Background: HSP90/70 inactivation reduces cancer cell invasion by unknown mechanisms.
Results: The WASF3 metastasis promoting gene stability and activation is regulated by HSP90/70 chaperones.
Conclusion: The ability of HSP90/70 to suppress invasion results from its regulation of WASF3 function.
Significance: Inhibiting HSP may provide an approach to prevent metastasis.
Keywords: Cell Migration, Heat Shock Protein, Metastasis, Proteomics, Signal Transduction, WASF3, Invasion
Abstract
Inactivation of HSP90 and HSP70 leads to loss of invasion in a variety of cancer cell types, presumably as a result of destabilization of, as yet, undefined clients of these molecular chaperones that influence this phenotype. The WASF3 gene has been shown to be up-regulated in high-grade tumors and its down-regulation leads to loss of invasion and metastasis. WASF3 phosphorylation by ABL kinase is essential for its ability to regulate invasion. Mass spectroscopy analysis now shows that HSP90 is present in the WASF3 immunocomplex from prostate cancer cells. Inactivation of HSP90 in these and other cell types does not affect WASF3 stability but prevents its phosphoactivation as a result of destabilization of ABL. HSP70 was also found in the WASF3 immunocomplex and inactivation of HSP70 results in destabilization of WASF3 through proteasome degradation. Knockdown of WASF3, HSP90, and HSP70 individually, all lead to loss of invasion but as knockdown of WASF3 in the presence of robust expression of HSP90/70 has the same effect, it seems that the influence these chaperone proteins have on invasion is mediated, at least in part, by their control over the critical invasion promoting capacity of the WASF3 protein. Overexpression of HSP70 in WASF3 null cells does not enhance invasion. These observations suggest that targeting HSP90/70 may have efficacy in reducing cancer cell invasion.
Introduction
The WASF3 protein is a member of the Wiskott-Aldridge syndrome family of proteins that have been implicated in actin polymerization and cell movement (1). We have demonstrated that knockdown of WASF3 in breast and prostate cancer cells has a profound effect on their ability to migrate and invade in vitro (2, 3) and to metastasize in vivo (3, 4). Specifically, WASF3 appears to coordinate the development of lamellipodia (2) at the leading edges of cells, and loss of its function prevents motility. This function of WASF3 is regulated through its phosphorylation by ABL kinase (4). In primary breast (4, 5) and prostate (3) cancer, WASF3 has been shown to be up-regulated in advanced stage tumors supporting a role in promoting metastasis. WASF3 has been shown to be in a complex with the p85 component of PI3K as well as ABL kinase (2), and other members of the WASF family also form a complex with at least four other proteins: Abi1/2, the Rac effector protein CYFIP1/2 (also known as PIR121), NCKAP1 (NAP1), and HSPC300 (6, 7), which appear to hold the protein in an inactive form. Upon activation, this protein complex is released, and the VCA domain is exposed allowing binding of ARP2-ARP3 complexes, which facilitate actin polymerization from globular to filamentous actin. This process is presumed to facilitate the generation of lamellipodia and membrane ruffles, which are used by different cells for their motility.
Because the function and stability of a protein is often regulated by the proteins it interacts with, important insights can be obtained by identifying interacting partners. We have now used immunoprecipitation and mass spectroscopy (MS) to demonstrate that WASF3 is in a complex with the HSP90 and HSP70 chaperone proteins, which are frequently involved in the folding of newly synthesized proteins; assembly of multi-protein complexes; translocation of proteins across cellular membranes; and channeling of misfolded proteins into the proteasomal degradation pathway (8, 9). Inactivation of HSP90 did not affect WASF3 stability but prevented its phosphoactivation, and inactivation of HSP70 resulted in WASF3 protein destabilization. Loss of either HSP90 or HSP70 led to reduced cell motility and invasion in breast cancer cells, demonstrating a dependence of WASF3 on these chaperone proteins for its function.
MATERIALS AND METHODS
Cell Cultures and Reagents
All cell lines were obtained from the American Type Culture Collection (ATCC, Rockville, MD) and were cultured in RPMI 1640 medium supplemented with 10% FBS (Invitrogen). Stock aliquots of 2 mm 17-allylamino-17-demethoxygeldanamycin (17-AAG,2 Sigma), 100 mm N-formyl-3,4-methylenedioxy-γ-butyrolactam (KNK437; EMD, San Diego, CA), and 20 mm 2-phenylethynesulfonamide (PES, EMD) were prepared in dimethyl sulfoxide. Stocks of 5 mm MG132 (EMD) and 100 mm novobiocin were prepared in ethanol or water, respectively. All drugs were stored at −20 °C in the dark. In all drug treatment assays, except for novobiocin, dimethyl sulfoxide was used as a control.
Molecular Reagents and Procedures
For HSP70 overexpression, the pcDNA-FLAG-HSP70 construct was generated by add an N-terminal FLAG tag to the human HSP70 full-length cDNA. To construct the WASF3 transgene lentiviral expression vector, WASF3 fragments were generated using the PCR from the template WASF3 cDNA clone BC050283 (Open Biosystems, Huntsville, AL), and the full-length (W3-FL) and truncated human WASF3 (W3-PRD+VCA, W3-PRD, W3-dVCA) with HA tags were subcloned into pCDH-CMV-MCS-EF1-PURO (System Biosciences, Mountain View, CA) using XhoI and SalI sites. All constructs were confirmed by sequencing. pLKO lentiviral vector containing a short hairpin RNA against WASF3 (shW3-1-RHS3979-98060783 and shW3-2-RHS3979-98060797; Open Biosystems) was used to generate knockdown WASF3 stable cells. Lentivirus production was performed by co-transfecting the lentiviral vectors and packaging plasmids (Viropower kit; Invitrogen) into HEK293FT cells (Invitrogen) with Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. The supernatants containing virus were collected and filtered after 48 and 72 h post-transfection and used to infect breast cancer cells at least two times in the presence of 4 μg/ml polybrene (Sigma). The stable cells were selected with 800–1000 ng/ml puromycin (Sigma) for 2 or 3 weeks. RNA interference-Ready pSIREN-RetroQ vector with a double-stranded oligonucleotide encoding a hairpin small interfering RNA (5′-GAAGGACGAGTTTGAGCACAA-3′) was used for knockdown of HSP70. The retroviral transfection and infection was carried out as described previously (10, 11). The puromycin-resistant stable cells were verified for knockdown or overexpression by RT-PCR and Western blotting. Quantitative RT-PCR analysis was carried out as described previously (3). Primer sequences were as follows: β-actin forward primer, 5′-TCCCTGGAGAAGAGCTACGA-3′ and β-actin reverse primer, 5′-AGCACTGTGTTGGCGTACAG-3′; HSP70 forward primer, 5′-TGTCGTCCAGCACCCAGGCCAGC-3′ and HSP70 reverse primer, 5′-GCTCTTGTTCAGGTCGCGCCCG-3′. In this assay, the specific primers for human WASF3 were obtained from Sabiosciences (Frederick, MD).
