Background: DypB, a Dyp-type peroxidase, oxidizes Mn(II) and transforms lignin.
Results: DypB forms a stable Compound I that rapidly decays to Compound II in the D153A and N246A but is undetectable in the R244L variant.
Conclusion: The requirement of Arg-244 but not Asp-153 to form Compound I indicates that DyPs modulate the peroxidative cycle differently than plant peroxidase.
Significance: Understanding DyPs helps harness their biotechnological potential.
Keywords: Enzyme Kinetics, Enzyme Mechanisms, Lignin Degradation, Peroxidase, X-ray Crystallography, Bacteria, Compound I
Abstract
DypB from Rhodococcus jostii RHA1 is a bacterial dye-decolorizing peroxidase (DyP) that oxidizes lignin and Mn(II). Three residues interact with the iron-bound solvent species in ferric DypB: Asn-246 and the conserved Asp-153 and Arg-244. Substitution of either Asp-153 or Asn-246 with alanine minimally affected the second order rate constant for Compound I formation (k1 ∼ 105 m−1s−1) and the specificity constant (kcat/Km) for H2O2. Even in the D153A/N246A double variant, these values were reduced less than 30-fold. However, these substitutions dramatically reduced the stability of Compound I (t½ ∼ 0.13 s) as compared with the wild-type enzyme (540 s). By contrast, substitution of Arg-244 with leucine abolished the peroxidase activity, and heme iron of the variant showed a pH-dependent transition from high spin (pH 5) to low spin (pH 8.5). Two variants were designed to mimic the plant peroxidase active site: D153H, which was more than an order of magnitude less reactive with H2O2, and N246H, which had no detectable peroxidase activity. X-ray crystallographic studies revealed that structural changes in the variants are confined to the distal heme environment. The data establish an essential role for Arg-244 in Compound I formation in DypB, possibly through charge stabilization and proton transfer. The principle roles of Asp-153 and Asn-246 appear to be in modulating the subsequent reactivity of Compound I. These results expand the range of residues known to catalyze Compound I formation in heme peroxidases.
Introduction
Dyp-type peroxidases (DyPs)2 are a recently identified family of heme proteins present in a wide range of bacteria and fungi (1–4). Based on phylogenetic analysis, DyPs have been classified into four subfamilies: A–C are predominantly bacterial, whereas subfamily D is mostly fungal (2–5). Although DyPs were initially characterized for their ability to degrade anthraquinone dyes (1, 5), the subfamilies have remarkably different specificities for reductive substrates, with some DyPs catalyzing the oxidation of methoxylated aromatic compounds (6) and carotenoids. More recently, DypB from Rhodococcus jostii RHA1 was shown to oxidize lignin and Mn(II) (2, 7). Accordingly, there has been burgeoning interest in developing the biotechnological potential of these enzymes. Nevertheless, the physiological role of these enzymes is unclear, with proposals ranging from bacterial oxidative stress response (8) to virulence factors in plant pathogens (9), iron removal from heme (10), and porphyrinogen oxidation (11). Regardless of their physiological role, the peroxidative catalytic cycle of these enzymes has not been experimentally substantiated.
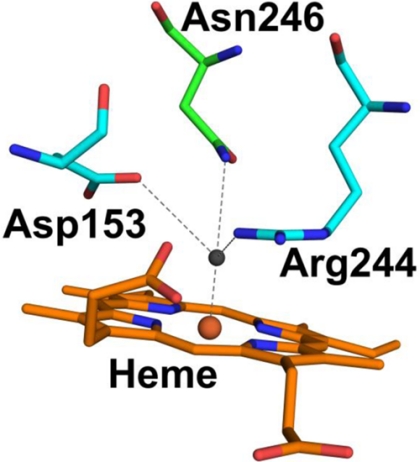
DyPs belong to the CDE (chlorite dismutases, DyPs, EfeB) superfamily (12), whose fold comprises two copies of a ferredoxin-like domain containing two β-sheets sandwiched between two α-helices. The heme is bound in the C-terminal domain (5, 7, 13–15). As in plant peroxidases, a histidine serves as the fifth ligand to the heme iron on the proximal face and is hydrogen-bonded to an acidic residue. The distal heme environment includes two conserved residues, an aspartate and an arginine, numbered 153 and 244 in DypB, respectively. The aspartate forms a hydrogen bond with the distal solvent species and has been proposed to be essential for catalysis, analogous to the distal histidine in plant peroxidases and distal glutamate of structurally distinct chloroperoxidases (5, 7, 13–16). The arginine guanidinium side chain interacts electrostatically with heme propionate A (5, 7, 14, 15). Finally, B-type DyPs also have an asparagine in the distal heme pocket (Asn-246 in DypB of RHA1) that interacts with the distal solvent ligand. In the well characterized D-type DypDec1, this residue is Ser-331 (5).
