Background: There is an urgent need to develop small molecule Mcl-1-specific inhibitors for the treatment of Mcl-1-dependent ABT-737/263-resistant cancers.
Results: Maritoclax binds to and induces Mcl-1 degradation, thereby leading to Mcl-1-dependent apoptosis and sensitizing leukemia/lymphoma cells to ABT-737.
Conclusion: Maritoclax is a novel Mcl-1-specific inhibitor.
Significance: Antagonizing Mcl-1 by maritoclax has the potential to prevent and overcome Mcl-1-mediated resistance to ABT-737/263.
Keywords: Anticancer Drug, Apoptosis, Bcl-2 Family Proteins, Drug Discovery, Drug Resistance, ABT-737, Marinopyrrole A, Maritoclax, Mcl-1, Obatoclax
Abstract
The anti-apoptotic Bcl-2 family of proteins, including Bcl-2, Bcl-XL and Mcl-1, are well-validated drug targets for cancer treatment. Several small molecules have been designed to interfere with Bcl-2 and its fellow pro-survival family members. While ABT-737 and its orally active analog ABT-263 are the most potent and specific inhibitors to date that bind Bcl-2 and Bcl-XL with high affinity but have a much lower affinity for Mcl-1, they are not very effective as single agents in certain cancer types because of elevated levels of Mcl-1. Accordingly, compounds that specifically target Mcl-1 may overcome this resistance. In this study, we identified and characterized the natural product marinopyrrole A as a novel Mcl-1-specific inhibitor and named it maritoclax. We found that maritoclax binds to Mcl-1, but not Bcl-XL, and is able to disrupt the interaction between Bim and Mcl-1. Moreover, maritoclax induces Mcl-1 degradation via the proteasome system, which is associated with the pro-apoptotic activity of maritoclax. Importantly, maritoclax selectively kills Mcl-1-dependent, but not Bcl-2- or Bcl-XL-dependent, leukemia cells and markedly enhances the efficacy of ABT-737 against hematologic malignancies, including K562, Raji, and multidrug-resistant HL60/VCR, by ∼60- to 2000-fold at 1–2 μm. Taken together, these results suggest that maritoclax represents a new class of Mcl-1 inhibitors, which antagonizes Mcl-1 and overcomes ABT-737 resistance by targeting Mcl-1 for degradation.
Introduction
Apoptosis is the best-characterized mode of physiological cell death, which plays an essential role in the development and homeostasis of multicellular organisms. Apoptosis is executed by caspases, a family of cysteine proteases, whose activation is initiated via two major pathways: the death receptor (extrinsic) pathway and the mitochondrial (intrinsic) pathway. The activated caspases cleave a number of cellular proteins to generate many of the hallmark morphological features of apoptosis, including DNA fragmentation and membrane blebbing.
The Bcl-2 family of proteins plays a pivotal role in apoptosis by regulating mitochondrial outer membrane permeabilization (MOMP).2 MOMP results in the release of apoptogenic factors (e.g. cytochrome c and Smac) from the mitochondria into the cytosol where they directly promote caspase activation and subsequent cell death. Members of the Bcl-2 family contain up to four evolutionarily conserved domains called Bcl-2 homology (BH) domains 1 to 4 and can be classified into three groups based on their domain architecture and function in apoptosis: multidomain (BH1–4) anti-apoptotic Bcl-2 proteins (e.g. Bcl-2, Bcl-XL, and Mcl-1), multidomain (BH1–3) pro-apoptotic Bcl-2 proteins (e.g. Bax and Bak), and BH3-only Bcl-2 proteins (e.g. Bad, Bid, Bim, Noxa, and Puma). Many of the Bcl-2 family proteins can interact with each other to determine cell fate. Three-dimensional structures reveal that the BH1–3 domains of anti-apoptotic Bcl-2 proteins form a hydrophobic surface groove to which the BH3 domains of pro-apoptotic Bcl-2 family members bind (1, 2). The multidomain pro-apoptotic Bcl-2 proteins Bax and Bak are two major effectors of MOMP, which homo-oligomerize and form pores in the mitochondrial outer membrane to induce MOMP upon apoptotic stimulation. The anti-apoptotic Bcl-2 proteins prevent MOMP by directly binding to both classes of pro-apoptotic Bcl-2 proteins. In contrast, the BH3-only proteins trigger Bax and Bak to induce MOMP. Based on their ability to interact with the multidomain anti- and pro-apoptotic Bcl-2 proteins, the BH3-only proteins are often further divided into two subgroups: direct activators and sensitizers/de-repressors. The direct activators, including Bid, Bim and Puma, are not only able to interact with and inhibit all the anti-apoptotic Bcl-2 proteins but also directly bind to and activate the effectors Bax and Bak. On the other hand, the sensitizers/de-repressors appear to function essentially as transdominant inhibitors by occupying the hydrophobic groove of anti-apoptotic Bcl-2 proteins, thereby displacing the direct activators to promote MOMP and prevent any future bindings of the direct activators or effectors to anti-apoptotic Bcl-2 proteins. Moreover, unlike the direct activators, the sensitizers/de-repressors are more selective in binding to the anti-apoptotic Bcl-2 members. For example, Bad binds and antagonizes Bcl-2 and Bcl-XL but not Mcl-1, whereas Noxa binds and antagonizes Mcl-1 but not Bcl-2 and Bcl-XL. This observation suggests that the BH3-only proteins provide a fine control of MOMP in a Bax/Bak-dependent manner and opportunities to design specific inhibitors for each of the anti-apoptotic Bcl-2 family members.
The evasion of apoptosis is considered to be a hallmark of cancers and a cause of resistance to radiation and chemotherapies. Consistently, high levels of the anti-apoptotic Bcl-2 family proteins are associated with the pathogenesis of cancer and resistance to therapy (3, 4). A recent analysis of somatic copy number alterations (SCNAs) showed that two anti-apoptotic Bcl-2 family genes (Bcl-XL and Mcl-1) undergo frequent somatic amplifications in multiple cancers and that cancer cells carrying Bcl-XL and Mcl-1 amplifications are dependent on the expression of these genes for survival (5). Thus, Bcl-XL and Mcl-1 are very attractive targets for the development of anticancer agents.
Over the last few years, several small molecule Bcl-2 inhibitors have been synthesized as BH3 mimetics and some of these molecules have entered clinical trials (6–8). Although Bcl-2 and Bcl-XL have been the primary focus for the design of small molecule inhibitors, recent studies have demonstrated that Mcl-1 also plays an important role for cancer cell survival and that it is necessary to neutralize both arms of the anti-apoptotic Bcl-2 family (Bcl-2/Bcl-XL and Mcl-1) for apoptosis to occur in many cell types (9).
To date, the most potent and selective small-molecule Bcl-2 inhibitors are ABT-737 and its orally active analog ABT-263, which inhibit Bcl-2 and Bcl-XL at subnanomolar concentrations but only weakly target Mcl-1 (10). Consequently, these agents generally lack efficacy in cancers with elevated Mcl-1 and in many instances this resistance can be overcome by down-regulation of Mcl-1 (10–16). Moreover, it has recently been shown that cancer cells can quickly acquire resistance to ABT-737 by up-regulation of Mcl-1 (17, 18), suggesting that a treatment regime combining ABT-737 with a Mcl-1-specific inhibitor may be necessary to overcome the resistance against ABT-737.
In this report, we report on the identification and characterization of marinopyrrole A (referred to as maritoclax) as a novel class of Mcl-1 inhibitors. Maritoclax is a natural product recently identified from a species of marine-derived streptomycetes and has been reported to exhibit excellent antimicrobial activity against methicillin-resistant Staphylococcus aureus (19–21). We found that maritoclax antagonizes Mcl-1 by directly binding to and targeting Mcl-1 for proteasomal degradation. Moreover, maritoclax induces apoptosis selectively in Mcl-1-dependent cells and synergistically sensitizes cancer cells to ABT-737 by down-regulation of Mcl-1. This study is the first to identify a small-molecule Mcl-1-specific inhibitor that binds Mcl-1 and induces its degradation in human cancer cells.