Immunoblotting and Immunoprecipitation
Cells were lysed in modified radioimmune precipitation assay lysis buffer (Pierce). Lysate (1500 μg total protein) was incubated with the antibodies followed by addition of protein A/G-agarose beads (Pierce) overnight at 4 °C on a rotating platform. Beads were washed extensively in lysis buffer, and proteins were eluted with 2× Laemmli buffer (100 mm Tris-HCl, pH 6.8, 20% glycerol, 4% SDS, 10% β-mercaptoethanol) and heated to 95 °C for 5 min before electrophoresis. For Western blotting, equal amounts of protein were resolved by 8–10% SDS-PAGE, transferred to nitrocellulose membranes, and probed with primary antibodies. Membranes were then incubated with species-specific horseradish peroxidase-conjugated secondary antibody (Pierce) followed by detection with ECL substrate (Pierce). In some cases, the protein levels were quantified from at least two independent experiments by NIH ImageJ software (version 1.41). The primary antibodies used in this study were as follows: HSP70 (SPA-810) and HSP90 (SPA-840) were from Assay Designs; c-ABL was from Calbiochem and Santa Cruz Biotechnology; WASF3, AKT, and CHIP were from Cell Signaling Technology (Danvers, MA); β-actin, HA, PY20, and FLAG were from Sigma.
Cycloheximide Chase Assay
Degradation of WASF3 proteins was assessed using a cycloheximide chase assay. MDA-MB-231 cells were transfected with either pcDNA-FLAG-HSP70 or the empty vector. After 48 h of transfection, cells were treated with 100 μg/ml of cycloheximide (Sigma) for the indicated time period, and Western blotting was then performed.
MS
Detailed procedures for protein preparation for MS analysis are provided in supplemental “Methods.”
Database Searching
All MS/MS data were analyzed using Sequest (Thermo Fisher Scientific, San Jose, CA; version 1.2.0.208) and X!Tandem (The GPM, version 2007.01.01.1). Sequest and X!Tandem were set up to search NCBInr_Homosapiens_05262011.fasta (221,863 entries) assuming trypsin digestion. Sequest and X!Tandem were searched with a fragment ion mass tolerance of 0.80 Da and a parent ion tolerance of 10.0 ppm. Iodoacetamide derivative of cysteine was specified in Sequest and X!Tandem as a fixed modification. Oxidation of methionine was specified in Sequest and X!Tandem as a variable modification.
Criteria for Protein Identification
Scaffold (version Scaffold_3.1.4.1, Proteome Software, Inc., Portland, OR) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at >95.0% probability as specified by the Peptide Prophet algorithm (12). Protein identifications were accepted if they could be established at >90.0% probability and contained at least one identified peptide. Protein probabilities were assigned by the Protein Prophet algorithm (13). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Single-peptide protein identification was accepted only if the protein was independently identified by both Sequest and X!Tandem.
Motility and Invasion Assays
The wound-closure assay was performed as described previously (3). Invasion assays were performed in Matrigel-coated modified Boyden chambers with 8-μm pore size filters (BD Biosciences). In brief, the serum-starved cells were added in the upper chamber (5 × 104 cells per insert) and RPMI 1640 medium with 5% FBS was used as a chemoattractant in the lower chamber. After 24 h of incubation, non-invading cells that remained on the upper surface of the filter were removed, and the cells that had passed through the filter and attached to the bottom of the membrane were stained with crystal violet cell stain solution (Millipore, Billerica, MA). The solution was eluted with 10% acetic acid extraction buffer (Millipore) and transferred to 96-well microplates. The absorbance was calculated at 590 nm in each well, and each sample was analyzed in triplicate.
RESULTS
WASF3 Interacts with HSP90
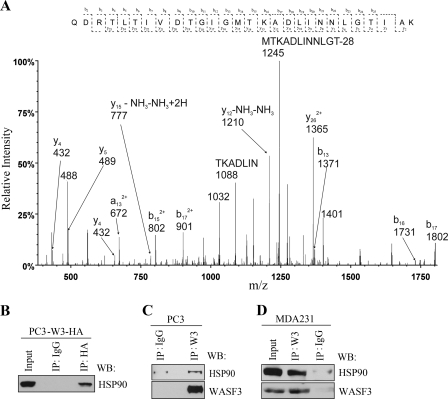
To identify proteins that interact with WASF3, an exogenous HA-tagged WASF3 gene was expressed in PC3 prostate cancer cells, and immunocomplexes were isolated using an anti-HA antibody. LTQ Orbitrap MS was then used to analyze tryptic peptides generated following this immunoprecipitation (IP). Among the highest ranking proteins (Table 1) in each of two independent experiments was the WASF3 bait protein as well as NCKAP1 (NAP1) and CYFIP2 (PIR121), which are known interacting proteins for other members of the WASF family. The MS analysis also identified HSP90 in the immunocomplex (Fig. 1A), which was confirmed using IP and Western blotting with either the HA antibody to immunoprecipitate the exogenous WASF3 (Fig. 1B) or a WASF3-specific antibody to immunoprecipitate the endogenous protein (Fig. 1C). These results demonstrate that the interaction between WASF3 and HSP90 is not specific to a system in which WASF3 is overexpressed. This interaction was also seen in MDA-MB-231 breast cancer cells using the WASF3-specific antibody (Fig. 1D).
TABLE 1.
Summary of selected MS identified proteins from the WASF3 immunocomplex
Lowercase letters represent modified amino acids.
| Protein | Accession no. | Mol. mass | Amino acid coverage | No. of peptides | Peptide sequence |
|---|---|---|---|---|---|
| Da | % | ||||
| WASF3 | gi 13699803 | 55, 293.8 | 43/502 amino acids (8.6%) | 2 | GALPEGITSELEcVTNSTLAAIIREQEAKREPVGNDVATILSR |
| HSP90 | gi 153792590 | 98, 165.1 | 139/854 amino acids (16.3%) | 6 | QDRTLTIVDTGIGMTKADLINNLGTIAK LSELLRYYTSASGDEMVSLK AFmEALQAGADISMIGQFGVGFYSAYLVAEK HIYYITGETKDQVANSAFVERLR KcLELFTELAEDKENYK mPPcSGGDGSTPPGPSLRDR |
| NCKAP1 | gi 119631358 | 129, 280.1 | 100/1132 amino acids (8.8%) | 5 | NLITDIcTEQcTLSDQLLPK VAMNVYELSSAAGLPcEIDPALVVALSSQK NNNQQLAQLQKEKSEILK EYPRLGQmIVDYENPLKK AINQIAAALFTIHK |
| CYFIP2 | gi 125490318 | 150, 789.6 | 184/1304 amino acids (14.1%) | 7 | YIEQATVHSSmNEmLEEGHEYAVMLYTWR cNEQPNRVEIYEKTVEVLEPEVTK LADQIFAYYKAmAGSVLLDKR YSNSEVVTGSGLDSQK mYLTPSEKHMLLKVmGFGLYLmDGNVSNIYK GLQVLmGRmESVFNQAIRNTIYAALQDFAQVTLR YIEQATVHSSMNEmLEEGHEYAVmLYTWR |
FIGURE 1.