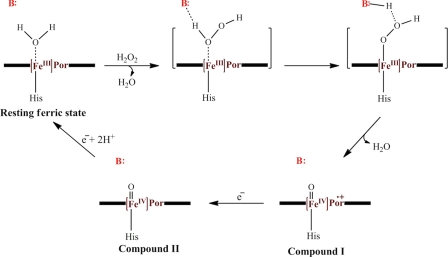
The peroxidative cycle of DyPs has been proposed to be similar to that of plant peroxidases (4, 5) whereby the distal aspartate serves as the base-acid catalyst. Accordingly, this aspartate transfers the proton from the proximal oxygen atom of the Fe(III)-bound H2O2 to the distal oxygen atom, facilitating heterolytic cleavage of the O-O bond to form Compound I, an Fe(IV) O intermediate with an additional oxidizing equivalent delocalized over either the porphyrin ([Fe(IV)
O intermediate with an additional oxidizing equivalent delocalized over either the porphyrin ([Fe(IV) O Por•]+) or an oxidizable amino acid. The distal arginine is proposed to stabilize and polarize the negative charge on the Fe(III)-OOH complex during formation of Compound I. The reaction of Compound I with one equivalent of the reducing substrate yields Compound II, an [Fe(IV)
O Por•]+) or an oxidizable amino acid. The distal arginine is proposed to stabilize and polarize the negative charge on the Fe(III)-OOH complex during formation of Compound I. The reaction of Compound I with one equivalent of the reducing substrate yields Compound II, an [Fe(IV) O] species, which further reacts with a second equivalent of the reducing substrate yielding the resting state Fe(III) peroxidase. Critical aspects of this proposed mechanism remain unsubstantiated, and the few studies to date indicate important differences both with the plant peroxidases and within the four DyP subfamilies. For example, the identity of Compound I in DyPs has not been definitively established, although the electron absorption spectrum (7) and the 35-GHz EPR spectrum3 of the green-colored intermediate are consistent with an [Fe(IV)
O] species, which further reacts with a second equivalent of the reducing substrate yielding the resting state Fe(III) peroxidase. Critical aspects of this proposed mechanism remain unsubstantiated, and the few studies to date indicate important differences both with the plant peroxidases and within the four DyP subfamilies. For example, the identity of Compound I in DyPs has not been definitively established, although the electron absorption spectrum (7) and the 35-GHz EPR spectrum3 of the green-colored intermediate are consistent with an [Fe(IV) OPor•]+ species. Regardless of its identity, B- and D-type DyPs stabilize Compound I (t½ ∼ 9 min), which then decays to the ferric enzyme without detectable Compound II formation (4, 5, 7). Moreover, substitution of the conserved aspartate in DyPDec1 abolished the enzyme peroxidase activity (5). By contrast, the equivalent substitution in EfeB/YcdB from Escherichia coli, an A-type DyP, retained ∼80% of the wild-type peroxidase activity (15), suggesting the residue is not essential for peroxidase activity in all DyPs. Further structure-function studies are required to substantiate the peroxidative catalytic cycle in DyPs and to identify their catalytic determinants.
OPor•]+ species. Regardless of its identity, B- and D-type DyPs stabilize Compound I (t½ ∼ 9 min), which then decays to the ferric enzyme without detectable Compound II formation (4, 5, 7). Moreover, substitution of the conserved aspartate in DyPDec1 abolished the enzyme peroxidase activity (5). By contrast, the equivalent substitution in EfeB/YcdB from Escherichia coli, an A-type DyP, retained ∼80% of the wild-type peroxidase activity (15), suggesting the residue is not essential for peroxidase activity in all DyPs. Further structure-function studies are required to substantiate the peroxidative catalytic cycle in DyPs and to identify their catalytic determinants.
Herein, we investigated the roles of each of the three distal residues (Fig. 1) in the peroxidative cycle of DypB of R. jostii RHA1. Asp-153 and Asn-246 were single- and double-substituted with alanine, whereas Arg-244 was substituted with leucine. In addition, the D153H and N246H variants were constructed to engineer histidine in the active site of DypB to more closely resemble the active site of plant peroxidases. The reaction of the variant proteins with H2O2 was analyzed using steady-state and transient state kinetics. X-ray crystal structures were solved for each of the single variants. The role of these residues in the peroxidative cycle of DypB is discussed.
FIGURE 1.
The distal residues of DypB from R. jostii RHA1. Non-covalent bonds are shown as dashed lines. The atoms are colored as follows: heme carbon, orange; residue carbons, cyan; nitrogen, blue; oxygen, red; iron, brown; solvent species oxygen, gray. The figure was generated using coordinates from PDB ID 3QNR.
EXPERIMENTAL PROCEDURES
Reagents and Chemicals
HPLC grade DMSO was from Alfa Aesar. All other reagents and chemicals were purchased from Sigma, ACROS, MP Bio-medicals, or Fisher and were used without further purification. Water was purified using a Barnstead NANO pure UV apparatus (Barnstead International, Dubuque, IA) to a resistivity of greater than 17 megaohms cm.
Recombinant DypB and Variants
Wild-type DypB (WT) was heterologously produced in E. coli BL21(DE3) using pETDYPB1 as described elsewhere (2). The variants were constructed using oligonucleotide-directed mutagenesis as described in the supplemental Experimental Procedures. The variants were produced, purified, and reconstituted with heme as described for the WT (2). For reconstitution, 20 mg/ml hemin chloride in DMSO was added dropwise with gentle stirring to 20 mg/ml protein in to a molar ratio of 2:1. Excess heme was removed via centrifugation and gel filtration. The protein samples were then dialyzed overnight at 4 °C in buffer containing 1 mm EDTA and further purified by anion-exchange chromatography. Fractions with Rz values > 2.5 were pooled, concentrated, and flash-frozen as beads in liquid nitrogen and stored at −80 °C until use. Protein concentration was measured using the Micro BCA assay (Thermo Scientific). The molar absorption coefficients for heme in the DypB variants was determined by using a pyridine hemochromogen assay (17) and were used to determine the enzyme concentrations in kinetic assays. Electronic absorption spectra were recorded using a Cary 5000 spectrophotometer (Varian) equipped with a thermostatted cuvette holder.
Steady-state Kinetic Analysis
Apparent steady-state kinetic parameters for H2O2 were determined using a previously described spectrophotometric assay containing 10 mm ABTS, appropriately diluted enzyme, and H2O2 (0.05–5 mm) in 1 ml of 50 mm sodium acetate (pH 5.5) at 25.0 ± 0.5 °C (7). Reactions were initiated by the addition of H2O2, and initial rates were monitored at 414 nm (ϵ414 = 36.6 mm−1cm−1 (18)). Steady-state kinetic equations were fit to the data using LEONORA (19).
Stopped-flow Kinetics
Transient state kinetics were performed using an SX18 stopped-flow spectrophotometer (Applied Photo Physics Ltd.) equipped with a diode array detector and a monochromator. A circulating water bath was used to maintain the temperature of the reactant syringes and the mixing cell at 25 °C. Typically, 10 μm enzyme was mixed with H2O2 at various concentrations at pH 5.5 and 7.5. Multiwavelength data were analyzed using singular value decomposition with the Pro-K.2000 Global Analysis program (Applied Photo Physics) to obtain the transition rates (kn) between intermediates. Reactions were then monitored at selected single wavelengths to follow the formation and decay of intermediates, and multiple exponential equations were fitted to the data to obtain pseudo-first order rate constants (kobs). Second order rate constants for the formation of Compound I were evaluated from plots of kobs versus H2O2.
