-CD at time 0 (green and red).
Solution Structure of Spir-Actin Complexes
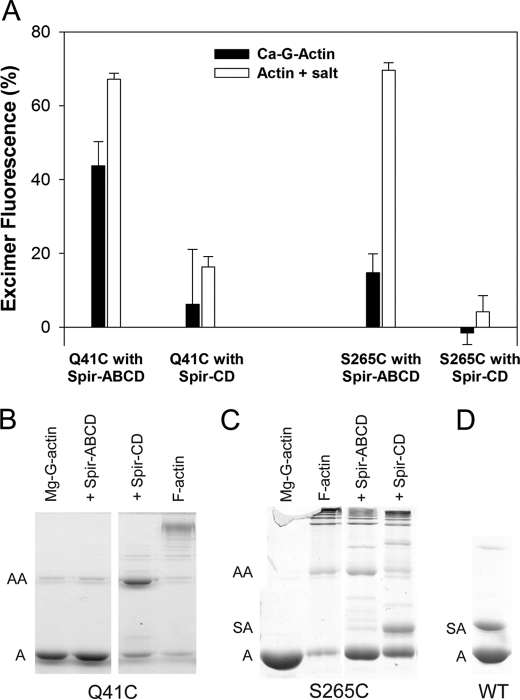
To examine the Spir-actin complexes in solution, we used spectroscopy, cross-linking, and analytical ultracentrifugation. We used yeast actin mutants to examine both longitudinal and lateral contacts between actin monomers associated with Spir. To study longitudinal contacts, we labeled the yeast actin mutant Q41C with pyrene (at Cys-41 and Cys-374). Residue 41 is within the DNase I binding loop, and pyrene probes bound to Cys-41 and Cys-374 from adjacent longitudinal monomers in F-actin will stack, producing excimer emission at 470 nm (22). We monitored excimer emission in the presence of stoichiometric concentrations of Spir-ABCD or Spir-CD under nonpolymerizing (in G buffer) and polymerizing (added MgCl2 and KCl) conditions. If four actin monomers bind Spir-ABCD, or two bind Spir-CD, in a filament-like configuration, we expect the amplitude of excimer fluorescence to be ∼75% or ∼50% that of polymerized actin, respectively. Excimer emission of stoichiometric actin and Spir-ABCD in G buffer was 44 ± 6% that of polymerized actin (Fig. 5A). The addition of salts increased the excimer emission (to 67 ± 2%), indicating that the orientation and/or affinity of actin and Spir are sensitive to solvent conditions. Increased excimer emission in the presence of salts was also observed for Spir-CD, but the excimer signal was much lower than could be expected for stoichiometric actin binding in a filament-like configuration (16 ± 3% versus 50%; Fig. 5A). The signal is also much lower than the expected two-thirds of that with Spir-ABCD, indicating structural differences in monomer stacking between Spir-ABCD-actin and Spir-CD-actin complexes. The addition of an excess of actin did not change the excimer emission levels, suggesting that the lower levels were not caused by insufficient actin binding and indicating that further structural changes were unlikely (data not shown).
FIGURE 5.
Longitudinal and lateral actin contacts induced by Spir. A, yeast actin mutants Q41C and S265C incubated with Spir-ABCD or Spir-CD were monitored for excimer formation under nonpolymerizing and polymerizing conditions. Excimer fluorescence is plotted relative to that of the corresponding F-actin controls (100%). Stoichiometric concentrations of Spir-ABCD and Q41C showed excimer fluorescence under nonpolymerizing conditions. Minimal excimer formation was visible in all other Ca-G-actin cases. Under polymerizing conditions, Spir-ABCD incubated with either Q41C or S265C produced high excimer fluorescence. B, cross-linking of Q41C in the presence of Spir-ABCD or Spir-CD is shown. 5 μm yeast actin mutant Q41C converted to Mg-G-actin was incubated with stoichiometric concentrations of either Spir-ABCD or Spir-CD and the cross-linker MTS1. In the presence of Spir-CD, actin dimer formation is strong compared with the Mg-G-actin (with a trace level of cross-linking) and F-actin (cross-linked into higher order oligomers) controls. In contrast, little cross-linking is evident when Q41C is incubated with Spir-ABCD. C, cross-linking of S265C in the presence of Spir-ABCD or Spir-CD. Conditions were the same as in B. Cross-linking with S265C and Spir-ABCD or Spir-CD both result in higher order oligomers. A band between actin monomer and dimer is detected only for Spir-CD and S265C. We interpret this as cross-linking between Spir and actin. D, cross-linking with Spir-CD and wild type yeast actin. A band above the actin monomers confirms that Spir-CD cross-links to actin Cys-374. A indicates molecular weight consistent with an actin monomer. AA indicates an actin dimer. SA indicates Spir-CD-actin.
