Background: The membrane attack complex (MAC) is an ancient component of immune defense that assembles lytic pores in pathogen membranes.
Results: Structural comparisons between C6 and C8 reveal the available conformations of MAC proteins.
Conclusion: We propose a critical role for the “auxiliary” domains in driving and regulating assembly.
Significance: The model rationalizes the sequential and unidirectional nature of assembly.
Keywords: Allosteric Regulation, Complement System, Crystal Structure, Host Defense, Host-Pathogen Interactions, Innate Immunity, Membrane Proteins, Protein Conformation, Protein-Protein Interactions, Structural Biology
Abstract
The complement membrane attack complex (MAC) is formed by the sequential assembly of C5b with four homologous proteins as follows: one copy each of C6, C7, and C8 and 12–14 copies of C9. Together these form a lytic pore in bacterial membranes. C6 through C9 comprise a MAC-perforin domain flanked by 4–9 “auxiliary” domains. Here, we report the crystal structure of C6, the first and longest of the pore proteins to be recruited by C5b. Comparisons with the structures of the C8αβγ heterodimer and perforin show that the central domain of C6 adopts a “closed” (perforin-like) state that is distinct from the “open” conformations in C8. We further show that C6, C8α, and C8β contain three homologous subdomains (“upper,” “lower,” and “regulatory”) related by rotations about two hinge points. In C6, the regulatory segment includes four auxiliary domains that stabilize the closed conformation, inhibiting release of membrane-inserting elements. In C8β, rotation of the regulatory segment is linked to an opening of the central β-sheet of its clockwise partner, C8α. Based on these observations, we propose a model for initiation and unidirectional propagation of the MAC in which the auxiliary domains play key roles: in the assembly of the C5b-8 initiation complex; in driving and regulating the opening of the β-sheet of the MAC-performin domain of each new recruit as it adds to the growing pore; and in stabilizing the final pore. Our model of the assembled pore resembles those of the cholesterol-dependent cytolysins but is distinct from that recently proposed for perforin.
Introduction
The complement system is an ancient component of vertebrate immune defense (1). One major end point of complement activation is the formation of the membrane attack complex (MAC),2 which is a circular multiprotein assembly that can embed into a phospholipid membrane of a target cell generating a large pore (2–5). Deficiencies in any of the MAC components cause enhanced susceptibility to Gram-negative bacterial infections such as those caused by Neisseria meningitidis (6–8).
The mature MAC includes single copies of C5b, C6, and C7 and the heterotrimeric C8αβγ complex and 12–18 copies of C9 (2–5). All MAC components, except C5b and C8γ (a small protein attached to the side of C8α), contain a homologous central region of ∼350 amino acids termed the membrane attack complex-perforin (MACPF) domain (9, 10). MAC assembly starts with proteolytic cleavage of complement component C5 (Mr ∼196,000), giving rise to a large fragment, C5b (Mr∼185,000), which forms an initial complex with C6 and C7 at the membrane surface (“C5b-7”) (3, 11). C5b-7 then binds C8αβγ to form the “C5b-8” complex, which then binds sequentially to multiple copies of C9 to form the mature MAC (3, 12), a circular SDS-stable complex with an internal diameter of ∼100 Å that constitutes the lytic pore (13, 14). Radiolabeling experiments indicate that in the mature MAC all MACPF components insert at least partially into the membrane, with C8α and C9 being the most highly labeled (15–17). Although the order in which the MAC components assemble is known, the structural and regulatory bases of initiation and propagation of pore assembly are not understood.
An unexpected structural similarity between the MACPF domain and a family of bacterial cholesterol-dependent cytolysins (CDCs) led to the suggestion that MACPF proteins function analogously by assembling into a ring on the target membrane and inserting β-hairpins to create a contiguous β-barrel that forms the lumen of the pore (18, 19). The dimensions of the mature MAC derived from EM images (20) are consistent with such a model, in which two helical clusters (which we call CH1 and CH2) from the MACPF domain unfurl to form two β-hairpins (referred to as trans-membrane hairpins in the CDCs) (18). It should be noted that the predicted hairpins are twice as long as in the CDCs, and there is no evidence that a circular “pre-pore” intermediate forms above the membrane prior to insertion (18, 21). Available evidence points instead to an assembly pathway in which each MAC protein attaches and inserts sequentially into the membrane (3, 13).
Perforin is the only other mammalian member of the MACPF family whose structure is known (22); its role is to form homo-oligomeric lytic pores in infected or transformed host cells (23, 24). A recent crystallographic and EM study of the perforin pore supported the β-barrel hypothesis, but the authors proposed that the MACPF domain had a reversed orientation with respect to models of the CDC pore (22). However, the packing between MACPF domains in the first crystal structure of a full-length MAC protein, the C8αβγ complex, supports a CDC-like organization for the MAC (25).
C6 is the longest of the MAC proteins. In its monomeric form, it is a single chain plasma glycoprotein of Mr ∼106,000, consisting of 913 residues folded as nine auxiliary/regulatory domains complementing the MACPF core. Although the precise functions of the auxiliary domains are unclear, their importance in MAC assembly is supported by several studies. For example, two independent studies (using deletion mutants) demonstrated that the N-terminal modules of C8α (TS2 and LR) are strictly required for MACPF formation and hemolytic activity, although deletion of the C-terminal TS3 domain greatly reduced activity (26, 27). A study on C9 provided evidence for regulatory roles for the N terminus and TS2 domain; thus, short deletions at the N terminus promoted MACPF formation, and deletions or mutations within the TS2 domain caused nonproductive C9 self-polymerization (28).
Here, we describe the crystal structure of full-length C6 at 2.85 Å resolution. Comparisons with complement C8 and perforin, as well as with the broader family of CDCs, led us to propose an atomic model of initiation, regulation, and propagation of assembly of the MAC, in which the key process is the sequential template-driven opening of the MACPF β-sheet of each new component as it is recruited to the growing pore. In this model, the auxiliary domains play critical roles in the initial assembly of the C5b-8 initiation complex, in driving and regulating sheet opening, and in stabilizing the final pore.
EXPERIMENTAL PROCEDURES
Protein Production
Complement C6 was purified from human plasma by a modification of a published procedure (29). The steps included barium citrate depletion of the vitamin K proteins, 4–12% polyethylene glycol precipitation, DEAE-Sephadex column chromatography, euglobulin precipitation, gel filtration on Sephacryl-300, dextran sulfate-Sepharose column chromatography, and rabbit anti-contaminant IgG-Sepharose 6B.
Crystallization and Heavy Atom Derivatives
Small crystals of C6 were prepared in batch by incubating protein at 11 mg/ml in 5 mm MES, pH 5.8, 80 mm NaCl, and 10 μm CdCl2 at 1 °C for 2 weeks and then at 6 °C for 3–4 weeks, resulting in large (∼300 μm) rhombic bipyramidal crystals. They were cryoprotected by addition of pure glycerol to the crystallization liquor to a final concentration of 28% v/v. Heavy atom derivatives were obtained by soaking crystals in mother liquor and heavy atoms at concentration/times as follows: Ta6Br12 (data set 1, 1.0 mm/4 h; data set 2, 0.5 mm/12 h; K2PtCl4 (0.1 mm/4 h); (NH4)2OsCl6 (0.5 mm/2 h); and K2IrCl6 (0.5 mm/2 h).
Data Collection and Structure Solution
All data were collected at SSRL beamline 9-2. Data were processed using IMOSFLM and CCP4i (30, 31). Several native data sets were collected to obtain the highest isomorphism with the heavy atom derivatives. Initial phases were calculated using a 3.5 Å native data set (Nat-2) and five derivatives (Table 1). The best diffracting crystals (Nat-2), which were less isomorphous, were used for the final model refinement. Data extended to 2.85 Å in the nest directions but were truncated ellipsoidally (32) because of significant anisotropy in the 3.0 to 2.85 Å resolution shell, reducing its completeness to ∼60%. Heavy atom sites were determined by SOLVE (33) and refined with SHARP (34). Initial phasing power was poor above 4 Å, but the high solvent content of the crystals (64%) allowed SHARP to extend the phases to 3.5 Å using density modification.
TABLE 1.