Crystallization, Data Collection, and Structure Solution
Crystals of each of the D153H, D153A, N246H, N246A, and R244L variants were grown at room temperature by hanging drop vapor diffusion. Drops were made from 2 μl of 18 mg/ml protein solution in 20 mm MOPS (pH 7.5) 50 mm NaCl, and 2 μl of well solution consisting of 0.1 m sodium acetate trihydrate, 3 m NaCl (pH 4.5). Crystals formed overnight. To prepare crystals for mounting, they were briefly soaked in well solution supplemented with 16% glycerol and then flash-frozen by immersion in liquid nitrogen. Data were collected at the Canadian Light Source on beamline 08B1-1 at 0.97952 Å wavelength. The crystals grew in the space group P3221 and crystallized with three molecules in the asymmetric unit as reported for the WT structure (7). Data were processed using HKL2000 (20). Structures were solved directly by refinement using the WT P3221 crystal structure (PDB ID 3QNR) with alanine replacement of active site residues. Structures were manually edited using Coot (21) and refined using Refmac (22) from the CCP4 program suite (23). Structure figures were generated using PyMol (24). Data collection and structure refinement statistics are summarized in Supplemental Table S2.
RESULTS
Electronic Structure of Variants
As with WT DypB, the variants were produced predominantly as apoproteins in E. coli (Rz < 0.1). Accordingly, purified proteins were reconstituted with heme as previously described (2). The spectra of the D153A, N246A, and D153A/N246A variants were remarkably similar to that of the WT, with a Soret band at 404 nm and the charge transfer band 1 at 632 nm (Fig. 2), consistent with the presence of high spin ferric heme. The molar extinction coefficients of these variants were similar to that of WT (supplemental Table S1). By contrast, the spectra of the D153H and N246H variants suggest the presence of low spin ferric heme complexes. Thus, the Soret band was red-shifted to 410 nm, the charge transfer band 1 was absent, and the α and β bands occurred between 530 and 580 nm (supplemental Fig. S1). The spectra of position 153 and 246 variants were not significantly affected by pH over the range of 4.5 to 8.0. By contrast, the spectrum of the R244L variant showed a strong pH dependence, with the Soret band shifting from 411 nm at pH 8 to 404 nm at pH 6 (supplemental Fig. S1). The species at low pH was further characterized by charge transfer bands at 503 and 632 nm. These spectral changes are consistent with a transition from high spin to low spin with increasing pH.
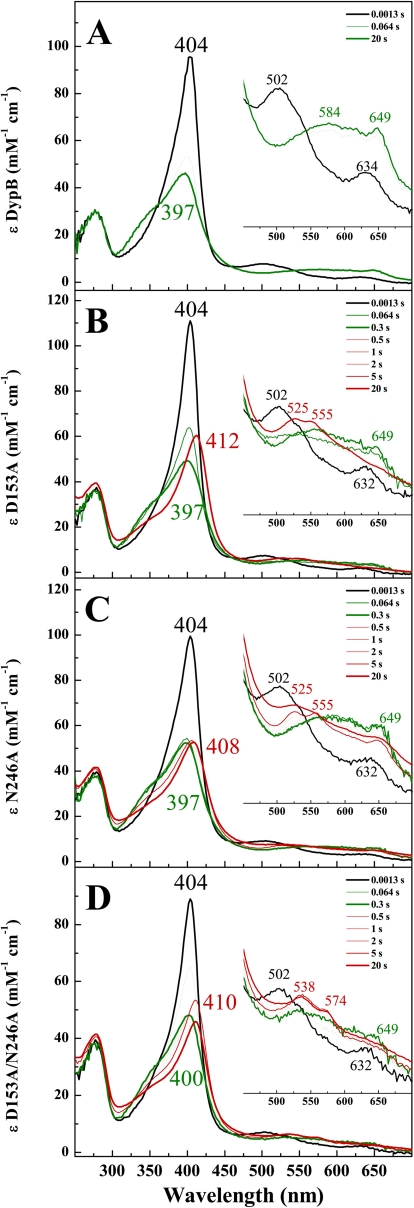
FIGURE 2.
Stopped-flow analyses of the reaction of DypB (A), D153A (B), N246A (C), and D153A/N246A (D) with H2O2 (20 mm MOPS, 50 mm NaCl (pH 7.5) at 25 °C. In each case, 10 μm concentrations of enzyme were mixed with an equal volume of 200 μm H2O2, except in D, where 1000 μm H2O2 was used. The black spectrum, recorded immediately after the mixing, corresponds to the ferric enzyme. The transitions to the first and second intermediates are shown in green and red, respectively. The inset shows the region between 450 and 700 nm.
Steady-state Kinetic Analyses
The steady-state kinetic parameters for H2O2 were determined for the WT and the variant proteins using a saturating amount of ABTS (10 mm) as the reducing substrate (Table 1). Substitution of Arg-244 with leucine had a much greater effect on the reactivity of DypB with H2O2 than did substitution of either Asp-153 or Asn-246 with alanine. Thus, the respective kcat/Km values of the D153A and N246A variants for H2O2 were very similar to that of WT, whereas the R244L variant possessed no detectable peroxidase activity. Nevertheless, both the kcat and Km values of D153A for H2O2 were ∼17-fold higher compared with the WT, whereas these values were relatively unchanged in N246A. The D153A/N246A double variant was also surprisingly reactive, possessing a slightly higher kcat value than the WT, although its Km for H2O2 was 200-fold greater. Finally, the introduction of histidine at either position 153 or 246 did not increase the peroxidase activity of DypB. Indeed, the N246H variant did not show any detectable peroxidase activity under the experimental conditions.
TABLE 1.
