Background: To investigate the role of miR-217 in mediating ethanol action in the liver.
Results: miR-217 promotes ethanol-induced fat accumulation through down-regulating SIRT1 in vitro and in vivo.
Conclusion: miR-217 is a specific target of ethanol action in the liver and contributes to development of alcoholic fatty liver.
Significance: miR-217 may present as a potential therapeutic target for treating human alcoholic fatty liver disease.
Keywords: Alcohol, Lipids, Liver, MicroRNA, Signal Transduction, SIRT1, Alcoholic Fatty Liver, Lipid Metabolism
Abstract
Ethanol-mediated inhibition of hepatic sirtuin 1 (SIRT1) plays a crucial role in the pathogenesis of alcoholic fatty liver disease. Here, we investigated the underlying mechanisms of this inhibition by identifying a new hepatic target of ethanol action, microRNA-217 (miR-217). The role of miR-217 in the regulation of the effects of ethanol was investigated in cultured mouse AML-12 hepatocytes and in the livers of chronically ethanol-fed mice. In AML-12 hepatocytes and in mouse livers, chronic ethanol exposure drastically and specifically induced miR-217 levels and caused excess fat accumulation. Further studies revealed that overexpression of miR-217 in AML-12 cells promoted ethanol-mediated impairments of SIRT1 and SIRT1-regulated genes encoding lipogenic or fatty acid oxidation enzymes. More importantly, miR-217 impairs functions of lipin-1, a vital lipid regulator, in hepatocytes. Taken together, our novel findings suggest that miR-217 is a specific target of ethanol action in the liver and may present as a potential therapeutic target for treating human alcoholic fatty liver disease.
Introduction
Alcoholic fatty liver disease is clinically characterized by excess fat accumulation in the liver in response to ethanol consumption. Excessive lipid accumulation in the liver can progress to more harmful forms of liver injury, such as hepatic inflammation, necrosis, progressive fibrosis, and hepatocellular carcinoma in humans (1). The underlying cellular and molecular mechanisms by which ethanol causes liver steatosis are complex and elusive.
In liver, SIRT1 (sirtuin 1) regulates lipid metabolism by deacetylation of modified lysine residues on transcription regulators, such as SREBP-1 (sterol regulatory element-binding protein 1) and PGC-1α (PPARγ2 co-activator-1 α) (2–4). In recent years, SIRT1 has been gaining recognition as one of the most important targets of ethanol action in the liver (5–11). Accumulating evidence has demonstrated that chronic ethanol exposure impairs hepatic lipid metabolism pathways largely mediated by SIRT1 and causes development of fatty liver in animals. Moreover, chronic ethanol exposure decreases SIRT1 gene and protein expression levels, induces SIRT1 nucleocytoplasmic shuttling, and ultimately inhibits SIRT1 deacetylase activity in the liver (11). Although the role of SIRT1 signaling in the development of alcoholic fatty liver is firmly established, little is known about the hepatic signaling molecules affected by ethanol, which result in altering the gene and protein expression and enzyme activity of SIRT1, leading to hepatic fat accumulation.
MicroRNAs (miRs) are small, non-coding RNAs that inhibit gene expression by binding to the 3′-untranslated region (UTR) of target mRNAs, causing mRNA cleavage or suppression of translation (12). A growing body of evidence suggests that aberrant expression of miRNAs contributes to the development of alcoholic liver injury (13–18). Thus, miRNAs have been suggested as novel therapeutic targets for alcoholic liver disease. However, the specific miRNAs that are involved in the development of alcoholic liver steatosis remain largely unknown.
In the present study, we have identified a particular miRNA, miR-217, as a target of ethanol in the liver. More importantly, we demonstrate that ethanol-mediated induction of miR-217 results in SIRT1 inhibition and subsequently excess fat accumulation in hepatocytes.
EXPERIMENTAL PROCEDURES
Plasmids, Antibodies, and Reagents
pcDNATM 6.2-GW/miR-217 and pcDNATM 6.2-GW/scmiR-217 expression plasmids were kind gifts from Dr. Gregory M. Vercellotti (University of Minnesota, Minneapolis, MN). miRIDIAN miR-217 mimic (miR-217) and miRIDIAN hairpin inhibitor miR-217 (anti-miR-217) were purchased from Thermo Scientific.
Wild-type SIRT1 (SIRT1) expression plasmid was a kind gift from Dr. Marty W. Mayo (University of Virginia, Charlottesville, VA). The luciferase vector, including 3′-UTR of human SIRT1 containing the SIRT1-miR-217 response elements (SIRT1-3′-UTR), was purchased from Addgene Inc. Plasmids for (UAS)3-TK luciferase, expression vector driving expression of a Gal4-PGC-1α fusion protein, and HA-lipin-1α were kind gifts from Dr. Brian Finck (Washington University School of Medicine, St. Louis, MO). SIRT1, phosphorylated AMP-activated kinase α (p-AMPKα), and AMPKα antibodies were purchased from Cell Signaling Technology. The lipin-1 antibody for immunostaining was purchased from Abnova. GAPDH antibody (Santa Cruz Biotechnology, Inc.) was used as a loading control. Most chemicals and supplies were purchased from Sigma.
Cell Culture
The mouse AML-12 hepatocyte cell line was obtained from American Type Culture Collection (Manassas, VA). The AML-12 cells were cultured in DMEM/F-12 medium supplemented with 10% fetal bovine serum (FBS), 100 μg/ml streptomycin, 63 μg/ml penicillin G, 0.1 μm dexamethasone, and insulin-transferrin-selenium (Invitrogen). Cells were grown at 37 °C in an atmosphere of 5% CO2.
Measurement of Intracellular Triglyceride (TG)
Whole cell lysates were prepared with radioimmune precipitation assay lysis buffer (Santa Cruz Biotechnology, Inc.), and total intracellular lipids were extracted from cell lysates using a chloroform/methanol mix (2:1, v/v). TGs in the total lipid fraction were determined using a commercially available enzymatic kit (Cayman Chemical) according to the manufacturer's protocol.
Nile Red Staining
Cells were fixed with 4% formaldehyde and stained with Nile Red solution (1 μg/ml) for 10 min at 37 °C. Lipid-bound Nile Red fluorescence was observed with a fluorescence microscope after counterstaining with DAPI (1 μg/ml) for 10 min.
Luciferase Reporter Assay
AML-12 cells were seeded in 12-well plates for 24 h. On the day of transfection, cells were washed with DPBS and switched to reduced serum medium (Invitrogen). For the SIRT1 luciferase activity assay, cells were transiently transfected with PGL3-SIRT1–3′-UTR (1 μg) and β-galactosidase (0.1 μg) as an internal control. For measuring the regulation of miR-217 on lipin-1α-mediated PGC-1α activity, AML-12 cells were transfected with Gal4-PGC-1α and (UAS)3-TK luciferase plasmid. HA-lipin-1α was transfected alone or together with pcDNATM 6.2-GW/miR-217, pcDNATM 6.2-GW/Sc-miR-217, anti-miR-217, or β-galactosidase. Cells were transfected using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's protocol. Luciferase assays were performed using a luciferase reporter assay system (Promega).