EXPERIMENTAL PROCEDURES
Antibodies and Compounds
Antibodies were obtained from the following sources: mouse monoclonal anti-Mcl-1 (BD Pharmingen 559027); rabbit polyclonal anti-phospho-Mcl-1 (Ser159/Thr163; Cell Signaling 4579); mouse monoclonal anti-Bcl-XL (Sigma B9429); rabbit polyclonal anti-Bim (Sigma B7929); mouse monoclonal anti-cytochrome c (BD Pharmingen 556432); rabbit polyclonal anti-Bak (Millipore 06-536); rabbit polyclonal anti-caspase-3 (22), rabbit polyclonal anti-PARP (Cell Signaling 9542); mouse monoclonal anti-β-actin (Sigma A5441); mouse monoclonal anti-GAPDH (Imgenex 5019A); rabbit polyclonal anti-Mcl-1 for immunoprecipitation (23); rat monoclonal anti-Bim (Millipore 17001); HRP-conjugated goat polyclonal anti-GST (Bethyl A190-121P). ABT-737 and dasatinib were obtained from Abbott Laboratories and Bristol-Myers Squibb, respectively, as described (24). Obatoclax and MG132 were purchased from Active Biochem and Sigma, respectively. Marinopyrrole A (maritoclax) was synthesized as racemic mixture as previously described (25).
Protein Expression and Purification
A cDNA fragment encoding mouse Mcl-1 with an N-terminal truncation of 151 residues and a C-terminal truncation of 23 residues (Mcl-1ΔNC23) (26) was amplified by PCR and cloned into the pGEX-6P-1 vector (GE Healthcare). A cDNA fragment encoding human Bcl-XL lacking the C-terminal transmembrane (TM) domain was released from the pGEX-4T-Bcl-XLΔTM plasmid (27) and subcloned into the pGEX-6P-1 vector. The GST-tagged Mcl-1 and Bcl-XL proteins were expressed in Escherichia coli BL21 (DE3) and purified as previously described (27). For NMR studies, the GST-Mcl-1 fusion protein was prepared by growth of the transformed E. coli in minimal media using 15NH4Cl as the sole source of nitrogen. The 15N-labeled Mcl-1 protein was cleaved off from GST with Prescission Protease (GE Healthcare) and further purified on a Superdex 200 column (GE Healthcare) in 50 mm Tris (pH 8.0) and 150 mm NaCl. NMR samples containing 0.5 mm 15N-labeled Mcl-1 protein were prepared in 50 mm sodium phosphate (pH 6.7), 70 mm NaCl, and 0.04% sodium azide in H2O:2H2O (95:5) as described (26).
Enzyme-linked Immunosorbent Assay (ELISA)
In brief, 100 μl of 100 nm biotinylated Bim BH3 peptide (biotin-(β)A(β)ADMRPEIWIAQELRRIGDEFNAYYARR-amide) in SuperBlock Blocking Buffer in PBS (Thermo Scientific) was added into Reacti-BindTM Streptavidin High Binding Capacity Coated Plates (8-well strips, Pierce #15501) and incubated for at least 2 h with gentle shaking at room temperature or at 4 °C overnight. After washing with PBS, 200 μl of SuperBlock Blocking Buffer in PBS was added and incubated for 1 h with gentle shaking at room temperature. The plate can be completely dried and stored at 4 °C until use or immediately used for the experiment after washing three times with 200 μl of PBS. At the time of each experiment, compounds were prepared at 2× concentration in PBS containing 4% DMSO and pre-incubated with 2× concentration of GST-tagged Mcl-1 (10 nm final concentration) or Bcl-XL (25 nm final concentration) proteins in 1.5 ml tubes for 1 h. Then, 100 μl of the protein/compound mixture was transferred to the ELISA plate and incubated for 2 h at room temperature with gentle shaking. After washing 5 times with 200 μl of wash buffer (PBS containing 0.05% Tween 20), the plate was incubated with 100 μl of HRP-conjugated anti-GST goat polyclonal antibody in SuperBlock Blocking Buffer in PBS for 1.5 h with gentle shaking at RT. Then, the plate was washed five times with 200 μl of wash buffer and rinsed with PBS to remove Tween-20. To develop the blue color, 100 μl of SureBlue TMB Microwell Peroxidase Substrate (Thermo Scientific) was added to each well and incubated for 10∼20 min at RT. The reaction was stopped by adding 100 μl of 1N HCl to each well, and the absorbance of each well was immediately measured (0.1 s) at 450 nm using a PerkinElmer 2030 multi-plate reader.
Nuclear Magnetic Resonance (NMR) Spectroscopy
A 0.5 mm 15N-labeled sample of Mcl-1ΔNC23 was prepared as described (2). A 15N-1H HSQC spectrum was collected on this sample. A 50 mm stock of maritoclax was prepared in deuterated DMSO. 12 μl of the maritoclax stock was added to the 15N-labeled Mcl-1 sample to achieve a final concentration of 1 mm. A 15N-1H HSQC spectrum was collected on this sample. This procedure was repeated five times for a total of six additions of maritoclax. This should have resulted in a final maritoclax concentration of 6 mm. In practice, the poor solubility of maritoclax made it impossible to determine the exact concentration after each addition. Precipitate was observed after each addition of maritoclax. This precipitation reduced the concentration of Mcl-1 and also reduced the volume of the sample. Following the last addition of maritoclax, the Mcl-1 concentration was estimated from UV absorption to be 0.215 mm. Binding was assumed to be saturated based on the minimal chemical shift differences observed between the 5th and 6th additions of maritoclax.
All of the 15N-1H HSQC spectra were recorded at 25 °C on Agilent VNMRS 800 spectrometer equipped with triple resonance cryogenic probe and Z-axis pulse field gradient. The sweep widths and complex points of the series of the HSQCs were 9615.4(t2)'2600(t1) Hz and 1024(t2)'256(t1). All NMR spectra were processed with nmrPipe and analyzed using nmrview software. Apodization was achieved in the 1H and 15N dimensions using a squared sine bell function shifted by 70º and followed by zero filling to twice the number of real data points. The 1H carrier frequency was set on the water peak, and 4.753 ppm of the water peak at 25 ºC was used as reference in this report. The amide 1H and 15N resonance assignments in the apo-Mcl-1 15N-1H HSQC were made based on the work by Day et al. (2). For the protein sequence G147-G308, 158 residues were assigned from a total 159 non-proline residues. Only the first residue, Gly-147, which is a remnant of the affinity tag, was not assigned. Using these assignments we were able to reliably identify 145 resonances in the 15N-1H HSQC spectrum of apo Mcl-1. Resonances for the other 13 residues were missing. Chemical shift changes were observed for specific residues following each addition of maritoclax. Very small chemical shift changes were observed between the final and penultimate additions, suggesting that binding was approaching saturation. The chemical shifts for all assigned residues at each titration point were tabulated and the average differences between the chemical shifts of apo Mcl-1 and the chemical shifts observed after the final addition of maritoclax were determined using the following relationship: Average Chemical Shift Change = ((DH2) + (DN2/5)/2)^0.5 where DH is the amide proton chemical shift difference in parts per million (ppm), and DN is the amide nitrogen chemical shift difference in ppm.
Molecular Modeling
Maritoclax docking was carried out using the GLIDE (Grid-based Ligand Docking from Energetics) program (28) from Schrödinger, L.L.C. OPLS-2005 force field (29) was applied in the GLIDE program. The optimal binding geometry for each model was obtained by utilization of Monte Carlo sampling techniques coupled with energy minimization. GLIDE uses a scoring method based on ChemScore (30) but with additional terms added for greater accuracy. The NMR solution structures of mouse Mcl-1 in complex with mouse NoxaB (2JM6.pdb) (26) were used for docking maritoclax to Mcl-1.