Data collection, phasing, and refinement statistics
| Nat-1 | Nat-2 | Ta6Br12-1 | Ta6Br12-2 | K2PtCl4 | (NH4)2OsCl6 | K2IrCl6 | |
|---|---|---|---|---|---|---|---|
| Data collection | |||||||
| Wavelength (Å) | 0.991 | 1.078 | 1.123 | 1.255 | 1.068 | 0.991 | 1.105 |
| Resolution (Å) | 49/2.85 | 20-3.5 | 20-3.9 | 20-3.5 | 20-3.8 | 20-3.5 | 20-3.9 |
| Space group | P21212 | P21212 | P21212 | P21212 | P21212 | P21212 | P21212 |
| Cell dimensions (Å) | a = 146.8 | 146.5 | 146.8 | 146.4 | 147.6 | 146.1 | 144.8 |
| b = 180.2 | 181.0 | 181.5 | 179.7 | 179.6 | 180.3 | 179.7 | |
| c = 60.5 | 60.5 | 60.3 | 60.7 | 60.0 | 60.5 | 59.9 | |
| RMERGEa (%) | 6.9 (50)b | 5.4 (35) | 9.3 (56) | 6.5 (35) | 6.7 (40) | 6.2 (28) | 6.1 (25) |
| I/σI | 6.4 (1.5)c | 9.8 (2.2) | 5.9 (1.3) | 7.1 (2.1) | 6.5 (1.9) | 6.9 (2.7) | 8.4 (3.0) |
| Completeness (%) | 99.6 (100) | 99.9 (100) | 99.6 (100) | 99.8 (100) | 99.9 (100) | 99.7 (100) | 99.7 (100) |
| Redundancy | 8.3 (8.5) | 7.3 (7.4) | 14.5 (14.8) | 6.1 (6.2) | 9.0 (9.0) | 8.1 (8.2) | 5.1 (5.2) |
| Phasing statistics | |||||||
| Power (Iso/Ano) | 3.3/1.1 | 2.2/1.3 | 0.6/0.6 | 0.7/0.8 | 0.3/0.3 | ||
| Resolution (Iso/Ano)d | 5.0/6.0 | 5.4/5.4 | 20.0/6 | 8.0/6.0 | 20.0/7.0 | ||
| Heavy atoms | 4 | 4 | 1 | 2 | 2 | ||
| Refinement statistics | |||||||
| Resolution (Å) | 40-2.85 | ||||||
| Completeness | 94.9 | ||||||
| RWORKe/RFREEf | 0.22/0.28 | ||||||
| Reflections | 35,745/2179 | ||||||
| Protein atoms | 6937 | ||||||
| Ions (Cd2+) | 1 | ||||||
| Glycosylation sites/atoms | 7/102 | ||||||
| Wilson B (Å2) | 81.7 | ||||||
| Root mean square deviation from ideality | |||||||
| Bond lengths (Å) | 0.01 | ||||||
| Bond angles (°) | 1.4 | ||||||
| Ramachandran plot | |||||||
| Most favored (%) | 77.5 | ||||||
| Additional allowed | 20.8 | ||||||
| Generously allowed | 1.6 | ||||||
| Disallowed | 0.1 | ||||||
a RMERGE = Σ|Ih − 〈Ih〉|/ΣIh, where 〈Ih〉 is the average intensity over symmetry equivalent reflection.
b All values in parentheses refer to the highest resolution shell (3 to 2.85 Å).
c For the high resolution native data set (Nat-1), because of anisotropic diffraction, data were truncated ellipsoidally in the 3.0 to 2.85 Å shell.
d This is the resolution at which the phasing power fell below 1.0.
e RWORK = Σ|Fobs − Fcalc|/ΣFobs, where the summation is over the 35,745 reflections used for refinement.
f RFREE was calculated using 5% of data (2179) excluded from refinement (70).
The initial map quality was reasonable for this resolution but did not permit automated chain tracing. Structural homologs were therefore identified for each domain using FFAS (35), and three-dimensional models were built by omitting insertions and substituting nonidentical amino acids with serines. The following structures were used as templates (PDB entry/% identity): MACPF-C8β (2RD7/25%); TS1–TS3 (1LSL/29–45%); LR (1CR8/50%); CCP1 (1HO4/30%); CCP2 (3KXV/25%), and FIM1–FIM2 (2WCY/30%).
The MACPF domain, TS1 domain, and CCPs were readily located in the 3.5-Å experimental map using real space molecular replacement with FFFEAR (36), and a distinct conformation for the MACPF was immediately apparent. These domains fixed the topology of the molecule, enabling the remaining domains to be identified in the experimental map. Because of the low sequence identity of many templates (sometimes with distinct disulfide linkages), the domains were built manually, using the templates as guides. Side chains were built first in the well resolved fragments and then in the less ordered domains as refinement progressed. Rigid-body refinement of domains was followed by torsion angle refinement using simulated annealing and refinement of individual B-factors using CNS (37) in the phased target function (maximum likelihood (amplitude and Hendrickson-Lattman coefficients)) mode with the 3.5-Å (Nat-2) data set. The model was then refined in PHENIX (38) using the 2.85-Å (Nat-1) data set with experimental phase restraints removed (ML mode). Reciprocal space refinement was iterated with manual model building and real space refinement in COOT (39).
The final cycles included the refinement of nine translation/libration/screw (TLS) groups (assigned to individual protein domains), bulk solvent/scale corrections, and individual atomic coordinates and B-factors. Final refinement statistics are presented in Table 1. Stereochemical quality was validated with PROCHECK (40). Figures were prepared with PyMOL and CHIMERA (41).
Residue B-factors are shown schematically in supplemental Fig. 1. Some domains had limited intramolecular or crystal lattice interactions, leading to high B-factors (100–200 Å2). Electron density for the last FIM domain (residues 834–913) was diffuse and fragmented, but rigid-body refinement of a homology-based model reduced the RFREE by 0.38%, supporting its presence at that location. Electron density is absent for interdomain linker residues 243–259, 591–600, 605–619, and 744–755. For illustrative purposes only, these fragments were built as extended coils to show the domain topology.
The map revealed seven glycosylation sites (N-glycosylation of Asn-303, O-glycosylation of Thr-17 and Thr-371, and α1-Cδ-linked mannosylation (42) of Trp-8, Trp-11, Trp-547, and Trp-550). The sugar moieties at Asn-303 were built as β1-OG1- and β1–4-linked N-acetylglucosamine. Two sugar moieties at Thr-17 were built as α1-OG1-linked fucose and β1–3-linked glucose by analogy with other thrombospondin-like repeat domains (e.g. PDB entry 3GHN). There is no information about glycosylation at Thr-371, but based on the density it was built as OG1-α1-linked fucose. N-Glycosylation is predicted at Asn-834 (43, 44), but we could not validate this because of the disorder in this region (FIM2). The LR module Ca2+-binding site is occupied by Cd2+, as judged by its 2Fo − Fc peak height and a strong anomalous peak in an anomalous difference Fourier. The ionic radii of Cd2+ and Ca2+ are very similar. Atomic coordinates and structure factors for C6 are deposited in the Protein Data Bank with accession number 3T5O.
RESULTS
Overall Structure of C6
The crystal structure of C6 was solved by a combination of experimental phasing and molecular replacement at ∼3 Å resolution (see “Experimental Procedures,” Table 1, and supplemental Fig. 1). Interpretable electron density exists for all domains except for FIM2 (see below) and some interdomain linkers. The nine auxiliary domains are generally small and rigid, ranging in size from 35 to 75 residues and containing 2–5 disulfide bonds. In contrast, the large MACPF domain contains only one intradomain disulfide bond.
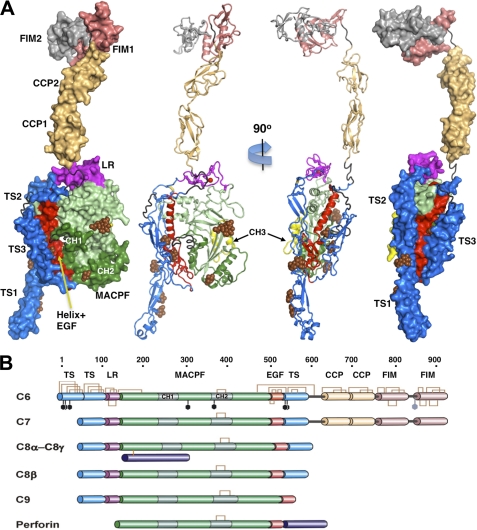
C6 has the overall shape of a seahorse, a rather flat molecule with a head-to-tail distance of 215 Å (Fig. 1). The MACPF domain (residues 160–501) itself is only 75 Å tall, and the additional height of C6 is accounted for by auxiliary domains as follows: in particular, four C-terminal domains (complement control protein (“CCP”) modules and factor I modules (“FIMs”) that extend from the upper body of the MACPF core. These auxiliary domains are found only in C6 and C7 among the late acting components of complement (Fig. 1 and supplemental Fig. 2). The overall shape is consistent with EM images of C6 and C7, although a more compact conformation is often observed for the C-terminal domains, in which the “head” folds back onto the main body (11, 43). In addition, C6 has a unique N-terminal thrombospondin-like domain (“TS1”) that forms a protrusion from its base that was observed in the earlier EM comparisons. TS1 also has an unusual helical insert at its base with amphipathic properties that may promote membrane targeting/specificity (supplemental Fig. 3). Intriguingly, perforin and the bacterial cytolysins have a similar membrane-binding domain at their base, although it is attached to the C terminus (Fig. 2).
FIGURE 1.