SIRT1 Deacetylase Activity
SIRT1 activity was measured by a SIRT1 fluorometric kit (AK-555, Biomol) (10). Briefly, the assays were performed by incubation with recombinant SIRT1 and Fluor de Lys substrate, including a fluorogenic acetylated Lys382 p53 peptide (50 μm) and NAD (100 μm) at 37 °C for 30 min. Fluorescent intensity was measured on a fluorometric reader with excitation set at 360 nm and emission detection set at 460 nm.
Western Blots
Twenty micrograms of protein of the whole cell extracts were separated by 8 or 10% SDS-PAGE, transferred to a nitrocellulose membrane, and immunoblotted with various antibodies.
Immunofluorescence Imaging
AML-12 cells were grown and transfected with HA-lipin-1α on Chamber BD FalconTM culture slides (BD Biosciences) (11). Following treatment, AML-12 cells were fixed for 10 min with 4% paraformaldehyde in PBS. Fixed cells were permeabilized for 10 min in 0.3% Triton X-100 in PBS and blocked for 1 h using 4% BSA in PBS at room temperature. Cells were incubated with a HA.11 clone 16 B12 monoclonal antibody overnight at 4 °C. Samples were then incubated with the secondary Cy5 antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) for 1 h at room temperature. After washing with PBS, cells were mounted with DAPI (ProLong Gold, Invitrogen). Samples without primary antibodies were used as negative controls. Images were obtained with a Leica DM4000 upright microscope and DFC350X camera. Images were processed with Leica LAS software.
Animal Studies
The detailed animal feeding protocol has been described previously (5, 8, 9). Liquid diets were based on the Lieber-DeCarli formulation (Dyets, Bethlehem, PA). Six- to eight-week-old male C57BL/6J mice (Jackson Laboratory, Bar Harbor, ME) were divided into two dietary groups: (a) control low fat diet (fat comprising 10% of total calories and 72% of calories as carbohydrate) and (b) ethanol-containing low fat diet (identical to the control diet except with ethanol added to account for 29% of total calories and the caloric equivalent of carbohydrate (maltose-dextrin) removed). The animals were pair-fed for 4 weeks. The experimental protocols were approved by the Institutional Animal Care and Use Committee of the University of South Florida. We have reported previously the relevant hepatic histology and lipid data in these ethanol-fed mice.
Real-time PCR
Total RNA was prepared from cells by use of the TRIzol reagent according to the manufacturer's protocol (Invitrogen) (8–10). Reverse transcription of total RNA to cDNA was performed using the High Capacity cDNA Archive kit (Applied Biosystems, Foster City, CA). Real-time quantitative polymerase chain reaction (qRT-PCR) amplification was performed in an iCycler spectrofluorometric thermal cycler (Bio-Rad) using RT2 SYBR Green qPCR Master Mix and gene-specific primers (Table 1). The relative amount of target mRNA was calculated by the comparative cycle threshold (Ct) method by normalizing target mRNA Ct values to those for GAPDH. For miRNA analysis, miRNA first-strand cDNA was synthesized from total RNA using an optimized mix of poly(A) polymerase and SMARTTM MMLV reverse transcriptase. RT2SYBR Green qPCR Master Mix and mRQ 3′ primer supplied with the Mir-X miRNA qRT-PCR SYBR kits (Clontech, Palo Alto, CA) were then used in qRT-PCR, along with miRNA-specific 5′-primer (Table 1). U6 small nuclear RNA was used as an internal control for miRNA analysis.
TABLE 1.
Primers used in this study
| Direction | Sequence (5′–3′) | |
|---|---|---|
| Gene-specific primers | ||
| Sirt1 | Forward | ATCGGCTACCGAGACAAC |
| Reverse | GTCACTAGAGCTGGCGTGT | |
| Acaca | Forward | TGAGGAGGACCGCATTTATC |
| Reverse | GAAGCTTCCTTCGTGACCAG | |
| Scd1 | Forward | CCTCATCATTGCCAACACCAT |
| Reverse | AGCCAACCCACGTGAGAGAA | |
| Ppargc1a | Forward | TTGCTAGCGGTTCTCACAGA |
| Reverse | GGCTCTTCTGCCTCCTGA | |
| Ppara | Forward | AGAGCCCCATCTGTCCTCTC |
| Reverse | ACTGGTAGTCTGCAAAACCAAA | |
| Acox1 | Forward | TGGTATGGTGTCGTACTTGAATGAC |
| Reverse | AATTTCTACCAATCTGGCTGAAC | |
| Lpin-1 | Forward | CCCTCGATTTCAACGTACCC |
| Reverse | GCAGCCTGTGGCAATTCA | |
| Lpin-1α | Forward | GGTCCCCCAGCCCCAGTCCTT |
| Reverse | GCAGCCTGTGGCAATTCA | |
| Lpin-1β | Forward | CAGCCTGGTAGATTGCCAGA |
| Reverse | GCAGCCTGTGGCAATTCA | |
| Sfrs10 | Forward | GTGGACAACCTGACCTACCG |
| Reverse | TCCTTGGTGTAGCGATCCC | |
| Gapdh | Forward | CTTCACCACCATGGAGAAGGC |
| Reverse | GGCATGGACTGTGGTCATGAG | |
| miRNA-specific primers | ||
| miR-33 | Forward | GTGCATTGTAGTTGCATTGCA |
| miR-34a | Forward | TGGCAGTGTCTTAGCTGGTTGT |
| miR-217 | Forward | TACTGCATCAGGAACTGACTGGA |
Statistical Analysis
Data are presented as means ± S.E. Multiple comparisons were evaluated by two-way analysis of variance followed by Tukey's multiple comparison procedure with p < 0.05 considered significant.
RESULTS
Ethanol Drastically Up-regulates miR-217 in Mouse AML-12 Hepatocytes
Mouse AML-12 hepatocytes efficiently metabolize ethanol through class I (low Km) alcohol dehydrogenase and aldehyde dehydrogenase 2 proteins (19). Furthermore, this cell line provides an adequate system in which to study how ethanol regulates several lipid metabolism signaling pathways (19). Thus, we investigated miR-217 expression in AML-12 cells in response to ethanol exposure.