Kinetic parameters for DypB and the distal residue variants
Steady-state kinetic parameters were determined using 10 mm ABTS, 50 mm sodium acetate (pH 5.5) at 25.0 ± 0.5 °C. Pseudo-first order and -second order rate constants were determined using 20 mm MOPS, 50 mm NaCl (pH 7.5) at 25.0 ± 0.5 °C.
| Enzyme | KmH2O2 | kcat | kcat/Km | k1 | k2 |
|---|---|---|---|---|---|
| μm | s−1 | ×105m−1s−1 | ×105m−1s−1 | s−1 | |
| DypB | 27 ± 5 | 3.2 ± 0.03 | 1.20 ± 0.08 | 2.0 ± 0.07 | –a |
| D153H | 87 ± 7 | 1.8 ± 0.04 | 0.20 ± 0.06 | 0.12 ± 0.05 | 1.2 ± 0.01 |
| D153A | 460 ± 30 | 51 ± 3 | 1.10 ± 0.04 | 0.94 ± 0.01 | 0.75 ± 0.06 |
| R244L | –b | ||||
| N246H | –b | ||||
| N246A | 15 ± 1 | 5.4 ± 0.2 | 3.60 ± 0.20 | 3.20 ± 0.01 | 0.37 ± 0.04 |
| D153A/N246A | 5000 ± 2000 | 19 ± 8 | 0.04 ± 0.002 | 0.13 ± 0.02 | –c |
a Not measured because Compound I did not transition to Compound II.
b No significant activity was detected (detection limit was 0.04 μmole/min/μmol of heme).
c Not measured because the high concentrations of H2O2 required to produced Compound I resulted in the conversion of the latter to Compound III without detectable formation of Compound II.
Compound I Formation in Variants
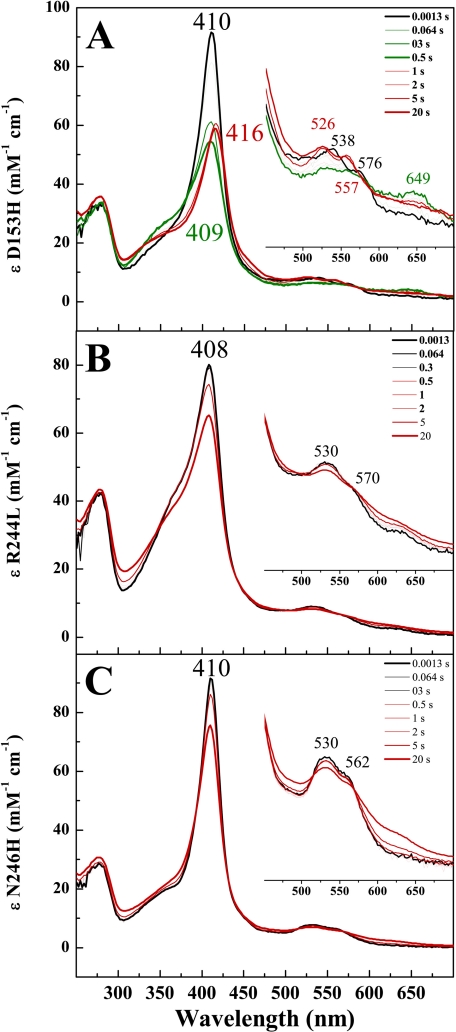
Transient intermediates in DypB and the variants were observed by following the variants reaction with 5–200 eq of H2O2 using a rapid-mixing, stopped-flow spectrophotometer. In the D153A and N246A variants at pH 7.5, the spectrum of the first intermediate, observed within ∼0.1 s upon mixing with H2O2, was almost identical to Compound I observed in WT (Fig. 2, A–C, thick green trace) with a hypochromatic, blue-shifted Soret at 397 nm and a hyperchromic CT band at 649 nm. Similar intermediates were observed in D153A/N246A and D153H, although the CT band of Compound I in the double variant was less hyperchromic (Fig. 2D, thick green trace), and >100 eq of H2O2 were required to form it, consistent with the high Km for H2O2. In the D153H variant, the Soret was less blue-shifted at 406 nm (Fig. 3A, thick green trace), and the visible region was less hypochromatic compared with the WT. Neither R244L nor N246H detectably reacted with H2O2 even at 100-fold greater concentrations of the latter (Fig. 3, B and C, thick red spectrum). Moreover, neither the high spin state (pH 6) nor pH 7.5 the low spin state (pH 7.5) of the R244L variant detectably reacted with H2O2 (Fig. 3B, thick red spectrum). However, the heme of this variant was more susceptible to degradation at pH 6 (supplemental Fig. S2) compared with pH 8.
FIGURE 3.
Stopped-flow analyses of the reaction of D153H (A), R244L (B), and N246H (C) with H2O2 (20 mm MOPS, 50 mm NaCl (pH 7.5) at 25 °C). In each case, 10 μm concentrations of enzyme were mixed with an equal volume of 2000 μm H2O2, except in A, where 200 μm H2O2 was used. The black spectrum, recorded immediately after the mixing, corresponds to the ferric enzyme. The spectra are represented as in Fig. 2.
The second order rate constants for Compound I formation (k1) by the WT and the variants were measured by following the changes in absorption at 404 and 647 nm. The kinetic traces were described by single order exponentials, and the determined pseudo-first order rate constants, kobs, were similar at the two wavelengths (supplemental Fig. S5). Plots of kobs versus H2O2 concentration were linear (supplemental Fig. S6). In each of the studied enzymes, the k1 values were comparable at pH 7.5 (Table 1) and pH 5.5 (data not shown) and were remarkably similar to the kcat/Km values for H2O2. Thus, k1 for D153A was about half that for WT, whereas that for N246A was 1.6-fold greater. Similarly, the k1 values for D153H and D153A/N246A were more than an order of magnitude less than that for WT.
Compound II Formation in Variants
Compound I was much shorter lived in each of the D153H, D153A, and N246A variants (t½ ∼ 0.13 s) than in WT (t½ ∼ 540 s). Moreover, in these variants, Compound I decayed to an intermediate whose spectra resemble Compound II of peroxidase (Fig. 2, B and C, and Fig. 3A, thick red spectrum). Thus, the Soret of Compound II was red-shifted to various extents in each of the three variants, whereas α and β bands appeared near 525 and 555 nm, respectively. Interestingly, the Soret band for Compound II is less red-shifted in the spectrum of N246A compared with D153A. In addition, the spectrum of N246A possessed a charge transfer band at 649 nm, suggesting the presence of a mixture of Compounds I and II (Fig. 2C, thick red spectrum).
In D153A/N246A, Compound I decayed to an intermediate whose spectrum resembled that of Compound III, with a Soret for this intermediate located at 410 nm and α/β bands at 538 and 574 nm, respectively (Fig. 2D, thick red spectrum). Formation of this intermediate was likely due to the use of higher concentrations of H2O2 required to produce detectable Compound I. Compound III was not observed in either D153A or N246A even upon reaction with 100 eq of H2O2 (supplemental Fig. S4).