Cell Culture and Transfection
K562, Raji, HL60, HL60/VCR, Jurkat, and JurkatΔBak cell lines were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) and 100 units/ml penicillin, 100 μg/ml streptomycin, 0.25 μg/ml amphotericin B (Cellgro) at 37 °C and 5% CO2. Primary large-granular lymphocyte leukemia (LGLL) cells were obtained from LGLL patients as previously described (31) and an informed consent was signed for sample collection according to a protocol approved by the Institutional Review Board of Penn State Hershey Cancer Institute. Patients received no treatment at the time of sample acquisition. Peripheral blood mononuclear cells (PBMCs) from healthy donors were used as a control. K562 cells stably expressing Bcl-2-IRES-BimEL, Bcl-XL-IRES-BimEL, or Mcl-1-IRES-BimEL were generated by retroviral transduction and puromycin selection as previously described (24).
Cell Viability Assay
Cell viability was determined by measuring intracellular ATP levels with the CellTiter Glo Luminescent Cell Viability Assay kit (Promega G7571). EC50 values were calculated by non-linear regression analysis using SigmaPlot.
Co-immunoprecipitation
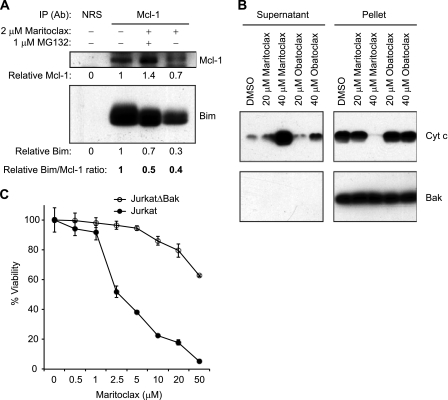
K562 cells expressing Mcl-1-IRES-BimEL were treated with DMSO, 2 μm maritoclax alone, or in combination with 1 μm MG132 for 12 h. Cells were lysed in 1% Chaps buffer (1% Chaps, 150 mm NaCl, 10 mm Hepes, pH7.4) containing protease inhibitors. Cell lysates containing 350 μg of protein were incubated with 4 μl of rabbit anti-Mcl-1 antiserum or control pre-immune serum in 250 μl of the same lysis buffer at 4 °C overnight on a rotator. Immunoprecipitates were collected by adding 20 μl of protein A-Sepharose beads (GE Healthcare) for 3 h at 4 °C, followed by centrifugation at 6,000 rpm for 30 s. The beads were washed five times with the same lysis buffer, boiled for 5 min in Laemmli sample buffer and analyzed by Western blotting.
Cytochrome c Release Assay
Mitochondria were isolated from K562 cells expressing Mcl-1-IRES-BimEL using Qproteome Mitochondria Isolation kit (Qiagen) in accordance with the manufacturer's instructions. The isolated mitochondrial pellet was resuspended in mitochondria assay buffer (210 mm d-mannitol, 70 mm sucrose, 5 mm KH2PO4, 4 mm MgCl2, 5 mm sodium succinate, 1 mm EGTA, 10 mm Hepes-NaOH, pH7.4) and the protein concentration was measured by BCA assay (Pierce). The purified mitochondria were incubated with DMSO, maritoclax (20 and 40 μm), or obatoclax (20 and 40 μm) for 1 h at 37 °C. After centrifugation at 13,000 × g for 10 min, the resulting supernatant and pellet were analyzed by Western blotting.
RESULTS
Maritoclax Binds to Mcl-1 and Inhibits the Binding of Bim-BH3 Peptide to Mcl-1 but Not Bcl-XL
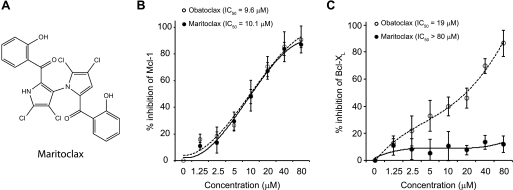
During the past several years, we have taken several approaches, including rational design (8), kinetic target-guided synthesis (27) and high throughput screening, to identify BH3-mimetic small-molecule inhibitors of Bcl-XL and Mcl-1. As part of this effort, we screened a small compound library comprised of marinopyrrole A (herein referred to as maritoclax; Fig. 1A) and 23 derivatives (25) for their ability to disrupt Bim binding to Mcl-1 and Bcl-XL using an ELISA assay. Of these, maritoclax blocked the interaction between a biotin-labeled Bim-BH3 peptide and GST-Mcl-1 in a dose-dependent manner with an IC50 value comparable to that of obatoclax (Fig. 1B). However, unlike obatoclax, which is a pan-Bcl-2 inhibitor (32), maritoclax did not inhibit the binding of Bim-BH3 peptide to GST-Bcl-XL at concentrations up to 80 μm (Fig. 1C).
FIGURE 1.
Maritoclax blocks the binding of Bim BH3 α-helix to Mcl-1 but not Bcl-XL. A, chemical structure of maritoclax. B, maritoclax and obatoclax are comparable in inhibiting the binding of a biotinylated Bim BH3 peptide to Mcl-1 protein as determined using an ELISA assay. C, in contrast to obatoclax, a pan Bcl-2 inhibitor, maritoclax does not block the binding of a biotinylated Bim BH3 peptide to Bcl-XL protein as determined using an ELISA assay.
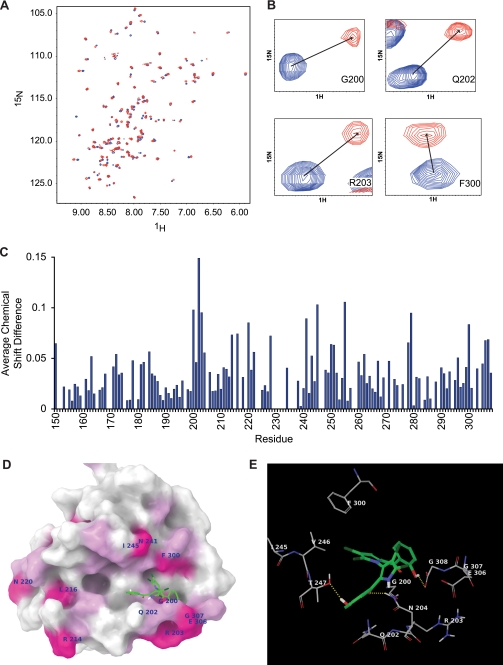
To determine whether maritoclax physically interacts with Mcl-1, NMR titration experiments were performed to monitor chemical shift perturbations in 1H-15N HSQC spectra of 15N-labeled Mcl-1 upon addition of maritoclax. Fig. 2A shows an overlay of the 1H-15N HSQC spectra of 15N-labeled Mcl-1 before (blue resonances) and after (red resonances) the addition of maritoclax. The 1H-15N HSQC spectrum of 15N-labeled Mcl-1 from the 6th titration point was used to make the figure. Fig. 2A shows that several residues experience significant chemical shift changes in the presence of maritoclax, strongly suggesting that maritoclax binds Mcl-1. Fig. 2B shows close-up views of selected residues that undergo large chemical shift changes in the presence of maritoclax. Fig. 2C shows a plot of the average chemical shift perturbations against the Mcl-1 protein primary sequence. The results show that the residues 150, 163, 172, 184, 200, 202–204, 214, 216, 220, 222, 228, 241, 243, 245, 250, 251, 255, 262, 278, 279, 300, 306, and 307 experience average chemical shift changes of at least 0.05 ppm and residues 200, 202, 203, 220, 241, 245, 255, 279, and 300 experience at changes of least 0.08 ppm. In addition, there were several residues that experienced intensity reductions associated with line broadening during the titration. In particular, residues 229, 235, 237, 244, and 246 have large intensity changes following the first addition of maritoclax, to a point where they are no longer detectable.
FIGURE 2.