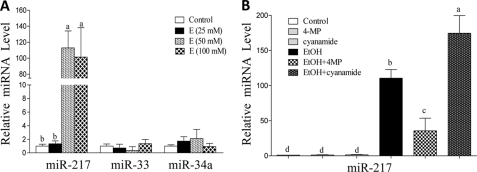
As shown in Fig. 1A, ethanol dramatically increased the miR-217 levels in a concentration-dependent manner in AML-12 hepatocytes up to nearly 100-fold compared with controls. There were no significant changes in miR-33 or miR-34a levels in AML-12 cells exposed to ethanol, indicating that ethanol-induced miR-217 is specific (Fig. 1A). Ethanol, at 50 mm, generated optimal effects and was used in subsequent experiments.
FIGURE 1.
Ethanol up-regulates miR-217 in mouse AML-12 hepatocytes. A, AML-12 cells were treated with various concentrations of ethanol for 24 h, and miRNA-217 levels were measured using qRT-PCR. B, AML-12 cells were treated with ethanol (50 mm), 4-methylpyrazole (4-MP) (0.1 mm), or cyanamide (Cya) (0.1 mm) for 24 h, and miR-217 levels were measured. All data are means ± S.E. (error bars) from at least 3–5 experiments. Means without a common letter differ; p < 0.05.
We used the alcohol dehydrogenase inhibitor 4-methylpyrazole and the aldehyde dehydrogenase 2 inhibitor cyanamide to determine whether ethanol metabolism was required for the ethanol-mediated miR-217 induction in AML-12 cells. As shown in Fig. 1B, 4-methylpyrazole largely abolished the effect of ethanol on miR-217, whereas cyanamide augmented the ethanol effect. Collectively, these results suggest that acetaldehyde generated from ethanol may be, at least in part, responsible for the ability of ethanol to induce miR-217 in AML-12 hepatocytes.
miR-217 Promotes Fat Accumulation in AML-12 Hepatocytes Exposed to Ethanol
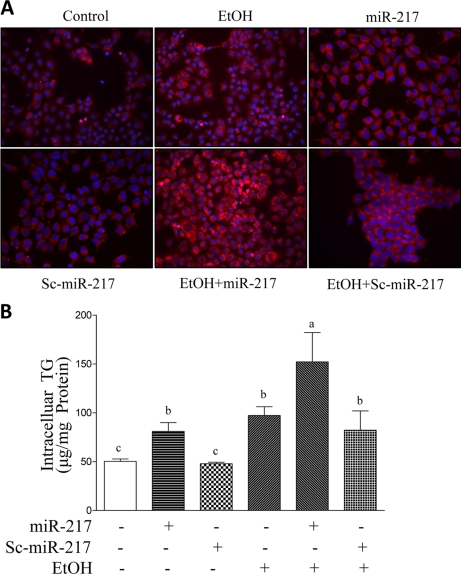
To determine whether the increased miR-217 levels induced by ethanol affect cellular TG accumulation, we examined the TG levels in AML-12 cells transfected with empty vector, miR-217, or a scrambled version of miR-217 (Sc-miR-217) with or without the presence of ethanol using Nile Red staining and enzymatic assays. Ethanol or overexpression of miR-217 significantly increased the TG accumulation in AML-12 cells compared with controls (Fig. 2). More importantly, miR-217 markedly augmented fat accumulation in AML-12 cells exposed to ethanol. These results were not seen when Sc-miR-217 was expressed. Our results clearly demonstrate that miR-217 promotes TG accumulation in AML-12 hepatocytes exposed to ethanol.
FIGURE 2.
miR-217 promotes TG accumulation in AML-12 hepatocytes exposed to ethanol. AML-12 cells were transfected with control vector, miR-217, or Sc-miR217. Forty-eight hours after transfection, ethanol (50 mm) was added. After incubation for 24 h, cellular TG content was measured by Nile Red staining (A) and a cellular enzymatic kit (B). All data are means ± S.E. (error bars) from at least 3–5 experiments. Means without a common letter differ; p < 0.05.
miR-217 Is Dramatically Up-regulated in Livers of Chronically Ethanol-fed Mice
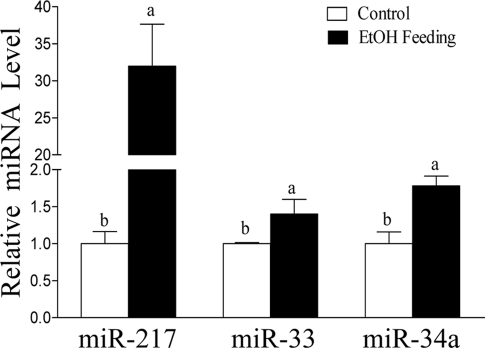
We assessed whether the effects of ethanol observed in vitro also occurred in vivo by using liver samples from previously studied ethanol-fed and pair-fed control mice. Feeding mice ethanol (29% of the total calories) via a modified low fat Lieber-DeCarli liquid diet for 4 weeks led to development of liver steatosis and mildly elevated liver enzymes (5, 8, 9). The in vivo effect of ethanol on hepatic miR-217 was determined using cryo-preserved liver samples from those mice. Indeed, the level of miR-217 was drastically increased nearly 30-fold by ethanol feeding compared with the pair-fed controls (Fig. 3). The levels of hepatic miR-33 and miR-34a were slightly but significantly increased by ethanol feeding in mice (Fig. 3).
FIGURE 3.
miR-217 is markedly up-regulated in the livers of chronically ethanol-fed mice. Relative levels of miRNA from liver of mice fed a control diet with or without ethanol. All data are means ± S.E. (error bars); n = 4–6 animals. Means without a common letter differ; p < 0.05.
miR-217 Promotes Ethanol-mediated Inhibition of SIRT1
MiR-217 functions as an endogenous inhibitor of SIRT1 in endothelial cells (20). We used a reporter vector containing a luciferase gene followed by the 3′-UTR of human SIRT1 mRNA (SIRT1-3′-UTR) to explore the role of miR-217 in the regulation of SIRT1 in AML-12 cells.