WASF3 is a novel HSP90-interacting protein. A, after overexpression of an HA-tagged WASF3 construct in PC3 cells, mass spectrometric analysis identified HSP90 in the WASF3 complex. An example of a representative MS/MS spectrum of a tryptic peptide derived from HSP90 is shown in A. IP of an exogenous HA-tagged WASF3 protein from PC3 cells (left panel) identified HSP90 in the immunocomplex (B). IP from parental PC3 cells using an anti-WASF3 antibody (C) also identified HSP90 in the immunocomplex. Preimmune IgG was used as a control. D, IP of WASF3 from MDA-MB-231 cells also identified HSP90 in the immunocomplex. WB, Western blot.
Inhibition of HSP90 Leads to WASF3 Inactivation
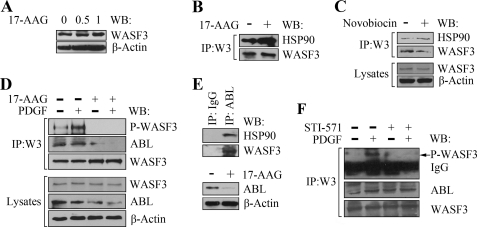
HSP90 has a central position in the network of molecular chaperones that promote necessary protein folding and degradation (14). HSP90 has emerged as a promising target for the treatment of cancer and inhibition of its function by the 17-AAG drug leads to decreased cancer cell motility and invasion (15–17). We have recently demonstrated that WASF3 is important in controlling cell motility and invasion in breast and prostate cancer cells (2, 3, 5). To determine whether the interaction between WASF3 and HSP90 maintains protein stability, we inactivated HSP90 function using 17-AAG, which binds directly to the essential ATP/ADP binding pocket in the N-terminal region of HSP90 (8, 18). When PC3 cells were treated with low and high doses of 17-AAG, there was no resultant change in WASF3 protein levels (Fig. 2A). The same was true for MDA-MB-231 and SkBr3 breast cancer cell lines as well as COS7 cells (supplemental Fig. S1). Interestingly, after 17-AAG treatment, increased levels of HSP90 were observed following IP of WASF3 in PC3 cells (Fig. 2B). A second ATP-binding site is located in the C terminus of HSP90 (19, 20), which can be inactivated using novobiocin, which binds to the HSP90 C-terminal nucleotide binding pocket. When we treated PC3 cells with 100 nm novobiocin, a slight decrease in WASF3 protein levels was observed, whereas increased levels of the HSP90 protein were found in the WASF3 immunocomplex following IP (Fig. 2C). These observations suggest that WASF3 is probably not an HSP90 client protein and that the interaction with HSP90 is not facilitated through the ATP/ADP binding domain.
FIGURE 2.
Inhibition of HSP90 leads to WASF3 inactivation. A, treatment of PC3 cells with varying concentrations (μm) of 17-AAG for 24 h does not affect intracellular WASF3 protein levels. B, IP of WASF3 from PC3 cells treated with 17-AAG for 24 h demonstrates increased HSP90 levels in the immunocomplex compared with untreated cells. C, treatment of PC3 cells with 100 nm novobiocin for 24 h shows a decrease in WASF3 levels and a slight increase in HSP90 levels in the immunocomplex recovered following IP with an anti-WASF3 antibody. D, Western blot (WB) analysis of WASF3 IPs (upper panel) shows increased levels of phosphoactivated WASF3 in serum-starved PC3 cells using anti-phosphotyrosine antibodies (PY20) following stimulation with 50 ng/ml PDGF for 10 min. After a 24-h exposure to 1 μm 17-AAG, phosphoactivated WASF3 is undetectable despite high levels of WASF3 protein. This loss of activated protein cannot be recovered following PDGF treatment. IP of WASF3 also results in decreased levels of ABL in the immunocomplex after 17-AAG treatment regardless of PDGF stimulation. E, Western blot analysis of the ABL immunocomplex shows the presence of HSP90 in PC3 cells. Following treatment with 17-AAG levels of ABL are reduced. F, IP of WASF3 from PC3 cells shows increased activation following treatment with PDGF. In the presence of 50 μm ABL inhibitor STI571, this activation is significantly reduced and cannot be recovered in the presence of PDGF.
Because HSP90 does not appear to affect the stability of the WASF3 protein, we investigated whether HSP90 played an important role in WASF3 activation. As shown in Fig. 2D, treatment of PC3 cells with PDGF leads to a dramatic increase in WASF3 phosphorylation levels. However, treatment with 17-AAG for 24 h completely prevents WASF3 activation, even in the presence of PDGF (Fig. 2D). The same effect was observed using SkBr3 breast cancer cells (data not shown). We have previously shown that ABL kinase can phospoactivate WASF3 (21) in MDA-MB-231 breast cancer cells, and others have shown that inhibition of HSP90 function decreases ABL protein levels (22). Treatment of PC3 cells with 17-AAG for 24 h (Fig. 2D) results in a dramatic reduction in the phosphorylation levels of WASF3 regardless of PDGF stimulation. IP of WASF3 from these cells shows reduced ABL protein levels in the immunocomplex. To confirm that ABL interacts with HSP90, we used an anti-ABL antibody to IP ABL from PC3 cells and Western blot analysis demonstrated the presence of HSP90 in the immunocomplex (Fig. 2E). When HSP90 function was inhibited in these cells using 17-AAG, ABL protein levels were reduced to almost beyond detection, compared with the untreated cells (Fig. 2E). These observations support the idea that ABL is a client protein for HSP90.3 When PC3 cells were treated with 50 μm STI-571 (Gleevec or imatinib), which inhibits ABL, for 24 h, the phosphorylation level of WASF3 was reduced significantly (Fig. 2F). PDGF stimulation could not rescue inactivation of WASF3 (Fig. 2F). These observations indicate that the loss of activation of WASF3 following inhibition of HSP90 results from destabilization of ABL, suggesting the basis of the indirect action of HSP90 loss on WASF3 phosphoactivation.