The transient observed in reactions of WT DypB and the variants with H2O2 were modeled using singular value decomposition, global analysis, and a three-step model (A → B → C) (supplemental Fig. S7). The simulated spectra of the intermediates were consistent with the formation of a single intermediate in the reaction with WT and two intermediates in the reactions with each of D153A, N246A, D153A/N246A, and D153H.
Structural Characterization of Variants
Crystal structures of each of the four single variants were determined to at least 2.64 Å resolution (supplemental Table S2). The average r.m.s.d for all Cα atoms is <0.25 Å when compared with the WT crystal structure (PDB ID 3QNS), indicating that the substitutions had minimal effect on the overall -fold. Generally, the active site residues are well defined and are similarly positioned relative to the wild-type structure (Fig. 4A). The only significant differences in the structures are observed on the distal side of the heme where Asp-153, Arg-244, and Asn-246 interact with the iron-coordinated solvent molecule in the wild-type structure.
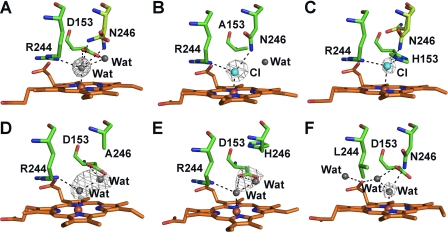
FIGURE 4.
X-ray crystal structures of DypB variants. Shown are wild-type (PDB ID 3QNS) (A), D153A (B), D153H (C), N246A (D), N246H (E), R244L (F). Active sites residues are shown as green (carbon), blue (nitrogen), and red (oxygen) sticks. Alternate conformations for Asn-246 (A and C) are highlighted in light green. Water (gray), iron (orange), and chloride (cyan) are shown as spheres. Hydrogen bonds are indicated by dashed lines. Gray mesh represents omit difference (Fo − Fc) maps for the distal ligand contoured at 3 σ for all panels except E (2.5 σ) and F (2.0 σ).
Difference electron density maps of the D153A and D153H variants revealed a strong positive peak on the distal side of the heme iron. Refinement of each site as a water molecule resulted in residual positive electron density and a low B-factor relative to surrounding atoms. Chloride, present at 3.0 m in the crystallization buffer, fully models the electron density and refines at full occupancy with B-factors comparable with those of the surrounding atoms (D153A, Fe(III) ∼45 Å2 and Cl− ∼58 Å2; D153H, Fe(III) ∼39 Å2 and Cl− ∼58 Å2). The presence of a chloride in the Asp-153 variants may be favored in the absence of the negatively charged carboxylate. In D153A, the chloride ion interacts with the heme iron (∼2.3 Å), Arg-244 Nη1 (3.2 Å), and Asn-246 Nδ2 (3.1 Å) (Fig. 4B). Arg-244 is in the same conformation as in the WT structure, and the orientation of Asn-246 corresponds to one of the two orientations modeled in the WT structure. A bound water molecule was observed to occupy part of the void created by the aspartate-to-alanine replacement. This solvent species is located ∼4.5 Å from the chloride ion and forms a hydrogen bond with Asp147. Similarly, in the D153H structure, the chloride also interacts with Arg-244 Nη1 (3.1 Å) and Asn-246 Nδ2 (2.4 Å in one conformation). However, it also interacts with His-153 (3.0 Å), and the chloride-iron bond is elongated (∼2.7 Å) (Fig. 4C). In contrast to D153A, two conformations are observed for Asn-246 in the D153H structure; one bound to chloride through Nδ2 and the other directed into the distal solvent access channel by an ∼135° rotation about χ1. The increased chloride-iron bond length in D153H appears to be a result of the imidazole side chain, which sterically prevents the chloride from occupying a coordination site closer to the iron. This amino acid replacement also causes a 0.6-Å shift of Asn-246 Nδ2 away from position 153 relative to its position in the structure of WT DypB.
The replacement of Asn-246 with either alanine or histidine removes the Nδ2 atom that is directly bound to the axially coordinated solvent molecule in the WT structure (Fig. 4A). Omit difference (Fo − Fc) electron density maps of N246A and N246H clearly display elongated density on the distal side of heme-iron, extending toward residue 246 (Figs. 4, D and E). Two water molecules may be refined into the density of each variant with one coordinating the iron (2.1–2.2 Å) and the second hydrogen-bonded to the first (2.5 Å). The B-factors for these water molecules are in line with those of heme iron (N246A, Fe(III) ∼35 Å2 and waters both ∼38 Å2; N246H, Fe(III) ∼41 Å2 and waters ∼41 and 50 Å2). Attempts to model the density by an acetate molecule (present at 0.1 m in the crystallization buffer) were unsuccessful, but the possibility that the structure contains a mixture of coordinated solvent and acetate cannot be excluded. A positionally equivalent second solvent molecule was observed in the high resolution WT crystal structure (Fig. 4A), where it is modeled at 40% occupancy, correlated with one of two conformations of Asn-246. In contrast to WT, in N246A and N246H the second solvent molecule is shifted ∼0.7 Å toward residues 246 and forms a hydrogen bond with Asp-153 (3.1–3.3 Å). The remaining residues in N246A active site overlay well with those of the WT structure, indicating that the mutation has little effect on the rest of the heme environment. In the N246H variant, the imidazole of His-246 is orientated away from the heme and projects into the distal solvent access channel, potentially blocking substrate access. The orientation of His-246 appears to perturb surrounding residues. Asp-153 is displaced toward the heme (maximal deviation ∼1.2 Å at Oδ1 relative to WT), and the side chain of Arg-244 is displaced ∼0.5 Å away from the distal coordination site (Fig. 4E). The remainder of the N246H structure closely resembles WT.
In R244L, the distal ligand is a solvent molecule coordinated at 2.0 Å from the iron (Fig. 4F), similar to what is observed in the WT structure. The coordinated solvent forms a H-bond with Asn-246 (3.1 Å) but is shifted ∼0.5 Å away from Asp-153 relative to its position in the WT structure. As a result, Asp-153 Oδ1 is located ∼4.2 Å from the solvent molecule, outside hydrogen bonding distance. The guanidinium group of Arg-244 is effectively replaced by two solvent species. One solvent molecule is positioned similarly to Arg-244 Nη1 in the WT structure, bridging the coordinating solvent molecule (2.6 Å) and heme propionate A (2.7 Å). This species also forms an additional hydrogen bond with Asp-153 Oδ1 (2.9 Å). The second additional solvent species occupies a position similar to that of Arg-244 Nη2 in the WT structure, forming another hydrogen bond with heme propionate A (2.8 Å). The remainder of the active site is unperturbed by the mutation.