Maritoclax directly binds to Mcl-1. A, comparison of the 1H-15N HSQC spectra of 15N-labeled Mcl-1 before (blue resonances) and after (red resonances) the addition of maritoclax. B, close-up of residues with large chemical shift changes. C, plot of average chemical shift changes (in ppm) in the spectra of 15N-labeled Mcl-1 upon titration with maritoclax. D, structure of maritoclax docked to Mcl-1. Mcl-1 is represented by its molecular surface colored according to NMR chemical shift data. Areas with average chemical shift greater than 0.06 are colored magenta with amino acid labels. Areas with an average chemical shift between 0.04 and 0.06 are colored light pink. A pyrrole group of maritoclax is docked in the p4 pocket with its phenol group extended to the tail region of Mcl-1 helix 8. The other phenol group is extended toward p3 pocket in the left. E, hydrogen bonding and amino acid residues with large chemical shifts are shown near the predicated binding site of maritoclax. Three hydrogen bonds between maritoclax and Mcl-1 are shown as yellow dotted lines. Carbon atoms of maritoclax are colored green. Carbon atoms of Mcl-1 are colored gray. Chlorine atoms are colored dark green, oxygen atoms red, nitrogen blue, and polar hydrogen white. Non-polar hydrogen atoms and amide hydrogen atoms are not shown.
The anti-apoptotic Bcl-2 family proteins share similar structural features of eight α-helices and a similar function of binding to BH3 domains (2). Mcl-1 has four hydrophobic pockets, namely p1, p2, p3 and p4 that interact with E74, L78, I81, and V85 of mouse NoxaB (mNoxaB) BH3 domain, respectively (2). When maritoclax is docked to Mcl-1, two binding regions are observed; one is centered at the p2 pocket between helices 4 and 5 and in contact with helix 3, and the other is centered at the p4 pocket between helices 5 and 8 and in contact with helix 2. The maritoclax binding site with the best docking score is located in the area centered at pocket p4, as shown in Fig. 2D. One pyrrole group of maritoclax is located in the p4 pocket with its chlorine atoms pointing toward Mcl-1. The connecting phenol group has its hydroxyl group forming a hydrogen bond with G308 of the Mcl-1 helix 8 tail region. Another pyrrole group is pointing toward the solvent and its connecting phenol group toward the p3 pocket with the hydroxyl group forming a hydrogen bond with T247 of Mcl-1 and the carbonyl group forming a hydrogen bond with N204. These hydrogen bonds are shown in Fig. 2E, together with the amino acids with large chemical shifts near the binding site. Among other residues with large chemical shifts, V255 is on helix 5 in contact with helix 3 and L279 is near the end of helix 6 in contact with helix 5. Their large chemical shifts could result from conformational changes upon ligand binding. L279 is not in a position of direct contact with ligand when the BH3 domain binds, but V255 is near the p2 pocket. Therefore, maritoclax binding site near pocket p4 may not be the only possible site, but it is the most probable site from the docking study, and it is consistent with the majority of the chemical shift data.
Maritoclax Kills Mcl-1-dependent but Not Bcl-2/Bcl-XL-dependent Cells
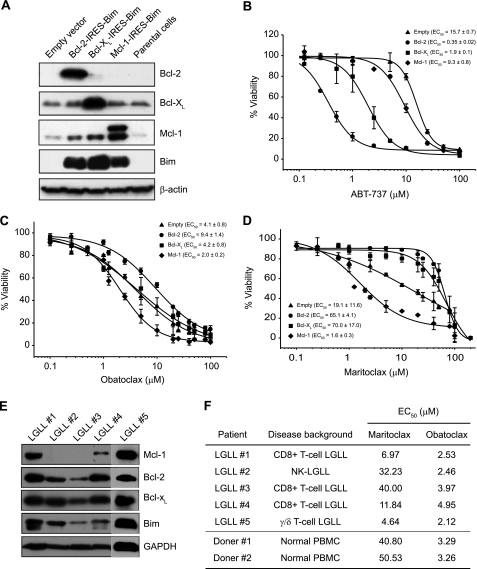
To study whether maritoclax selectively induces apoptosis in cells that are dependent on Mcl-1 for survival, we stably transfected human myeloid leukemia K562 cells with Bcl-2-IRES-BimEL, Bcl-XL-IRES-BimEL, Mcl-1-IRES-BimEL or control empty vector (Fig. 3A) and treated with increasing concentrations of ABT-737 (Fig. 3B), obatoclax (Fig. 3C) or maritoclax (Fig. 3D). The survival of these cells is dependent on a balanced association of BimEL with the respective pro-survival Bcl-2 family proteins (primed for death). Thus, these stable cell lines should enable us to test the specificity of BH3 mimetics for Bcl-2, Bcl-XL or Mcl-1. As expected, the Bcl-2/Bcl-XL specific inhibitor ABT-737 potently induced cell death in K562 cells expressing Bcl-2-IRES-BimEL (EC50 = 0.35 μm) or Bcl-XL-IRES-BimEL (EC50 = 1.9 μm) compared with Mcl-1-IRES-BimEL (EC50 = 9.3 μm) or empty vector (EC50 = 15.7 μm) transfectants (Fig. 3B), whereas the pan-Bcl-2 inhibitor obatoclax was able to kill all four cell lines with EC50 values ranging from 2.0 to 9.4 μm (Fig. 3C). In contrast, treatment with maritoclax markedly inhibited the viability of Mcl-1-IRES-BimEL cells (EC50 = 1.6 μm) with a selectivity greater than 40-fold over Bcl-2-IRES-BimEL (EC50 = 65.1 μm) and Bcl-XL-IRES-BimEL (EC50 = 70.0 μm) cells (Fig. 3D), consistent with its affinity for Mcl-1 over Bcl-XL (Figs. 1B and 1C). Moreover, maritoclax was able to kill primary human large granular lymphocyte leukemia (LGLL) cells and a negative correlation was observed between Mcl-1 protein levels and EC50 values (Fig. 3, E and F). In contrast, normal PBMCs were resistant to maritoclax (Fig. 3F). However, obatoclax induced similar levels of cell death in normal PBMCs and LGLL cells regardless of their expression of Bcl-2 family proteins (Fig. 3, E and F).
FIGURE 3.
Maritoclax induces cell death selectively in Mcl-1-dependent but not Bcl-2- or Bcl-XL-dependent leukemia cells. A, K562 cells were retrovirally transduced with empty, Bcl-2-IRES-BimEL, Bcl-XL-IRES-BimEL, or Mcl-1-IRES-BimEL and analyzed for expression of the proteins by immunoblot. The parental K562 cells were used as a control. B–D, stably transfected K562 cells were treated with increasing concentrations of ABT-737, obatoclax, or maritoclax for 24 h. Cell viability was determined by measuring intracellular ATP levels with the CellTiter Glo assay. All experiments were repeated at least three times to determine the EC50 values (mean ± S.D.). E, five primary human LGLL samples were analyzed by immunoblotting for expression of Bcl-2 family proteins. F, LGLL cells and normal PBMCs were treated with various concentrations of maritoclax for 24 h to determine EC50 values with the CellTiter Glo assay.
Maritoclax Induces Caspase-3 Activation by Degradation of Mcl-1 Protein
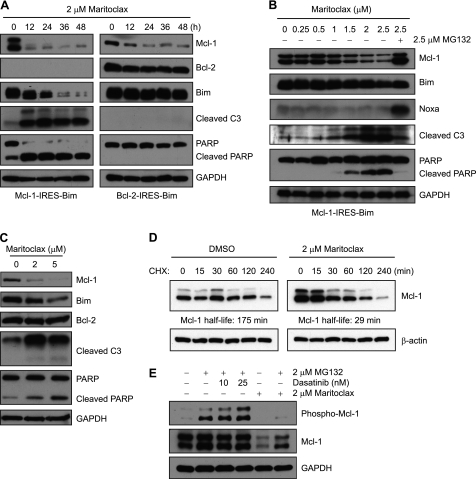
In agreement with the cell viability data, maritoclax induced robust caspase-3 activation in K562 cells expressing Mcl-1-IRES-BimEL but not Bcl-2-IRES-BimEL as demonstrated by procaspase-3 processing and PARP cleavage (Fig. 4A). Notably, maritoclax treatment led to a rapid and marked decrease in Mcl-1 protein levels. As shown in Fig. 4B, maritoclax induced a dose-dependent decrease in Mcl-1 expression, which was accompanied by an increase in cleavage of caspase-3 and PARP, in K562 cells stably transfected with Mcl-1-IRES-BimEL. Similar results were also observed in primary human LGLL cells (Fig. 4C). Interestingly, addition of the proteasome inhibitor MG132 attenuated maritoclax-mediated Mcl-1 degradation, caspase-3 activation and PARP cleavage (Fig. 4B), suggesting that the pro-apoptotic effect of maritoclax is attributed to its ability to induce proteasomal degradation of Mcl-1. Treatment with maritoclax markedly reduced the half-life of Mcl-1 to ∼0.5 h as compared with nearly 3 h in control cells (Fig. 4D). It has been shown that phosphorylation of Mcl-1 at S159 as well as expression of Noxa promotes the turnover of Mcl-1 protein (26, 33). However, neither Noxa expression (Fig. 4B) nor Mcl-1 phosphorylation (Fig. 4E) was induced by maritoclax. In contrast, dasatinib, a tyrosine kinase inhibitor, induced a dose-dependent phosphorylation of Mcl-1 (Fig. 4E) and obatoclax, a pan-Bcl-2 inhibitor, up-regulated Noxa expression (not shown), which are consistent with previous studies (24, 34).