HSP70 Is Important Regulator of WASF3 Protein Stability
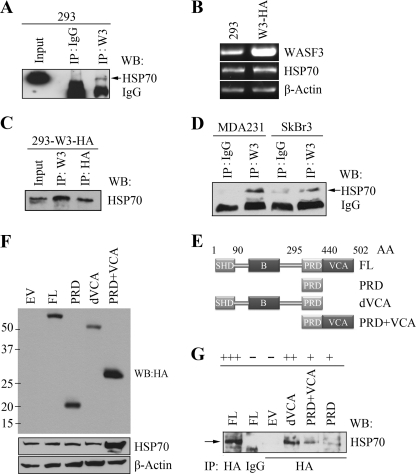
HSP70, also called HSP70A1A or HSP72, is the main member of the 70-kDa heat shock protein family. As a molecular chaperone, HSP70 guides the pathways of protein folding (23), protein translocation (24), and protein degradation (25, 26) at the molecular level. Because the observations described above suggest that HSP90 does not appear to affect the stability of WASF3, we next investigated whether HSP70 can affect its stability. Using the WASF3 antibody to immunoprecipitate the WASF3 protein complex from 293 cells, we identified HSP70 in the immunocomplex (Fig. 3A). We then generated 293 cells expressing an exogenous HA-tagged WASF3 gene (Fig. 3B) and, in an IP of WASF3 using anti-HA antibodies, HSP70 was identified in the immunocomplex (Fig. 3C). We have recently demonstrated that WASF3 is important in controlling cell motility and invasion in breast cancer cells (2, 3) and IP of WASF3 from MDA-MB-231 and SkBr3 also identified HSP70 in the immunocomplex (Fig. 3D). To determine where HSP70 binds within the WASF3 protein, we created a series of HA-tagged truncations (Fig. 3E) of WASF3 and expressed them in MDA-MB-231 cells (Fig. 3F). IP from these cells using the anti-HA antibody, demonstrated that HSP70 is present in the immunocomplex with the full-length protein as expected and was also present in the immunocomplex with WASF3 in which the VCA domain was deleted (Fig. 3G). Using the cell lines expressing the PRD only, minimal participation in the immunocomplex was seen. The same results were observed in cells expressing the PRD-VCA domain although HSP70 is elevated remarkably (Fig. 3, F and G). Taken together, therefore, it appears that HSP70 likely interacts with the N-terminal end of WASF3.
FIGURE 3.
HSP70 is present in the WASF3 protein complex. A, IP of WASF3 from HEK293 cells shows the presence of HSP70 in the immunocomplex but not in the nonspecific IgG control. An asterisk represents the IgG heavy chain. RT-PCR analysis of stable 293 clones expressing HA-tagged WASF3 (B) show high levels of WASF3. IP of WASF3 using either the anti-HA or anti-WASF3 antibodies show co-IP of HSP70 (C). IP of WASF3 from MDA-MB-231 and SkBr3 breast cancer cells (D) also shows the presence of HSP70 in the immunocomplex. HA-tagged full-length (FL) or truncated WASF3 (W3) deletion constructs were generated to map the WASF3 domains required for HSP70 binding (summarized in E). Western blot (WB) analysis confirmed overexpression of these constructs in MDA-MB-231 cells (F). When WASF3 was recovered from these cells using anti-HA antibodies, Western blot analysis showed the presence of appreciable levels only when the N-terminal part of the protein was present (G). Preimmune IgG was used as negative or positive controls. + and −, relative ability of the variants to co-precipitate HSP70.
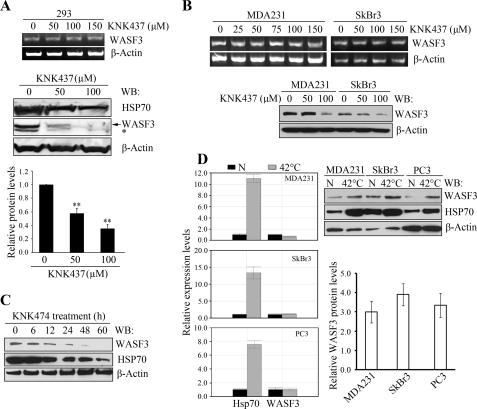
To investigate the functional inter-relationship between HSP70 and WASF3, we treated 293 cells for 24 h with increasing concentrations of KNK437, which has been shown to interfere with transcriptional activation of HSP70 (27). This treatment showed no effect on WASF3 mRNA levels in the cells (Fig. 4A) regardless of concentration but showed a dose-dependent reduction in HSP70 protein levels (Fig. 4A). This reduction in HSP70 was accompanied by a more dramatic reduction in WASF3 (Fig. 4A). The same effect was also seen in the MDA-MB-231 and SkBr3 breast cancer cell lines (Fig. 4B). In a time course experiment (Fig. 4C), KNK437 was shown to exert its effect beyond 6 h. Thus, WASF3 appears to be a client protein for HSP70, which stabilizes it.
FIGURE 4.
HSP70 stabilizes the WASF3 protein. Treatment of HEK293 cells with varying concentrations of KNK437 for 24 h (A) demonstrates no effect on WASF3 mRNA levels by RT-PCR (upper panel). WASF3 protein levels determined by Western blot analysis (lower panel), demonstrates significantly reduced WASF3 protein levels (middle panel). Quantification of the Western blot data for WASF3 using ImageJ software is shown in the lower panel. **, p = 0.001. The same effects of KNK437 treatment was seen for MDA-MB-231 and SkBr3 cells (B) using RT-PCR (upper panel) and Western blot (WB) analysis (lower panel). C, when MDA-MB-231 cells were treated with 100 μm KNK437 for varying times (0–60 h), Western blot analysis of the WASF3 protein using an anti-WASF3 antibody, shows an increasing reduction in both WASF3 and HSP70 proteins. D, quantitative RT-PCR analysis (left panel) of MDA-MB-231, SkBr3, and PC3 cells exposed to heat shock (42 °C for 1 h) show large increases in HSP70 expression levels but not WASF3 (N, 37 °C treatment). Western blot analysis of the same extracts (right panel) shows that the increased HSP70 protein levels are accompanied by increased levels of WASF3. Quantification of the 37 °C/42 °C ratios using ImageJ software for WASF3 protein levels from two independent experiments are shown on the lower right panel.
The more common effect of HSP70 binding is to target client proteins for degradation, although there are examples where proteins are stabilized as seen for WASF3 above. The HSP70 mediated degradation process typically involves recruitment of CHIP E3 ubiquitin ligase. To investigate the relationship between CHIP and WASF3, we first demonstrated that AKT, a protein known to be degraded through HSP70-CHIP, shows increased expression following knockdown of HSP70 in our system, whereas WASF3 shows a decrease in protein levels (supplemental Fig. S2). IP from parental MDA231 cells using anti-CHIP antibodies demonstrated that although AKT was present in the immunocomplex, WASF3 was not (supplemental Fig. S2). These observations support the idea that stabilization of proteins by HSP70 is related to the inability of proteins such as CHIP to engage the complex, although the mechanism behind this observation is not known.