DISCUSSION
The current study establishes that of the three distal polar residues, only Arg-244 is essential for peroxidase activity in DypB. Indeed, the replacement of Asp-153 and/or Asn-246 with an alanine had a remarkably small effect on the enzyme reactivity with H2O2, as evaluated by the apparent specificity constant (kcat/Km) and the first order rate constant for the formation of Compound I (k1). Nevertheless, substitution of these residues dramatically decreased the stability of Compound I in DypB. In addition, the data from the D153H and N246H variants suggest that DypB cannot efficiently utilize imidazole as an acid-base catalyst on the distal face of the heme. Overall, this study indicates that in DypB, the peroxidative cycle is modulated very differently than in plant peroxidases.
The ability of D153A to form Compound I and to react with peroxide almost as efficiently as WT DypB is surprising considering that substitution of the equivalent residue, Asp-171, in DyPDec1 reduced the enzyme peroxidase activity to undetectable levels (5). The latter result led to the proposal that the distal aspartate functions as an acid-base catalyst, similar to the distal histidine of plant peroxidases and the distal glutamate of chloroperoxidase (B: in Fig. 5). Substitution of the distal acid-base catalyst in plant peroxidases reduces catalytic efficiency by four to six orders of magnitude (25, 26). The distal aspartate also does not appear to be essential for peroxidase activity in A-type DyPs. Thus, WT EfeB and the D235N variant oxidized guaiacol at similar rates (15). Interestingly, the variant-oxidized catechol at ∼20% that of the rate of WT indicated that the aspartate has a role in substrate specificity. The lack of the steady-state and transient state kinetic data from the DyPDec1 and EfeB variants makes comparison with DypB difficult. Nevertheless, the available data suggest that although the distal aspartate is the essential acid-base catalyst in d-type DyPs, it is not in either A- or B-type DyPs. This may reflect the different physiological substrates and/or roles of the different subfamilies of enzymes as discussed below.
FIGURE 5.
The proposed mechanism of peroxidases. The steps leading to Compound I formation are in parentheses. An acid-base catalyst, B:, assists in transferring a proton from the proximal O to the distal O of the iron-bound H2O2, accelerating Compound I formation. This residue is histidine and glutamate in plant peroxidases and chloroperoxidase, respectively. In D- and B-type DyPs, this residue appears to be aspartate and arginine, respectively.
Although the current data indicate that Asn-246 plays a minor role in Compound I formation, it is positioned remarkably similarly to the conserved asparagine in catalases (27). In bovine liver catalase the conserved asparagine (Asn-147) and histidine (His-74) on the distal face of the heme assist in the binding of H2O2 to the heme iron (27). In resting state bovine liver catalase, the two residues are hydrogen-bonded to a shared solvent molecule, much as Asn-246 and Asp-153 are in DypB. Although the kinetic parameters of the N246A variant (Table 1) establish that Asp-246 is not required for catalysis, those of the D153A/N246A double variant indicate that these two residues interact with the H2O2. Further insight into the interaction of Asn-246 and Asp-153 was obtained by inverse analysis. This analysis uses the double variant as a reference point to evaluate whether the effect of substituting two residues is additive, cooperative, anti-cooperative, or antagonistic (28). The effect of the Asp-153 and Asn-246 substitutions was anti-cooperative (supplemental Fig. S9), indicating that the two residues function synergistically with respect to peroxidase activity.
The inability of the R244L variant to form Compound I further distinguishes DypB from HRP and cytochrome c peroxidase. Although the distal arginine is similarly located in these enzymes, it forms an electrostatic interaction with heme propionate A (supplemental Fig. S8) in DyPs (5, 7, 14). In plant peroxidases, the propionate is orientated away from the arginine guanidinium group (29). Substitution of this residue with leucine in HRP or cytochrome c peroxidase lowers the rate of Compound I formation by 2–3 orders of magnitude, although this intermediate is still formed (25, 26). Nevertheless, the pH-dependent iron spin state transition of R244L is similar to that of the R48L variant of cytochrome c peroxidase (26). This transition suggests that the pKa of Asp-153 in R244L is ∼7, much higher than the expected value of 3.5. Indeed, it would be interesting to determine the pKa values of Asp-153, which is 0.7 Å further away from the distal solvent species in R244L than in WT.
The large effect of substituting Arg-244 in DypB indicates that this residue has a major catalytic role in Compound I formation and may be the requisite acid-base catalyst (B: in Fig. 5). It is unclear what other residue could fulfill this role; other than the three residues targeted in this study, the closest residue with the requisite functionality is Asp-147, located up to 7 Å away from the heme iron. Although the pKa of the acid-base catalyst is expected to be ∼5, much lower than that of an arginine residue (12.5), the pKa of catalytically relevant arginines can be significantly shifted. For example, Arg-82 of bacteriorhodopsin has a pKa of 5.8 in the M state (deprotonated retinal) (30), and Arg-418 of inosine 5′-monophosphate dehydrogenase has a pKa of ∼8 (31). In both of these cases, the arginine is in close proximity to an aspartate, as observed in DypB. Moreover, in all three cases, the arginine is part of a hydrogen bonding network that connects the residue to the protein surface. In DypB, the network involves the iron-bound solvent species, Arg-244, heme propionate A, a structurally conserved solvent molecule, and the highly conserved Arg-208 on the protein surface (supplemental Fig. S8). This network is conserved in DyPs and may mediate proton transfer as has been proposed in ascorbate peroxidase and other enzymes (32, 33). Disruption of this network may help explain the lack of peroxidase activity in R244L.