FIGURE 4.
Maritoclax induces proteasome-mediated Mcl-1 degradation without induction of Mcl-1 phosphorylation and Noxa expression. A, Mcl-1-IRES-BimEL and Bcl-2-IRES-BimEL K562 cells were treated with 2 μm Maritoclax for the indicated times and analyzed by Western blotting. B, Mcl-1-IRES-BimEL K562 cells were treated with increasing concentrations of maritoclax (up to 2.5 μm) alone or a combination of 2.5 μm maritoclax and 2.5 μm MG132 for 12 h and subjected to immunoblot analysis. C, primary LGLL cells were treated with DMSO or maritoclax (2 or 5 μm) for 12 h and analyzed by Western blotting. D, Mcl-1-IRES-BimEL K562 cells were treated with 2 μm maritoclax for 1 h, followed by addition of 10 μg/ml CHX to block protein synthesis. Cells were harvested at the indicated time points and subjected to immunoblot analysis. The intensity of Mcl-1 bands was quantified by densitometry and normalized to β-actin. The half-life of Mcl-1 was calculated by linear regression equations. E, Mcl-1-IRES-BimEL K562 cells were left untreated or were treated as indicated for 6 h and analyzed by Western blotting for Ser159/Thr163 phosphorylation of Mcl-1.
Interestingly, a time- and dose-dependent decrease in the protein expression of BimEL was also observed following Mcl-1 degradation in cells treated with maritoclax (Fig. 4, A and B). Because free Mcl-1 and Bim were reported to be more susceptible to proteasomal degradation (35), the decreases in Mcl-1 and BimEL protein levels in maritoclax-treated cells are probably due to the disruption of the Bim-Mcl-1 complex by maritoclax. In fact, treatment with maritoclax disrupted the interaction between BimEL and Mcl-1 by 50–60%, as demonstrated by co-immunoprecipitation (Fig. 5A). Consistent with its ability to disrupt BimEL/Mcl-1 heterodimerization, maritoclax was able to induce cytochrome c release in vitro from mitochondria isolated from K562 cells stably expressing Mcl-1-IRES-BimEL with a much higher potency than obatoclax (Fig. 5B), suggesting that maritoclax directly engages MOMP. To further support this notion, we tested whether maritoclax induces cell death in a Bax/Bak-dependent manner using a subclone (JurkatΔBak) of Jurkat human acute T cell leukemia cell line that constitutively lacks Bax and Bak (36). As expected, maritoclax was much less effective against JurkatΔBak cells than it was against wild type Jurkat cells (Fig. 5C).
FIGURE 5.
Maritoclax inhibits Mcl-1 interaction with Bim in intact cells and triggers cytochrome c release from isolated mitochondria. A, Mcl-1-IRES-BimEL K562 cells were treated with DMSO or 2 μm maritoclax with or without 1 μm MG132 for 12 h and subjected to immunoprecipitation with anti-Mcl-1 rabbit antiserum or pre-immune rabbit serum (NRS). The resulting immune complexes were analyzed by immunoblotting with anti-Mcl-1 and anti-Bim monoclonal antibodies. The amounts of Mcl-1 and BimEL in the immunocomplexes were quantified and are listed relative to those of untreated cells, which were set as 1.0. B, isolated mitochondria from Mcl-1-IRES-BimEL K562 cells were incubated with DMSO, maritoclax (20 or 40 μm) or obatoclax (20 or 40 μm) for 1 h at 37 °C. After centrifugation, the resulting supernatant and pellet fractions were analyzed by Western blotting with antibodies specific for cytochrome c or Bak as a mitochondrial control marker. C, wild type and ΔBak Jurkat cells were treated with increasing concentrations of maritoclax for 24 h and subjected to cell viability assay (mean ± S.D.; n = 3).
Maritoclax Sensitizes Cancer Cells to ABT-737
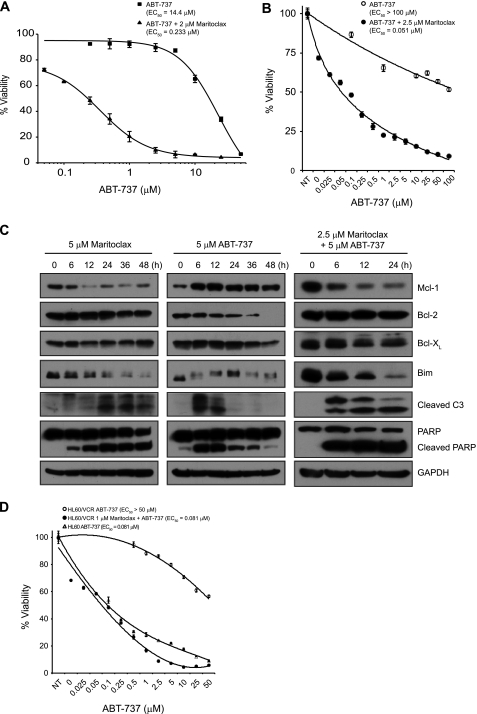
Although ABT-737 and its orally active analog, ABT-263, are the most potent and selective small-molecule Bcl-2 inhibitors described to date, they are not very effective against cancer cells that express high levels of Mcl-1, such as K562 and Raji cells (7, 10). To determine whether maritoclax sensitizes cancer cells to ABT-737, we treated K562 and Raji cells with increasing doses of ABT-737 alone or in combination with 2 or 2.5 μm maritoclax for K562 or Raji cells, respectively. As a single agent, ABT-737 was not efficient in killing K562 and Raji cells with EC50 values of 14.4 μm and >100 μm, respectively (Fig. 6, A and B). However, in combination with a sub-optimal dose of maritoclax, the efficacy of ABT-737 in K562 and Raji cells was enhanced by ∼60- and 2000-fold, respectively. In accordance with the results in K562 cells (Fig. 4A), treating Raji cells with maritoclax resulted in degradation of Mcl-1, which was followed by BimEL degradation, procaspase-3 processing, and PARP cleavage (Fig. 6C). In contrast, exposure of Raji cells to ABT-737 led to a rapid increase in Mcl-1 and a decrease in Bcl-2 protein levels. Interestingly, caspase-3 activation was observed only at early time points after ABT-737 treatment, suggesting that Raji cells can rapidly develop resistance to ABT-737 probably by up-regulating Mcl-1 expression. In fact, addition of maritoclax to ABT-737 attenuated the protein expression of Mcl-1 and enhanced the cleavage of caspase-3 and PARP in Raji cells. These data suggest that the pro-apoptotic synergy between maritoclax and ABT-737 is, at least in part, due to the degradation of Mcl-1 induced by maritoclax.
FIGURE 6.