Expression of HSP70 is either constitutively elevated, or inducible in response to heat shock stress (28). To determine whether stress-induced increases in HSP70 levels affect WASF3 protein levels, quantitative RT-PCR and Western blot analysis was performed before and after a 42 °C heat shock for 60 min in various cell lines. As shown in Fig. 4D, in MDA-MB-231 and SkBr3, as well as PC3 cells, the increase in HSP70 levels resulting from the heat shock led to a concomitant increase in WASF3 protein levels, which was not a result of increased WASF3 expression. These observations support the suggestion that stabilization of the WASF3 protein is facilitated by HSP70.
WASF3 Is Protected from Proteasome Degradation by HSP70
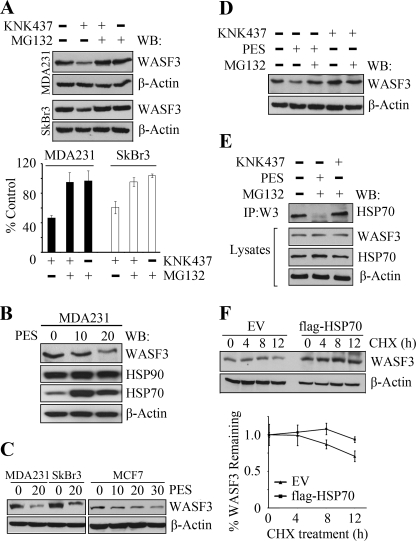
To determine whether WASF3 can be degraded through the proteasome following down-regulation of HSP70, we used the MG132 proteasome inhibitor in combination with KNK437 to investigate its stability. As shown earlier, treatment with KNK437 alone results in reduced WASF3 levels (Fig. 5A). Treatment with MG132 alone or in combination with KNK437 in both MDA-MB-231 and SkBr3 cells, shows no significant effect on WASF3 protein levels in either cell line, demonstrating that WASF3 is likely protected from proteasome degradation by HSP70. These observations further confirm that HSP70, not HSP90, is essential for the stable presence of WASF3.
FIGURE 5.
WASF3 is protected from proteasome degradation by HSP70. A, MDA-MB-231 and SkBr3 cells were treated with KNK437 and MG132 in various combinations for 24 h. Western blot analysis (upper panel) shows significantly increased WASF3 protein levels in the presence of the proteasome inhibitor. Quantification (lower panel) of protein levels as a percentage of the untreated control cells supports this observation. When MDA-MB-231 cells were treated with varying concentrations of PES (B) for 24 h, WASF3 protein levels were reduced but not HSP70. The same effect was seen in SkBr3 and MCF7 breast cancer cells (C). After various combinations of treatment for 6 h, WASF3 levels are decreased following PES treatment (D) but unaffected by the presence of KNK437 or MG132. IP from MDA-MB-231 cells (E) shows increased HSP70 levels following MG132 treatment and reduced HSP70 levels when PES was present. F, in MDA-MB-231 cells, the half-life of WASF3 was longer in HSP70-overexpressing cells, compared with that in empty vector-transfected cells. Upper panel, inhibition of protein synthesis by cycloheximide (CHX) and Western blotting (WB) at the indicated times. Lower panel, quantification of WASF3 half-life from the Western blots, normalized to β-actin.
KNK437 is a specific inhibitor of HSP70 transcription and to further confirm that WASF3 binding to HSP70 affected protein stabilization, we used another HSP70 inhibitor, PES. PES interacts with HSP70 and has been shown to disrupt its association with some of its client proteins (29). In MDA-MB-231 cells, WASF3 protein levels were decreased remarkably when treated with 20 μm PES for 24 h (Fig. 5B). The same result was found in SkBr3 cells (Fig. 5C). Interestingly, in the noninvasive MCF7 epithelial human breast cancer cell line, which expresses low WASF3 levels, higher doses of PES (30 μm) were required to deplete WASF3 levels compared with the two invasive breast cancer cell lines described above (Fig. 5C). To compare the different effect of the various HSP70 inhibitors, we treated MDA-MB-231 with PES and KNK437, alone or in combination with MG132, for 6 h (Fig. 5D). As described above, KNK437 did not affect WASF3 levels after 6 h of treatment, whereas PES treatment leads to a distinct reduction in WASF3 protein levels. Analysis of the interaction between HSP70 and WASF3 using IP, showed no appreciable change with or without KNK437 treatment (Fig. 5E). Most strikingly, PES disrupted the association between HSP70 and WASF3, in the presence of MG132, suggesting that PES leads to depletion of WASF3 protein once the interaction with HSP70 is lost. These observations demonstrate that it is the HSP70-WASF3 complex formation that facilitates WASF3 stability, rather than endogenous HSP70 levels. To investigate the half-life of WASF3, we treated MDA231 cells with cycloheximide to prevent de novo protein synthesis and measured protein levels in cells overexpressing HSP70 compared with cells expressing the empty vector. In empty vector cells, WASF3 is relatively stable for 8 h, but protein levels are reduced by ∼30% after 12 h (Fig. 5F). In cells overexpressing HSP70, however, protein levels were relatively unaffected and, over the same 12-h period, were only reduced by 7% (Fig. 5F). These data further confirm the importance of HSP70 in the stabilization of WASF3.