Although Asp-153 and Asn-246 have relatively minor roles in Compound I formation, they appear to have important roles in stabilizing this intermediate; substitution of either residue decreased the half-life of Compound I by >3 orders of magnitude. This is consistent with their close proximity to the heme iron: ∼5.0 Å for Asn-246Nδ2 and 5.1 Å for Asp-153Oδ1. Although this is similar to the corresponding distances for His-42Nϵ2 in HRP (5.8 Å (29)) and Glu-183Oϵ2 in chloroperoxidase (5.1 Å (16)), neither of the latter has an equivalent of Asn-246. At this time, it is unclear whether the stabilization of Compound I by Asp-153 and Asn-246 is due to kinetic or thermodynamic effects. Thus, these residues contribute to both the electrostatic environment and the hydrogen bonding network of the active site. The precise role of Asp-153 in stabilizing Compound I clearly depends on the residue protonation state. Although the iron spin-state transition in R244L suggests that the pKa value of Asp-153 is ∼7, this is unlikely to be the case in WT DypB.
The poor catalytic efficiency of the D153H and N246H variants is likely due in part to the suboptimal positioning of the imidazole in the distal heme pocket of DypB. In the case of D153H, the Nϵ2 atom of His-153 is 1.8 Å closer to the heme iron than His-42 is in HRP (29). This close proximity also likely favors the low spin state of the D153H variant. In the case of N246H, the imidazole is 7.8 Å from the iron. However, it is positioned at the center of the distal solvent access channel, potentially limiting the access of H2O2 to the heme iron. In addition, hydrogen bonding (3.3 Å) between His-246 Nδ1 and the main chain carbonyl of Asp-153 would also influence the catalytic role of His-246. Overall, this result highlights how protein active sites are fine-tuned for efficient utilization of specific functional groups. Additional amino acid substitutions are required to transform DypB into a more efficient peroxidase.
This work highlights the differences in the modulation of the active site residues in the peroxidative cycle of DypB as compared with that of D-type DyPs and plant peroxidases. Our data rule out Asp-153 as the acid-base catalyst in the formation of Compound I in DypB and identifies Arg-244 as best candidate for this role. It is possible that the different modulation of the peroxidative cycle in the DyPs reflects their different physiological roles. For example, C- and D-type DyPs may oxidize a variety of substrates such as dyes (5) and melanin (14), whereas A- and B-type DyPs may oxidize a specific substrate, such as porphyrinogen (11). Thus, although the lignin-and Mn2+-oxidizing activities of DypB from R. jostii RHA1 may be biotechnologically useful, it may not be physiologically relevant. This possibility is consistent with the ∼100-fold lesser activity of A- and B-type DyPs with classic peroxidase substrates (7). Further studies should elucidate the structural bases of the diverse activities of DyPs and facilitate developing the biotechnological potential of this family of microbial enzymes.
Supplementary Material
Acknowledgments
Part of this work was performed at the Canadian Light Source, which is supported by Natural Sciences and Engineering Research Council of Canada, the National Research Council of Canada, the Canadian Institutes of Health Research, the Province of Saskatchewan, Western Economic Diversification Canada, and the University of Saskatchewan.
This work was supported by Natural Sciences and Engineering Research Council of Canada Discovery grants (to L. D. E. and M. E. P. M.), respectively.

This article contains supplemental Experimental Procedures, Figs. S1–S9, and Tables S1 and S2.
R. Davydov and B. M. Hoffman, personal communication.
- DyP
- dye-decolorizing peroxidase
- ABTS
- 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulphonic acid.
REFERENCES
- 1. Sugano Y., Sasaki K., Shoda M. (1999) cDNA cloning and genetic analysis of a novel decolorizing enzyme, peroxidase gene dyp from Geotrichum candidum Dec 1. J. Biosci. Bioeng. 87, 411–417 [DOI] [PubMed] [Google Scholar]
- 2. Ahmad M., Roberts J. N., Hardiman E. M., Singh R., Eltis L. D., Bugg T. D. (2011) Identification of DypB from Rhodococcus jostii RHA1 as a lignin peroxidase. Biochemistry 50, 5096–5107 [DOI] [PubMed] [Google Scholar]
- 3. Ogola H. J., Kamiike T., Hashimoto N., Ashida H., Ishikawa T., Shibata H., Sawa Y. (2009) Molecular characterization of a novel peroxidase from the cyanobacterium Anabaena sp. strain PCC 7120. Appl. Environ. Microbiol. 75, 7509–7518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sugano Y. (2009) DyP-type peroxidases comprise a novel heme peroxidase family. Cell. Mol. Life. Sci. 66, 1387–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sugano Y., Muramatsu R., Ichiyanagi A., Sato T., Shoda M. (2007) DyP, a unique dye-decolorizing peroxidase, represents a novel heme peroxidase family. Asp-171 replaces the distal histidine of classical peroxidases. J. Biol. Chem. 282, 36652–36658 [DOI] [PubMed] [Google Scholar]
- 6. Liers C., Bobeth C., Pecyna M., Ullrich R., Hofrichter M. (2010) DyP-like peroxidases of the jelly fungus Auricularia auricula-judae oxidize nonphenolic lignin model compounds and high redox potential dyes. Appl. Microbiol. Biotechnol. 85, 1869–1879 [DOI] [PubMed] [Google Scholar]
- 7. Roberts J. N., Singh R., Grigg J. C., Murphy M. E., Bugg T. D., Eltis L. D. (2011) Characterization of dye-decolorizing peroxidases from Rhodococcus jostii RHA1. Biochemistry 50, 5108–5119 [DOI] [PubMed] [Google Scholar]
- 8. Kaur A., Van P. T., Busch C. R., Robinson C. K., Pan M., Pang W. L., Reiss D. J., DiRuggiero J., Baliga N. S. (2010) Coordination of frontline defense mechanisms under severe oxidative stress. Mol. Syst. Biol. 6, 393–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kong L., Guo D., Zhou S., Yu X., Hou G., Li R., Zhao B. (2010) Cloning and expression of a toxin gene from Pseudomonas fluorescens GcM5-1A. Arch. Microbiol. 192, 585–593 [DOI] [PubMed] [Google Scholar]