Crystal structure and domain organization of C6. A, surface and secondary structure presentations of C6 (orthogonal orientations). The disordered residues between modules TS3 and CCP1, CCP2 and FIM1, and the entire FIM2 are colored gray-black; the sugars of the glycosylation sites are shown as brown spheres. B, schematic presentation of the primary structural features of the complement MAC components and perforin. The disulfide bonds and the glycosylation sites of C6 are shown as brown brackets and black hexagons. Perforin contains a distinct membrane-binding C-terminal domain.
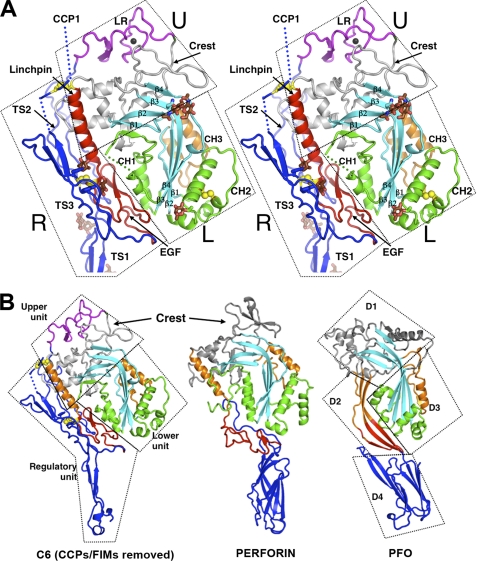
FIGURE 2.
Structure of C6 core and its interaction with auxiliary domains. A, stereo view of the core fragment of C6 presented as a secondary structure ribbon. The rigid-body units are enclosed with boxes and labeled U (upper), L (lower), and R (regulatory). The regulatory unit consists of EGF (red), TS1, TS2, and TS3 modules (blue). The upper unit contains the LR module (magenta) and the upper fragment of MACPF, including the linchpin helix (red). The lower unit contains the lower fragment of MACPF including CH1-CH2 (green) and CH3 (orange). Glycosylation sites are shown as brown sticks. Two disulfide bonds linking TS3 to MACPF and EGF are shown as yellow balls. B, comparison of C6 (lacking CCPs and FIMs) with perforin (PDB code 2NSJ) and a member of the CDC family, PFO (PDB code 1PFO). The domains of PFO are designated D1 to D4. D1 and D3 are analogous to the upper and lower domains of C6. The linchpin helices (in orange) and the EGF domains (in red) of C6 and perforin have some functional analogy with domain D2 of PFO, but PFO and perforin lack the regulatory functions provided by the auxiliary domains of C6. D4 may be identified with TS1 of C6 on structural and possibly functional grounds. β-Sheets are in cyan; CH1 and CH2 are in green; CH3 is orange, and the rest of the domain is gray. TS1-TS3 of C6 and the membrane-binding domains of perforin and PFO are in blue.
C6 Adopts a Default Closed Autoinhibited State
The central core of C6 adopts the typical MACPF organization, built around a central four-stranded β-sheet with up-down-up-down topology (18, 22, 25). The β-sheet bends abruptly in the middle and has extensive elaborations between and flanking the β-strands (Fig. 2). The two ∼50-residue helical clusters (CH1 and CH2) that are hypothesized to unfurl upon activation connect the β1-β2 and β3-β4 strands at the bottom of the central β-sheet. In our crystal structure, conformational rearrangements of CH1 are inhibited by a module (colored red in Fig. 2) comprising a long α-helix (which we call the “linchpin,” residues 478–498) and a rigid, disulfide-rich epidermal growth factor (EGF) domain. The module connects the upper and lower ends of the β-sheet, creating a central D-shaped enclosure in which the two helices of CH1 are packed. CH2, however, is located on the external (convex) face of the β-sheet and is stabilized by interactions with the β-sheet and another helical cluster, CH3, which is an insert in strand β4.
Most full-length monomeric MACPF/CDC proteins, including perforin and perfringolysin O (PFO) (Fig. 2) (19, 22, 45), contain a highly bent β-sheet that is held shut by an analogous module, creating a similar enclosure that locks the CH1 helices in place. We consider this to be an auto-inhibited or “closed” conformation. By contrast, the C8 complex (25) adopts a much more open conformation than in C6 and perforin (see below).
Auxiliary Domains (EGF and TS1–3) Form a Y-shaped Module Attached to the Linchpin Helix
At the base of the MACPF, N- and C-terminal auxiliary domains pack tightly around the EGF module, forming a Y shape (colored blue in Fig. 3). TS1 and TS2 form a tandem linear dimer (very similar to a dimeric module of thrombospondin-1 (46), see supplemental Fig. 3) that forms two arms of the “Y”; the module is stabilized by an inter-domain disulfide bridge and O-linked glycosylation. The amphipathic N terminus of TS1 lies 50 Å below the body of the MACPF, whereas the TS1–2 interface packs tightly against the EGF module. TS2 then proceeds up the side of MACPF, without making further direct contacts. C-terminal to MACPF and the EGF domain (also at the base of MACPF), the chain makes an abrupt turn, folding as a third thrombospondin domain (TS3) that forms the third arm of the Y. TS3 behaves similarly to TS2, packing tightly against the (opposite face) EGF domain, before proceeding to the top of MACPF, but making few contacts along the way. Both ends of TS3 (in C6 and C7) are, however, disulfide-bonded to cysteines located N- and C-terminal to the linchpin helix. A close-knit array of disulfide-bridged elements connects the EGF domain (in contact with CH1) through the base of the linchpin to the mid-section of TS3 on the exterior face of the molecule (distal to the presumed channel lumen). TS2 is not disulfide-linked to the EGF domain but is nevertheless connected via an extensive interface.
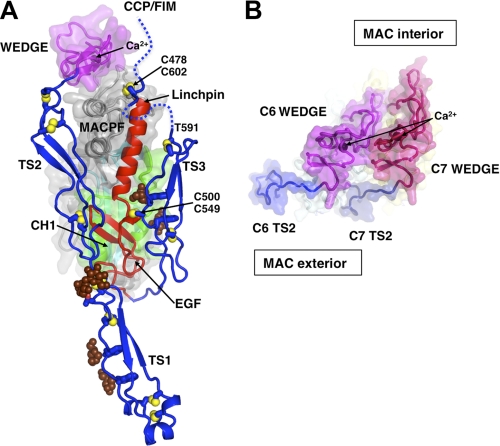
FIGURE 3.
Interactions between the Y-shaped regulatory module and MACPF and a model for packing of two wedge domains. A, regulatory module is colored in blue (TS1–3) and orange (EGF domains). The MACPF core is covered with a semi-transparent surface, except for the linchpin helix. The LR domain and Crest domain of MACPF, which collectively form a wedge-shaped unit, are covered with a magenta surface. The mannose rings of C1-glycosylated Trp-547 and Trp-550 and the O-glycosylation site (glucose-fucose) of Thr-17 are shown as brown balls. Selected intra- and interdomain (numbered) disulfide bonds are shown as yellow balls. Hydrophobic side chains of TS2 and TS3 that contact the EGF domain are labeled. The TS3-CCP1 linker (residues 591–620, shown as dashed lines) is disordered everywhere except for three amino acids around the Cys-478–Cys-602 disulfide bond. The second disulfide bond Cys-500–Cys-549 attaches TS3 to EGF. CCP modules and FIMs are omitted. B, model of two “wedge” domains (C6 and C7, viewed from the “top”), forming two putative building blocks that create the upper surface of the growing pore. Note the shape complementarity and curvature, which places the TS2 modules on the MAC exterior. Ca2+ ions are labeled. The view includes only the tops of the subunits sliced near to the ends of the TS2 modules.
LR Domain Creates a Wedge-shaped Building Block at the Top of MACPF
Downstream of TS2, the polypeptide chain continues across the top of the MACPF, adopting a “low density lipoprotein receptor class A repeat” (LR module), which is stabilized by disulfide bonding and a divalent cation site. By analogy with homologous domains (47), Ca2+ is likely to occupy this site in vivo (in our crystals, the site is occupied by Cd2+ that was added to the crystallization buffer) (Fig. 3). The module is very similar in C8 (25), and sequence alignments suggest that identical binding sites will be found in all MAC MACPF domains (supplemental Fig. 2).
The LR module augments the first segment of the consensus MACPF domain (a long loop that lies across the top of the upper β-sheet and includes two β-hairpins, one of which inserts into the central enclosure). Together, these elements form a wedge-shaped “building block.” Thus, we found that the C6 wedge packs well against the same region of a model of C7 (applying the same transformation that relates homologous segments of C8α and C8β (25)), forming a tandem dimer with a curvature appropriate for the mature MAC. Importantly, this model places the convex face of the β-sheet on the inside of the pore, as proposed for the CDCs (Fig. 3B).
