Background: Nα-Acetylation is catalyzed by N-terminal acetyltransferases (NATs). The reaction mechanisms of NATs are unknown. hNaa50p is a member of the human NAT family.
Results: Kinetic parameters and product inhibition patterns were determined. Acetyl-CoA binding induced conformational changes facilitating peptide binding.
Conclusion: hNaa50p most likely utilizes the Theorell-Chance mechanism.
Significance: Bisubstrate inhibitors, mimicking a ternary complex, should function as specific inhibitors of human NATs.
Keywords: Acetyl Coenzyme A, Enzyme Kinetics, Enzyme Mechanisms, NMR, Post-translational Modification, Protein Acylation
Abstract
Nα-Acetylation is a common protein modification catalyzed by different N-terminal acetyltransferases (NATs). Their essential role in the biogenesis and degradation of proteins is becoming increasingly evident. The NAT hNaa50p preferentially modifies peptides starting with methionine followed by a hydrophobic amino acid. hNaa50p also possesses Nϵ-autoacetylation activity. So far, no eukaryotic NAT has been mechanistically investigated. In this study, we used NMR spectroscopy, bisubstrate kinetic assays, and product inhibition experiments to demonstrate that hNaa50p utilizes an ordered Bi Bi reaction of the Theorell-Chance type. The NMR results, both the substrate binding study and the dynamic data, further indicate that the binding of acetyl-CoA induces a conformational change that is required for the peptide to bind to the active site. In support of an ordered Bi Bi reaction mechanism, addition of peptide in the absence of acetyl-CoA did not alter the structure of the protein. This model is further strengthened by the NMR results using a catalytically inactive hNaa50p mutant.
Introduction
Acetylation is one of the most common covalent modifications, occurring on the majority of eukaryotic proteins, in which an acetyl group is transferred from acetyl coenzyme A either to the α-amino group of protein N termini (Nα-acetylation) or to the ϵ-amino group of specific lysine residues (Nϵ-acetylation) (1, 2). So far, six different N-terminal acetyltransferase (NAT)4 complexes have been identified in eukaryotes (NatA–NatF) (3, 4), and interestingly, subunits of the human NatA complex, i.e. hNaa10p and hNaa15p, have been increasingly linked to cancer development and prognosis. For example, the genes encoding hNaa10p and hNaa15p are up-regulated in several types of cancer (5–7). Functional studies indicate that hNaa10p and hNaa15p are essential for growth and survival of cancer cell lines (8–11). Antitumorigenic roles have also been proposed (12, 13). Naa50p is physically associated with NatA (14, 15) but has its own catalytic activity defined as NatE (16, 17). From fruit flies to humans, Naa50p is essential for proper sister chromatid cohesion and chromosome resolution (18–20).
These observations highlight the biological significance of the human NATs, and mark them as potential targets for cancer (21). Detailed knowledge of the catalytic and kinetic mechanisms of the NATs will undoubtedly aid our efforts to develop inhibitors targeting these enzymes. Two types of kinetic mechanisms are observed for acetyl transfer reactions: a ping-pong mechanism (22) and the ternary complex/sequential mechanism (23–26). The former mechanism is typically associated with an acetyl-enzyme intermediate, whereas the latter mechanism requires that both substrates bind to the enzyme to form a ternary complex prior to acetyl transfer. Our data demonstrate that the acetyl transfer reaction catalyzed by hNaa50p involves the formation of a ternary complex. Thus, bisubstrate inhibitors, mimicking a ternary complex molecule, should function as highly specific and efficient inhibitors of hNaa50p and possibly other human NATs.
EXPERIMENTAL PROCEDURES
Prokaryotic Expression and Purification of Recombinant Proteins
GST-hNaa50p and the H112A mutant were expressed and purified as described previously (16).
In Vitro Acetylation/Kinetic Assays
All enzyme kinetic experiments were performed essentially as described previously (27). The steady-state kinetic parameters and the enzyme inhibition patterns were analyzed by global fit analysis using GraFit 7 software (Erithacus Software). See supplemental material for detailed information. The peptide substrate used was 1MLGPEGGRWGRPVGRRRRPVRVYP24 (denoted 1MLGP–RRR24), as it is the optimal in vitro substrate for hNaa50p (17). The C-terminal 17 amino acids of this peptide correspond to the sequence of ACTH, except that all Lys residues have been replaced with Arg to minimize the potential interference from Nϵ-acetylation. The positively charged Arg residues also facilitate peptide solubility.
NMR Spectroscopy
Resonances were assigned for hNaa50p samples (250 μm in 600 μl, 15N- and 13C-labeled) in 90% H2O, 10% D2O, 1.0 mm acetyl-CoA, 100 mm NaCl, and 50 mm NaH2PO4 (pH 7.4). A two-dimensional 1H-15N heteronuclear single-quantum correlation spectrum (28, 29) was collected at 600.13 MHz (1H) on a Bruker BioSpin AV600 spectrometer equipped with a superconducting actively shielded magnet. A 5-mm triple-resonance (1H, 13C, 15N) inverse cryogenic probe head with z-gradient coils and cold 1H and 13C preamplifiers was used. The sample temperature was kept at 310 K. The data were processed using Bruker BioSpin TopSpin 1.3 software. Sequential backbone assignment was achieved using the standard three-dimensional experiments (HNCA, HN(CO)CA, HNCO, HN(CA)CO, HNCACB, and CBCA(CO)NH) (30) on a Varian Inova 800 NMR spectrometer with a 5-mm triple-resonance (1H, 13C, 15N) probe at 310 K (Swedish NMR Centre, University of Gothenburg). The spectra were processed with NMRPipe (31) and analyzed using Cara (32). For further diminishing of side chain ambiguity, three-dimensional CC(CO)NH (33, 34) and several two-dimensional 1H-15N MUSIC (multiplicity selective in-phase coherence transfer) (35–37) experiments were conducted on the Bruker BioSpin AV600 spectrometer. The 1H and 13C chemical shifts were referenced to 4,4-dimethyl-4-silapentane-1-sulfonic acid as an internal standard. The 15N chemical shifts were calculated from the adjusted 1H frequency (38). In spectra without 4,4-dimethyl-4-silapentane-1-sulfonic acid, the signal of the solvent HDO was set to 4.68 ppm, as this was the measured shift value of HDO in the 4,4-dimethyl-4-silapentane-1-sulfonic acid sample. Furthermore, 4.68 ppm is the calculated shift for HDO at this temperature, pH, and ionic strength (39). All experiments with important acquisition parameters are listed in supplemental Table S1.