WASF3 Contributes to HSP70-induced Cancer Cell Migration and Invasion
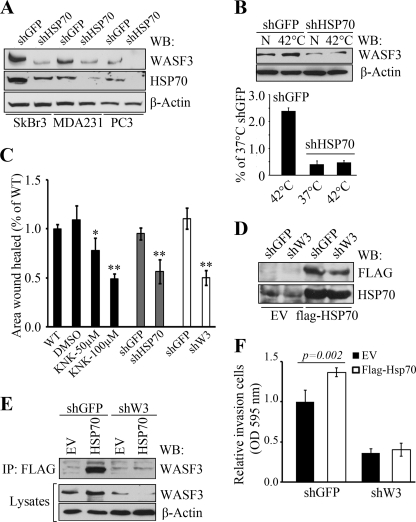
The HSP70 inhibitors described above, while targeting HSP70, also affect HSP90 and, although loss of invasion is seen when treated with these agents, these could also be due to off target effects. To investigate this possibility, we used a highly specific shRNA targeting strategy to further investigate the relationship between HSP70 and WASF3. As shown in Fig. 6A, we identified shRNA constructs that caused significant knockdown of the HSP70 levels in PC3, MDA-MB-231, and SkBr3 cells, resulting in a concomitant reduction in WASF3 protein levels (Fig. 6A). In contrast, when WASF3 is knocked down in the high WASF3-expressing cell lines MDA-MB-231 and PC3, or in low WASF3-expressing MCF7 and T47D cells overexpressing an exogenous WASF3 gene, there is no change in HSP70 levels (supplemental Fig. S3). As expected, in HSP70-silenced MDA-MB-231 cells, no change was found in WASF3 protein level before or after heat shock (Fig. 6B). Most interestingly, using wound healing assays, we found that both pharmacological inhibition of HSP70 as well as HSP70-specific shRNA knockdown produced the same reduction in cell migration seen following shRNA knockdown of WASF3 (Fig. 6C). Next, to determine whether WASF3 contributes to HSP70-induced cancer cell migration and invasion, we transfected the stable WASF3 knockdown MDA-MB-231 cells, as well as control cells, with a FLAG-tagged HSP70 construct (Fig. 6D). Western blot analysis showed that overexpression of HSP70 leads to elevated WASF3 protein levels in the control cells but had no effect on WASF3 levels in WASF3 knockdown cells (Fig. 6E). Using an anti-FLAG antibody, exogenous HSP70 was immunoprecipitated from these modified MDA-MB-231 cells and WASF3 was only identified in the immunocomplex in control cells that overexpressed HSP70 (Fig. 6E), which further confirms the association between these two proteins. Finally, we used transwell assays to determine whether knockdown of WASF3 attenuated HSP70-induced cancer cell invasion. In response to HSP70 overexpression, cell invasion was increased in the control cells, simultaneously up-regulating the WASF3 protein (Fig. 6, E and F). No significant change was found in WASF3-silenced cells with or without HSP70 overexpression (Fig. 6F), although the large reduction in invasion potential seen as a result of WASF3 knockdown may have masked any effect of overexpressing HSP70 in this system. When we analyzed all WASF3 knockdown cells transfected with different shRNAs targeting different sequences within the WASF3 gene (supplemental Fig. S4), it became clear that to see an effect on invasion, >50% knockdown of WASF3 was required. As such, overexpressing HSP70 in partial knockdown cells is unlikely to reveal any difference in invasion potential either. Taken together, however, these data support the concept that WASF3 mediates HSP70-assocaited cell migration and invasion in breast cancer cells.
FIGURE 6.
HSP70 stabilization of WASF3 is required for cell invasion. SkBr3, MDA-MB-231, and PC3 cells stably expressing shRNAs against HSP70 (shHSP70) show highly reduced levels of HSP70 protein (A) compared with cells expressing a control shRNA against GFP (shGFP). Knockdown of HSP70 also leads to knockdown of WASF3. When parental MDA-MB-231 cells were exposed to 42 °C for 1 h (B), WASF3 protein levels increase but when HSP70 knockdown cells are heat-shocked, there is no change in WASF3 levels (upper panel). The lower panel shows normalized quantification of WASF3 protein levels from the Western blots (WB). C, MDA-MB-231 cells treated with KNK437 show reduced cell motility, which was comparable with the reduced motility seen in cells in which either HSP70 or WASF3 were knocked down. When WASF3 stable knockdown MDA-MB-231 cells and shGFP control cells overexpressed FLAG-HSP70 (D), increased HSP70 protein levels were detected using anti-FLAG and anti-HSP70 antibodies. When WASF3 stable knockdown MDA-MB-231 cells (shW3) and the control cells (shGFP) were forced to express FLAG-HSP70 (E), increased levels of WASF3 protein was seen in control cells overexpressing HSP70 but not in the WASF3 knockdown cells using both anti-FLAG and anti-WASF3 antibodies. F, the stable WASF3 knockdown MDA-MB-231 cells and the control cells with or without HSP70 overexpression were used to determine the invasion potential using transwell assays. In response to HSP70 overexpression, the control cells increase their invasion potential after simultaneously up-regulating the WASF3 protein, whereas no significant change was found in WASF3 silenced cells with or without HSP70 overexpression.
DISCUSSION
The WASF3 protein appears to be important for in vitro invasion in many cell types and knockdown in breast (2) and prostate (3) almost completely suppresses metastasis in vivo. The association with HSP90/70 has been demonstrated clearly in this study and potentially provides a basis for other observations involving the influences of HSP70/90 over invasion. In a number of cases, involving, for example, cervical carcinoma (30) and bladder carcinoma (31), the HSP70 protein was shown to be highly expressed in advanced stage tumors suggesting a role in progression (30, 31). When HSP70 was inactivated using shRNA knockdown in cell lines from cervical and bladder cancer, invasion and migration was also suppressed, in the same way these phenotypes are affected by WASF3 knockdown. In other studies of breast cancer (17), treatment with 17-AAG led to decreased filopodia and lamellipodia formation, as well as actin bundles and actin polymerization. These changes were associated with decreased invasion, as we demonstrated for different cell lines in this study. In highly invasive PC3 prostate cancer cells, HSP90 was localized to the leading edge of migrating cells and in this cell line, knockdown of HSP90 led to reduced invasion as well (32). It is likely, therefore, that the effect of HSP70/90 inhibitors on invasion in these systems is affected by reducing WASF3 protein stability and activation, which as we have shown is the mechanism of suppression of invasion. In our early studies, we demonstrated that knockdown of WASF3 led to reduced expression of matrix metalloproteinases such as matrix metalloproteinase 2. Furthermore, matrix metalloproteinase 2 activity was also suppressed when HSP90/70 was targeted, which is correlated with decreased invasion, as these matrix metalloproteinases facilitate degradation of basement membrane matrix, allowing increased cell movement and invasion (33, 34). In one recent study (35), inactivation of the HSP70/90 organizing protein, which is a co-chaperone for HSP70/90, led to the reduction of invasion in pancreatic cells. It was suggested that this was due to client protein destabilization involving HSP90-dependent complexes. Knockdown of HSP70/90 organizing protein also leads to reduced expression of matrix metalloproteinase 2 (35), as well as other client proteins such as HER2, MET, and SRC, which have also been implicated in invasion and metastasis (36). One function of WASF family members is to assemble at receptor tyrosine kinases in response to growth factor stimulation, and so, it is potentially implicated in signaling through these receptors as well. Indeed, stimulation of breast cancer cells with PDGF leads to phosphactivation of WASF3 and increased lamellipodia formation and invasion (2).