C-terminal CCP Modules and FIMs Form Tandem Modules That Extend Above the Body of MACPF
Downstream of TS3, the C-terminal 300 residues fold as tandem pairs of CCP modules and FIMs. In our crystal structure, these extend from the top of MACPF and make no contact with other modules. The CCPs form a tandem module with a small but significant interface whose sequence is conserved in C7 (supplemental Fig. 4). Their fold and organization are similar to other complement proteins (factor H, CR1, and C4b-binding protein) that mediate protein-protein and protein-heparin interactions (48–50). The electron density for FIM1 is clear, and a reliable model has been built. Although the density for C6 FIM2 is fragmented, the domain location is clear and displays a very different FIM1/2 organization from that observed in the solution structure of the C7 pair, which forms a tightly packed pseudo-symmetric dimer (51). The difference likely arises from an insertion between the two FIMs of C6 (a helix and disulfide-linked hairpin) that is absent in C7. Another difference is a positively charged face of C7 FIM1 implicated in binding to C5/C5b (51), which is negatively charged in C6 (supplemental Fig. 5).
There is evidence that C5 binds the FIMs of both C6 and C7 as part of the initial activation process leading to C5b-7 (52–54). A naturally occurring variant of C6 lacking the FIM domains does, however, retain some activity (bactericidal efficiency is reduced ∼10-fold (55)), suggesting that additional interactions occur. In our crystals, the extended organization for the two C-terminal module pairs has been selected, at least in part, by the lattice environment (supplemental Fig. 1); and it is possible that the structure mimics an early activated state of C6 in which the FIMs and CCP modules have been released from inhibitory binding sites on the upper surface of the MACPF that were inferred from EM images (11, 43). This possibility is also supported by modeling studies of C6 and C7 linker sequences (supplemental Fig. 6).
C6, C8α, and C8β Contain Three Homologous Segments Related by Rigid-body Rotations about Two Distinct Axes
In the case of the CDCs, manual fitting of crystallographic models into EM reconstructions of the pneumolysin pore (56) led to a model in which the β-sheets opened during the prepore-to-pore transition, promoting the reorganization of the helical clusters (equivalent to CH1 and CH2) into membrane-spanning β-hairpins. However, no such analysis of MACPF proteins has previously been described. Notably, sheet opening was not considered in the recent model of the perforin pore (22). We therefore analyzed C6 and C8 using an objective procedure (DYNDOM (57)) that compares homologous molecules and evaluates the extent to which quaternary differences can be described in terms of rigid-body motions of analogous segments/subdomains.
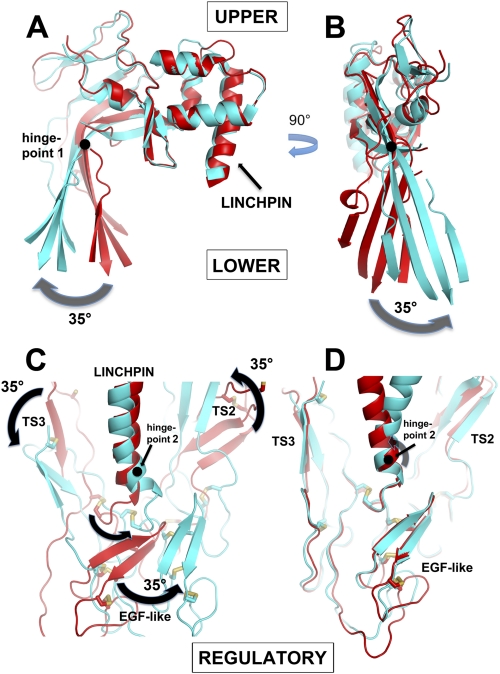
This procedure identified three homologous segments (“upper,” lower,” and “regulatory”) common to all three proteins (C6, C8α, and C8β) that are related by rigid-body rotations about two distinct axes (Fig. 4). These segments are broadly equivalent to the D1–D3 domains defined for the CDCs. The upper segment includes the top part of the β-sheet (above its bend) that includes the β2-β3 turn, the wedge-shaped unit that includes the LR module, and sequences downstream of β4, ending at the linchpin helix. The lower segment includes the bottom half of the β-sheet (below the bend), together with the CH1–3 helical clusters. The regulatory segment includes TS1–2, the EGF-like domain, and TS3. The conserved structures of these segments are revealed when the analogous segments are overlaid, giving root mean square main chain differences in the 1-Å range for pairwise comparisons (Fig. 4D and supplemental Fig. 7); note that the closed conformation observed in C6 is conserved in perforin (supplemental Fig. 7C).
FIGURE 4.
Comparison of C6 with C8α and C8β illustrating the existence of three rigid-body units connected by two hinge points. A and B, two orthogonal views (“side” and “inside” views) of MACPFs of C6 (red) and C8α (cyan) after superposition of their UPPER segments. The arrows indicate the large rigid-body rotation (by ∼35°) of the LOWER segment, about an axis (hinge point 1) shown as a black circle, represented by the lower half of the β-sheet (i.e. the CH1-CH3 clusters follow this movement but have been omitted for clarity; see also supplemental Fig. 7). Note that the sheet both “opens” (A) and “twists” (B). When C6 is compared with C8β (data not shown), there is a small opening of the sheet, but it does not twist. C, comparison of C6 (red) and C8β (cyan) after superposition of their upper segments, illustrating the ∼35° rotation of the REGULATORY segment (TS2+EGF+TS3) around hinge point 2, at the end of the linchpin helix. The view is the same as in B but is from the outside of the predicted pore. Note how the motion of the REGULATORY domain correlates with the twist of the β-sheet in B. When C6 is compared with C8α (data not shown), the motions are similar but smaller in magnitude. D, same view as in C, but here the overlay is on the REGULATORY domains of C6 and C8β, showing their close superposition, and thus demonstrating that they behave as a rigid body.
Evidence for Sheet Rotation (“Opening” and “Twisting”) in MACPF Domains (Hinge Point 1)
Overlay of the upper segments of C6 and C8α shows that their upper and lower segments are related by a rigid-body rotation of ∼35° about an axis (“Hinge Point 1”) that passes through the bend in the β-sheet (Fig. 4). This rotation may be further resolved into two components as follows: the first along the ridge formed by the bend in the sheet, leading to an opening of the sheet similar to that modeled for the CDCs (Fig. 4A); the second component is orthogonal to the first, leading to a “twist” of the sheet of a similar magnitude (counterclockwise, when viewed from the side presumed to form the channel lumen) (Fig. 4B). The rotation equates to a relative translation of the lower edge of the β-sheet of C8α by as much as 20 Å. Because CH1 shifts in concert with the lower part of the sheet, whereas the linchpin, EGF domain, and β-hairpin do not, the CH1 enclosure is substantially opened in C8α. This opening relaxes constraints on the CH1 helices, which accordingly show greater disorder and higher B-factors than in C6. The straightening of the sheet also appears to weaken the bonds between CH2 and CH3, which are packed against the outer surface of the sheet, because even greater disorder is observed in the CH2-CH3 interface. Indeed, in the case of C9, antibodies raised against CH3 provide evidence that the domain undergoes an order-disorder transition upon recruitment to the MAC (58).
Thus, the opening and twisting of the sheet may facilitate unfurling of both CH1 and CH2. Overlay of C6 with C8β shows that the sheet in C8β also opens, but to a smaller extent than in C8α; and the sheet does not twist (not shown).
Regulatory Segment Rotates about a Distinct Axis at Base of Linchpin Helix (Hinge Point 2)
The second rigid-body motion is illustrated in Fig. 4C. When overlaid on their upper segments, compared with C6, the regulatory segments (TS2-EGF-TS3) of C8α and C8β rotate as a rigid body about hinge point 2, which is located near the end of the linchpin helix (centered at Phe-497 in C6). The rotations are similar in nature in C8α and C8β but twice as large in the latter, leading to shifts of >20 Å in parts of the C8β EGF domain. The direction of movement is nearly orthogonal to that of sheet opening and is parallel to (and commensurate with) the sheet twisting observed in C8α (compare the movements in Fig. 4, B and C).
In our model, both TS2 and TS3 lie on the outer surface of the MAC pore. Observe that in following the rotation of the EGF domain TS3 appears to be “pulled down” the side of C6 MACPF (compared with C8), whereas TS2 is “pushed up.” In C6 and C7, an analogous downward shift of TS3 is restrained by a disulfide connection at the top of the linchpin helix; however, modeling suggests that the flexible linker segment (residues 591–602) provides just enough slack to allow TS3 to adopt the position analogous to that seen in C8β, and hence enable a similar movement of its EGF domain.
DISCUSSION
The discovery in the early 1970s of “neo-epitopes,” antigenic surfaces present on the MAC but not on the monomeric components, led Kolb and Müller-Eberhard to conclude that “It is likely that the quaternary structure of the complex imposes conformational changes on the subunits” (59). Our crystal structure of C6, together with a detailed comparison with the recently determined structure of C8 (25), allowed us for the first time to propose the nature of this quaternary-tertiary linkage and how it controls MAC assembly.