The interpretation of the above excimer results must be qualified by the assay dependence on the stacking of pyrene probes. Thus, in our case, excimer emission does not report directly and strictly on filament-like positions of the labeled residues. Instead, it reports on their longitudinal or lateral stacking, which requires proximities, within ∼3–18 Å. A useful example is that of cofilin-decorated actin filaments. Cofilin binding enhances excimer emission above that of unbound filaments. However, in contrast to excimer increase, FRET measurements show an increased distance between Cys-41 and Cys-374 probe pairs upon cofilin binding (22). In agreement with FRET results, cofilin inhibits the normal cross-linking of actin monomers via the same cysteines that are used for excimer experiments. With this in mind, we also probed the complexes of Spir with the Q41C actin mutant with cross-linking reagents. As expected, Q41C F-actin alone was cross-linked by MTS-1, a homo-bifunctional cysteine cross-linker, but Mg-G-actin was not (Fig. 5B). When we treated Spir-CD and Q41C with MTS-1, the major species detected was ∼84 kDa, consistent with an actin dimer (Fig. 5B). However, when we added MTS-1 to Q41C in the presence of Spir-ABCD, no cross-linking was observed. Other homo-bifunctional cross-linkers of varying lengths (CuCl2, MTS-3, and MTS-6) produced similar results. These data confirm that the Spir-ABCD-actin structures are distinct from Spir-CD-actin structures and actin filaments. Actin bound to Spir-ABCD is stabilized in a structure that holds the neighboring pyrene rings close enough to produce excimer emission but not close enough to be cross-linked. Thus, the excimer emission is not reflecting precise filament-like configuration in this case. In contrast, the Spir-CD-actin complex is flexible, allowing cross-linkers to capture monomers when they are close to a filament-like configuration, although they are not close enough to each other or oriented correctly on average to give eximer emission levels proportional to Spir-ABCD-actin.
We performed analogous experiments with yeast actin mutated to Cys at Ser-265. Pyrene excimer emission from labeled Cys-265 and Cys-374 reflects lateral proximity within the actin filament (23), which is not expected if Spir-ABCD and Spir-CD bind actin in strictly longitudinal structures (1, 8). Excimer emission was negligible for Spir-CD and actin, regardless of the buffer conditions (Fig. 5A). Surprisingly, we detected high excimer emission when stoichiometric amounts of Spir-ABCD and actin were combined under polymerizing conditions (Fig. 5A). We also performed cross-linking experiments with S265C actin. Consistent with excimer emission data, we observed lateral actin cross-linking in the presence of Spir-ABCD (Fig. 5C). This cross-linking was weak but reproducible (with ∼80% of the actin remaining in the monomer form), confirming lateral contacts between actin monomers in the presence of Spir-ABCD. This result differs from Spir-ABCD+Q41C actin, in which no cross-linking is observed. Perhaps, the lateral contacts approximate filamentous orientations more closely than the longitudinal contacts. As with Q41C actin, S265C actin was cross-linked in the presence of Spir-CD, despite low excimer emission. We also note that, despite low overall cross-linking of lateral actin contacts, higher order oligomers were generated with Spir-CD in contrast to the dimer observed when longitudinal contacts were cross-linked.
We also detected a band between actin monomers and dimers in the Spir-CD/S265 actin cross-linking experiment (Fig. 5C). We interpret this cross-linking as another indication of the flexibility of the Spir-CD-actin structures. To learn more about the orientation of contacts being detected, we performed cross-linking with Spir-CD and wild type actin. We observed a Spir-actin band in this case as well. Because Spir-CD has only one cysteine (Cys-459 within linker 3) we conclude that Spir Cys-459 can be cross-linked to actin through its sole reactive cysteine, Cys-374.