Maritoclax synergistically sensitizes lymphoma/leukemia cells to ABT-737. A, parental K562 cells were treated with increasing concentrations of ABT-737 alone or in combination with 2 μm maritoclax for 24 h. Cell viability was determined by measuring intracellular ATP levels with the CellTiter Glo assay (mean ± S.D.; n = 3). B, Raji cells were treated with increasing doses of ABT-737 alone or in combination with 2.5 μm maritoclax for 24 h and subjected to cell viability assay (mean ± S.D.; n = 3). C, Raji cells were treated with 5 μm maritoclax, 5 μm ABT-737, or 2.5 μm maritoclax and 5 μm ABT-737 for the indicated times and analyzed by Western blotting. D, parental HL60 and HL60/VCR cells were treated with increasing concentrations of ABT-737 without or with 1 μm maritoclax for 48 h and subjected to cell viability assay (mean ± S.D.; n = 3).
It was suggested that up-regulation of Mcl-1 may contribute to the multidrug-resistance phenotype in human leukemia (39). Consistently, while parental HL60 cells were sensitive to ABT-737 (EC50 = 81.1 nm), the multidrug resistant variant HL60/VCR, which has elevated levels of Mcl-1, was resistant to ABT-737 (EC50 > 50 μm), as shown in Fig. 6D. Importantly, however, the efficacy of ABT-737 in HL60/VCR cells was fully restored when combined with 1 μm maritoclax; the EC50 value decreased ∼600-fold (Fig. 6D), suggesting that combination treatment of maritoclax with ABT-737 may overcome multidrug resistance.
DISCUSSION
Mcl-1 is an anti-apoptotic member of the Bcl-2 family of proteins and is highly expressed in a variety of human cancers (40). Mcl-1 overexpression has been demonstrated to be an important contributor to cancer cell survival and resistance to chemotherapy. To date, numerous strategies, including small-molecule BH3 mimetics, stapled BH3 peptides, and down-regulation of Mcl-1 by kinase inhibitors, deubiquitinase inhibitors, and antisense oligonucleotides, have been attempted to target Mcl-1 for cancer treatment (40, 41). However, none of the small molecule BH3 mimetics reported so far, including ABT-737, obatoclax, BH3-M6, gossypol, and its derivatives, are specific for Mcl-1.
In this study, we have identified the natural product marinopyrrole A as a novel Mcl-1-specific inhibitor and named it maritoclax. Our results suggest that maritoclax antagonizes Mcl-1 by binding to and targeting Mcl-1 for proteasome-mediated degradation. The Mcl-1 protein has a relatively short half-life (i.e. 2–3 h) and its stability is regulated at multiple levels. It has been demonstrated that ERK-mediated phosphorylation of Mcl-1 at Thr163 prolongs the half-life of Mcl-1 (42), whereas phosphorylation of Ser159 by GSK3 leads to increased ubiquitination and degradation of Mcl-1 (33). However, maritoclax had no apparent effect on Mcl-1 (Ser159/Thr163) phosphorylation (Fig. 4E), suggesting that maritoclax induces phosphorylation-independent Mcl-1 degradation. Moreover, Noxa overexpression has been shown to promote Mcl-1 degradation (26), probably by increasing the binding of the E3 ligase Mule and decreasing the binding of the deubiquitinase USP9X to Mcl-1 (43). However, maritoclax alone did not significantly affect Noxa protein levels (Fig. 4B). Noxa binding to Mcl-1 induces Mcl-1 degradation, and the amino acid sequence LRQKLL in the tail region of mNoxaB BH3 helix is considered to be necessary for this effect (26). Amino acid residues D83, K84, R88, and N93 of mNoxaB form hydrogen bonds with Mcl-1 residues R244, G308, E306, E298, and F299. These hydrogen bonds attach the C terminus of mNoxaB BH3 helix to the unfolded tail of Mcl-1 helix 8 (2, 26). Interestingly, the docking study predicted that maritoclax is likely to bind at the p4 pocket of Mcl-1 (Fig. 2D), mimicking the tail region of human Noxa BH3 or mNoxaB BH3 domain, which is important for inducing Mcl-1 degradation (26). The close interaction of maritoclax with the tail region of Mcl-1 helix 8, similar to that of mNoxaB, suggests that binding to the Mcl-1 helix 8 tail region may be related to Mcl-1 degradation in cells. Taken together, we speculate that maritoclax binds to Mcl-1 in a similar manner as Noxa, resulting in a conformational change that makes Mcl-1 more susceptible to destruction by the proteasome.
While certain cancers rely on Bcl-2/Bcl-XL for survival, others are more heavily dependent on Mcl-1. Accordingly, specific inhibitors for individual Bcl-2 family members are expected to be effective with less undesired side effects for a particular cancer based on such dependence. Indeed, ABT-737 and its orally active analog, ABT-263, have a single agent activity in a subset of cancers (including multiple myeloma and small-cell lung cancer) that rely on Bcl-2/Bcl-XL but not Mcl-1 for survival (7). However, recent studies demonstrated that cancer cells rapidly develop resistance to ABT-737 through up-regulation of Mcl-1 and that down-regulation of Mcl-1 restores the sensitivity to ABT-737 (38, 44). This suggests that the anti-apoptotic members of the Bcl-2 family can functionally compensate for each other. Similarly, in this study, we observed that ABT-737 as a single agent resulted in an increase in Mcl-1 protein levels, which was associated with the acquired resistance to ABT-737; however, co-treatment with a suboptimal dose of maritoclax (1∼2.5 μm) abolished Mcl-1 accumulation and markedly enhanced ABT-737 sensitivity (Fig. 6). Moreover, maritoclax was able to completely restore the sensitivity of multidrug resistant leukemia cells to ABT-737 (Fig. 6D). Taken together, these results support the notion that combination or sequential treatment with Mcl-1 inhibitors may be necessary for ABT-737 to achieve maximum therapeutic efficacy and avoid or overcome drug resistance. Accordingly, one might argue that it would be preferable to employ a pan rather than a more specific Bcl-2 inhibitor for the treatment of cancer. However, pan-Bcl-2 inhibitors have a higher likelihood of inducing toxicities than more specific inhibitors in normal cells or tissues (Fig. 3F). Thus, it will be important to investigate whether sequential treatment with ABT-737 followed by Mcl-1 inhibitors (e.g. maritoclax) has a synergistic effect on cancer cell death with much less toxicity to normal tissues when compared with pan Bcl-2 inhibitors.
In summary, maritoclax represents a novel class of Mcl-1 inhibitors, which binds to Mcl-1, presumably at the p4 binding site in a manner similar to Noxa, and displaces Bim from Mcl-1, thereby leading to Mcl-1 degradation via the proteasome system. This mode of action is different from that of obatoclax, which is predicted to bind at the p1 and p2 binding sites of Mcl-1 (37) and does not destabilize Mcl-1 (34). While obatoclax up-regulates Noxa (34), maritoclax has no effect on Noxa protein levels (Fig. 4B). Notably, maritoclax is much more efficient than obatoclax in inducing cytochrome c release from mitochondria isolated from cells co-transfected with BimEL and Mcl-1 (Fig. 5B). These results suggest that maritoclax is a specific Mcl-1 inhibitor. Consistently, maritoclax selectively kills Mcl-1-dependent but not Bcl-2- or Bcl-XL-dependent cells as a single agent and markedly synergizes with ABT-737 to induce apoptosis in ABT-737-resistant cancer cells by targeting Mcl-1 for proteasome-mediated degradation.
Acknowledgments
We thank Dr. Mark G. Hinds (Walter and Eliza Hall Institute of Medical Research) for the backbone and side-chain resonance assignments of the mouse Mcl-1 fragment; Dr. Myles Cabot (John Wayne Cancer Institute) for HL60/VCR cells; Dr. Hannah Rabinowich (University of Pittsburgh) for JurkatΔBak cells. NMR data were collected in the Florida Center of Excellence for Drug Discovery and Innovation.
This work was supported, in whole or in part, by National Institutes of Health Grant CA118210, by Moffitt Cancer Center startup funds (to R. L.), and by the National Science Foundation (Award No. MCB0939014) and the American Cancer Society (RSG-07-89-01-GMC) (to G. W. D.).
- MOMP
- mitochondrial outer membrane permeabilization
- SCNA
- somatic copy number alteration
- PBMC
- peripheral blood mononuclear cell
- CHX
- cycloheximide
- BH
- Bcl-2 homology.