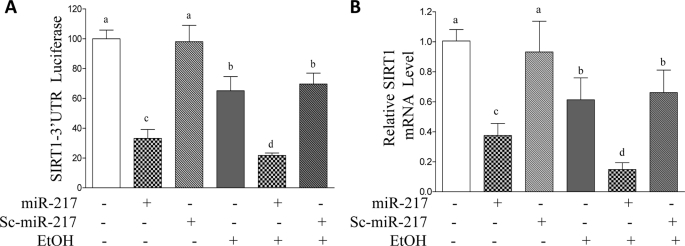
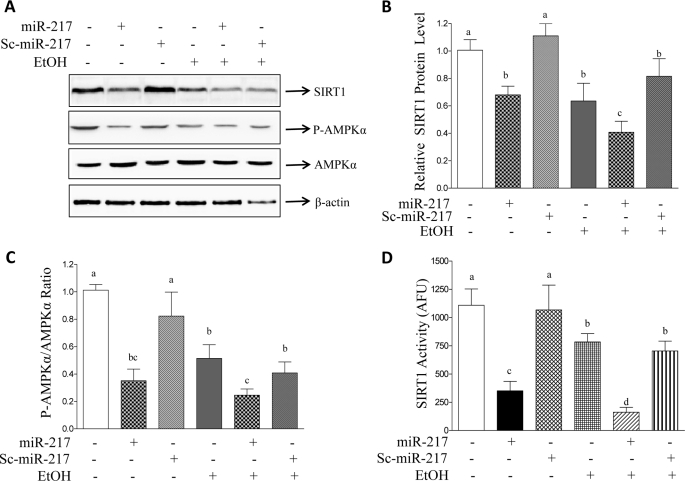
Overexpression of miR-217 significantly inhibited SIRT1-3′-UTR reporter activity in AML-12 cells (Fig. 4A). Accordingly, AML-12 cells transfected with miR-217 showed a significant reduction in SIRT1 mRNA and protein levels compared with controls (Figs. 4B and 5). SIRT1 deacetylase activity was also significantly diminished in AML-12 cells transfected with miR-217 compared with controls (Fig. 5D). Note that expression of Sc-miR-217 showed no significant effects on SIRT1.
FIGURE 4.
miR-217 promotes ethanol-mediated inhibition of SIRT1 in AML-12 hepatocytes. A, AML-12 cells were transfected with a SIRT1-3′-UTR reporter and expression plasmids of control vector, miR-217, Sc-miR-217, and β-galactosidase (internal control). Ethanol (50 mm) was added for 24 h. Forty-eight hours after transfection, cells were harvested, and luciferase and β-galactosidase activities were determined. B, AML-12 cells were transfected with plasmids of control vector, miR-217, or Sc-miR-217. Ethanol (50 mm) was then added. qRT-PCR was used to estimate relative mRNA levels of SIRT1. All data are means ± S.E. (error bars) from at least 3–5 experiments. Means without a common letter differ; p < 0.05.
FIGURE 5.
miR-217 promotes ethanol-mediated inhibition of SIRT1 protein and activity in AML-12 hepatocytes. A, cell extracts of AML-12 cells transfected with plasmids of control vector, miR-217, or Sc-miR-217 with or without ethanol (50 mm) were immunoblotted with a SIRT1, p-AMPKα, AMPKα, or β-actin antibody. B, relative protein levels of SIRT1. C, relative levels of p-AMPKα/AMPKα ratio. D, SIRT1 deacetylase activity was measured in AML-12 cells transfected with plasmids of control vector, miR-217, or Sc-miR-217 with or without ethanol (50 mm). SIRT1 deacetylase activity is represented as arbitrary fluorescence units (AFU). All data are means ± S.E. (error bars) from at least 3–5 experiments. Means without a common letter differ; p < 0.05.
As expected, ethanol exposure to AML-12 cells significantly decreased SIRT1-3′-UTR reporter activity, SIRT1 mRNA and protein expression, and SIRT1 activity (Figs. 4 and 5). Remarkably, expression of miR-217 significantly exacerbated the ethanol-mediated suppression of SIRT1 (Figs. 4 and 5).
SIRT1 regulates AMPK activity, and ethanol causes fat accumulation by inhibition of AMPK in hepatocytes (21, 22). Thus, we further explored the role of miR-217 in regulating AMPK activity with or without ethanol in AML-12 cells. As shown in Fig. 5, A and C, miR-217 or ethanol substantially inhibited AMPK activity, demonstrated by a reduced p-AMPKα/AMPKα ratio in AML-12 cells, and ethanol-mediated AMPK inhibition was significantly augmented by miR-217 expression compared with Sc-miR-217 controls. Taken together, our data suggest that inhibition of SIRT1 by ethanol may be mediated, at least in part, through miR-217 induction in AML-12 hepatocytes.
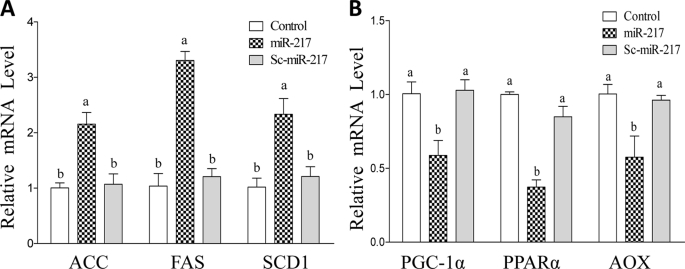
miR-217 Regulates Expression of Genes Encoding Lipogenic Enzymes or Fatty Acid Oxidation Enzymes in AML-12 Cells
To verify the consequence of the miR-217-mediated inhibition of SIRT1, we transfected miR-217 into AML-12 cells and evaluated the mRNA expression levels of several known SIRT1-targeted enzymes involved in lipogenesis or fatty acid oxidation (2–4). As shown in Fig. 6A, overexpression of miR-217 significantly increased mRNA expression levels of several lipogenic enzymes (acetyl-CoA carboxylase (ACC), fatty acid synthase (FAS), and stearoyl-coenzyme A desaturase 1 (SCD1)) compared with controls. Conversely, the mRNA expression levels of enzymes involved in fatty acid oxidation (PPARα, PGC-1α, and acyl-CoA oxidase (AOX)) were significantly reduced in AML-12 cells transfected with miR-217 (Fig. 6B). Overexpression of Sc-miR-217 did not affect those enzymes. Collectively, our findings clearly suggest that miR-217 regulates genes encoding lipogenic or fatty acid oxidation enzymes in AML-12 cells.
FIGURE 6.
miR-217 regulates expression of genes encoding lipogenic enzymes or fatty acid oxidation enzymes in AML-12 cells. A, AML-12 cells were transfected with plasmids of control vector, miR-217, or Sc-miR-217. The qRT-PCR was used to estimate relative mRNA levels of acetyl-CoA carboxylese (ACC), stearoyl-coenzyme A desaturase 1 (SCD1), and fatty acid synthase (FAS). B, the relative mRNA levels of PGC-1α, PPARα, and acyl-CoA oxidase (AOX) in AML-12 cells transfected with plasmids of miR-217 and Sc-miR-217. All data are means ± S.E. (error bars) from at least 3–5 experiments. Means without a common letter differ; p < 0.05.