To determine the substrate binding order, two-dimensional 1H-15N SOFAST-heteronuclear multiple-quantum correlation (HMQC) experiments (40) were collected at 600 MHz on the following samples: 100 μm 15N-labeled hNaa50p, hNaa50p with 500 μm 1MLGP–RRR24, hNaa50p with 500 μm acetyl-CoA, hNaa50p with 500 μm CoA, hNaa50p with 500 μm CoA and 500 μm 1MLGP–RRR24, and hNaa50p with 500 μm CoA and 500 μm acetylated 1MLGP–RRR24. To study the interaction between the enzyme and both substrates simultaneously, HMQC spectra of ∼100 μm 15N-labeled hNaa50p H112A mutant with 500 μm acetyl-CoA and mutant with 500 μm acetyl-CoA and 1 mm 1MLGP–RRR24 were collected. The temperature was lowered to 298 K to further slow down the enzyme reaction. All samples were dissolved in the buffer that was used for the resonance assignment, with exception of the H112A mutant, for which the pH in the buffer was raised to 8.0.
15N T1 and T2 relaxation rates and 15N{1H} heteronuclear NOEs (41) at 298 K were measured using two-dimensional 1H-15N heteronuclear single-quantum correlation-based methods at 600 MHz. T1 and T2 values were obtained using three parallel series of 10 randomized delays in the range of 50–1850 ms for T1 and the range of 0–123.2 ms for T2. Four interleaved heteronuclear NOE spectra were recorded. In the spectra with NOE, a proton saturation time of 3 s and a recycling delay of 7 s were used, whereas in the spectra without NOE, the recycling delay was 10 s. No saturation was included in the latter case. All spectra were processed using the NMRPipe software package. Peak heights were measured in all spectra of a relaxation series and fitted with a two-parameter single-exponential function to extract the relaxation rates, except for the heteronuclear NOE experiments, in which the result was determined by the intensity ratio from the spectrum with NOE and the reference spectrum without NOE, averaged for the four experiments. Errors in T1 and T2 were obtained by Monte Carlo simulations (42), whereas the errors in heteronuclear NOE are the root mean squares of ratios derived from the four sets of measurements. The T1, T2, and heteronuclear NOE values were used to calculate the order parameter (S2) for a completely anisotropic model in the TENSOR2 program (43).
RESULTS
Bisubstrate Kinetics
Because the hNaa50p reaction involves two substrates, acetyl-CoA and peptide, bisubstrate kinetic experiments were performed to begin to distinguish between the requirement for an acetyl-enzyme intermediate (ping-pong) or a direct transfer mechanism (ternary complex) reaction, as well as to determine kinetic parameters. A summary of the inhibition patterns and values of kinetic parameters is presented in Table 1. Initial velocity experiments were performed by varying the concentration of acetyl-CoA at several fixed concentrations of peptide (supplemental Fig. S1). The data were globally fit to the ternary complex model and the ping-pong model. Double-reciprocal plots of the initial velocity experiments generated an intersecting line pattern, indicating that hNaa50p follows a ternary complex mechanism (supplemental Fig. S1).
TABLE 1.
Summary of results from kinetic experiments on hNaa50p with various combinations of inhibitors and substrates (supplemental Figs. S1–S3)
| Inhibitor and type of inhibition | Variable substrate | Fixed substrate (subsaturated) | Kinetic parametersa |
|---|---|---|---|
| Control | Acetyl-CoA (2–25 μm) | Peptide (300 μm) | Km = 5.0 ± 1.5 μm; Vmax = 6.0 ± 0.56b,c |
| CoA (0–80 μm), competitive | Acetyl-CoA (2–25 μm) | Peptide (80 μm) | KI = 2.27 ± 0.16 μm; Km = 4.7 ± 0.4 μm; Vmax = 5.7 ± 0.2b,c |
| Desulfo-CoA (0–60 μm), competitive | Acetyl-CoA (2–25 μm) | Peptide (80 μm) | KI = 67 ± 9 μm; Km = 17.4 ± 1.7 μm; Vmax = 21.2 ± 1.1b,c |
| CoA (0–80 μm), non-competitive | Peptide (30–500 μm) | Acetyl-CoA (100 μm) | KI = 27.7 ± 1.7 μm; Km 59 ± 5 μm; Vmax = 7.4 ± 0.3c,d |
a The kinetic parameters Vmax and Km are apparent values calculated for the experiments without added inhibitor. The specific activity of different lots of the purified enzyme may vary and thus contribute to variation in the values of these parameters.
b Apparent Km and Vmax for acetyl-CoA in the absence of inhibitor.
c Vmax is given as picomoles of acetylated peptide × min−1 × pmol hNaa50p−1.
d Apparent Km and Vmax for unacetylated peptide substrate.
Product Inhibition
To further investigate the kinetic mechanism, product inhibition experiments were performed using several fixed concentrations of the reaction product CoA and the peptide substrate while varying the concentration of acetyl-CoA. Double-reciprocal plots of acetyl-CoA versus the initial velocities at different concentrations of CoA produced an intersecting line pattern consistent with CoA functioning as a competitive inhibitor for acetyl-CoA (supplemental Fig. S2A). KI for the competitive inhibition of CoA was calculated to be 2.27 ± 0.16 μm. To verify the product inhibition observations, the experiments were repeated with desulfo-CoA, a dead-end analog inhibitor of acetyl-CoA. Desulfo-CoA showed the same competitive inhibition pattern as CoA (supplemental Fig. S2B), with a KI of 67 ± 9 μm.