Thus, we have shown how the conformational differences between crystal structures of C6, C8α, and C8β (see Fig. 4 and supplemental Fig. 7) may be described in terms of rigid-body rotations of three conserved segments (upper, lower, and regulatory) about two distinct axes, leading to three distinct quaternary arrangements. We postulate that these conformational differences reflect distinct quaternary states of MAC proteins on the pathway to activation/assembly; and the C8αβγ complex has evolved to adopt a partially activated but stable (in the absence of the C5b-7 complex) MACPF dimer.
Our initial model of MAC pore is based on the proposal of Lovelace et al. (25), who found that iterating the tandem packing of C8α and C8β observed in the C8αβγ complex led to a circular assembly that resembled poly(C9). In support of this model, we have shown how the LR domains on the crest of the upper segments of C6 and C8 create wedge-shaped building blocks. In addition to shape complementarity, we note that the “leading” and “trailing” faces of the wedge-shaped segments of each successive MACPF pair have complementary/opposite charges (supplemental Fig. 8). The model orients the concave faces of the MACPF β-sheets toward the center of the pore (consistent with models of CDC pores), with the CH3 and C8γ domains contained within the inner lumen, and the TS1-TS3 domains on the outer surface of the MAC.
This topological model of the assembled MAC does not by itself address the mechanisms of pore formation, but it does provide a structural framework for developing such models, which must include the steps of initiation, propagation, and the sequential, unidirectional recruitment of protomers that lead to the mature membrane-bound MACPF.
Model for MAC Propagation
What is the underlying mechanism that enables each monomeric recruit to spontaneously attach to the nascent pore and undergo a major conformational change leading to membrane insertion? The comparisons between C6 and C8αβγ give us many clues. Thus, in C8β, a large rotation of its TS2 domain (compared with C6 and C8α) creates a new interface both with its own MACPF domains and with its clockwise neighbor (C8α) that augments the binding between their upper segments. But our analysis suggests that a necessary consequence of this is a linked rotation of the C8β EGF domain that thrusts it toward the CH1 enclosure of C8α.
In the C8 crystal structure, C8α responds to this motion in several ways, most notably via a commensurate (30°) opening/twisting of its β-sheet. However, this motion significantly reduces favorable interactions between its own EGF domain and CH1 (the latter moves in concert with the β-sheet, because it is part of the lower segment). Thus, the EGF-CH1 interface in C8α is 360 Å2 versus 750 Å2 in C6, resulting in decreased order and weak or nonexistent electron density for parts of C8α and its EGF domain. The opening of the C8α sheet also necessitates a repacking (and weakening) of the CH2-CH3 interface as noted above, i.e. the transformation from a closed auto-inhibited state seen in our C6 structure to a more “open” and more activated conformation as seen in C8α leads to a weakening of the restraints that stabilize the helical conformations of both CH1 and CH2 (thus promoting their unfolding and transformation into β-hairpins).
A further key observation here is that the rotation of the regulatory segment of C8β drives the opening and twisting of the β-sheet of its clockwise partner (C8α), but it has little effect on its own sheet, i.e. it is the rotation of the regulatory segment that rationalizes the directionality (clockwise) of pore formation (C8β will only open when it gains a counter-clockwise partner).
Why is the C8αβγ module, with its tandem MACPF domains, required for regulated MAC assembly? Two possible reasons are as follows. (i) It may have evolved its dimeric form to provide a pair of rigid building blocks to propagate pore assembly with the correct curvature. (ii) Once the critical high energy task of assembling the membrane-bound C5b-7 complex has been accomplished, the partly activated conformation of C8 may provide a fast/low energy pathway enabling facile binding to both C5b-7 and the first C9, resulting in rapid propagation of the nascent pore.
Model for MAC Initiation
The binding of C6 and C7 to C5b via their C-terminal modules (FIMs and/or CCPs) primes them initially to form a reversible complex, which then reorganizes into an irreversible complex, C5b-7, that attaches to the target membrane (3, 60, 61). C5b may promote this process in several ways as follows: (i) simply by bringing the partners into close apposition; (ii) by releasing constraints (e.g. removing the CCPs and FIMs) on the upper segments, thereby promoting formation of an initial encounter complex; and (iii) by binding to the top of the C6-C7 pair (as it does in the mature MAC), a process that could push down on the TS3 domains of C6 and/or C7, thereby promoting rotation/activation of their regulatory segments. We note that a role for the TS3 domain in regulating assembly is supported by studies of an inhibitory antibody that maps to TS3 of C6 (62).
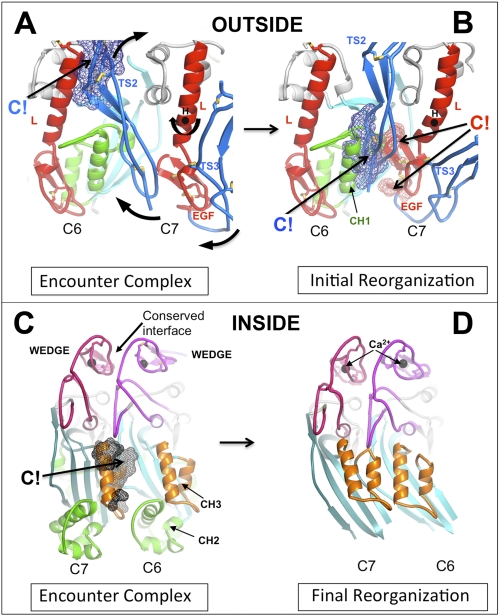
Nevertheless, the bimolecular association of C6 and C7 is rate-limiting for membrane association, with an activation energy estimated at 35 ± 2 kcal/mol (60, 63), consistent with the need for large conformational changes. We assume that C5b-bound C6 and C7 first form an encounter complex by binding via their wedge domains, as described above. We will further (arbitrarily) assume that C6 is the clockwise partner of C7 (see Fig. 7 and supplemental Fig. 9). Although there is good shape and charge complementarity between the wedge domains, computational modeling suggests that steric clashes would occur at several other points, notably between the CH3 domain of C7 and the β-sheet of C6 (on the inner surface of the nascent pore, see Fig. 5A) and between TS2 of C7 and the linchpin helix of C6 on the outer surface (Fig. 5B). These clashes would be relieved by rotation of the regulatory segment, with TS2 repositioned to augment the C6-C7 interface, and the EGF domain of C7 driven into the C6 enclosure, forcing open its neighbor's β-sheet.
FIGURE 7.
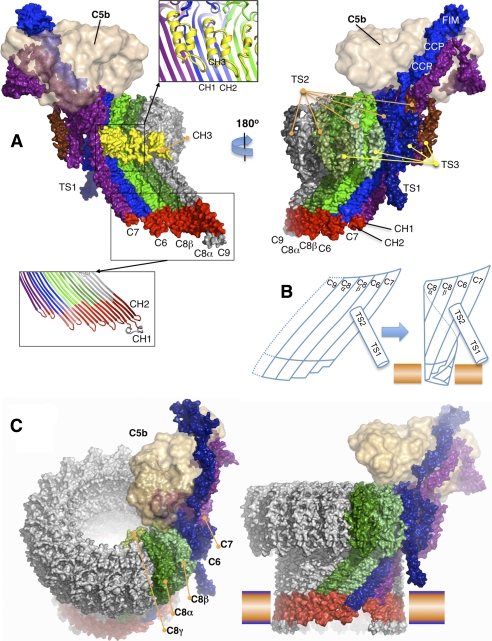
Molecular model of the C5b-8 complex extended by one C9 (C5b-9). A, two views of the C5b-9 complex, seen from the inside (left) and outside (right) of the pore. C6 through C9 were initially modeled from the C6 and C8α crystal structures. C5b was modeled from C3b and placed on top of MAC at a position consistent with EM images. The complex is shown as a solvent-accessible surface, colored primarily by subunit, except that the amphipathic transmembrane regions are red, and the CH3 elements are yellow. Two inserted panels show the selected regions as the secondary structure ribbons. B, schematic of the C7-C6-C8α-C8β complex, hypothesized to form a small membrane-spanning pore that is facilitated by the unusually short hairpins of C7. The upper segments maintain low curvature, so that the leading edge of the growing β-sheet remains available for binding the next recruit and promoting its membrane insertion. C, two views of an atomic model for the mature MAC, viewed from different directions. The blue-orange bar represents a membrane bilayer.
FIGURE 5.