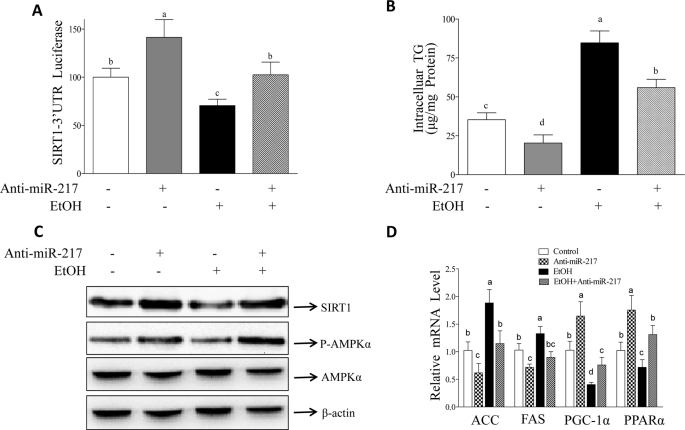
The Inhibition of miR-217 Largely Alleviates Ethanol-mediated Inhibition of SIRT1 Signaling in AML-12 Cells
To further determine whether miR-217 is important in ethanol-mediated attenuation of SIRT1 signaling, we performed experiments inhibiting miR-217 in AML-12 cells using a miR-217 antisense oligonucleotide inhibitor (anti-miR-217). As shown in Fig. 7, reduction of miR-217 levels by transfection with anti-miR-217 significantly enhanced SIRT1 signaling in AML-12 cells. More importantly, anti-miR-217 restored SIRT1 signaling and largely prevented TG accumulation in AML-12 cells exposed to ethanol, suggesting a causal role of miR-217 in ethanol-mediated excess fat accumulation via down-regulation of SIRT1.
FIGURE 7.
Inhibition of miR-217 alleviates ethanol-mediated inhibition of SIRT1 signaling in AML-12 cell. A, AML-12 cells were transfected with a SIRT1-3′-UTR reporter and expression plasmids of control vector, anti-miR-217, and β-galactosidase (internal control). Ethanol (50 mm) was added for 24 h. 48 h after transfection, cells were harvested, and luciferase and β-galactosidase activities were determined. B, AML-12 cells were transfected with control vector, miR-217, or anti-miR-217. 48 h after transfection, ethanol (50 mm) was added. After incubation for 24 h, cellular TG content was measured by an enzymatic kit. C, cell extracts of AML-12 cells transfected with plasmids of control vector or anti-miR-217 were immunoblotted with a SIRT1, p-AMPKα, AMPKα, or β-actin antibody. D, AML-12 cells were transfected with plasmids of control vector or anti-miR-217. qRT-PCR was used to estimate relative mRNA levels of acetyl-CoA carboxylase (ACC), fatty acid synthase (FAS), PGC-1α, and PPARα. All data are means ± S.E. (error bars) from at least 3–5 experiments. Means without a common letter differ; p < 0.05.
miR-217 Impairs Lipin-1 Signaling in AML-12 Hepatocytes
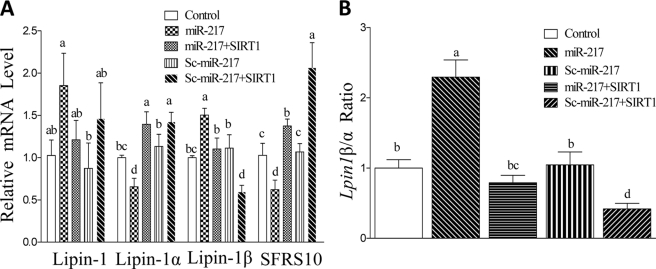
Lipin-1 has emerged as a vital lipid regulator in the development of both alcoholic and nonalcoholic fatty liver disease (9, 19, 23). We therefore examined the effects of miR-217 on lipin-1 in AML-12 cells.
Overexpression of miR-217 in AML-12 cells significantly induced total lipin-1 mRNA levels by nearly 2-fold compared with the control (Fig. 8A). The lipin-1 gene (Lpin1) has two major alternatively spliced isoforms, lipin-1α and lipin-1β (23–26). The Lpin-1 β/α ratio was substantially increased by miR-217 expression compared with controls (Fig. 8B). In parallel, the mRNA levels of SFRS10, a known splicing factor that regulates lipin-1 mRNA alternative splicing (26), were significantly inhibited by miR-217 compared with the controls (Fig. 8A). Importantly, SIRT1wt expression largely relieved the effects of miR-217 on lipin-1, clearly suggesting the involvement of SIRT1 (Fig. 8).
FIGURE 8.
miR-217 impairs lipin-1 signaling in AML-12 hepatocytes. A, relative mRNA levels of total lipin-1, lipin-1α, lipin-1β, and SFRS10 in AML-12 cells transfected with plasmids of control vector, miR-217, Sc-miR-217, or SIRT1. B, relative Lpin1 β/α ratio levels in AML-12 cells transfected with plasmids of control vector, miR-217, Sc-miR-217, or SIRT1. All data are means ± S.E. from at least 3–5 experiments. Means without a common letter differ; p < 0.05.
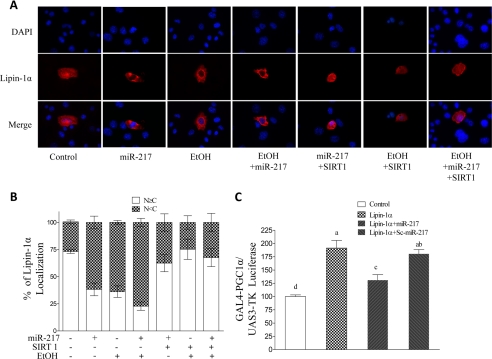
Lipin-1α is a nucleocytoplasmic shuttling protein (19, 27–31). To investigate whether miR-217 governs lipin-1α nucleocytoplasmic shuttling, AML-12 cells were co-transfected with control vector or HA-lipin-1α plasmid with or without the presence of ethanol. The localization of the HA-tagged lipin-1α was determined by immunofluorescence with anti-HA antibody (red). Nuclei were visualized with DAPI staining (blue). Consistent with the findings reported by others (19, 27–31), lipin-1α was localized in both the cytoplasm and the nucleus of AML-12 cells (Fig. 9, A and B). However, overexpression of miR-217, ethanol treatment, or the combination of miR-217 and ethanol resulted in lipin-1α localized to the cytoplasm. Co-transfection of SIRT1 largely blocked lipin-1α nucleocytoplasmic shuttling induced by ethanol, miR-217, or a combination of both (Fig. 9, A and B).
FIGURE 9.
miR-217 blocks lipin-1α nuclear entry and impairs lipin-1 nuclear function. A, representative photomicrographs of HA-lipin-1 (red) or DAPI (blue) immunofluorescence in AML-12 cells. AML-12 cells were transfected with control vector, miR-217, Sc-miR-217, HA-lipin-1α, or SIRT1 for 72 h and then treated with or without ethanol (50 mm) for 18 h. The cells were subjected to immunostaining with anti-HA (lipin-1) antibody (red), and nuclei were stained with DAPI (blue). Original magnification was ×200. B, the distribution patterns of HA-lipin-1α were scored for nearly 100 cells and classified into two categories: nuclear-dominant distribution (N [mteq] C) and cytoplasmic-dominant distribution (N < C). C, AML-12 cells were transfected with plasmids for (UAS)3-TK luciferase and expression vector driving expression of a Gal4-PGC-1α fusion protein along with control vector, miR-217, or Sc-miR-217 for 72 h. Cells then were harvested for assay of luciferase activity. All data are means ± S.E. from at least 3–5 experiments. Means without a common letter differ; p < 0.05.