Next, CoA was used as product inhibitor in experiments in which the concentration of acetyl-CoA was maintained at a constant level and the concentration of the peptide substrate was varied (supplemental Fig. S3). A non-competitive inhibition pattern was obtained (KI = 27.7 ± 1.7 μm), well in line with a ternary complex mechanism.
In a typical ternary complex mechanism, the reaction product, acetylated peptide, should also show an inhibitory effect on both substrates. To study the possible product inhibition patterns of acetylated peptides, experiments were performed using fixed levels of N-terminally acetylated peptide as the product inhibitor and acetyl-CoA or peptide as the variable substrate. Several experiments using saturating or subsaturating concentrations of substrates were performed. Surprisingly, even very high concentrations (up to 1 mm) of acetylated peptide failed to show any inhibition against any of the substrates (supplemental Fig. S4, A and B, and data not shown). This indicates that the acetylated peptide is an extremely weak product inhibitor. Unfortunately, a dead-end analog of the acetylated product peptide does not exist; thus, additional dead-end analog inhibition experiments could not be performed to further refine the kinetic mechanism of hNaa50p. Nevertheless, these data indicate that hNaa50p forms a very unstable ternary complex and suggest that hNaa50p does not follow a classical ternary complex mechanism.
NMR Resonance Assignment and Secondary Structures
140 of the 164 amino acid backbone NH correlations (∼85%) could be assigned to the hNaa50p sequence using three-dimensional heteronuclear NMR experiments combined with amino acid-specific techniques (Biological Magnetic Resonance Data Bank accession number 18202) (supplemental Table S2). Missing residues are due to the protein size, leading to signal broadening and spectral overlap in some regions and to conformational exchange in flexible parts of the protein, like the unstructured C terminus.
The 13C shift deviations from their random coil values (39, 44) were used to determine the secondary structural elements of hNaa50p (supplemental Fig. S5). The results are overall in agreement with available information on the secondary structures obtained from x-ray crystallography of hNaa50p in complex with acetyl-CoA (Protein Data Bank code 2OB0).
Determining Order of Substrate Binding by NMR
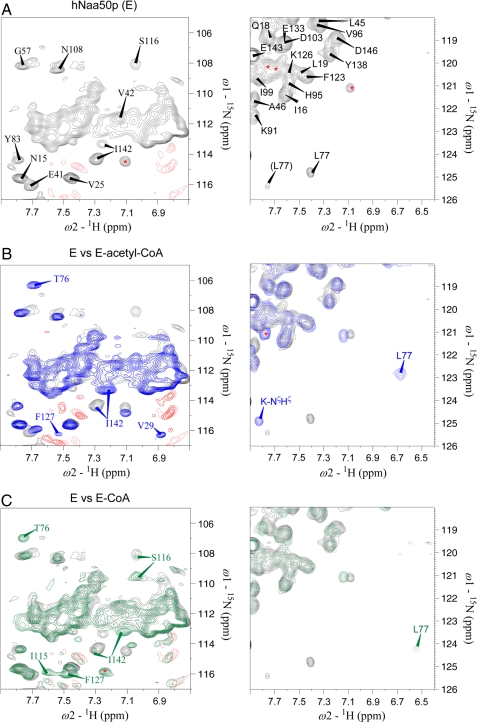
The changes in the backbone NH correlations were used to investigate the order of substrate binding. First, a HMQC experiment was performed on hNaa50p in the absence of substrates. Next, recordings were made in the presence of substrates and products in various combinations. The regions of the spectra presented in Fig. 1 comprise the majority of peaks that were consistently and strongly affected by substrate or product binding.
FIGURE 1.
Two-dimensional 1H-15N HMQC spectra of 15N-labeled hNaa50p in absence or presence of acetyl-CoA or CoA. A, two expanded regions of the two-dimensional 1H-15N HMQC spectrum of 15N-labeled free hNaa50p. B, overlay of the HMQC spectra of hNaa50p (black) and hNaa50p with acetyl-CoA (blue). C, overlay of the HMQC spectra of hNaa50p (black) and hNaa50p with CoA (green). For clarity, the complete peak identity is shown only in A, whereas peaks with significant chemical shift changes upon acetyl-CoA or CoA binding are shown in B and C. The side chain signals of Asn and Gln present in the expansions in the left panels (severely overlapped peaks) are not assigned. Negative signals (red) are either noise or side chain signals of Arg. There was no effect observed on these peaks. Peaks with unknown identity are indicated with red asterisks. Note that the extraction in the right panel in B includes one of the autoacetylated lysine side chains, the peak marked K-NζHζ (not assigned).
The spectral positions of Val-29, Thr-76, Leu-77, Ser-116, Phe-127, and Ile-142 show significant changes upon adding acetyl-CoA to hNaa50p (Fig. 1B), whereas most of the remaining NH signals in the spectrum were either unaffected or only slightly altered (Fig. 1, A and B). This spectral repositioning indicates that acetyl-CoA is capable of binding to hNaa50p in the absence of peptide substrate and that probably a conformational change occurs upon binding this substrate.
Because the product inhibition experiments (supplemental Fig. S2A) indicated that CoA is a competitive inhibitor of acetyl-CoA, NMR spectroscopy was used to determine whether or not CoA binds directly to hNaa50p in the absence of acetyl-CoA (Fig. 1C). The spectrum of hNaa50p in the presence of CoA shows several similarities to the spectrum of hNaa50p in complex with acetyl-CoA (Fig. 1B); both spectra differ from that of the substrate-free protein (Fig. 1A). The spectral positions of Thr-76, Leu-77, Phe-127, and Ile-142 change similarly to that observed in the spectrum with acetyl-CoA. On the other hand, the NH shifts of Val-29, Ile-115, and Ser-116 (all situated close to the acetyl group of acetyl-CoA) differ between the two spectra (Fig. 1, B and C). This indicates that CoA binds free hNaa50p at the same structural moiety and has a similar structural effect on hNaa50p as acetyl-CoA, but with a differing “induced fit” response.