To further examine the solution structure of Spir-actin complexes, we measured their sedimentation velocity coefficients. As before, we combined Spir-ABCD and Spir-CD with stoichiometric concentrations of actin under polymerizing (with MgCl2 and KCl) and nonpolymerizing (in G buffer) conditions. In all cases, the sedimentation coefficient(s) were larger in the presence of salts, consistent with the increase in actin contacts observed under these conditions in the excimer experiments described above (Table 1). In the presence of salts, the observed sedimentation coefficient for Spir-CD and actin (4.65 S) approached the theoretical sedimentation coefficient of an actin dimer (4.94 S). In contrast, the sedimentation coefficient for stoichiometric ratios of actin and Spir-ABCD was less than that of an actin trimer. Asymmetry in the g(s) plot for Spir-ABCD indicates a mixture of species (supplemental Fig. S4). This mixture of species could represent a variable number of actin monomers bound to Spir-ABCD, as well as a mixture of orientations of monomers within the structure. In either case, we do not observe full occupancy of WH2 domains of Spir-ABCD in an elongated structure.
TABLE 1.
Comparison of observed and theoretical sedimentation coefficients (in Svedbergs) of Spir constructs incubated with actin at stoichiometric ratios under polymerizing and nonpolymerizing conditions
The theoretical S values for actin oligomers were calculated as described under “Experimental Procedures” (+ salt indicates 2 mm MgCl2 and 50 mm KCl).
| Observed sedimentation coefficients | S | Theoretical sedimentation coefficients | S |
|---|---|---|---|
| Actin | 3.25 | Actin | 3.25 |
| Actin + Spir-CD | 3.80 | 2 actin prolated ellipsoid | 4.94 |
| Actin + Spir-CD (+ salt) | 4.65 | 3 actin prolated ellipsoid | 6.08 |
| Actin + Spir-ABCD | 5.80 | 3 actin oblate ellipsoid | 6.49 |
| Actin + Spir-ABCD (+salt) | 6.25 | 3 actin spherical | 6.76 |
| 4 actin prolated ellipsoid | 6.93 | ||
| 4 actin oblate ellipsoid | 7.86 | ||
| 4 actin spherical | 8.19 |
Crystal Structure of Spir-Actin Complex
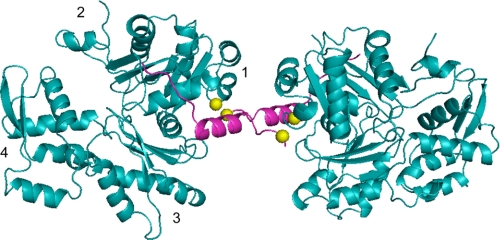
We co-crystallized Spir-CD with nonpolymerizing ECP-cleaved actin (24) to obtain high resolution data about the Spir-actin complex. Crystals were acquired with a starting ratio of 1 Spir-CD:1 actin, and the complex we observed contained only one actin monomer per Spir-CD. At 2.8 Å resolution, we determined that actin was consistently bound to Spir-D (Fig. 6). As expected, the WH2 domains forms an α-helix that binds in the hydrophobic cleft between subdomains 1 and 3. The C-terminal half of WH2-D extends along subdomain 1 toward subdomain 2. The configuration of Spir-D is similar to that in other reported WH2-actin co-crystal structures (e.g. root mean square = 0.472 Å with respect to the WH2 domain of WASP (17)) and similar to the previously published structure of Spir-D-actin (which did not include linker 3 or Spir-C; root mean square = 0.563 Å (16)). Our structure suggests that linker 3 does not fold back to bind the same monomer as Spir-D, instead favoring a conformation in which it is reaching away, leaving Spir-C more able to bind an actin monomer. However, Spir-C and most of the linker between C and D were disordered in the structure and could not be mapped.
FIGURE 6.