REFERENCES
- 1. Sattler M., Liang H., Nettesheim D., Meadows R. P., Harlan J. E., Eberstadt M., Yoon H. S., Shuker S. B., Chang B. S., Minn A. J., Thompson C. B., Fesik S. W. (1997) Structure of Bcl-xL-Bak peptide complex: recognition between regulators of apoptosis. Science 275, 983–986 [DOI] [PubMed] [Google Scholar]
- 2. Day C. L., Smits C., Fan F. C., Lee E. F., Fairlie W. D., Hinds M. G. (2008) Structure of the BH3 domains from the p53-inducible BH3-only proteins Noxa and Puma in complex with Mcl-1. J. Mol. Biol. 380, 958–971 [DOI] [PubMed] [Google Scholar]
- 3. Reed J. C., Miyashita T., Takayama S., Wang H. G., Sato T., Krajewski S., Aimé-Sempé C., Bodrug S., Kitada S., Hanada M. (1996) BCL-2 family proteins: regulators of cell death involved in the pathogenesis of cancer and resistance to therapy. J. Cell Biochem. 60, 23–32 [DOI] [PubMed] [Google Scholar]
- 4. Reed J. C. (1997) Bcl-2 family proteins: strategies for overcoming chemoresistance in cancer. Adv. Pharmacol. 41, 501–532 [DOI] [PubMed] [Google Scholar]
- 5. Beroukhim R., Mermel C. H., Porter D., Wei G., Raychaudhuri S., Donovan J., Barretina J., Boehm J. S., Dobson J., Urashima M., Mc Henry K. T., Pinchback R. M., Ligon A. H., Cho Y. J., Haery L., Greulich H., Reich M., Winckler W., Lawrence M. S., Weir B. A., Tanaka K. E., Chiang D. Y., Bass A. J., Loo A., Hoffman C., Prensner J., Liefeld T., Gao Q., Yecies D., Signoretti S., Maher E., Kaye F. J., Sasaki H., Tepper J. E., Fletcher J. A., Tabernero J., Baselga J., Tsao M. S., Demichelis F., Rubin M. A., Janne P. A., Daly M. J., Nucera C., Levine R. L., Ebert B. L., Gabriel S., Rustgi A. K., Antonescu C. R., Ladanyi M., Letai A., Garraway L. A., Loda M., Beer D. G., True L. D., Okamoto A., Pomeroy S. L., Singer S., Golub T. R., Lander E. S., Getz G., Sellers W. R., Meyerson M. (2010) The landscape of somatic copy number alteration across human cancers. Nature 463, 899–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yip K. W., Reed J. C. (2008) Bcl-2 family proteins and cancer. Oncogene 27, 6398–6406 [DOI] [PubMed] [Google Scholar]
- 7. Vogler M., Dinsdale D., Dyer M. J., Cohen G. M. (2009) Bcl-2 inhibitors: small molecules with a big impact on cancer therapy. Cell Death Differ. 16, 360–367 [DOI] [PubMed] [Google Scholar]
- 8. Kazi A., Sun J., Doi K., Sung S. S., Takahashi Y., Yin H., Rodriguez J. M., Becerril J., Berndt N., Hamilton A. D., Wang H. G., Sebti S. M. (2011) The BH3 α-helical mimic BH3-M6 disrupts Bcl-X(L), Bcl-2, and MCL-1 protein-protein interactions with Bax, Bak, Bad, or Bim and induces apoptosis in a Bax- and Bim-dependent manner. J. Biol. Chem. 286, 9382–9392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Willis S. N., Chen L., Dewson G., Wei A., Naik E., Fletcher J. I., Adams J. M., Huang D. C. (2005) Proapoptotic Bak is sequestered by Mcl-1 and Bcl-xL, but not Bcl-2, until displaced by BH3-only proteins. Genes Dev. 19, 1294–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tse C., Shoemaker A. R., Adickes J., Anderson M. G., Chen J., Jin S., Johnson E. F., Marsh K. C., Mitten M. J., Nimmer P., Roberts L., Tahir S. K., Xiao Y., Yang X., Zhang H., Fesik S., Rosenberg S. H., Elmore S. W. (2008) ABT-263: a potent and orally bioavailable Bcl-2 family inhibitor. Cancer Res. 68, 3421–3428 [DOI] [PubMed] [Google Scholar]
- 11. Oltersdorf T., Elmore S. W., Shoemaker A. R., Armstrong R. C., Augeri D. J., Belli B. A., Bruncko M., Deckwerth T. L., Dinges J., Hajduk P. J., Joseph M. K., Kitada S., Korsmeyer S. J., Kunzer A. R., Letai A., Li C., Mitten M. J., Nettesheim D. G., Ng S., Nimmer P. M., O'Connor J. M., Oleksijew A., Petros A. M., Reed J. C., Shen W., Tahir S. K., Thompson C. B., Tomaselli K. J., Wang B., Wendt M. D., Zhang H., Fesik S. W., Rosenberg S. H. (2005) An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature 435, 677–681 [DOI] [PubMed] [Google Scholar]
- 12. van Delft M. F., Wei A. H., Mason K. D., Vandenberg C. J., Chen L., Czabotar P. E., Willis S. N., Scott C. L., Day C. L., Cory S., Adams J. M., Roberts A. W., Huang D. C. (2006) The BH3 mimetic ABT-737 targets selective Bcl-2 proteins and efficiently induces apoptosis via Bak/Bax if Mcl-1 is neutralized. Cancer Cell 10, 389–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen S., Dai Y., Harada H., Dent P., Grant S. (2007) Mcl-1 down-regulation potentiates ABT-737 lethality by cooperatively inducing Bak activation and Bax translocation. Cancer Res. 67, 782–791 [DOI] [PubMed] [Google Scholar]
- 14. Konopleva M., Contractor R., Tsao T., Samudio I., Ruvolo P. P., Kitada S., Deng X., Zhai D., Shi Y. X., Sneed T., Verhaegen M., Soengas M., Ruvolo V. R., McQueen T., Schober W. D., Watt J. C., Jiffar T., Ling X., Marini F. C., Harris D., Dietrich M., Estrov Z., McCubrey J., May W. S., Reed J. C., Andreeff M. (2006) Mechanisms of apoptosis sensitivity and resistance to the BH3 mimetic ABT-737 in acute myeloid leukemia. Cancer Cell 10, 375–388 [DOI] [PubMed] [Google Scholar]
- 15. Lin X., Morgan-Lappe S., Huang X., Li L., Zakula D. M., Vernetti L. A., Fesik S. W., Shen Y. (2007) 'Seed' analysis of off-target siRNAs reveals an essential role of Mcl-1 in resistance to the small-molecule Bcl-2/Bcl-XL inhibitor ABT-737. Oncogene 26, 3972–3979 [DOI] [PubMed] [Google Scholar]
- 16. Tahir S. K., Yang X., Anderson M. G., Morgan-Lappe S. E., Sarthy A. V., Chen J., Warner R. B., Ng S. C., Fesik S. W., Elmore S. W., Rosenberg S. H., Tse C. (2007) Influence of Bcl-2 family members on the cellular response of small-cell lung cancer cell lines to ABT-737. Cancer Res. 67, 1176–1183 [DOI] [PubMed] [Google Scholar]
- 17. Yecies D., Carlson N. E., Deng J., Letai A. (2010) Acquired resistance to ABT-737 in lymphoma cells that up-regulate MCL-1 and BFL-1. Blood. 115, 3304–3313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hikita H., Takehara T., Shimizu S., Kodama T., Shigekawa M., Iwase K., Hosui A., Miyagi T., Tatsumi T., Ishida H., Li W., Kanto T., Hiramatsu N., Hayashi N. (2010) The Bcl-xL inhibitor, ABT-737, efficiently induces apoptosis and suppresses growth of hepatoma cells in combination with sorafenib. Hepatology 52, 1310–1321 [DOI] [PubMed] [Google Scholar]