We further used NMR spectroscopy to investigate whether the peptide substrate interacts with hNaa50p in the absence of acetyl-CoA. Evidence of such an interaction would support a random ternary complex mechanism (random Bi Bi). When hNaa50p was mixed with saturating concentration of peptide, the only residue that appeared to be affected was Ile-142 (supplemental Fig. S6A), suggesting that the peptide substrate does not bind significantly to free enzyme to form a stable complex. Thus, a random ternary complex mechanism is unlikely. The lack of observable inhibition by acetylated peptide (supplemental Fig. S3) was further investigated. Acetylated 1MLGP–RRR24 was added to the hNaa50p-CoA complex, and a HMQC spectrum was recorded (supplemental Fig. S6, B and C). The spectrum did not differ from the one with hNaa50p and CoA (Fig. 1C).This result is consistent with a model in which acetylated 1MLGP–RRR24 does not form a stable interaction with the hNaa50p-CoA complex, concordant with the lack of product inhibition observed in the kinetic experiments (supplemental Fig. S4).
The different ternary complexes of hNaa50p and both substrates were investigated using the wild-type hNaa50p-CoA complex in the presence of peptide and the enzymatically inactive but natively folded hNaa50p H112A mutant (45). Asp-38, Tyr-73, Leu-77, Ala-81, and Tyr-110 showed small chemical shift changes when 1MLGP–RRR24 instead of acetylated 1MLGP–RRR24 was added to the hNaa50p-CoA complex. (Tyr-73 and Tyr-110 are shown in supplemental Fig. S7.) Ten additional amino acids (Arg-71, Leu-72, Gly-87, Thr-90, Gly-104, Asn-108, Leu-111, Phe-123, Phe-129, and Ile-142) were affected when the peptide was added to the mutant-acetyl-CoA complex. These results suggest that the affinity between hNaa50p and peptide is stronger when the enzyme is in complex with acetyl-CoA instead of CoA.
Protein Dynamics
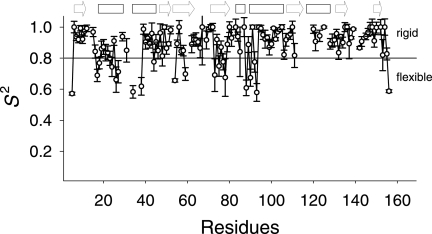
To elucidate the flexible parts in hNaa50p, the dynamics of the protein in the presence of acetyl-CoA were analyzed by NMR spectroscopy. The experimental T1, T2, and heteronuclear NOE values were used to calculate the order parameters (S2) (Fig. 2) in TENSOR2 (43) utilizing the simple Lipari-Szabo approach (46, 47). The most flexible parts in the structure, defined by an S2 value below 0.8 (Fig. 2), are the regions comprising the first two helices in the molecule (residues 17–26 and 33–38) in addition to the stretches 73YIMTLG78 and 87GIGTKML93. According to the crystal structure of hNaa50p in complex with acetyl-CoA, the former stretch is close to the cysteamine moiety of acetyl-CoA and appears to be involved in enzyme catalysis. The direct involvement of Tyr-73 in catalysis is strongly supported by a recent structural (Protein Data Bank code 3TFY) and functional investigation (45). The latter stretch, which is close to the pantothenic acid moiety, is part of the hNaa50p variant of the conserved motif A responsible for acetyl-CoA binding, (Q/R)XXGX(G/A), common to the GCN5 superfamily (48, 49). Overall, our NMR results suggest that acetyl-CoA, in accordance with the kinetic data (supplemental Fig. S1–S3), is the first substrate to enter the active site, causing structural changes required for peptide substrate interaction as indicated by cross-peak splittings and the dynamic data.
FIGURE 2.
Order parameters (S2) of hNaa50p determined from NMR relaxation data. The order parameters (S2) with their S.D. are plotted against the amino acid positions. Highly ordered residues have S2 > 0.8 (solid line), whereas residues below this border belong to more flexible parts of the protein. Missing residues in the plot are either unassigned or severely overlapping. The unstructured C terminus of the protein was excluded from the S2 calculation. The arrows and boxes above the plot indicate strands and helices, respectively. The secondary structures are according to the crystal structure of hNaa50p in complex with acetyl-CoA.
DISCUSSION
In this study, we have presented enzyme kinetic and NMR data that point to an ordered ternary complex mechanism for the human Naa50p NAT activity. The initial velocity experiments, as well as the competitive product inhibition pattern observed for hNaa50p using CoA as product inhibitor and acetyl-CoA as variable substrate (supplemental Fig. S2A), rule out a ping-pong type of mechanism. Also, results from experiments using desulfo-CoA as a dead-end acetyl-CoA analog (supplemental Fig. S2B) are inconsistent with a ping-pong mechanism. The non-competitive inhibition pattern observed when CoA was tested against peptide as variable substrate and at fixed concentrations of acetyl-CoA also rules out a ping-pong mechanism (supplemental Fig. S3). In a ping-pong mechanism, one expects the acetyl group to be transferred to an enzyme residue, with the subsequent release of CoA. However, in the crystal structure of the hNaa50p-acetyl-CoA complex, the acetyl group remains covalently bound to CoA. Thus, the enzyme kinetic experiments strongly support a ternary complex mechanism, which is consistent with other GCN5-like acetyltransferases (23, 50–52). Although the results of our kinetic experiments cannot completely distinguish between a random and an ordered sequential mechanism, the data are inconsistent with both the steady-state and equilibrium ordered mechanisms (see Table 1 for a summary of inhibition patterns and kinetic parameters).