Crystal structure of Spir-D (magenta) with ECP-cleaved actin (cyan) forming an anti-parallel dimer. Spir-D is positioned next to the W-loop of actin between subdomains 1 and 3. Subdomains are indicated on the left actin monomer. Part of linker 3 between Spir-C and -D is shown. Spir-C and part of actin subdomain 2 are disordered in the crystal. Actin Cys-374 and Spir Cys-459 (shown as yellow balls) form a disulfide bond, creating an anti-parallel dimer.
In fact pairs of Spir-CD-actin complexes were cross-linked to each other through a disulfide bond between Cys-374 in actin and Cys-459 in Spir (Fig. 6). The cross-linking is most likely driven by crystal packing, capturing the same transient interaction we detected in chemical cross-linking experiments. This structure could reflect a severing mechanism. If linker 3 can insert between subunits of a filament it would destabilize the structure. However, we note that we do not detect significant Spir-actin cross-linking with Spir-ABCD. Thus, we believe that the anti-parallel dimer reflects the flexibility inherent in the association of Spir-CD with actin. Potentially arguing against this is the fact that we do not observe cross-linking between Spir-CD and Q41C actin. Instead, we interpret this as evidence that the Q41C actin dimers form rapidly, preventing significant Spir-actin cross-linking.
DISCUSSION
Spir Severing
Spir can nucleate, sever, and depolymerize filaments in vitro. We found that a construct containing only two WH2 domains, Spir-CD, is sufficient to sever, consistent with recent findings for the WH2 nucleator Cobl (9). As is the case for nucleation, the severing activity of a construct containing four WH2 domains, Spir-ABCD, is more potent than one with two WH2 domains. Some of the difference may be due to the obvious fact that Spir-ABCD has twice as many WH2 domains, but our data show that the constructs actually behave differently when bound to actin. By combining the spectroscopic and cross-linking data with the solution and crystal structure data, we infer that Spir-CD-actin structures are more flexible than Spir-ABCD-actin. The relative stiffness of Spir-ABCD-actin compared with Spir-CD-actin may explain its stronger severing activity.
Both bulk assays and single filament assays show that the severing activity of Spir is quite weak. Under the conditions used here, it is approximately 2 orders of magnitude weaker than the classical severing protein, cofilin. We only observed significant amounts of severing at concentrations of Spir close to stoichiometric with actin, conditions that are unlikely to exist in the cell. Furthermore, severing is even weaker when actin monomers are present. This result explains our earlier observation that Spir does not sever at measurable levels in seeded polymerization assays (1). We therefore conclude that Spir is not a significant in vivo severing factor.
Spir Depolymerization
The severing activity of Spir alone is insufficient to explain the rapid depolymerization observed in bulk assays. TIRF experiments show that filaments also shrink in the presence of Spir (Fig. 3). Depolymerization occurred predominantly at one end of the filament, which we determined is the barbed end. Spir has been shown to sequester actin monomers (1, 8). Because bulk depolymerization rates were significantly higher for Spir than the sequestering agent, LatA, we are intrigued by the possibility that there is a difference in depolymerization mechanisms. However, data from TIRF experiments and pyrene bulk assays do not support this idea. Filaments shorten in control TIRF experiments as fast as they do when Spir is added. The major difference was in the number of filaments shortening, which may be a function of surface interactions and severing. Filaments depolymerize faster in pyrene bulk assays when sheared prior to the addition of LatA. Thus, the enhanced depolymerization rates observed in bulk experiments may reflect how even weak severing activity can markedly accelerate overall depolymerization rates.
Spir-Actin Complexes
Although Spir-CD binds two actin monomers (in the presence of MgCl2 and KCl) as expected, it is noteworthy that Spir-ABCD did not bind four actin monomers (on average) in sedimentation velocity experiments. Tight cooperative binding of four actin monomers was reported for the N-terminal construct of hSpir-1 (8). Perhaps the difference is due to the constructs used. However, Dm Spir-NT and Dm Spir-ABCD have equivalent nucleation activity and the only domain in the N terminus of Spir outside of the WH2 cluster, the KIND domain, does not bind actin (25, 26). Furthermore, earlier EM studies showed little difference between Spir-NT and Spir-ABCD in complex with actin (lengths of 22 ± 4 versus 26 ± 5 nm, respectively; (1)). Likewise, rigid body modeling of small angle x-ray scattering data by Sitar et al. (27) produced little difference between the WH2 domains and actin of Spir-NT and Spir-ABCD.