Steric clashes at the C6-C7 interfaces drive reorganization of the dimer. A, initial encounter complex was modeled by overlaying the upper segments of C8α and C8β in the C8αβγ crystal structure with C6 and C7 (modeled on C6). Note that we do not know which of C6 and C7 is the clockwise partner, but our arbitrary choice of C6 does not affect the underlying mechanism. A close-up of the C6-C7 dimers, viewed from the outer face of the presumptive pore, shows where the TS2 domain of C7 clashes with C6, principally at the linchpin helix of C7. B, rotation of the regulatory segment (TS1-TS2-EGF-TS3) of the C7 structure about the axis marked (H) relieves the steric clash in A and creates a new (favorable) interface between C7 and C6, but a concerted rotation of the EGF domain creates new clashes between the EGF domain of C7 and the CH1 enclosure of C6. The lower part of TS2 also makes substantial clashes with this region. We hypothesize that these clashes are relieved by the opening of the C6 β-sheet and release of the CH1 helices to form β-hairpins (see schematic in Fig. 6). C, same dimer as in A but viewed from the opposite side (from inside the presumptive pore), illustrating a major clash of the CH3 element of C6 with the β-sheet of C7. C6 and C7 are both in the closed conformation. D, open structure of the C7-C6 dimer modeled on the C8α structure (for clarity, their CH1 and CH2 helices are not shown). Opening and twisting of the β-sheets removes all steric clashes, allowing the formation of an 8-stranded contiguous β-sheet. CH3 is present in all MAC components, and its displacement may contribute to unidirectional assembly.
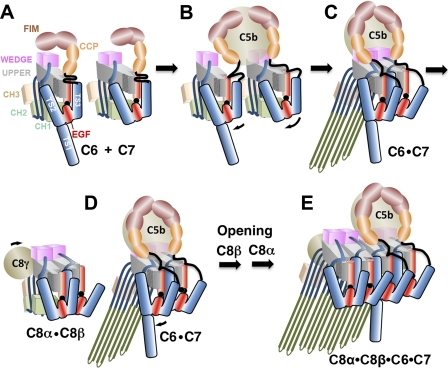
We do not know what provides the final trigger for the creation of the membrane-bound C5b-7, but we hypothesize that, while transiently resembling the C8αβ complex, C5b-7 lacks its additional (evolved) stability, such that the opening of the sheet and rotation of the regulatory segments are sufficient to trigger dislocation and unraveling of the CH1/2 elements to form β-hairpins (Fig. 6).
FIGURE 6.
Schematic of the hypothetical assembly pathway of the C5b-8 initiation complex. A, C6 and C7 in solution are maintained in monomeric states by the packing of their regulatory segments (TS1–3+EGF) against the back of the β-sheet. The FIMs may also fold back onto the upper segment of the MACPF. B, C5b engages C6 and C7, initially via their FIMs, bringing them into apposition. An initial encounter complex between the wedge modules triggers rotation of the C7 regulatory module about the linchpin hinge (hinge point 2) to relieve steric clashes with C6. C, EGF domain of C7 rotates in concert with TS2 and TS3, inserting into the CH1 enclosure of C6, whereas TS2 forms a new C7-C6 interface. These processes open and twist the β-sheet of C6 (rotation about hinge point 2), enabling the release and unfurling of CH1 and CH2 to form β-hairpins that associate with the outer leaflet of the membrane, supported by the TS1 domain of C6. D and E, following encounter with the C8αβγ complex, a similar process occurs, in which the regulatory element of C6 inserts its EGF domain into the C8β enclosure. The opening and twisting of the β-sheets allows the formation of a contiguous 16-stranded β-sheet. The amphipathic hairpins of C8α and C8β insert through the membrane bilayer. Typically, 12–14 C9 molecules will then add sequentially to the growing pore and insert into membrane until a complete circular MAC is formed.
Modeling of the β-hairpins of C6 and C7 suggests that they are long (∼50 Å) and amphipathic enough to insert partially into membrane, but not span it, consistent with radiolabeling experiments (11, 17) and the lack of pore formation (60) at this stage. We also note that the TS1 domain has the appropriate length to provide the third leg of a tripod to support the body of the MACPF domains at the correct height above the membrane surface for pore formation. The height (∼50 Å) is also consistent with the predicted hairpins of C8 and C9, which have ∼30 hydrophilic residues at the starts and ends of the amphipathic membrane-spanning sequences adopting extended conformations above the membrane.
Intriguingly, rotation of the regulatory segment of C6 produces a large shift of TS1, such that is brought into close contact with the beginning of the nascent β-hairpins, where it might provide the final trigger to release the CH1 domains and/or create a local disruption of the membrane to promote insertion of the β-hairpins. Whatever its precise role(s) in promoting MAC initiation, it is intriguing that the TS1-TS2 tandem pair is conserved in the most ancient characterized C6 MAC-like component from cartilaginous fish (64) (whose earliest common ancestor with humans existed about 500 million years ago), as well as in a C6-like molecule from the chordate, amphioxus (65).
Final Steps in MAC Assembly
We propose that our general model of unidirectional transmission of conformational changes applies to the addition of each new protomer to the nascent/growing MAC (Fig. 6). The next step is the encounter between membrane-bound C5b-7 and solute phase C8β within the C8αβγ complex (66). C6 should at this point resemble C8α, with an open twisted sheet in contrast to the more closed untwisted sheet of C8β. Iterating the process described above, C8β approaches C6, forming an encounter complex. C6 then rotates its TS2 domain to complete the new C6-C8β interface, but in a concerted motion thrusts its EGF domain into the CH1 enclosure of C8β. This drives the opening and untwisting of the C8β sheet, so that it closely resembles C8α. In so doing, it also disrupts the CH3 elements of C8α, causing them to release their grip on CH2. These motions would then bring the β-hairpins of four MACPF domains into close alignment and proximity, creating the possibility of forming a 16-stranded contiguous β-sheet (Fig. 7A). The predicted hairpins of C8α and C8β are amphipathic and long enough to traverse the bacterial membrane. As noted above, the β-hairpins of C6 and C7 hairpins are only long enough to insert their tips into the membrane, but this may create a local disturbance of the membrane that lowers the activation barrier for the (energetically demanding) insertion of the C8 hairpins into and across the membrane.
Once C8α is activated and inserted into the membrane, sequential recruitment of C9 molecules can presumably ensue. Note that the mechanism at this point involves both the “trigger” for opening the β-sheet by the insertion of the EGF module of the “receiver” into the CH1 enclosure of the incoming recruit as well as a template in the form of an open membrane-inserted β-sheet. This pathway rationalizes the directionality (the trigger operates in a clockwise direction as viewed from above) of assembly as well as its sequential nature. Therefore, in contrast to the CDCs, membrane insertion does not proceed via the assembly of a pre-pore above the membrane. Rather, a pore begins to form once C5b-8 assembles at the membrane, and the pore grows in size in a stepwise fashion as each new C9 is added, with each new recruit inserting two further β-hairpins, sequentially enlarging the pore (12, 21, 67, 68). Indeed, modeling suggests that it is sterically feasible to build a circular assembly starting with just four MACPF elements (Fig. 7B), and experiments suggest that the addition of only a single C9 is sufficient to create a transmembrane pore (69). The second β-hairpin of C9 (residues 200–260) has a large hydrophilic loop at its tip (residues 225–240) that may provide a strong anchor that is key to the formation of a stable membrane-permeating pore. However, the pore does not become SDS-stable until the MAC is complete (14). Based on our assembly model, we have built hypothetical atomic models of the MAC (Fig. 7C) as well as poly(C9) (supplemental Fig. 10).
Summary and Future Directions
In summary, although sequential assembly of the MAC pore via a series of distinct intermediates appears to be unique to the MAC, our model shares the following two major features with those of CDC pores (56): (i) opening of the β-sheets as a key step in assembly that releases the membrane-inserting elements; (ii) the orientation of the MACPF/CDC domain within the pore (which contrasts with the model proposed for perforin (22)). Furthermore, our detailed comparisons between C6 and C8 have allowed us to propose a novel mechanism of pore initiation and propagation, one that emphasizes roles for the auxiliary domains in this process. Thus, we propose what drives sheet opening, why assembly is unidirectional and sequential, and how a contiguous barrel is formed. It seems likely that all MACPF-based pores will have a similar architecture, although the mechanistic details of assembly will necessarily be influenced by the nature of the auxiliary domains.
Finally, although we are aware of the speculative nature of our model for MAC assembly, we note that it is readily testable. For example, crystal structures of stable subassemblies on the pathway to MAC formation, such as the C5b-6 and C5b-7 complexes, should reveal how C5b activates C6 and C7, and whether C6-C7 in the context of C5 does indeed resemble C8αβ. To define the orientation of the MACPF domains within the assembled pore, EM studies of the MAC (and also poly(C9)) are required at a resolution that allows the orientation of the constituent domains to be defined unambiguously; this may also require antibody labeling of defined epitopes on the predicted lumen (e.g. the CH3 and C8γ domains) and outer surface (e.g. TS3 domain) of the pore. Our results would also suggest that further modeling of the perforin pore be performed, allowing for the possibility of sheet opening.
Supplementary Material
Acknowledgments
We thank the outstanding beamline support team at the Stanford Synchrotron Radiation Laboratory (SSRL) for data collection facilities. The SSRL is a national synchrotron user facility operated by Stanford University on behalf of the United States Department of Energy, Office of Basic Energy Sciences, for synchrotron access.