From the NMR studies, it appears that the residues that show the greatest chemical shift changes upon addition of acetyl-CoA/CoA and 1MLGP–RRR24 are all either involved in the binding of the substrates or part of flexible structures of hNaa50p undergoing conformational changes similar to an induced fit response. Leu-77, the amino acid closest to the cysteamine moiety of acetyl-CoA, shows the greatest difference, shifting 2 ppm upfield in NH and 0.8 ppm upfield in HN upon acetyl-CoA binding (Fig. 1B). The change in the NH chemical shift of Leu-77 is less pronounced upon binding of CoA (Fig. 1C). The preceding amino acid, Thr-76, shows a similar effect. Remarkably, the peak strength of Leu-77 increases significantly when 1MLGP–RRR24 is added to either the hNaa50p-CoA or H112A mutant-acetyl-CoA complex, indicating a more stable structural conformation in the region of Leu-77 upon binding of the second substrate. In the dynamic data, the S2 values for hNaa50p in complex with acetyl-CoA indicate that amino acid region 73–78 is more flexible than expected for a β-strand (Fig. 2). This flexibility might be necessary for the second substrate, the peptide, to enter and bind the active site. In accordance with this suggestion, it was observed that Arg-71, Leu-72, and probably Ala-81 change spectral positions when both substrates are added to the hNaa50p H112A mutant.
Val-29 and Ile-142 show very interesting chemical shift changes compared with all of the other amino acids. These residues are positioned opposite to each other in two loops that, according to recent results (45), are part of the hydrophobic pocket in which the N-terminal methionine of the peptide substrate binds (supplemental Fig. S9). Thus, it is likely that these loops alter positions relative to each other in an induced fit response upon acetyl-CoA binding.
It was possible to assign Val-29 only in the spectrum of the hNaa50p-acetyl-CoA complex at 310 K, suggesting that Val-29 undergoes intermediate-rate conformational exchange both in the absence of acetyl-CoA and at lower temperatures, leading to signal broadening. This α-helix has already previously proven to be of special interest: we demonstrated that Lys-34 and Lys-37 are autoacetylated and important for catalytic activity and specificity (17). Introducing conservative K34R and K37R mutations results in a 4-fold decreased Nα-acetyltransferase activity and specificity toward 1MLGP–RRR24. In addition, the dynamic data (Fig. 2) suggest that amino acid region 20–40 is more flexible than expected for a sequence that contains two α-helices, whereas the chemical shift indices indicate that the second helix is shorter than observed in the crystal structure (Protein Data Bank code 2OB0) (supplemental Fig. S5). This region is likely to be prone to conformational changes that are important for optimal catalytic activity and specificity. This notion is supported by the fact that Asp-38 is one of the amino acids that change spectral position upon addition of peptide to hNaa50p in complex with both CoA and acetyl-CoA.
The NH signal of Ile-142, on the other side of the hydrophobic pocket, is split in two in the spectrum of free hNaa50p, indicating a slow conformational change between two states, where the strongest peak corresponds to the dominating conformation (Fig. 1A). Surprisingly, this splitting is not observed in the spectrum of hNaa50p in combination with 1MLGP–RRR24 (supplemental Fig. S6A). After binding of acetyl-CoA/CoA, the ratio between the two states changes, and the former dominating signal almost disappears; however, the chemical shifts of Ile-142 in the two spectra are not identical (Fig. 1, B and C). Because both Val-29 and Ile-142 appear to take part in peptide binding, the chemical shift differences of these two residues upon addition of acetyl-CoA or CoA indicate that binding of acetyl-CoA leads to the formation of a more rigid hydrophobic pocket for 1MLGP–RRR24 than CoA does. This idea is supported by the observation that Ile-142 changes spectral position again upon binding of the peptide to the mutant-acetyl-CoA complex. Also, the dynamic data and the 13C chemical shifts suggest that Ile-142 in the hNaa50p-acetyl-CoA complex is situated in a more rigid environment than is expected for a loop (Fig. 2).
No major chemical shift changes are observed in the sequence 84RRLGIG89, which is the hNaa50p variant of the acetyl-CoA-binding motif A, (Q/R)XXGX(G/A), common to the GCN5 superfamily (48, 49), upon binding of the first substrate. However, the dynamic data presented in Fig. 2 strongly indicate that amino acid stretch 87–93 is far more flexible than is expected for the start of a long α-helix. Again, this flexibility might be important for the binding of the peptide as the second substrate: both Gly-87 and Thr-90 show small chemical shift changes upon addition of peptide to the mutant-acetyl-CoA complex.
Interestingly, Tyr-124 (supplemental Fig. S8A) shows chemical shift changes in the spectrum of the hNaa50p-acetyl-CoA complex, which may indicate that Tyr-124 is involved in substrate binding. On the basis of the crystal structure (supplemental Fig. S8B), we hypothesize that a water molecule mediates contact between the hydroxyl group of Tyr-124 and the nitrogen atom of the cysteamine moiety of acetyl-CoA. The same water molecule may also be coordinated to the backbone of Leu-77, which presents significant chemical shift changes upon substrate binding (as discussed above and in Fig. 1B). The Tyr-124 chemical shift change is consistent with our previous observation that Tyr-124 is essential for both the Nα-acetylation activity and Nϵ-autoacetylation function of hNaa50p (17). Furthermore, the mutant shows chemical shift changes of the neighboring Phe-123.