Although cross-linking of actin in the presence of Spir-CD captures a filament-like orientation, Spir-ABCD brings more actins into close proximity, including some lateral interactions, both of which may make it the stronger nucleator. The absence of strong longitudinal actin cross-linking in the presence of Spir-ABCD suggests that the contacts are not precisely filament-like. Sitar et al. (27) consistently predict elongated structures for Dm Spir bound to actin from small angle x-ray scattering data. They also conclude that these complexes are not strictly filament-like. They do not observe lateral interactions, but our data suggest that they are a small fraction of all interactions. Deviations between their models and measured values are well explained if there is a mixture of species in these experiments.
It is interesting to speculate whether Spir can form distinct complexes with actin: a nucleus and a sequestration complex. Perhaps fully occupied Spir is stable and effectively sequesters actin, as described for the SA4 complex (8). Here we present evidence of alternate, perhaps less stable, Spir-actin complexes with actin making lateral contacts in addition to longitudinal contacts. Such structures are good candidates for filament nucleation. Indeed, we observed accelerated actin polymerization under these same conditions, consistent with nuclei being present. However, the number of filament seeds formed by Spir-actin must be relatively small, judging by the level of actin polymerization acceleration. Together these data suggest that the dominant species observed is not the nucleus, adding to the challenge of understanding how Spir nucleates.
Ducka et al. (16) observed enhanced pyrene fluorescence when at least 4 mol of actin were added per mole of Spir-BCD (3 WH2 domains). They interpreted this as evidence that lateral monomer interactions are necessary to stabilize longitudinal contacts between monomers bound to Spir in a filament-like structure. However, we observed lateral interactions between actin monomers when stoichiometric concentrations were added to Spir-ABCD in multiple experiments, and our sedimentation velocity data suggest a distribution of the number of monomers binding to Spir-ABCD. The pyrene-actin experiments of Ducka et al. (16) must be reconsidered in light of these results. Given the mixture of species that we detect, it is unlikely that the enhancement of pyrene fluorescence that Ducka et al. (16) observe is solely the product of adding a monomer in a lateral position to fully occupied WH2 domains. Instead, increasing concentrations of actin probably lead to greater occupation of WH2 domains and a concomitant change in the pyrene environment.
Concluding Remarks
The importance of the ability of Spir to nucleate and sever needs to be examined further in vivo. Identifying residues that are essential to severing versus nucleation, if possible, would be the ideal way to test which of these activities is essential. Short of this, we argue that nucleation by Spir is likely to be a critical function because it has been shown to play a positive role in the establishment of an actin-based mesh that is essential for Drosophila and mammalian oogenesis (13, 14). Genetic and biochemical evidence indicates that Spir coordinates with the formin Capu (Fmn2 in mammals) (11, 14, 25, 28). Capu binds two molecules of Spir and enhances its nucleation activity. Our preliminary results indicate that Capu does not influence depolymerization or lateral cross-linking by Spir. This suggests that if Spir, in fact, has multiple activities in vivo, regulation by factors other than Capu may play a role.
Finally, it will be important to study other WH2 nucleators with the approaches taken here to learn more about how this disparate class of nucleators works and what specific factors create the differences in their activities. To date, one common theme among this class of nucleators is that WH2 domains alone are not sufficient to create a nucleator. Instead an extra domain, such as the MBL domain of Spir, the K domain, and linker 2 of Cobl and the C-terminal dimerization domain of VopL, works with the WH2 domains (1, 2, 6, 9, 29, 30).
Supplementary Material
Acknowledgments
We thank Corie Ralston for data collection at the Advanced Light Source, Alex Shvetsov for helping with crystal conditions, Dima Kudryashov for initial work on this project, Ron Lin for help with TIRF and slide preparation, and Christina Vizcarra for sharing reagents.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM096133, a Burroughs-Wellcome Fund Career Award in the Biomedical Sciences, and a March of Dimes Foundation, Basil O'Connor Starter Scholar Award (#5-FY10-81) (to M. E. Q). This work was also supported by U.S. Public Health Service Grant GM077190 (to E. R.).

This article contains supplemental text, references, Table S1, and Figs. S1–S4.