This work was supported, in whole or in part, by National Institutes of Health Grant R21 HL094878 (to I. U. S.). This work was also supported by United States Army Medical Research and Materiel Command Grant DAMD17-03-2-0038 (to R. C. L.) and Multiple Sclerosis National Research Institute Grant 4061 (to R. G. D.).
This article was selected as a Paper of the Week.

This article contains supplemental Figs. 1–10.
The atomic coordinates and structure factors (code 3T5O) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- MAC
- membrane attack complex
- CCP
- complement control protein
- CDC
- cholesterol-dependent cytolysin
- FIM
- factor I module
- LR
- low density lipoprotein receptor class A repeat
- MACPF
- membrane attack complex perforin (domain)
- PFO
- perfringolysin O
- PDB
- Protein Data Bank.
REFERENCES
- 1. Kimura A., Ikeo K., Nonaka M. (2009) Evolutionary origin of the vertebrate blood complement and coagulation systems inferred from liver EST analysis of lamprey. Dev. Comp. Immunol. 33, 77–87 [DOI] [PubMed] [Google Scholar]
- 2. Dourmashkin R. R. (1978) The structural events associated with the attachment of complement components to cell membranes in reactive lysis. Immunology 35, 205–212 [PMC free article] [PubMed] [Google Scholar]
- 3. Podack E. R., Esser A. F., Biesecker G., Müller-Eberhard H. J. (1980) Membrane attack complex of complement: a structural analysis of its assembly. J. Exp. Med. 151, 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. DiScipio R. G. (1998) in The Complement System (Rother K., Till G. O., Hansch G. M., eds) 2 Ed., pp. 50–68, Springer-Verlag, New York [Google Scholar]
- 5. Müller-Eberhard H. J. (1986) The membrane attack complex of complement. Annu. Rev. Immunol. 4, 503–528 [DOI] [PubMed] [Google Scholar]
- 6. Ducret F., Decoux M., Pointet P., Lambert C., Grosperrin E., Sédaillan A. (1988) Hereditary C5 deficiency and recurrent Neisseria meningitidis meningitis. Rev. Med. Intern. 9, 534–537 [DOI] [PubMed] [Google Scholar]
- 7. Corvini M., Randolph C., Aronin S. I. (2004) Complement C7 deficiency presenting as recurrent aseptic meningitis. Ann. Allergy Asthma Immunol. 93, 200–205 [DOI] [PubMed] [Google Scholar]
- 8. Orren A., Potter P. C. (2004) Complement component C6 deficiency and susceptibility to Neisseria meningitidis infections. S. Afr. Med. J. 94, 345–346 [PubMed] [Google Scholar]
- 9. Shinkai Y., Takio K., Okumura K. (1988) Homology of perforin to the ninth component of complement (C9). Nature 334, 525–527 [DOI] [PubMed] [Google Scholar]
- 10. Rosado C. J., Kondos S., Bull T. E., Kuiper M. J., Law R. H., Buckle A. M., Voskoboinik I., Bird P. I., Trapani J. A., Whisstock J. C., Dunstone M. A. (2008) The MACPF/CDC family of pore-forming toxins. Cell. Microbiol. 10, 1765–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. DiScipio R. G., Chakravarti D. N., Muller-Eberhard H. J., Fey G. H. (1988) The structure of human complement component C7 and the C5b-7 complex. J. Biol. Chem. 263, 549–560 [PubMed] [Google Scholar]
- 12. Tschopp J. (1984) Ultrastructure of the membrane attack complex of complement. heterogeneity of the complex caused by different degree of C9 polymerization. J. Biol. Chem. 259, 7857–7863 [PubMed] [Google Scholar]
- 13. Podack E. R., Tschoop J., Müller-Eberhard H. J. (1982) Molecular organization of C9 within the membrane attack complex of complement. Induction of circular C9 polymerization by the C5b-8 assembly. J. Exp. Med. 156, 268–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Podack E. R. (1984) Molecular composition of the tubular structure of the membrane attack complex of complement. J. Biol. Chem. 259, 8641–8647 [PubMed] [Google Scholar]
- 15. Hu V. W., Esser A. F., Podack E. R., Wisnieski B. J. (1981) The membrane attack mechanism of complement. Photolabeling reveals insertion of terminal proteins into target membrane. J. Immunol. 127, 380–386 [PubMed] [Google Scholar]
- 16. Podack E. R., Stoffel W., Esser A. F., Müller-Eberhard H. J. (1981) Membrane attack complex of complement: distribution of subunits between the hydrocarbon phase of target membranes and water. Proc. Natl. Acad. Sci. U.S.A. 78, 4544–4548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Steckel E. W., Welbaum B. E., Sodetz J. M. (1983) Evidence of direct insertion of terminal complement proteins into cell membrane bilayers during cytolysis. Labeling by a photosensitive membrane probe reveals a major role for the eighth and ninth components. J. Biol. Chem. 258, 4318–4324 [PubMed] [Google Scholar]
- 18. Hadders M. A., Beringer D. X., Gros P. (2007) Structure of C8α-MACPF reveals mechanism of membrane attack in complement immune defense. Science 317, 1552–1554 [DOI] [PubMed] [Google Scholar]
- 19. Rosado C. J., Buckle A. M., Law R. H., Butcher R. E., Kan W. T., Bird C. H., Ung K., Browne K. A., Baran K., Bashtannyk-Puhalovich T. A., Faux N. G., Wong W., Porter C. J., Pike R. N., Ellisdon A. M., Pearce M. C., Bottomley S. P., Emsley J., Smith A. I., Rossjohn J., Hartland E. L., Voskoboinik I., Trapani J. A., Bird P. I., Dunstone M. A., Whisstock J. C. (2007) A common fold mediates vertebrate defense and bacterial attack. Science 317, 1548–1551 [DOI] [PubMed] [Google Scholar]
- 20. Bhakdi S., Tranum-Jensen J. (1978) Molecular nature of the complement lesion. Proc. Natl. Acad. Sci. U.S.A. 75, 5655–5659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Czajkowsky D. M., Hotze E. M., Shao Z., Tweten R. K. (2004) Vertical collapse of a cytolysin prepore moves its transmembrane β-hairpins to the membrane. EMBO J. 23, 3206–3215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Law R. H., Lukoyanova N., Voskoboinik I., Caradoc-Davies T. T., Baran K., Dunstone M. A., D'Angelo M. E., Orlova E. V., Coulibaly F., Verschoor S., Browne K. A., Ciccone A., Kuiper M. J., Bird P. I., Trapani J. A., Saibil H. R., Whisstock J. C. (2010) The structural basis for membrane binding and pore formation by lymphocyte perforin. Nature 468, 447–451 [DOI] [PubMed] [Google Scholar]
- 23. Lichtenheld M. G., Olsen K. J., Lu P., Lowrey D. M., Hameed A., Hengartner H., Podack E. R. (1988) Structure and function of human perforin. Nature 335, 448–451 [DOI] [PubMed] [Google Scholar]
- 24. Voskoboinik I., Smyth M. J., Trapani J. A. (2006) Perforin-mediated target-cell death and immune homeostasis. Nat. Rev. Immunol. 6, 940–952 [DOI] [PubMed] [Google Scholar]
- 25. Lovelace L. L., Cooper C. L., Sodetz J. M., Lebioda L. (2011) Structure of human C8 protein provides mechanistic insight into membrane pore formation by complement. J. Biol. Chem. 286, 17585–17592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Scibek J. J., Plumb M. E., Sodetz J. M. (2002) Binding of human complement C8 to C9. Role of the N-terminal modules in the C8 α subunit. Biochemistry 41, 14546–14551 [DOI] [PubMed] [Google Scholar]
- 27. Plumb M. E., Scibek J. J., Barber T. D., Dunlap R. J., Platteborze P. L., Sodetz J. M. (1999) Chimeric and truncated forms of human complement protein C8 α reveal binding sites for C8β and C8γ within the membrane attack complex/perforin region. Biochemistry 38, 8478–8484 [DOI] [PubMed] [Google Scholar]
- 28. Taylor K. M., Trimby A. R., Campbell A. K. (1997) Mutation of recombinant complement component C9 reveals the significance of the N-terminal region for polymerization. Immunology 91, 20–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. DiScipio R. G., Sweeney S. P. (1994) The fractionation of human plasma proteins. II. The purification of human complement proteins C3, C3u, and C5 by application of affinity chromatography. Protein Expr. Purif. 5, 170–177 [DOI] [PubMed] [Google Scholar]
- 30. Leslie A. G. (2006) The integration of macromolecular diffraction data. Acta Crystallogr. D Biol. Crystallogr. 62, 48–57 [DOI] [PubMed] [Google Scholar]
- 31. Potterton E., Briggs P., Turkenburg M., Dodson E. (2003) A graphical user interface to the CCP4 program suite. Acta Crystallogr. D Biol. Crystallogr. 59, 1131–1137 [DOI] [PubMed] [Google Scholar]