Recent results (45) suggest that the residues likely to be involved in enzyme catalysis are Tyr-73 and His-112. Tyr-73 shows chemical shift changes upon binding of CoA to the protein, whereas this chemical shift is not affected by acetyl-CoA binding (data not shown). Interestingly, the spectrum of hNaa50p-CoA in complex with 1MLGP–RRR24 contains both peaks (supplemental Fig. S7A). On the other hand, the addition of peptide to the hNaa50p H112A mutant-acetyl-CoA complex changes the intensity distribution of the three Tyr-73 peaks that are found in the HMQC spectra of the mutant (supplemental Fig. S7B). Asn-108, Tyr-110, Leu-111 (supplemental Fig. S8A), Ile-115, and Ser-116 all present changes upon acetyl-CoA/CoA and peptide binding whereas, the catalytically important residue His-112 (supplemental Fig. S9) was not possible to assign. Similar to Tyr-73, Tyr-110 shows chemical shift changes only upon binding of CoA, but not acetyl-CoA (data not shown), and the spectra (Tyr-110) of hNaa50p-CoA-1MLGP–RRR24 and hNaa50p H112A mutant-acetyl-CoA-1MLGP–RRR24 differ slightly from the spectra of both the hNaa50p-CoA and mutant-acetyl-CoA complexes with or without acetylated peptide (supplemental Fig. S7). In hNaa50p H112A, Asn-108 and Leu-111 are also affected. The latter appears to show conformational exchange between two states, in both the presence and absence of 1MLGP–RRR24. On the other hand, Ile-115 and Ser-116 are detected only in spectra of hNaa50p in complex with CoA (Fig. 1C). Combined, these data all support an ordered Bi Bi sequential mechanism in which the peptide is the second substrate to enter the active site. Significant NMR signal shifts indicating an interaction between acetylated peptide and enzyme were not identified.
Finally, two residues change spectral position only upon addition of peptide to the mutant-acetyl-CoA complex: Gly-104, which shows conformational exchange between two states, and Phe-129. Neither of these amino acids is in direct contact with the substrates, indicating that some long-range effects occur upon substrate binding.
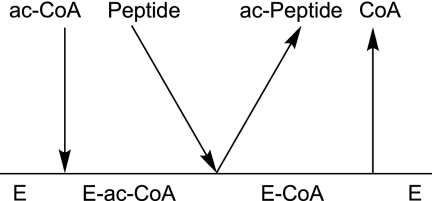
Taken together, the NMR results indicate an essential difference in interactions between the protein and the substrate acetyl-CoA and the product CoA around the active site. Furthermore, the affinity between the enzyme and the peptide substrate is very small in the absence of acetyl-CoA. Overall, the NMR data suggest that hNaa50p utilizes an ordered mechanism of substrate binding, with acetyl-CoA binding to the enzyme prior to the peptide substrate. Given that the product and dead-end analog inhibition studies ruled out both the steady-state and equilibrium ordered mechanisms, the sum of the data suggests that hNaa50p utilizes a special type of Bi Bi sequential mechanism, possibly of the Theorell-Chance type as outlined in Fig. 3. This type of reaction mechanism has been reported for the human lysine acetyltransferase p300 (26) and, as such differs, from other protein acetyltransferases (25). A Theorell-Chance mechanism is supported by the following observations. (a) Only one chemical shift in the HMQC spectrum of hNaa50p slightly changes when the enzyme is incubated in the presence of peptide, which indicates that the affinity of the free enzyme for the peptide substrate in the absence of acetyl-CoA is very low (supplemental Fig. S6A). At the same time, close to 20 NH correlations change when the peptide is added to the hNaa50p H112A mutant incubated with acetyl-CoA. Similar but more subtle effects are observed when the peptide is added to the wild-type hNaa50p-CoA complex. (b) Upon binding of either acetyl-CoA or CoA, the enzyme undergoes a dramatic conformational change, which is observable in different NMR experiments. It is likely that this change is required for the formation of the hNaa50p-acetyl-CoA-1MLGP–RRR24 complex. (c) There is no observable effect on the NMR spectrum when acetylated peptide product is added to the enzyme-CoA complex. This indicates that the affinity between the hNaa50p-CoA complex and acetylated peptide is significantly lower than would be expected for a classical ordered Bi Bi kinetic mechanism. This work represents the first mechanistic study of a eukaryotic NAT and provides important information that will potentially aid the generation of specific inhibitors of this group of GCN5 enzymes.
FIGURE 3.
Proposed kinetic mechanism of hNaa50p NAT activity.
Supplementary Material
Acknowledgments
We thank L. Vikebø, M. Algrøy, and N. Glomnes for technical assistance and K. J. Bjuland for assistance in protein modeling. We also thank the Swedish NMR Centre at the University of Gothenburg.

This article contains supplemental Figs. S1–S9, Tables S1 and S2, and Equations 1–4.
The NMR assignments have been deposited in the Biological Magnetic Resonance DataBank (accession number 18202).
- NAT
- N-terminal acetyltransferase
- HMQC
- heteronuclear multiple-quantum correlation.