To further determine the ability of miR-217 to regulate lipin-1α-mediated PGC-1α transcriptional activity, AML-12 cells were transfected with a Gal4-responsive (UAS)3-TK luciferase (UAS.TKLuc) construct with expression vector driving expression of a Gal4-PGC-1α fusion protein either alone (control vector) or together with lipin-1α. As shown in Fig. 9C, whereas the transactivation mediated by the PGC-1α fusion protein was significantly increased in AML-12 cells expressing lipin-1α, co-expression of miR-217 largely inhibited the lipin-1α-mediated effects.
DISCUSSION
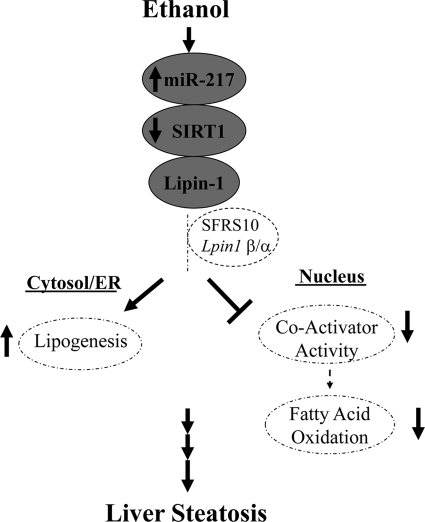
In the present study, we have identified miR-217 as a novel target of ethanol action in the liver. We demonstrated that chronic ethanol exposure dramatically and specifically induced miR-217 levels and caused excess fat accumulation in AML-12 hepatocytes and in mouse livers. We further showed that overexpression of miR-217 in AML-12 cells promoted ethanol-mediated impairments of SIRT1 expression and SIRT1-regulated genes encoding lipogenic or fatty acid oxidation enzymes. Moreover, we found that miR-217 disturbed the functions of lipin-1, a vital lipid regulator, in AML-12 hepatocytes. Taken together, our findings suggest for the first time that ethanol-mediated up-regulation of hepatic miR-217 may play an essential role in the development of alcoholic fatty liver in mice (Fig. 10).
FIGURE 10.
Proposed role of miR-217 in alcoholic fatty liver.
In recent years, several miRNAs, such as miR-155 and miR-212, involved in alcoholic liver disease have been identified (13–18). Although our present study further supports the concept that miRNAs contribute to the development of alcoholic liver injury in mice, further work is warranted to identify more specific hepatic miRNAs and their functions involved in the pathogenesis of alcoholic fatty liver disease. More importantly, a causal role of miR-217 in the development of alcoholic fatty liver needs to be further established. Whether down-regulation of elevated hepatic miR-217 using anti-miR-217 approaches would alleviate liver steatosis in ethanol-fed mice is currently under investigation in our laboratory.
The exact mechanism by which ethanol induces miR-217 in hepatocytes remains to be determined. Our data suggest that acetaldehyde generated from ethanol metabolism via alcohol dehydrogenase may be responsible for the ability of ethanol to induce miR-217 levels in AML-12 cells. Recently, we and several other groups have shown that both acetaldehyde and acetate, two major metabolites of ethanol metabolism via alcohol dehydrogenase and aldehyde dehydrogenase 2, are required for the effect of ethanol on several hepatic signaling molecules, such as SIRT1, lipin-1, and histone H3 (5, 19, 32). This discrepancy may be due to different pathways involved in the regulation of those signaling molecules. On the other hand, hepatic ethanol metabolism induces a shift in the ratio of [NAD+] to [NADH] and releases reactive oxygen species (1). Therefore, it is tempting to speculate that hepatic miR-217 may be dramatically elevated in response to altered redox state or excessive reactive oxygen species production in the livers of ethanol-fed mice.
miRNAs exert their actions by targeting specific mRNAs and inhibiting their protein expression (12). We demonstrated that SIRT1 is a direct target of miR-217 in response to ethanol exposure in AML-12 cells. In recent years, SIRT1 has emerged as a central molecule controlling the pathways of hepatic lipid metabolism and in the development of alcoholic fatty liver (5–11). However, the mechanism by which ethanol induces loss of function of SIRT1 in liver is unknown. Our present study demonstrates that ethanol dramatically up-regulates miR-217, which affects SIRT1 expression, leading to loss of SIRT1 function, causing hepatic fat accumulation.
SIRT1 expression can be regulated by multiple mechanisms. For instance, tumor suppressor p53 can bind to the SIRT1 promoter and form a complex with SIRT1, leading to inhibition of SIRT1 transcription (33). Moreover, the RNA-binding protein HuR stabilizes SIRT1 mRNA through 3′-UTR interactions, leading to increased SIRT1 levels (34). Given that both p53 and HuR have been implicated in alcoholic liver disease, it is logical to speculate that ethanol-mediated up-regulation of miR-217 may affect p53 or HuR that bind to the 3′-UTR of SIRT1 and hence may lead to suppression of SIRT1 expression (35, 36). On the other hand, our data cannot rule out the possibility that other miRNAs may also play roles in governing SIRT1 expression in response to ethanol treatment. For example, miR-34a functions as a negative regulator of SIRT1 (37). Indeed, we observed that there was a mild but significant increase in miR-34a in the livers of ethanol-fed mice, suggesting that miR-34a may also be involved in ethanol-mediated SIRT1 inhibition in mouse livers.
Another novel finding of our present study is that miR-217 regulates lipin-1, a vital lipid regulator. Lipin-1 is a bifunctional molecule that regulates lipid metabolism as a phosphatidic acid phosphohydrolase type enzyme in the cytosol and a transcriptional co-activator in the nucleus (23–31). In recent years, lipin-1 has emerged as a major player in the development of both alcoholic and non-alcoholic fatty liver diseases (9, 19, 23–31). Accumulating evidence suggests that lipin-1β primarily functions in the cytoplasm as a phosphatidic acid phosphohydrolase type enzyme in lipid synthesis, whereas lipin-1α functions in the nucleus as a transcriptional co-activator (19, 23–31). Our present data showed that in accordance with miR-217-mediated up-regulation of total lipin-1 mRNA levels and Lpin β/α ratio, miR-217 sequesters lipin-1α to the cytosol, preventing its proper functioning in the nucleus of AML-12 cells. Remarkably, activation of SIRT1 signaling largely alleviates the effects of miR-217 on lipin-1. Although there is no report on the role of a potential SIRT1-lipin-1 axis, our group has recently suggested that SIRT1 inhibition by ethanol may impair acetylation/SUMOylation modifications of lipin-1 that ultimately affect nuclear lipin-1 activity in mouse livers (19). Therefore, it is logical to speculate that ethanol-mediated impairment of the miR-217-SIRT1 axis may be associated with lipin-1 in an acetylation- or SUMOylation-dependent manner, blocking lipin-1α nuclear entry, which, in turn, reduces its nuclear functions.