- 32. Strong M., Sawaya M. R., Wang S., Phillips M., Cascio D., Eisenberg D. (2006) Toward the structural genomics of complexes. Crystal structure of a PE-PPE protein complex from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 103, 8060–8065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Terwilliger T. C. (2003) SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzymol. 374, 22–37 [DOI] [PubMed] [Google Scholar]
- 34. Bricogne G., Vonrhein C., Flensburg C., Schiltz M., Paciorek W. (2003) Generation, representation, and flow of phase information in structure determination. Recent developments in and around SHARP 2.0. Acta Crystallogr. D Biol. Crystallogr. 59, 2023–2030 [DOI] [PubMed] [Google Scholar]
- 35. Jaroszewski L., Rychlewski L., Li Z., Li W., Godzik A. (2005) FFAS03. A server for profile-profile sequence alignments. Nucleic Acids Res. 33, W284–W288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cowtan K. (1998) Modified phased translation functions and their application to molecular-fragment location. Acta Crystallogr. D Biol. Crystallogr. 54, 750–756 [DOI] [PubMed] [Google Scholar]
- 37. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., Warren G. L. (1998) Crystallography & NMR system. A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 [DOI] [PubMed] [Google Scholar]
- 38. Adams P. D., Afonine P. V., Bunkoczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hun L. W., Kapral G. J., Grosse-Kunstleve R. W. (2010) PHENIX. A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 2123–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Laskowski R. A., MacAuthor M. W., Moss D. S., Thornton J. M. (1993) PROCHECK. A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 41. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF Chimera. A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 42. Hofsteenge J., Blommers M., Hess D., Furmanek A., Miroshnichenko O. (1999) The four terminal components of the complement system are C-mannosylated on multiple tryptophan residues. J. Biol. Chem. 274, 32786–32794 [DOI] [PubMed] [Google Scholar]
- 43. DiScipio R. G., Hugli T. E. (1989) The molecular architecture of human complement component C6. J. Biol. Chem. 264, 16197–16206 [PubMed] [Google Scholar]
- 44. Haefliger J. A., Tschopp J., Vial N., Jenne D. E. (1989) Complete primary structure and functional characterization of the sixth component of the human complement system. Identification of the C5b-binding domain in complement C6. J. Biol. Chem. 264, 18041–18051 [PubMed] [Google Scholar]
- 45. Rossjohn J., Feil S. C., McKinstry W. J., Tweten R. K., Parker M. W. (1997) Structure of a cholesterol-binding, thiol-activated cytolysin and a model of its membrane form. Cell 89, 685–692 [DOI] [PubMed] [Google Scholar]
- 46. Tan K., Duquette M., Liu J. H., Dong Y., Zhang R., Joachimiak A., Lawler J., Wang J. H. (2002) Crystal structure of the TSP-1 type 1 repeats. A novel layered fold and its biological implication. J. Cell Biol. 159, 373–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Arias-Moreno X., Cuesta-Lopez S., Millet O., Sancho J., Velazquez-Campoy A. (2010) Thermodynamics of protein-cation interaction. Ca2+ and Mg2+ binding to the fifth binding module of the LDL receptor. Proteins 78, 950–961 [DOI] [PubMed] [Google Scholar]
- 48. Kirkitadze M. D., Barlow P. N. (2001) Structure and flexibility of the multiple domain proteins that regulate complement activation. Immunol. Rev. 180, 146–161 [DOI] [PubMed] [Google Scholar]
- 49. Schmidt C. Q., Herbert A. P., Mertens H. D., Guariento M., Soares D. C., Uhrin D., Rowe A. J., Svergun D. I., Barlow P. N. (2010) The central portion of factor H (modules 10–15) is compact and contains a structurally deviant CCP module. J. Mol. Biol. 395, 105–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Soares D. C., Barlow P. N. (2005) in Structural Biology of the Complement System (Lambris J. D., Morikis D., eds) pp. 21–59, CRC Press, Inc., Boca Raton, FL [Google Scholar]
- 51. Phelan M. M., Thai C. T., Soares D. C., Ogata R. T., Barlow P. N., Bramham J. (2009) Solution structure of factor I-like modules from complement C7 reveals a pair of follistatin domains in compact pseudosymmetric arrangement. J. Biol. Chem. 284, 19637–19649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Thai C. T., Ogata R. T. (2004) Complement components C5 and C7. Recombinant factor I modules of C7 bind to the C345C domain of C5. J. Immunol. 173, 4547–4552 [DOI] [PubMed] [Google Scholar]
- 53. Thai C. T., Ogata R. T. (2005) Recombinant C345C and factor I modules of complement components C5 and C7 inhibit C7 incorporation into the complement membrane attack complex. J. Immunol. 174, 6227–6232 [DOI] [PubMed] [Google Scholar]
- 54. DiScipio R. G., Linton S. M., Rushmere N. K. (1999) Function of the factor I modules (FIMS) of human complement component C6. J. Biol. Chem. 274, 31811–31818 [DOI] [PubMed] [Google Scholar]
- 55. Würzner R., Hobart M. J., Fernie B. A., Mewar D., Potter P. C., Orren A., Lachmann P. J. (1995) Molecular basis of subtotal complement C6 deficiency. A carboxyl-terminally truncated but functionally active C6. J. Clin. Invest. 95, 1877–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tilley S. J., Orlova E. V., Gilbert R. J., Andrew P. W., Saibil H. R. (2005) Structural basis of pore formation by the bacterial toxin pneumolysin. Cell 121, 247–256 [DOI] [PubMed] [Google Scholar]
- 57. Hayward S., Lee R. A. (2002) Improvements in the analysis of domain motions in proteins from conformational change. DynDom version 1.50. J. Mol. Graph. Model. 21, 181–183 [DOI] [PubMed] [Google Scholar]
- 58. Laine R. O., Esser A. F. (1989) Detection of refolding conformers of complement protein C9 during insertion into membranes. Nature 341, 63–65 [DOI] [PubMed] [Google Scholar]
- 59. Kolb W. P., Müller-Eberhard H. J. (1975) Neoantigens of the membrane attack complex of human complement. Proc. Natl. Acad. Sci. U.S.A. 72, 1687–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Silversmith R. E., Nelsestuen G. L. (1986) Interaction of complement proteins C5b-6 and C5b-7 with phospholipid vesicles. Effects of phospholipid structural features. Biochemistry 25, 7717–7725 [DOI] [PubMed] [Google Scholar]
- 61. DiScipio R. G. (1992) Formation and structure of the C5b-7 complex of the lytic pathway of complement. J. Biol. Chem. 267, 17087–17094 [PubMed] [Google Scholar]
- 62. Würzner R., Mewar D., Fernie B. A., Hobart M. J., Lachmann P. J. (1995) Importance of the third thrombospondin repeat of C6 for terminal complement complex assembly. Immunology 85, 214–219 [PMC free article] [PubMed] [Google Scholar]
- 63. Podack E. R., Kolb W. P., Müller-Eberhard H. J. (1978) The C5b-6 complex. Formation, isolation, and inhibition of its activity by lipoprotein and the S-protein of human serum. J. Immunol. 120, 1841–1848 [PubMed] [Google Scholar]
- 64. Kimura A., Nonaka M. (2009) Molecular cloning of the terminal complement components C6 and C8β of cartilaginous fish. Fish Shellfish Immunol. 27, 768–772 [DOI] [PubMed] [Google Scholar]
- 65. Suzuki M. M., Satoh N., Nonaka M. (2002) C6-like and C3-like molecules from the cephalochordate, amphioxus, suggest a cytolytic complement system in invertebrates. J. Mol. Evol. 54, 671–679 [DOI] [PubMed] [Google Scholar]
- 66. Brannen C. L., Sodetz J. M. (2007) Incorporation of human complement C8 into the membrane attack complex is mediated by a binding site located within the C8β MACPF domain. Mol. Immunol. 44, 960–965 [DOI] [PubMed] [Google Scholar]
- 67. Zalman L. S., Müller-Eberhard H. J. (1990) Comparison of channels formed by poly C9, C5b-8, and the membrane attack complex of complement. Mol. Immunol. 27, 533–537 [DOI] [PubMed] [Google Scholar]
- 68. Tschopp J., Podack E. R., Müller-Eberhard H. J. (1985) The membrane attack complex of complement. C5b-8 complex as accelerator of C9 polymerization. J. Immunol. 134, 495–499 [PubMed] [Google Scholar]
- 69. Bhakdi S., Tranum-Jensen J., Klump O. (1980) The terminal membrane C5b-9 complex of human complement. Evidence for the existence of multiple protease-resistant polypeptides that form the trans-membrane complement channel. J. Immunol. 124, 2451–2457 [PubMed] [Google Scholar]
- 70. Kleywegt G. J., Brünger A. T. (1996) Checking your imagination. Applications of the free R value. Structure 4, 897–904 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.