REFERENCES
- 1. Polevoda B., Sherman F. (2002) The diversity of acetylated proteins. Genome Biol. 3, reviews0006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Arnesen T., Van Damme P., Polevoda B., Helsens K., Evjenth R., Colaert N., Varhaug J. E., Vandekerckhove J., Lillehaug J. R., Sherman F., Gevaert K. (2009) Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans. Proc. Natl. Acad. Sci. U.S.A. 106, 8157–8162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Van Damme P., Hole K., Pimenta-Marques A., Helsens K., Vandekerckhove J., Martinho R. G., Gevaert K., Arnesen T. (2011) NatF contributes to an evolutionary shift in protein N-terminal acetylation and is important for normal chromosome segregation. PLoS Genet. 7, e1002169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Polevoda B., Arnesen T., Sherman F. (2009) A synopsis of eukaryotic Nα-terminal acetyltransferases: nomenclature, subunits, and substrates. BMC Proc. 3, S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ren T., Jiang B., Jin G., Li J., Dong B., Zhang J., Meng L., Wu J., Shou C. (2008) Generation of novel monoclonal antibodies and their application for detecting ARD1 expression in colorectal cancer. Cancer Lett. 264, 83–92 [DOI] [PubMed] [Google Scholar]
- 6. Yu M., Gong J., Ma M., Yang H., Lai J., Wu H., Li L., Li L., Tan D. (2009) Immunohistochemical analysis of human arrest-defective-1 expressed in cancers in vivo. Oncol. Rep. 21, 909–915 [DOI] [PubMed] [Google Scholar]
- 7. Fluge Ø., Bruland O., Akslen L. A., Varhaug J. E., Lillehaug J. R. (2002) NATH, a novel gene overexpressed in papillary thyroid carcinomas. Oncogene 21, 5056–5068 [DOI] [PubMed] [Google Scholar]
- 8. Arnesen T., Gromyko D., Pendino F., Ryningen A., Varhaug J. E., Lillehaug J. R. (2006) Induction of apoptosis in human cells by RNAi-mediated knockdown of hARD1 and NATH, components of the protein Nα-acetyltransferase complex. Oncogene 25, 4350–4360 [DOI] [PubMed] [Google Scholar]
- 9. Lim J. H., Park J. W., Chun Y. S. (2006) Human arrest-defective 1 acetylates and activates β-catenin, promoting lung cancer cell proliferation. Cancer Res. 66, 10677–10682 [DOI] [PubMed] [Google Scholar]
- 10. Fisher T. S., Etages S. D., Hayes L., Crimin K., Li B. (2005) Analysis of ARD1 function in hypoxia response using retroviral RNA interference. J. Biol. Chem. 280, 17749–17757 [DOI] [PubMed] [Google Scholar]
- 11. Gromyko D., Arnesen T., Ryningen A., Varhaug J. E., Lillehaug J. R. (2010) Depletion of the human Nα-terminal acetyltransferase A induces p53-dependent apoptosis and p53-independent growth inhibition. Int. J. Cancer 127, 2777–2789 [DOI] [PubMed] [Google Scholar]
- 12. Yi C. H., Sogah D. K., Boyce M., Degterev A., Christofferson D. E., Yuan J. (2007) A genome-wide RNAi screen reveals multiple regulators of caspase activation. J. Cell Biol. 179, 619–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kuo H. P., Lee D. F., Chen C. T., Liu M., Chou C. K., Lee H. J., Du Y., Xie X., Wei Y., Xia W., Weihua Z., Yang J. Y., Yen C. J., Huang T. H., Tan M., Xing G., Zhao Y., Lin C. H., Tsai S. F., Fidler I. J., Hung M. C. (2010) ARD1 stabilization of TSC2 suppresses tumorigenesis through the mTOR signaling pathway. Sci. Signal. 3, ra9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gautschi M., Just S., Mun A., Ross S., Rücknagel P., Dubaquié Y., Ehrenhofer-Murray A., Rospert S. (2003) The yeast Nα-acetyltransferase NatA is quantitatively anchored to the ribosome and interacts with nascent polypeptides. Mol. Cell. Biol. 23, 7403–7414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Arnesen T., Anderson D., Torsvik J., Halseth H. B., Varhaug J. E., Lillehaug J. R. (2006) Cloning and characterization of hNAT5/hSAN: an evolutionarily conserved component of the NatA protein Nα-acetyltransferase complex. Gene 371, 291–295 [DOI] [PubMed] [Google Scholar]
- 16. Van Damme P., Evjenth R., Foyn H., Demeyer K., De Bock P. J., Lillehaug J. R., Vandekerckhove J., Arnesen T., Gevaert K. (2011) Proteome-derived peptide libraries allow detailed analysis of the substrate specificities of Nα-acetyltransferases and point to hNaa10p as the post-translational actin Nα-acetyltransferase. Mol. Cell. Proteomics 10, M110.004580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Evjenth R., Hole K., Karlsen O. A., Ziegler M., Arnesen T., Lillehaug J. R. (2009) Human Naa50p (NAT5/SAN) displays both protein Nα- and Nϵ-acetyltransferase activity. J. Biol. Chem. 284, 31122–31129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hou F., Chu C. W., Kong X., Yokomori K., Zou H. (2007) The acetyltransferase activity of San stabilizes the mitotic cohesin at the centromeres in a shugoshin-independent manner. J. Cell Biol. 177, 587–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pimenta-Marques A., Tostões R., Marty T., Barbosa V., Lehmann R., Martinho R. G. (2008) Differential requirements of a mitotic acetyltransferase in somatic and germ line cells. Dev. Biol. 323, 197–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Williams B. C., Garrett-Engele C. M., Li Z., Williams E. V., Rosenman E. D., Goldberg M. L. (2003) Two putative acetyltransferases, san and deco, are required for establishing sister chromatid cohesion in Drosophila. Curr. Biol. 13, 2025–2036 [DOI] [PubMed] [Google Scholar]
- 21. Arnesen T., Thompson P. R., Varhaug J. E., Lillehaug J. R. (2008) The protein acetyltransferase ARD1: a novel cancer drug target? Curr. Cancer Drug Targets 8, 545–553 [DOI] [PubMed] [Google Scholar]
- 22. Yan Y., Harper S., Speicher D. W., Marmorstein R. (2002) The catalytic mechanism of the ESA1 histone acetyltransferase involves a self-acetylated intermediate. Nature Struct. Biol. 9, 862–869 [DOI] [PubMed] [Google Scholar]
- 23. Berndsen C. E., Albaugh B. N., Tan S., Denu J. M. (2007) Catalytic mechanism of a MYST family histone acetyltransferase. Biochemistry 46, 623–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Berndsen C. E., Denu J. M. (2008) Catalysis and substrate selection by histone/protein lysine acetyltransferases. Curr. Opin. Struct. Biol. 18, 682–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rojas J. R., Trievel R. C., Zhou J., Mo Y., Li X., Berger S. L., Allis C. D., Marmorstein R. (1999) Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide. Nature 401, 93–98 [DOI] [PubMed] [Google Scholar]