In mouse models of diabetic nephropathy, miR-217 is induced by transforming growth factor-β (TGF-β), leading to an inhibition of PTEN and ultimately activation of Akt (38). Moreover, miR-217 attenuates heme oxygenase-1 protein expression and enzyme activity (39). Conceivably, in addition to suppression of hepatic SIRT1 or lipin-1 signaling, ethanol-mediated miR-217 up-regulation may also affect signaling pathways mediated by AKT-PTEN, heme oxygenase-1, or AMPK, which, in turn, contribute to the development of alcoholic fatty liver in mice.
In summary, we have identified miR-217 as a specific target of ethanol in the liver. Our study further demonstrates that ethanol induces miR-217-dependent SIRT1 loss of function and ultimately induces fat accumulation in hepatocytes. Our novel findings shed light on the mechanism underlying the development of alcoholic fatty liver. miR-217 has been implicated in several pathological conditions, including diabetes and aging, via targeting SIRT1 (20). Our study suggests that hepatic miR-217 inhibition by nutritional or pharmacological modulation could be an attractive therapeutic approach for treating human alcoholic fatty liver disease.
This work was supported, in whole or in part, by National Institutes of Health, National Institute on Alcoholism and Alcohol Abuse, Grants AA-015951 and AA-013623 (to M. Y.).
- PPAR
- peroxisome proliferator-activated receptor
- AMPK
- AMP-activated kinase
- p-AMPKα
- phosphorylated AMPKα
- Ct
- cycle threshold
- miR
- microRNA
- qRT-PCR
- quantitative RT-PCR
- TG
- triglyceride
- TK
- thymidine kinase.
REFERENCES
- 1. Gao B., Bataller R. (2011) Alcoholic liver disease. Pathogenesis and new therapeutic targets. Gastroenterology 141, 1572–1585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ponugoti B., Kim D. H., Xiao Z., Smith Z., Miao J., Zang M., Wu S. Y., Chiang C. M., Veenstra T. D., Kemper J. K. (2010) SIRT1 deacetylates and inhibits SREBP-1C activity in regulation of hepatic lipid metabolism. J. Biol. Chem. 285, 33959–33970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Walker A. K., Yang F., Jiang K., Ji J. Y., Watts J. L., Purushotham A., Boss O., Hirsch M. L., Ribich S., Smith J. J., Israelian K., Westphal C. H., Rodgers J. T., Shioda T., Elson S. L., Mulligan P., Najafi-Shoushtari H., Black J. C., Thakur J. K., Kadyk L. C., Whetstine J. R., Mostoslavsky R., Puigserver P., Li X., Dyson N. J., Hart A. C., Näär A. M. (2010) Conserved role of SIRT1 orthologs in fasting-dependent inhibition of the lipid/cholesterol regulator SREBP. Genes Dev. 24, 1403–1417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sugden M. C., Caton P. W., Holness M. J. (2010) PPAR control. It's SIRTainly as easy as PGC. J. Endocrinol. 204, 93–104 [DOI] [PubMed] [Google Scholar]
- 5. You M., Liang X., Ajmo J. M., Ness G. C. (2008) Involvement of mammalian sirtuin 1 in the action of ethanol in the liver. Am. J. Physiol. Gastrointest. Liver Physiol. 294, G892–G898 [DOI] [PubMed] [Google Scholar]
- 6. Lieber C. S., Leo M. A., Wang X., Decarli L. M. (2008) Effect of chronic alcohol consumption on Hepatic SIRT1 and PGC-1α in rats. Biochem. Biophys. Res. Commun. 370, 44–48 [DOI] [PubMed] [Google Scholar]
- 7. You M., Cao Q., Liang X., Ajmo J. M., Ness G. C. (2008) Mammalian sirtuin 1 is involved in the protective action of dietary saturated fat against alcoholic fatty liver in mice. J. Nutr. 138, 497–501 [DOI] [PubMed] [Google Scholar]
- 8. Ajmo J. M., Liang X., Rogers C. Q., Pennock B., You M. (2008) Resveratrol alleviates alcoholic fatty liver in mice. Am. J. Physiol. Gastrointest. Liver Physiol. 295, G833–G842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shen Z., Liang X., Rogers C. Q., Rideout D., You M. (2010) Involvement of adiponectin-SIRT1-AMPK signaling in the protective action of rosiglitazone against alcoholic fatty liver in mice. Am. J. Physiol. Gastrointest. Liver Physiol. 298, G364–G374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shen Z., Ajmo J. M., Rogers C. Q., Liang X., Le L., Murr M. M., Peng Y., You M. (2009) Role of SIRT1 in regulation of LPS- or two ethanol metabolites-induced TNF-α production in cultured macrophage cell lines. Am. J. Physiol. Gastrointest. Liver Physiol. 296, G1047–G1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liang X., Hu M., Rogers C. Q., Shen Z., You M. (2011) Role of SIRT1-FoxO1 signaling in dietary saturated fat-dependent up-regulation of liver adiponectin receptor 2 in ethanol-administered mice. Antioxid. Redox Signal. 15, 425–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sayed D., Abdellatif M. (2011) MicroRNAs in development and disease. Physiol. Rev. 91, 827–887 [DOI] [PubMed] [Google Scholar]
- 13. Dolganiuc A., Petrasek J., Kodys K., Catalano D., Mandrekar P., Velayudham A., Szabo G. (2009) MicroRNA expression profile in Lieber-DeCarli diet-induced alcoholic and methionine choline-deficient diet-induced nonalcoholic steatohepatitis models in mice. Alcohol Clin. Exp. Res. 33, 1704–1710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bala S., Marcos M., Kodys K., Csak T., Catalano D., Mandrekar P., Szabo G. (2011) Up-regulation of microRNA-155 in macrophages contributes to increased tumor necrosis factor α (TNFα) production via increased mRNA half-life in alcoholic liver disease. J. Biol. Chem. 286, 1436–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhang Y., Jia Y., Zheng R., Guo Y., Wang Y., Guo H., Fei M., Sun S. (2010) Plasma microRNA-122 as a biomarker for viral-, alcohol-, and chemical-related hepatic diseases. Clin. Chem. 56, 1830–1838 [DOI] [PubMed] [Google Scholar]
- 16. Miranda R. C., Pietrzykowski A. Z., Tang Y., Sathyan P., Mayfield D., Keshavarzian A., Sampson W., Hereld D. (2010) MicroRNAs. Master regulators of ethanol abuse and toxicity? Alcohol Clin. Exp. Res. 34, 575–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yeligar S., Tsukamoto H., Kalra V. K. (2009) Ethanol-induced expression of ET-1 and ET-BR in liver sinusoidal endothelial cells and human endothelial cells involves hypoxia-inducible factor-1α and microRNA-199. J. Immunol. 183, 5232–5243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tang Y., Banan A., Forsyth C. B., Fields J. Z., Lau C. K., Zhang L. J., Keshavarzian A. (2008) Effect of alcohol on miR-212 expression in intestinal epithelial cells and its potential role in alcoholic liver disease. Alcohol Clin. Exp. Res. 32, 355–364 [DOI] [PubMed] [Google Scholar]