The atomic coordinates and structure factors (code 3UE5) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- WH2
- Wasp homology 2
- Dm
- D. melanogaster
- TIRF
- total internal reflection fluorescence
- LatA
- latrunculin A.
REFERENCES
- 1. Quinlan M. E., Heuser J. E., Kerkhoff E., Mullins R. D. (2005) Drosophila Spire is an actin nucleation factor. Nature 433, 382–388 [DOI] [PubMed] [Google Scholar]
- 2. Ahuja R., Pinyol R., Reichenbach N., Custer L., Klingensmith J., Kessels M. M., Qualmann B. (2007) Cordon-Bleu is an actin nucleation factor and controls neuronal morphology. Cell 131, 337–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liverman A. D., Cheng H. C., Trosky J. E., Leung D. W., Yarbrough M. L., Burdette D. L., Rosen M. K., Orth K. (2007) Arp2/3-independent assembly of actin by Vibrio type III effector VopL. Proc. Natl. Acad. Sci. U.S.A. 104, 17117–17122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tam V. C., Serruto D., Dziejman M., Brieher W., Mekalanos J. J. (2007) A type III secretion system in Vibrio cholerae translocates a formin/spire hybrid-like actin nucleator to promote intestinal colonization. Cell Host Microbe 1, 95–107 [DOI] [PubMed] [Google Scholar]
- 5. Chereau D., Boczkowska M., Skwarek-Maruszewska A., Fujiwara I., Hayes D. B., Rebowski G., Lappalainen P., Pollard T. D., Dominguez R. (2008) Leiomodin is an actin filament nucleator in muscle cells. Science 320, 239–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Zuchero J. B., Coutts A. S., Quinlan M. E., Thangue N. B., Mullins R. D. (2009) p53-cofactor JMY is a multifunctional actin nucleation factor. Nat. Cell Biol. 11, 451–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tam V. C., Suzuki M., Coughlin M., Saslowsky D., Biswas K., Lencer W. I., Faruque S. M., Mekalanos J. J. (2010) Functional analysis of VopF activity required for colonization in Vibrio cholerae. mBio 1, e00289–10 doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bosch M., Le K. H., Bugyi B., Correia J. J., Renault L., Carlier M. F. (2007) Analysis of the function of Spire in actin assembly and its synergy with formin and profilin. Mol. Cell 28, 555–568 [DOI] [PubMed] [Google Scholar]
- 9. Husson C., Renault L., Didry D., Pantaloni D., Carlier M. F. (2011) Cordon-Bleu uses WH2 domains as multifunctional dynamizers of actin filament assembly. Mol. Cell 43, 464–477 [DOI] [PubMed] [Google Scholar]
- 10. Bamburg J. R. (1999) Proteins of the ADF/cofilin family. Essential regulators of actin dynamics. Annu. Rev. Cell Dev. Biol. 15, 185–230 [DOI] [PubMed] [Google Scholar]
- 11. Manseau L. J., Schüpbach T. (1989) Cappuccino and Spire. Two unique maternal-effect loci required for both the anteroposterior and dorsoventral patterns of the Drosophila embryo. Genes Dev. 3, 1437–1452 [DOI] [PubMed] [Google Scholar]
- 12. Le Goff C., Laurent V., Le Bon K., Tanguy G., Couturier A., Le Goff X., Le Guellec R. (2006) pEg6, a spire family member, is a maternal gene encoding a vegetally localized mRNA in Xenopus embryos. Biol. Cell 98, 697–708 [DOI] [PubMed] [Google Scholar]
- 13. Pfender S., Kuznetsov V., Pleiser S., Kerkhoff E., Schuh M. (2011) Spire-type actin nucleators cooperate with Formin-2 to drive asymmetric oocyte division. Curr. Biol. 21, 955–960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dahlgaard K., Raposo A. A., Niccoli T., St Johnston D. (2007) Capu and Spire assemble a cytoplasmic actin mesh that maintains microtubule organization in the Drosophila oocyte. Dev. Cell 13, 539–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Schuh M. (2011) An actin-dependent mechanism for long-range vesicle transport. Nat. Cell Biol. 13, 1431–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ducka A. M., Joel P., Popowicz G. M., Trybus K. M., Schleicher