- 26. Wang L., Tang Y., Cole P. A., Marmorstein R. (2008) Structure and chemistry of the p300/CBP and Rtt109 histone acetyltransferases: implications for histone acetyltransferase evolution and function. Curr. Opin. Struct. Biol. 18, 741–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Evjenth R., Hole K., Ziegler M., Lillehaug J. R. (2009) Application of reverse-phase HPLC to quantify oligopeptide acetylation eliminates interference from unspecific acetyl-CoA hydrolysis. BMC Proc. 3, S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Grzesiek S., Bax A. (1993) The importance of not saturating H2O in protein NMR: application to sensitivity enhancement and NOE measurements. J. Am. Chem. Soc. 115, 12593–12594 [Google Scholar]
- 29. Davis A. L., Keeler J., Laue E. D., Moskau D. (1992) Experiments for recording pure-absorption heteronuclear correlation spectra using pulsed field gradients. J. Magn. Reson. 98, 207–216 [Google Scholar]
- 30. Sattler M., Schleucher J., Griesinger C. (1999) Heteronuclear multidimensional NMR experiments for the structure. Prog. Nucl. Magn. Reson. Spectrosc. 34, 93–158 [Google Scholar]
- 31. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 32. Keller R. L. J. (2005) Optimizing the Process of Nuclear Magnetic Resonance Spectrum Analysis and Computer Aided Resonance Assignment. Doctoral dissertation ETH No. 15947, ETH Zürich, Zürich, Switzerland [Google Scholar]
- 33. Grzesiek S., Anglister J., Bax A. (1993) Correlation of backbone amide and aliphatic side chain resonances in 13C/15N-enriched proteins by isotropic mixing of 13C magnetization. J. Magn. Reson. Ser. B 101, 114–119 [Google Scholar]
- 34. Montelione G. T., Lyons B. A., Emerson S. D., Tashiro M. (1992) An efficient triple-resonance experiments using C-13 isotropic mixing for determining sequence-specific resonance assignments of isotopically enriched proteins. J. Am. Chem. Soc. 114, 10974–10975 [Google Scholar]
- 35. Schubert M., Oschkinat H., Schmieder P. (2001) MUSIC and aromatic residues: amino acid type-selective 1H-15N correlations, III. J. Magn. Reson. 153, 186–192 [DOI] [PubMed] [Google Scholar]
- 36. Schubert M., Oschkinat H., Schmieder P. (2001) Amino acid type-selective backbone 1H-15N correlations for Arg and Lys. J. Biomol. NMR 20, 379–384 [DOI] [PubMed] [Google Scholar]
- 37. Schubert M., Smalla M., Schmieder P., Oschkinat H. (1999) MUSIC in triple-resonance experiments: amino acid type-selective 1H-15N correlations. J. Magn. Reson. 141, 34–43 [DOI] [PubMed] [Google Scholar]
- 38. Harris R. K., Becker E. D., De Menezes S. M., Goodfellow R., Granger P. (2001) Further conventions for NMR shielding and chemical shifts (IUPAC recommendations 2008). Pure Appl. Chem. 73, 1795–1818 [Google Scholar]
- 39. Wishart D. S., Bigam C. G., Yao J., Abildgaard F., Dyson H. J., Oldfield E., Markley J. L., Sykes B. D. (1995) 1H, 13C, and 15N chemical shift referencing in biomolecular NMR. J. Biomol. NMR 6, 135–140 [DOI] [PubMed] [Google Scholar]
- 40. Schanda P., Kupce E., Brutscher B. (2005) SOFAST-HMQC experiments for recording two-dimensional heteronuclear correlation spectra of proteins within a few seconds. J. Biomol. NMR 33, 199–211 [DOI] [PubMed] [Google Scholar]
- 41. Kay L. E., Torchia D. A., Bax A. (1989) Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry 28, 8972–8979 [DOI] [PubMed] [Google Scholar]
- 42. Kamath U., Shriver J. W. (1989) Characterization of thermotropic state changes in myosin subfragment-1 and heavy meromyosin by UV difference spectroscopy. J. Biol. Chem. 264, 5586–5592 [PubMed] [Google Scholar]
- 43. Dosset P., Hus J. C., Blackledge M., Marion D. (2000) Efficient analysis of macromolecular rotational diffusion from heteronuclear relaxation data. J. Biomol. NMR 16, 23–28 [DOI] [PubMed] [Google Scholar]
- 44. Wishart D. S., Sykes B. D., Richards F. M. (1991) Relationship between nuclear magnetic resonance chemical shift and protein secondary structure. J. Mol. Biol. 222, 311–333 [DOI] [PubMed] [Google Scholar]
- 45. Liszczak G., Arnesen T., Marmorstein R. (2011) Structure of a ternary Naa50p (NAT5/SAN) N-terminal acetyltransferase complex reveals the molecular basis for substrate-specific acetylation. J. Biol. Chem. 286, 37002–37010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lipari G., Szabo A. (1982) Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 1. Theory and range of validity. J. Am. Chem. Soc. 104, 4546–4559 [Google Scholar]
- 47. Lipari G., Szabo A. (1982) Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 2. Analysis of experimental results. J. Am. Chem. Soc. 104, 4559–4570 [Google Scholar]
- 48. Neuwald A. F., Landsman D. (1997) GCN5-related histone N-acetyltransferases belong to a diverse superfamily that includes the yeast Spt10 protein. Trends Biochem. Sci 22, 154–155 [DOI] [PubMed] [Google Scholar]
- 49. Clements A., Rojas J. R., Trievel R. C., Wang L., Berger S. L., Marmorstein R. (1999) Crystal structure of the histone acetyltransferase domain of the human PCAF transcriptional regulator bound to coenzyme A. EMBO J. 18, 3521–3532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vetting M. W., de Carvalho L. P., Roderick S. L., Blanchard J. S. (2005) A novel dimeric structure of the RimL Nα-acetyltransferase from Salmonella typhimurium. J. Biol. Chem. 280, 22108–22114 [DOI] [PubMed] [Google Scholar]
- 51. Liu X., Wang L., Zhao K., Thompson P. R., Hwang Y., Marmorstein R., Cole P. A. (2008) The structural basis of protein acetylation by the p300/CBP transcriptional coactivator. Nature 451, 846–850 [DOI] [PubMed] [Google Scholar]
- 52. Albaugh B. N., Kolonko E. M., Denu J. M. (2010) Kinetic mechanism of the Rtt109-Vps75 histone acetyltransferase-chaperone complex. Biochemistry 49, 6375–6385 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.