- 10. Létoffé S., Heuck G., Delepelaire P., Lange N., Wandersman C. (2009) Bacteria capture iron from heme by keeping tetrapyrrol skeleton intact. Proc. Natl. Acad. Sci. 106, 11719–11724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dailey H. A., Septer A. N., Daugherty L., Thames D., Gerdes S., Stabb E. V., Dunn A. K., Dailey T. A., Phillips J. D. (2011) The Escherichia coli protein YfeX functions as a porphyrinogen oxidase, not a heme dechelatase. MBio 2, e00248–00211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Goblirsch B., Kurker R. C., Streit B. R., Wilmot C. M., DuBois J. L. (2011) Chlorite dismutases, DyPs, and EfeB. three microbial heme enzyme families comprise the CDE structural superfamily. J. Mol. Biol. 408, 379–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zubieta C., Krishna S. S., Kapoor M., Kozbial P., McMullan D., Axelrod H. L., Miller M. D., Abdubek P., Ambing E., Astakhova T., Carlton D., Chiu H. J., Clayton T., Deller M. C., Duan L., Elsliger M. A., Feuerhelm J., Grzechnik S. K., Hale J., Hampton E., Han G. W., Jaroszewski L., Jin K. K., Klock H. E., Knuth M. W., Kumar A., Marciano D., Morse A. T., Nigoghossian E., Okach L., Oommachen S., Reyes R., Rife C. L., Schimmel P., van den Bedem H., Weekes D., White A., Xu Q., Hodgson K. O., Wooley J., Deacon A. M., Godzik A., Lesley S. A., Wilson I. A. (2007) Crystal structures of two novel dye-decolorizing peroxidases reveal a β-barrel-fold with a conserved heme binding motif. Proteins 69, 223–233 [DOI] [PubMed] [Google Scholar]
- 14. Zubieta C., Joseph R., Krishna S. S., McMullan D., Kapoor M., Axelrod H. L., Miller M. D., Abdubek P., Acosta C., Astakhova T., Carlton D., Chiu H. J., Clayton T., Deller M. C., Duan L., Elias Y., Elsliger M. A., Feuerhelm J., Grzechnik S. K., Hale J., Han G. W., Jaroszewski L., Jin K. K., Klock H. E., Knuth M. W., Kozbial P., Kumar A., Marciano D., Morse A. T., Murphy K. D., Nigoghossian E., Okach L., Oommachen S., Reyes R., Rife C. L., Schimmel P., Trout C. V., van den Bedem H., Weekes D., White A., Xu Q., Hodgson K. O., Wooley J., Deacon A. M., Godzik A., Lesley S. A., Wilson I. A. (2007) Identification and structural characterization of heme binding in a novel dye-decolorizing peroxidase, TyrA. Proteins 69, 234–243 [DOI] [PubMed] [Google Scholar]
- 15. Liu X., Du Q., Wang Z., Zhu D., Huang Y., Li N., Wei T., Xu S., Gu L. (2011) Crystal structure and biochemical features of EfeB/YcdB from Escherichia coli O157. Asp-235 plays divergent roles in different enzyme-catalyzed processes. J. Biol. Chem. 286, 14922–14931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sundaramoorthy M., Terner J., Poulos T. L. (1995) The crystal structure of chloroperoxidase. A heme peroxidase; cytochrome P450 functional hybrid. Structure 3, 1367–1377 [DOI] [PubMed] [Google Scholar]
- 17. Falk J. E. (1964) Porphyrins and Metalloporphyrins pp. 804–807, Elsevier Science Publishing Co., Inc., Amsterdam [Google Scholar]
- 18. Childs R. E., Bardsley W. G. (1975) The steady-state kinetics of peroxidase with 2,2′-azino-di-(3-ethyl-benzthiazoline-6-sulphonic acid) as chromogen. Biochem. J. 145, 93–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cornish-Bowden A. (1995) Analysis of Enzyme Kinetic Data, Oxford University Press, New York [Google Scholar]
- 20. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 21. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. Sect. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 23. Collaborative Computational Project, Number 4 (1994) The CCP4 suite. Programs for protein crystallography. Acta Crystallogr. D. Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 24. DeLano W. L. (2008) The PyMOL Molecular Graphics System, DeLano Scientific LLC, Palo Alto, CA [Google Scholar]
- 25. Rodriguez-Lopez J. N., Smith A. T., Thorneley R. N. (1996) Role of arginine 38 in horseradish peroxidase. A critical residue for substrate binding and catalysis. J. Biol. Chem. 271, 4023–4030 [DOI] [PubMed] [Google Scholar]
- 26. Vitello L. B., Erman J. E., Miller M. A., Wang J., Kraut J. (1993) Effect of arginine 48 replacement on the reaction between cytochrome c peroxidase and hydrogen peroxide. Biochemistry 32, 9807–9818 [DOI] [PubMed] [Google Scholar]
- 27. Fita I., Rossmann M. G. (1985) The active center of catalase. J. Mol. Biol. 185, 21–37 [DOI] [PubMed] [Google Scholar]
- 28. Mildvan A. S. (2004) Inverse thinking about double mutants of enzymes. Biochemistry 43, 14517–14520 [DOI] [PubMed] [Google Scholar]
- 29. Gajhede M., Schuller D. J., Henriksen A., Smith A. T., Poulos T. L. (1997) Crystal structure of horseradish peroxidase C at 2.15 A resolution. Nat. Struct. Biol. 4, 1032–1038 [DOI] [PubMed] [Google Scholar]
- 30. Xiao Y., Hutson M. S., Belenky M., Herzfeld J., Braiman M. S. (2004) Role of arginine 82 in fast proton release during the bacteriorhodopsin photocycle. A time-resolved FT-IR study of purple membranes containing 15N-labeled arginine. Biochemistry 43, 12809–12818 [DOI] [PubMed] [Google Scholar]
- 31. Guillén Schlippe Y. V., Hedstrom L. (2005) Is Arg-418 the catalytic base required for the hydrolysis step of the IMP dehydrogenase reaction? Biochemistry 44, 11700–11707 [DOI] [PubMed] [Google Scholar]
- 32. Efimov I., Badyal S. K., Metcalfe C. L., Macdonald I., Gumiero A., Raven E. L., Moody P. C. (2011) Proton delivery to ferryl heme in a heme peroxidase. Enzymatic use of the Grotthuss mechanism. J. Am. Chem. Soc. 133, 15376–15383 [DOI] [PubMed] [Google Scholar]
- 33. Sharp K. H., Mewies M., Moody P. C., Raven E. L. (2003) Crystal structure of the ascorbate peroxidase-ascorbate complex. Nat. Struct. Biol. 10, 303–307 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.