M., Noegel A. A., Huber R., Holak T. A., Sitar T. (2010) Structures of actin-bound Wiskott-Aldrich syndrome protein homology 2 (WH2) domains of Spire and the implication for filament nucleation. Proc. Natl. Acad. Sci. U.S.A. 107, 11757–11762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chereau D., Kerff F., Graceffa P., Grabarek Z., Langsetmo K., Dominguez R. (2005) Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly. Proc. Natl. Acad. Sci. U.S.A. 102, 16644–16649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rebowski G., Namgoong S., Boczkowska M., Leavis P. C., Navaza J., Dominguez R. (2010) Structure of a longitudinal actin dimer assembled by tandem W domains. Implications for actin filament nucleation. J. Mol. Biol. 403, 11–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ha T., Rasnik I., Cheng W., Babcock H. P., Gauss G. H., Lohman T. M., Chu S. (2002) Initiation and re-initiation of DNA unwinding by the Escherichia coli Rep helicase. Nature 419, 638–641 [DOI] [PubMed] [Google Scholar]
- 20. Pavlov D., Muhlrad A., Cooper J., Wear M., Reisler E. (2007) Actin filament severing by cofilin. J. Mol. Biol. 365, 1350–1358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chan A. Y., Bailly M., Zebda N., Segall J. E., Condeelis J. S. (2000) Role of cofilin in epidermal growth factor-stimulated actin polymerization and lamellipod protrusion. J. Cell Biol. 148, 531–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bobkov A. A., Muhlrad A., Kokabi K., Vorobiev S., Almo S. C., Reisler E. (2002) Structural effects of cofilin on longitudinal contacts in F-actin. J. Mol. Biol. 323, 739–750 [DOI] [PubMed] [Google Scholar]
- 23. Bobkov A. A., Muhlrad A., Shvetsov A., Benchaar S., Scoville D., Almo S. C., Reisler E. (2004) Cofilin (ADF) affects lateral contacts in F-actin. J. Mol. Biol. 337, 93–104 [DOI] [PubMed] [Google Scholar]
- 24. Khaitlina S. Y., Moraczewska J., Strzelecka-Gołaszewska H. (1993) The actin/actin interactions involving the N-terminus of the DNase-I-binding loop are crucial for stabilization of the actin filament. Eur. J. Biochem. 218, 911–920 [DOI] [PubMed] [Google Scholar]
- 25. Quinlan M. E., Hilgert S., Bedrossian A., Mullins R. D., Kerkhoff E. (2007) Regulatory interactions between two actin nucleators, Spire and Cappuccino. J. Cell Biol. 179, 117–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ito T., Narita A., Hirayama T., Taki M., Iyoshi S., Yamamoto Y., Maéda Y., Oda T. (2011) Human spire interacts with the barbed end of the actin filament. J. Mol. Biol. 408, 18–25 [DOI] [PubMed] [Google Scholar]
- 27. Sitar T., Gallinger J., Ducka A. M., Ikonen T. P., Wohlhoefler M., Schmoller K. M., Bausch A. R., Joel P., Trybus K. M., Noegel A. A., Schleicher M., Huber R., Holak T. A. (2011) Molecular architecture of the Spire-actin nucleus and its implication for actin filament assembly. Proc. Natl. Acad. Sci. U.S.A. 108, 19575–19580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vizcarra C. L., Kreutz B., Rodal A. A., Toms A. V., Lu J., Zheng W., Quinlan M. E., Eck M. J. (2011) Structure and function of the interacting domains of Spire and Fmn-family formins. Proc. Natl. Acad. Sci. U.S.A. 108, 11884–11889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Namgoong S., Boczkowska M., Glista M. J., Winkelman J. D., Rebowski G., Kovar D. R., Dominguez R. (2011) Mechanism of actin filament nucleation by Vibrio VopL and implications for tandem W domain nucleation. Nat. Struct. Mol. Biol. 18, 1060–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yu B., Cheng H. C., Brautigam C. A., Tomchick D. R., Rosen M. K. (2011) Mechanism of actin filament nucleation by the bacterial effector VopL. Nat. Struct. Mol. Biol. 18, 1068–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.