- 19. Hu M., Wang F., Li X., Rogers C. Q., Liang X., Finck B. N., Mitra M. S., Zhang R., Mitchell D. A., You M. (2012) Regulation of hepatic lipin-1 by ethanol: Role of AMPK-activated protein kinase/sterol regulatory element binding protein 1 signaling in mice. Hepatology 55, 437–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Menghini R., Casagrande V., Cardellini M., Martelli E., Terrinoni A., Amati F., Vasa-Nicotera M., Ippoliti A., Novelli G., Melino G., Lauro R., Federici M. (2009) MicroRNA 217 modulates endothelial cell senescence via silent information regulator 1. Circulation 120, 1524–1532 [DOI] [PubMed] [Google Scholar]
- 21. You M., Matsumoto M., Pacold C. M., Cho W. K., Crabb D. W. (2004) The role of AMP-activated protein kinase in the action of ethanol in the liver. Gastroenterology 127, 1798–1808 [DOI] [PubMed] [Google Scholar]
- 22. Hou X., Xu S., Maitland-Toolan K. A., Sato K., Jiang B., Ido Y., Lan F., Walsh K., Wierzbicki M., Verbeuren T. J., Cohen R. A., Zang M. (2008) SIRT1 regulates hepatocyte lipid metabolism through activating AMP-activated protein kinase. J. Biol. Chem. 283, 20015–20026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Harris T. E., Finck B. N. (2011) Dual function lipin proteins and glycerolipid metabolism. Trends Endocrinol. Metab. 22, 226–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Han G. S., Carman G. M. (2010) Characterization of the human LPIN1-encoded phosphatidate phosphatase isoforms. J. Biol. Chem. 285, 14628–14638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Péterfy M., Phan J., Reue K. (2005) Alternatively spliced lipin isoforms exhibit distinct expression pattern, subcellular localization, and role in adipogenesis. J. Biol. Chem. 280, 32883–32889 [DOI] [PubMed] [Google Scholar]
- 26. Pihlajamäki J., Lerin C., Itkonen P., Boes T., Floss T., Schroeder J., Dearie F., Crunkhorn S., Burak F., Jimenez-Chillaron J. C., Kuulasmaa T., Miettinen P., Park P. J., Nasser I., Zhao Z., Zhang Z., Xu Y., Wurst W., Ren H., Morris A. J., Stamm S., Goldfine A. B., Laakso M., Patti M. E. (2011) Expression of the splicing factor gene SFRS10 is reduced in human obesity and contributes to enhanced lipogenesis. Cell Metab. 14, 208–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu G. H., Gerace L. (2009) Sumoylation regulates nuclear localization of lipin-1α in neuronal cells. PLoS One 4, e7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bou Khalil M., Sundaram M., Zhang H. Y., Links P. H., Raven J. F., Manmontri B., Sariahmetoglu M., Tran K., Reue K., Brindley D. N., Yao Z. (2009) The level and compartmentalization of phosphatidate phosphatase-1 (lipin-1) control the assembly and secretion of hepatic VLDL. J. Lipid Res. 50, 47–58 [DOI] [PubMed] [Google Scholar]
- 29. Haller J. F., Krawczyk S. A., Gostilovitch L., Corkey B. E., Zoeller R. A. (2011) Glucose-6-phosphate isomerase deficiency results in mTOR activation, failed translocation of lipin 1α to the nucleus, and hypersensitivity to glucose. Implications for the inherited glycolytic disease. Biochim. Biophys. Acta 1812, 1393–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Valdearcos M., Esquinas E., Meana C., Gil-de-Gómez L., Guijas C., Balsinde J., Balboa M. A. (2011) Subcellular localization and role of lipin-1 in human macrophages. J. Immunol. 186, 6004–6013 [DOI] [PubMed] [Google Scholar]
- 31. Peterson T. R., Sengupta S. S., Harris T. E., Carmack A. E., Kang S. A., Balderas E., Guertin D. A., Madden K. L., Carpenter A. E., Finck B. N., Sabatini D. M. (2011) mTOR complex 1 regulates lipin 1 localization to control the SREBP pathway. Cell 146, 408–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Park P. H., Lim R. W., Shukla S. D. (2005) Involvement of histone acetyltransferase (HAT) in ethanol-induced acetylation of histone H3 in hepatocytes. Potential mechanism for gene expression. Am. J. Physiol. Gastrointest. Liver Physiol. 289, G1124–G1136 [DOI] [PubMed] [Google Scholar]
- 33. Nemoto S., Fergusson M. M., Finkel T. (2004) Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science 306, 2105–2108 [DOI] [PubMed] [Google Scholar]
- 34. Abdelmohsen K., Pullmann R., Jr., Lal A., Kim H. H., Galban S., Yang X., Blethrow J. D., Walker M., Shubert J., Gillespie D. A., Furneaux H., Gorospe M. (2007) Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol. Cell 25, 543–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. McMullen M. R., Cocuzzi E., Hatzoglou M., Nagy L. E. (2003) Chronic ethanol exposure increases the binding of HuR to the TNFα 3′-untranslated region in macrophages. J. Biol. Chem. 278, 38333–38341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Derdak Z., Lang C. H., Villegas K. A., Tong M., Mark N. M., de la Monte S. M., Wands J. R. (2011) Activation of p53 enhances apoptosis and insulin resistance in a rat model of alcoholic liver disease. J. Hepatol. 54, 164–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yamakuchi M., Ferlito M., Lowenstein C. J. (2008) miR-34a repression of SIRT1 regulates apoptosis. Proc. Natl. Acad. Sci. U.S.A. 105, 13421–13426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kato M., Putta S., Wang M., Yuan H., Lanting L., Nair I., Gunn A., Nakagawa Y., Shimano H., Todorov I., Rossi J. J., Natarajan R. (2009) TGF-β activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nat. Cell Biol. 11, 881–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Beckman J. D., Chen C., Nguyen J., Thayanithy V., Subramanian S., Steer C. J., Vercellotti G. M. (2011) Regulation of heme oxygenase-1 protein expression by miR-377 in combination with miR-217. J. Biol. Chem. 286, 3194–3202 [DOI] [PMC free article] [PubMed] [Google Scholar]