- 19. Hughes C. C., Prieto-Davo A., Jensen P. R., Fenical W. (2008) The marinopyrroles, antibiotics of an unprecedented structure class from a marine Streptomyces sp. Org. Lett. 10, 629–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hughes C. C., Yang Y. L., Liu W. T., Dorrestein P. C., La Clair J. J., Fenical W. (2009) Marinopyrrole A target elucidation by acyl dye transfer. J. Am. Chem. Soc. 131, 12094–12096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Haste N. M., Hughes C. C., Tran D. N., Fenical W., Jensen P. R., Nizet V., Hensler M. E. (2011) Pharmacological properties of the marine natural product marinopyrrole A against methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 55, 3305–3312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Krajewska M., Wang H. G., Krajewski S., Zapata J. M., Shabaik A., Gascoyne R., Reed J. C. (1997) Immunohistochemical analysis of in vivo patterns of expression of CPP32 (Caspase-3), a cell death protease. Cancer Res. 57, 1605–1613 [PubMed] [Google Scholar]
- 23. Krajewski S., Bodrug S., Gascoyne R., Berean K., Krajewska M., Reed J. C. (1994) Immunohistochemical analysis of Mcl-1 and Bcl-2 proteins in normal and neoplastic lymph nodes. Am. J. Pathol. 145, 515–525 [PMC free article] [PubMed] [Google Scholar]
- 24. Woods N. T., Yamaguchi H., Lee F. Y., Bhalla K. N., Wang H. G. (2007) Anoikis, initiated by Mcl-1 degradation and Bim induction, is deregulated during oncogenesis. Cancer Res. 67, 10744–10752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cheng C., Pan L., Chen Y., Song H., Qin Y., Li R. (2010) Total synthesis of (+/-)-marinopyrrole A and its library as potential antibiotic and anticancer agents. J. Combinatorial Chem. 12, 541–547 [DOI] [PubMed] [Google Scholar]
- 26. Czabotar P. E., Lee E. F., van Delft M. F., Day C. L., Smith B. J., Huang D. C., Fairlie W. D., Hinds M. G., Colman P. M. (2007) Structural insights into the degradation of Mcl-1 induced by BH3 domains. Proc. Natl. Acad. Sci. U.S.A. 104, 6217–6222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hu X., Sun J., Wang H. G., Manetsch R. (2008) Bcl-XL-templated assembly of its own protein-protein interaction modulator from fragments decorated with thio acids and sulfonyl azides. J. Am. Chem. Soc. 130, 13820–13821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Friesner R. A., Banks J. L., Murphy R. B., Halgren T. A., Klicic J. J., Mainz D. T., Repasky M. P., Knoll E. H., Shelley M., Perry J. K., Shaw D. E., Francis P., Shenkin P. S. (2004) Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J. Med. Chem. 47, 1739–1749 [DOI] [PubMed] [Google Scholar]
- 29. Jorgensen W. L., Maxwell D. S., Tirado-Rives J. (1996) Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 118, 11225–11236 [Google Scholar]
- 30. Eldridge M. D., Murray C. W., Auton T. R., Paolini G. V., Mee R. P. (1997) Empirical scoring functions: I. The development of a fast empirical scoring function to estimate the binding affinity of ligands in receptor complexes. J. Comput. Aided. Mol. Des. 11, 425–445 [DOI] [PubMed] [Google Scholar]
- 31. Liao A., Broeg K., Fox T., Tan S. F., Watters R., Shah M. V., Zhang L. Q., Li Y., Ryland L., Yang J., Aliaga C., Dewey A., Rogers A., Loughran K., Hirsch L., Jarbadan N. R., Baab K. T., Liao J., Wang H. G., Kester M., Desai D., Amin S., Loughran T. P., Jr., Liu X. (2011) Therapeutic efficacy of FTY720 in a rat model of NK-cell leukemia. Blood 118, 2793–2800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nguyen M., Marcellus R. C., Roulston A., Watson M., Serfass L., Murthy Madiraju S. R., Goulet D., Viallet J., Bélec L., Billot X., Acoca S., Purisima E., Wiegmans A., Cluse L., Johnstone R. W., Beauparlant P., Shore G. C. (2007) Small molecule obatoclax (GX15–070) antagonizes MCL-1 and overcomes MCL-1-mediated resistance to apoptosis. Proc. Natl. Acad. Sci. USA 104, 19512–19517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Maurer U., Charvet C., Wagman A. S., Dejardin E., Green D. R. (2006) Glycogen synthase kinase-3 regulates mitochondrial outer membrane permeabilization and apoptosis by destabilization of MCL-1. Molecular cell 21, 749–760 [DOI] [PubMed] [Google Scholar]
- 34. Albershardt T. C., Salerni B. L., Soderquist R. S., Bates D. J., Pletnev A. A., Kisselev A. F., Eastman A. (2011) Multiple BH3 mimetics antagonize antiapoptotic MCL1 protein by inducing the endoplasmic reticulum stress response and up-regulating BH3-only protein NOXA. J. Biol. Chem. 286, 24882–24895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wuillème-Toumi S., Trichet V., Gomez-Bougie P., Gratas C., Bataille R., Amiot M. (2007) Reciprocal protection of Mcl-1 and Bim from ubiquitin-proteasome degradation. Biochem. Biophys. Res. Commun. 361, 865–869 [DOI] [PubMed] [Google Scholar]
- 36. Wang G. Q., Gastman B. R., Wieckowski E., Goldstein L. A., Gambotto A., Kim T. H., Fang B., Rabinovitz A., Yin X. M., Rabinowich H. (2001) A role for mitochondrial Bak in apoptotic response to anticancer drugs. J. Biol. Chem. 276, 34307–34317 [DOI] [PubMed] [Google Scholar]
- 37. Acoca S., Cui Q., Shore G. C., Purisima E. O. (2011) Molecular dynamics study of small molecule inhibitors of the Bcl-2 family. Proteins 79, 2624–2636 [DOI] [PubMed] [Google Scholar]
- 38. Bhat U. G., Pandit B., Gartel A. L. (2010) ARC synergizes with ABT-737 to induce apoptosis in human cancer cells. Mol. Cancer Therap. 9, 1688–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ji M., Li J., Yu H., Ma D., Ye J., Sun X., Ji C. (2009) Simultaneous targeting of MCL1 and ABCB1 as a novel strategy to overcome drug resistance in human leukaemia. Br. J. Haematol. 145, 648–656 [DOI] [PubMed] [Google Scholar]
- 40. Quinn B. A., Dash R., Azab B., Sarkar S., Das S. K., Kumar S., Oyesanya R. A., Dasgupta S., Dent P., Grant S., Rahmani M., Curiel D. T., Dmitriev I., Hedvat M., Wei J., Wu B., Stebbins J. L., Reed J. C., Pellecchia M., Sarkar D., Fisher P. B. (2011) Targeting Mcl-1 for the therapy of cancer. Expert. Opin. Investig. Drugs 20, 1397–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Stewart M. L., Fire E., Keating A. E., Walensky L. D. (2010) The MCL-1 BH3 helix is an exclusive MCL-1 inhibitor and apoptosis sensitizer. Nat. Chem. Biol. 6, 595–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Domina A. M., Vrana J. A., Gregory M. A., Hann S. R., Craig R. W. (2004) MCL1 is phosphorylated in the PEST region and stabilized upon ERK activation in viable cells, and at additional sites with cytotoxic okadaic acid or taxol. Oncogene 23, 5301–5315 [DOI] [PubMed] [Google Scholar]
- 43. Gomez-Bougie P., Ménoret E., Juin P., Dousset C., Pellat-Deceunynck C., Amiot M. (2011) Noxa controls Mule-dependent Mcl-1 ubiquitination through the regulation of the Mcl-1/USP9X interaction. Biochem. Biophys. Res. Commun. 413, 460–464 [DOI] [PubMed] [Google Scholar]
- 44. Yecies D., Carlson N. E., Deng J., Letai A. (2010) Acquired resistance to ABT-737 in lymphoma cells that up-regulate MCL-1 and BFL-1. Blood 115, 3304–3313 [DOI] [PMC free article] [PubMed] [Google Scholar]