The heat shock proteins have numerous effects on their client proteins, most notably ensuring proper folding and promoting stability of the folded protein. In this study, even though IP of WASF3 identified HSP90 in the immunocomplex, it does not seem to act by stabilizing WASF3, as loss of HSP90 does not affect WASF3 protein levels. Thus, WASF3 does not appear to be a client protein for HSP90. Strikingly, however, loss of HSP90 almost completely prevents phosphoactivation of WASF3 as a result of destabilizing ABL kinase, which is a client of HSP90 (22). We have shown previously that ABL can activate WASF3 and greatly enhance cell migration and adhesion following growth factor stimulation (2, 37). However, it is not yet clear whether other kinases can also activate WASF3, and a large number of other tyrosine kinases are known to also be clients for HSP90 (36), including several receptor tyrosine kinases. In contrast, WASF3 has now been shown to be a client protein for HSP70, which directly influences its stability and hence its function. In this context, because knockdown of WASF3 alone leads to loss of invasion even in the presence of robust expression of HSP90/70, it appears that a functional WASF3 protein is required to facilitate this phenotype and that the observations of association between invasion and HSP90/70 is mediated through WASF3. Furthermore, overexpression of HSP70 in the absence of WASF3 function cannot rescue the invasion phenotype.
In our recent gene expression studies, we demonstrated that knockdown of WASF3 led to significant changes in gene expression profiles (5). One significant change involved the up-regulation of the KISS1 metastasis suppressor gene and down-regulation of matrix metalloproteinase 9. These changes were associated with increased IκBα levels in the cytoplasm and reduced NFκB in the nucleus. WASF3, therefore, appears to exert its influence on invasion through stimulating NFκB signaling. Treatment of colon cancer cells (38) with anti-HSP90 antibodies was also shown to inhibit invasion, and addition of recombinant HSP90 to these colon cancer cells induced the activity of a number of signaling pathways, including NFκB. In our previous studies (5), knockdown of WASF3 reduced NFκB signaling, leading to reduced invasion as a result of matrix metalloproteinase 9 inactivation. Suppression of secreted HSP90 function leads to reduced signaling presumed to be due to binding to essential receptor-mediated signaling in the same way that inhibition of intracellular signaling was prevented by disruption of HSP90-HSP70-WASF3 complex formation. These observations support the idea that WASF3 has a central role in regulating the invasion phenotype and that its interaction with the HSP90/70 proteins is required for this function. This observation raises the question whether HSP90/70 inhibitors would have an influence on metastasis.
Supplementary Material
Acknowledgments
We thank Dr. V. L. Gabai (Boston University School of Medicine, Boston, MA) for kindly providing HSP70 knockdown retroviral vectors and Novartis Pharma for providing STI-571. We also thank Dr. Suiquan Wang for helpful suggestions during the course of these experiments.
This work was supported, in whole or in part, by National Institutes of Health Grant CA120510 from NCI.

This article contains supplemental “Methods” and Figs. S1–S4.
Y. Teng, L. Ngoka, Y. Mei, L. Lesoon, and J. K. Cowell, unpublished data.
- 17-AAG
- 17-allylamino-17-demethoxygeldanamycin
- KNK437
- N-formyl-3,4-methylenedioxy-γ-butyrolactam
- PES
- 2-phenylethynesulfonamide
- IP
- immunoprecipitation
- PRD
- proline-rich domain
- VCA
- verprolin-cofilin-acidic.
REFERENCES
- 1. Suetsugu S., Miki H., Takenawa T. (1999) Identification of two human WAVE/SCAR homologues as general actin regulatory molecules that associate with the Arp2/3 complex. Biochem. Biophys. Res. Commun. 260, 296–302 [DOI] [PubMed] [Google Scholar]
- 2. Sossey-Alaoui K., Li X., Ranalli T. A., Cowell JK. (2005) WAVE3-mediated cell migration and lamellipodia formation are regulated downstream of phosphatidylinositol 3-kinase. J. Biol. Chem. 280, 21748–21755 [DOI] [PubMed] [Google Scholar]
- 3. Teng Y., Ren M. Q., Cheney R., Sharma S., Cowell J. K. (2010) Inactivation of the WASF3 gene in prostate cancer cells leads to suppression of tumorigenicity and metastases. Br. J. Cancer 103, 1066–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sossey-Alaoui K., Safina A., Li X., Vaughan M. M., Hicks D. G., Bakin A. V., Cowell J. K. (2007) Down-regulation of WAVE3, a metastasis promoter gene, inhibits invasion and metastasis of breast cancer cells. Am. J. Pathol. 170, 2112–2121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Teng Y., Liu M., Cowell J. K. (2011) Functional interrelationship between the WASF3 and KISS1 metastasis-associated genes in breast cancer cells. Int. J. Cancer 129, 2825–2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Eden S., Rohatgi R., Podtelejnikov A. V., Mann M., Kirschner M. W. (2002) Mechanism of regulation of WAVE1-induced actin nucleation by Rac1 and Nck. Nature 418, 790–793 [DOI] [PubMed] [Google Scholar]
- 7. Steffen A., Rottner K., Ehinger J., Innocenti M., Scita G., Wehland J., Stradal T. E. (2004) Sra-1 and Nap1 link Rac to actin assembly driving lamellipodia formation. EMBO J. 23, 749–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hartl F. U., Hayer-Hartl M. (2002) Molecular chaperones in the cytosol: From nascent chain to folded protein. Science 295, 1852–1858 [DOI] [PubMed] [Google Scholar]
- 9. Mayer M. P., Bukau B. (2005) Hsp70 chaperones: Cellular functions and molecular mechanism. Cell Mol. Life Sci. 62, 670–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zaarur N., Gabai V. L., Porco J. A., Jr., Calderwood S., Sherman M. Y. (2006) Targeting heat shock response to sensitize cancer cells to proteasome and Hsp90 inhibitors. Cancer Res. 66, 1783–1791 [DOI] [PubMed] [Google Scholar]
- 11. Gabai V. L., Yaglom J. A., Waldman T., Sherman M. Y. (2009) Heat shock protein Hsp72 controls oncogene-induced senescence pathways in cancer cells. Mol. Cell Biol. 29, 559–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 13. Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 [DOI] [PubMed] [Google Scholar]
- 14. Whitesell L., Lindquist S. L. (2005) HSP90 and the chaperoning of cancer. Nat. Rev. Cancer 5, 761–772 [DOI] [PubMed] [Google Scholar]
- 15. Sanderson S., Valenti M., Gowan S., Patterson L., Ahmad Z., Workman P., Eccles S. A. (2006) Benzoquinone ansamycin heat shock protein 90 inhibitors modulate multiple functions required for tumor angiogenesis. Mol. Cancer Ther. 5, 522–532 [DOI] [PubMed] [Google Scholar]
- 16. Kim M. S., Kwak H. J., Lee J. W., Kim H. J., Park M. J., Park J. B., Choi K. H., Yoo H., Shin S. H., Shin W. S., Song E. S., Lee S. H. (2008) 17-Allylamino-17-demethoxygeldanamycin down-regulates hyaluronic acid-induced glioma invasion by blocking matrix metalloproteinase-9 secretion. Mol. Cancer Res. 6, 1657–1665 [DOI] [PubMed] [Google Scholar]
- 17. Taiyab A., Rao ChM. (2011) HSP90 modulates actin dynamics: Inhibition of HSP90 leads to decreased cell motility and impairs invasion. Biochim. Biophys. Acta 1813, 213–221 [DOI] [PubMed] [Google Scholar]
- 18. Guo F., Rocha K., Bali P., Pranpat M., Fiskus W., Boyapalle S., Kumaraswamy S., Balasis M., Greedy B., Armitage E. S., Lawrence N., Bhalla K. (2005) Abrogation of heat shock protein 70 induction as a strategy to increase antileukemia activity of heat shock protein 90 inhibitor 17-allylamino-demethoxy geldanamycin. Cancer Res. 65, 10536–10544 [DOI] [PubMed] [Google Scholar]
- 19. Marcu M. G., Chadli A., Bouhouche I., Catelli M., Neckers L. M. (2000) The heat shock protein 90 antagonist novobiocin interacts with a previously unrecognized ATP-binding domain in the carboxyl terminus of the chaperone. J. Biol. Chem. 275, 37181–37186 [DOI] [PubMed] [Google Scholar]
- 20. Soti C., Vermes A., Haystead T. A., Csermely P. (2003) Comparative analysis of the ATP-binding sites of Hsp90 by nucleotide affinity cleavage: a distinct nucleotide specificity of the C-terminal ATP-binding site. Eur. J. Biochem. 270, 2421–2428 [DOI] [PubMed] [Google Scholar]
- 21. Sossey-Alaoui K., Li X., Cowell J. K. (2007) c-Abl-mediated phosphorylation of WAVE3 is required for lamellipodia formation and cell migration. J. Biol. Chem. 282, 26257–26265 [DOI] [PubMed] [Google Scholar]
- 22. An W. G., Schulte T. W., Neckers L. M. (2000) The heat shock protein 90 antagonist geldanamycin alters chaperone association with p210bcr-abl and v-src proteins before their degradation by the proteasome. Cell Growth Differ. 11, 355–360 [PubMed] [Google Scholar]
- 23. Beckmann R. P., Mizzen L. E., Welch W. J. (1990) Interaction of Hsp 70 with newly synthesized proteins: Implications for protein folding and assembly. Science 248, 850–854 [DOI] [PubMed] [Google Scholar]
- 24. Chirico W. J., Waters M. G., Blobel G. (1988) 70K heat shock related proteins stimulate protein translocation into microsomes. Nature 332, 805–810 [DOI] [PubMed] [Google Scholar]
- 25. Chiang H. L., Terlecky S. R., Plant C. P., Dice J. F. (1989) A role for a 70-kilodalton heat shock protein in lysosomal degradation of intracellular proteins. Science 246, 382–385 [DOI] [PubMed] [Google Scholar]
- 26. Saliba R. S., Munro P. M., Luthert P. J., Cheetham M. E. (2002) The cellular fate of mutant rhodopsin: Quality control, degradation, and aggresome formation. J. Cell Sci. 115, 2907–2918 [DOI] [PubMed] [Google Scholar]
- 27. Yokota S., Kitahara M., Nagata K. (2000) Benzylidene lactam compound, KNK437, a novel inhibitor of acquisition of thermotolerance and heat shock protein induction in human colon carcinoma cells. Cancer Res. 60, 2942–2948 [PubMed] [Google Scholar]
- 28. Hightower L. E. (1991) Heat shock, stress proteins, chaperones, and proteotoxicity. Cell 66, 191–197 [DOI] [PubMed] [Google Scholar]
- 29. Leu J. I., Pimkina J., Frank A., Murphy M. E., George D. L. (2009) A small molecule inhibitor of inducible heat shock protein 70. Mol. Cell 36, 15–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Garg M., Kanojia D., Saini S., Suri S., Gupta A., Surolia A., Suri A. (2010) Germ cell-specific heat shock protein 70-2 is expressed in cervical carcinoma and is involved in the growth, migration, and invasion of cervical cells. Cancer 116, 3785–3796 [DOI] [PubMed] [Google Scholar]
- 31. Garg M., Kanojia D., Seth A., Kumar R., Gupta A., Surolia A., Suri A. (2010) Heat shock protein 70-2 (HSP70-2) expression in bladder urothelial carcinoma is associated with tumor progression and promotes migration and invasion. Eur. J. Cancer 46, 207–215 [DOI] [PubMed] [Google Scholar]
- 32. Liu X., Yan Z., Huang L., Guo M., Zhang Z., Guo C. (2011) Cell surface heat shock protein 90 modulates prostate cancer cell adhesion and invasion through the integrin-β1/focal adhesion kinase/c-Src signaling pathway. Oncol. Rep. 25, 1343–1351 [DOI] [PubMed] [Google Scholar]
- 33. Eustace B. K., Sakurai T., Stewart J. K., Yimlamai D., Unger C., Zehetmeier C., Lain B., Torella C., Henning S. W., Beste G., Scroggins B. T., Neckers L., Ilag L. L., Jay D. G. (2004) Functional proteomic screens reveal an essential extracellular role for hsp90α in cancer cell invasiveness. Nat. Cell Biol. 6, 507–514 [DOI] [PubMed] [Google Scholar]
- 34. Sims J. D., McCready J., Jay D. G. (2011) Extracellular heat shock protein (Hsp)70 and Hsp90α assist in matrix metalloproteinase-2 activation and breast cancer cell migration and invasion. PLoS One 6, e18848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Walsh N., Larkin A., Swan N., Conlon K., Dowling P., McDermott R., Clynes M. (2011) RNAi knockdown of Hop (Hsp70/Hsp90 organizing protein) decreases invasion via MMP-2 down-regulation. Cancer Lett. 306, 180–189 [DOI] [PubMed] [Google Scholar]
- 36. Koga F., Kihara K., Neckers L. (2009) Inhibition of cancer invasion and metastasis by targeting the molecular chaperone heat shock protein 90. Anticancer Res. 29, 797–807 [PubMed] [Google Scholar]
- 37. Leng Y., Zhang J., Badour K., Arpaia E., Freeman S., Cheung P., Siu M., Siminovitch K. (2005) Abelson-interactor-1 promotes WAVE2 membrane translocation and Abelson-mediated tyrosine phosphorylation required for WAVE2 activation. Proc. Natl. Acad. Sci. U.S.A. 102, 1098–10103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen J. S., Hsu Y. M., Chen C. C., Chen L. L., Lee C. C., Huang T. S. (2010) Secreted heat shock protein 90α induces colorectal cancer cell invasion through CD91/LRP-1 and NF-κB-mediated integrin αV expression. J. Biol. Chem. 285, 25458–25466 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.